Introduction
Liver cancer is the third highest contributor to
cancer mortality worldwide (1,2). A
high rate of tumor metastasis and recurrence are major factors that
contribute to the poor prognosis associated with liver cancer.
Therefore, the efficacy of radiotherapy and chemotherapy is limited
for the majority of patients with liver cancer following diagnosis,
and surgical treatment is considered as the only available therapy
method at the initial stage of liver cancer (3,4).
Therefore, an improved understanding of the mechanism of liver
cancer is required to identify novel prognostic molecular markers,
as well as potential effective therapeutic targets, to improve the
effect of therapy and patient survival rate.
It has previously been demonstrated that the
accumulation of epigenetic and genetic alterations in hepatocytes,
and uncontrolled cell proliferation and death, are essential for
the progression and initiation of liver cancer (5). In liver cancer, chromosomal
abnormalities are the most common form of genetic mutation, and
numerous chromosomal regions that are frequently unstable have been
identified in liver cancer (6,7). For
example, a 13q34 amplification has been detected in several liver
cancer cell lines, and this region comprises five genes, including
transcription factor Dp-1, cullin 4A (CUL4A) and cell division
cycle protein 16 (8). These genes
may have potential as novel therapeutic targets for liver cancer;
however, further investigation into the particular roles of these
genes is required.
CUL4A is a single-copy gene and encodes an 87-kDa
protein that belongs to the cullin family. High expression levels
of CUL4A have been reported in the spleen and testis, with poor
expression in the liver, lung and thymus (9). The CUL4A protein is able to bind to
ring-box protein 1 and DNA damage-binding protein 1, forming the
ubiquitin ligase E3 complex. This complex mediates the
ubiquitination and degradation of particular substrates and has an
important function in the maintenance of cellular physiology
(10). The role of CUL4A in
oncogenesis has received increased interest; amplification or
overexpression of the CUL4A gene has been detected in various
cancer types, including liver cancer (8), adrenocortical carcinoma (11) and pituitary adenomas (12). In 2015, Pan et al (13) reported an inverse correlation
between the expression of the CUL4A gene and patient survival,
while a positive correlation with lymphatic and venous invasion was
identified. Additionally, the expression of CUL4A in liver cancer
tissues was associated with hepatitis B virus (HBV) e-antigen
(HBeAg) status in patients and may be upregulated by HBV in liver
cancer cell lines in vitro (13). Furthermore, knockdown of CUL4A
ameliorated the motility of liver cancer cell lines by regulating
the expression of epithelial-mesenchymal transition
(EMT)-associated genes (13).
Recently, another group identified that a novel long noncoding
(lnc)RNA, uc.134, repressed liver cancer progression by inhibiting
the CUL4A-mediated ubiquitination of the large tumor suppressor
kinase 1 (LATS1) protein, indicating that the application of uc.134
lncRNA may offer a promising treatment approach for liver cancer,
and that CUL4A may serve an important role in liver cancer
progression (14).
In the present study, the clinical relevance of
CUL4A in liver cancer was primarily investigated. The results
demonstrated that the expression levels of CUL4A in human liver
cancer tissues were markedly increased compared with paracancerous
tissues. CUL4A overexpression in liver cancer cell lines led to
enhanced liver cancer cell proliferation, migration and invasion,
while CUL4A knockdown suppressed the proliferation and motility of
liver cancer cells, and significantly induced cell apoptosis,
indicating that CUL4A may have the potential to serve as a novel
therapeutic target for liver cancer.
Materials and methods
Ethical approval and consent
The present study was approved by the Committee on
the Ethics of Animal Experiments and Human Subject Research of the
First People's Hospital of Kunming (Kunming, China). All volunteers
involved in the present study provided written informed
consent.
Liver cancer samples
In the present study, liver cancer tissues and
paracancerous tissues from 3 different patients (obtained from the
First People's Hospital of Kunming, Kunming, China) were used to
analyze the importance of CUL4A in liver cancer treatment. All
samples originated from primary tumors and were collected from
April-December 2016. The cancer tissue from patient 1 (age, 59;
sex, male;) and patient 2 (age, 56; sex, female) were diagnosed as
infiltrating liver cancer and the cancer tissue from patient 3
(age, 52; sex, male) was superficial liver cancer.
Cell culture
The liver cancer cell lines HEPG2 (hepatoblastoma
cell line) (15) and MHCC97-H
(hepatocellular carcinoma cell line) were employed in the present
study and were purchased from the American Type Culture Collection
(Manassas, VA, USA). The cells were cultured in high-glucose
Dulbecco's modified Eagle's medium (HG-DMEM; Hyclone; GE Healthcare
Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), 100 U/ml penicillin and 0.1 g/ml streptomycin (Hyclone; GE
Healthcare Life Sciences) in a humidified incubator at 37°C with 5%
CO2. Medium was replaced every other day and adherent
cells were passaged by 1:4 dilution every 5–7 days.
Overexpression and knockdown of CUL4A
in liver cancer cell lines
To generate the CUL4A overexpression vector,
CUL4A-coding sequences were obtained by reverse
transcription-polymerase chain reaction (RT-PCR) with the following
primer sequences: CUL4A forward,
5′-CGGAATTCATGGCGGACGAGGCCCCGCGGAA-3′ and reverse,
5′-ACGGTACCTCAGGCCACGTAGTGGTACTGAT-3′. The sequences were amplified
using the following parameters: Initial denaturation at 95°C for 5
min; 35 cycles of 95°C for 35 sec, 60°C for 35 sec and 72°C for 90
sec; followed by a final extension at 72°C for 5 min. Coding
sequences were cloned into a pCI-based overexpression plasmid
(Addgene, Inc., Cambridge, MA, USA). Human CUL4A small interfering
(si)RNA (sequence, AAGAAGAUUAACACGUGCUGG) was purchased from Santa
Cruz Biotechnology, Inc. (Dallas, TX, USA; cat. no. sc-44355). For
the overexpression and knockdown of CUL4A, liver cancer cell lines
(1×106 cells/well) were cultured in a 6-well plate
overnight at 37°C and were subsequently transfected with pCI-CUL4A
vector (2 µg/ml) and CUL4A-siRNA (10 µM/ml), respectively, using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), while cells transfected with an empty pCI vector
(2 µg/ml) or control-siRNA (10 µM/ml; sequence,
AACAGUCGCGUUUGCGACUGGdTdT) served as the control groups. The
detailed operation was performed according to the
Lipofectamine® 2000 protocol. At 36 h post-transfection,
cells were harvested for the subsequent experiments.
RT-quantitative PCR (RT-qPCR)
RT-qPCR was performed as previously described
(16,17). Briefly, the total RNA was extracted
from HEPG2 cells and converted into cDNA. The high capacity cDNA
Reverse Transcription Kit (Thermo Fisher Scientific, Inc.) and
PrimeScript™ II High Fidelity One Step RT-qPCR Kit (Takara
Biotechnology Co., Ltd., Dalian China) were used for reverse
transcription and qPCR. respectively. Following an initial
polymerase activation and denaturation step at 50°C for 2 min and
95°C for 5 min, respectively, the samples in each group underwent
40 amplification cycles of 95°C for 20 sec, 65°C for 10 sec and
72°C for 30 sec in the Light Cycler 480 instrument (Roche
Diagnostics, Basel, Switzerland). Three independent experiments
were performed. Results were quantified using the 2−ΔΔCq
method (18). In the present
study, 18s ribosomal (r)RNA was used for normalization and all
measurements were performed in triplicate. The primer sequences
(5′-3′) were as follows: CUL4A, 5′-TCCTGTTCTTGGACCGCACCT-3′
(forward) and 5′-ACCTGCAGGTCAGACAGCATGC-3′ (reverse); and 18s rRNA,
5′-CCTGGATACCGCAGCTAGGA-3′ (forward) and
5′-GCGGCGCAATACGAATGCCCC-3′ (reverse).
Western blotting
Both tissue samples and cell samples were harvested
with radioimmunoprecipitation assay lysis buffer (CST Biological
Reagents Co., Ltd., Dalian, China) and the protein content of cell
lysates in different groups was further detected with a
bicinchoninic acid protein estimation kit (Pierce; Thermo Fisher
Scientific, Inc.). Western blotting was performed as previously
described (19). Briefly, Protein
(15 µg/lane) was separated on 10% polyacrylamide gel, followed by
transfer to a nitrocellulose membrane. The membrane was blocked
with 5% bovine serum albumin (BSA; Thermo Fisher Scientific, Inc.)
for 2 h at room temperature and subsequently incubated with the
following primary antibodies overnight at 4°C: Anti-CUL4A (cat no.
ab72548; 1:1,000; Abcam, Cambridge, UK), anti-E-cadherin (cat no.
ab1416; 1:500; Abcam), anti-N-cadherin (cat no. ab18203; 1:500;
Abcam), anti-claudin 3 (Cldn3; cat no. ab15102; 1:1,000; Abcam),
anti-occludin (Ocln; cat no. ab31721; 1:1,000; Abcam),
anti-epithelial cell adhesion molecule (Epcam; cat no. ab71916;
1:800; Abcam), anti-Snail (cat no. ab53519; 1:500; Abcam),
anti-Slug (cat no. ab27568; 1:500; Abcam), anti-vimentin (cat no.
ab8978; 1:500; Abcam) and anti-GAPDH (cat no. ab8245; 1:10,000;
Abcam). Subsequently, the membrane was incubated with horseradish
peroxidase (HRP)-conjugated anti-mouse (cat no. sc-2005) or rabbit
(cat no. sc-2357) immunoglobulin G secondary antibodies (1:5,000;
Santa Cruz Biotechnology, Inc.) for 1 h at room temperature. Bands
were visualized with an Amersham ECL kit (GE Healthcare, Chicago,
IL, USA) and relative protein expression was quantified using
Quantity One software (version 4.6.2; Bio-Rad Laboratories, Inc.,
Hercules, CA, USA).
Immunohistochemical staining
Prior to immunohistochemical staining of patient
tissues, all tissue samples were fixed in 4% paraformaldehyde in
PBS at room temperature for 36 h, and sectioned to 5 µm thickness
for staining, as previously described (20,21).
For immunohistochemistry, sections were blocked with 5% BSA for 2 h
at room temperature, and endogenous peroxidase activity was
quenched with 3% H2O2 for 30 min at room
temperature. A polyclonal primary antibody against CUL4A (cat no.
ab72548; 1:200; Abcam) was employed. After 12 h of incubation at
4°C, the sections were washed three times with PBS and processed
with a HRP-conjugated Streptavidin-Biotin complex kit (cat no.
SA1040; Boster Biological Technology, Pleasanton, CA, USA) and
3′,3′-diaminobenzidine solution, according to the manufacturer's
protocol. Finally, the sections were observed using Axio Scope A1
(Carl Zeiss AG, Oberkochen, Germany) with AxioCAM MRc5 (Carl Zeiss
AG) and the relative staining intensity of each group was processed
with AxioVision software (version 4.7; Carl Zeiss AG).
Cell proliferation and cell cycle
analysis
To evaluate cell proliferation ability, cells
(1×105 cells/well) were seeded into a 96-well plate and
the proliferation index of each group was detected with the Cell
Counting Kit-8 (CCK-8) method (Dojindo Molecular Technologies,
Inc., Kumamoto, Japan) as previously described (22,23).
The following equation was used to measure cell proliferation
ability: Proliferation index = absorbance of the experimental
group-absorbance of blank group. The proliferation index of each
group was measured at 0 h (Day 0), 24 h (Day 1), 48 h (Day 2) and
72 h (Day 3) after seeding. To analyze the cell cycle,
5×106 cells were harvested and fixed with 70% ethanol
for 30 min at 4°C. The cell samples were stained with 200 µl
propidium iodide (PI; Beyotime Institute of Biotechnology, Haimen,
China) in the presence of RNase A (Beyotime Institute of
Biotechnology) for 10 min at room temperature. Finally, the samples
were analyzed using a FACSCalibur flow cytometer (BD Biosciences,
Franklin Lakes, NJ, USA) and Flowjo software (version 7.6.1;
Flowjo, LLC, Ashland, OR, USA).
Cell migration and invasion assay
In the present study, the migration and invasion of
liver cancer cells were measured with Transwell plates (8 µm pore
filter, Costar; Corning Incorporated, Corning, NY, USA) as
previously described (16,17). Briefly, the liver cancer cells were
seeded onto the upper insert at a concentration of 1×105
cells per insert in serum-free medium (HG-DMEM). The insert covered
with Matrigel was used for the invasion assay, while a normal
insert was used for the migration assay. Lower chambers were filled
with HG-DMEM containing 10% FBS as a chemoattractant; cells were
incubated for 48 h at 37°C. Non-invading cancer cells were removed
by swabbing the top layer and cancer cells that had migrated
through the gel and attached to the lower surface of the membrane
were stained with 0.5% crystal violet for 20 min at 37°C. The
number of cancer cells in four randomly selected microscopy fields
under a light microscope (magnification, ×100) was counted for each
group.
Cell apoptosis assay
In the present study, a cell apoptosis assay was
performed using a fluorescein isothiocyanate (FITC)-Annexin V/PI
cell apoptosis assay kit (Thermo Fisher Scientific, Inc.) according
to the manufacturer's protocol. Briefly, 5×106 cancer
cells in each group were dissociated into single cells with trypsin
and washed with PBS, followed by incubation with 200 µl
FITC-Annexin V and PI solution. Cancer cells incubated without the
addition of any reagents were used as the negative control group.
Finally, all cell samples were analyzed using a FACSCalibur
cytometer (BD Biosciences) and Flowjo software (version 7.6.1;
Flowjo, LLC).
Statistical analysis
In the present study, the results are presented as
the mean ± standard error of the mean and statistical analysis was
performed using SPSS 17.0 (SPSS, Inc., Chicago, IL, USA). Unpaired
Student's t-tests were used to compare the means of two groups.
One-way analysis of variance with Bonferroni's correction was used
to compare the means of three or more groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of CUL4A in liver cancer
tissue and paracancerous tissue
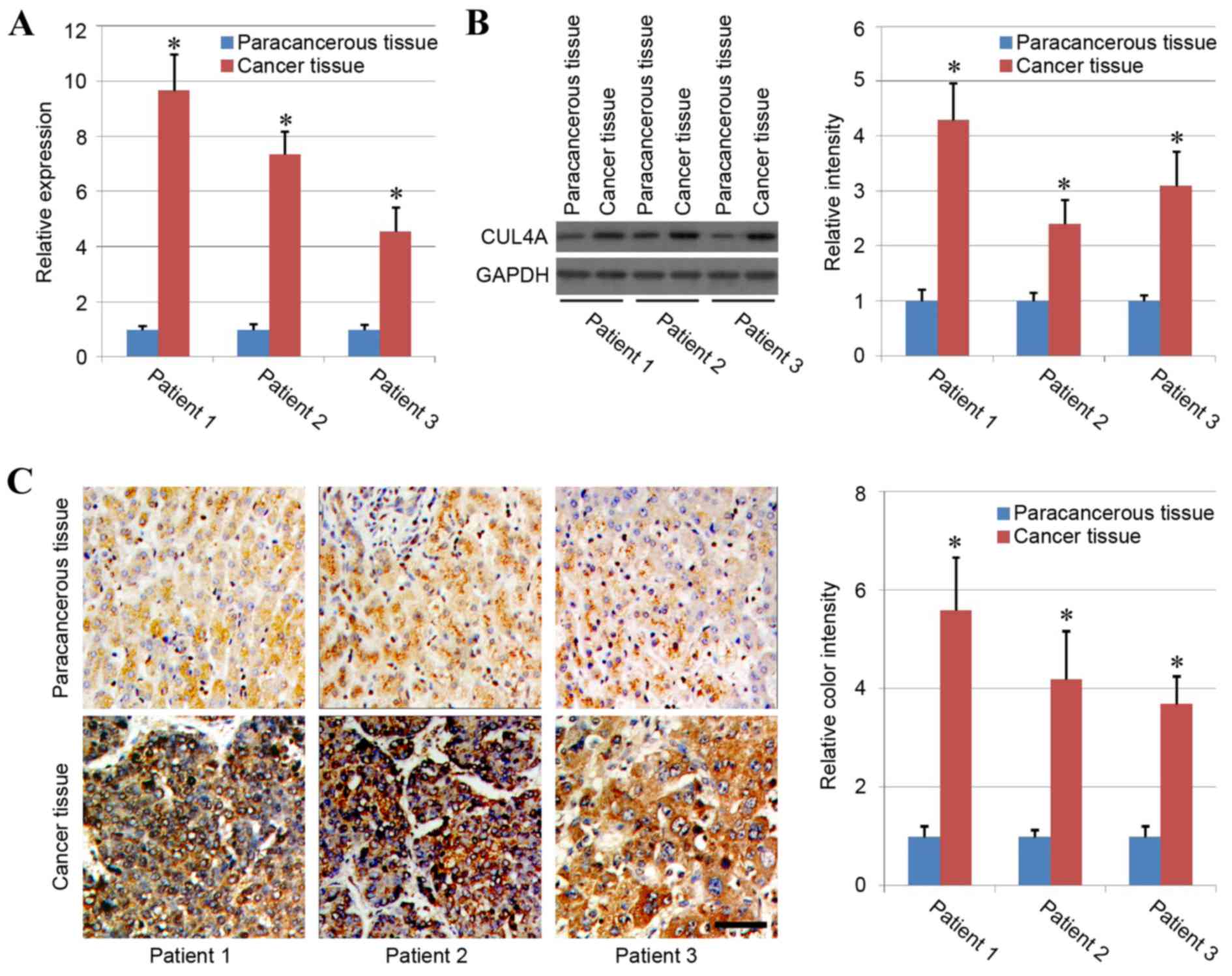
In the present study, liver cancer tissues and
paracancerous tissues from 3 different patients were harvested to
analyze the association between CUL4A and liver cancer. The results
of RT-qPCR and western blotting demonstrated that the expression
levels of CUL4A were significantly higher in liver cancer tissues
compared with in paracancerous tissues (P<0.05), with similar
patterns observed in the 3 different patients (Fig. 1A and B). The phenomenon was further
confirmed by immunohistochemical staining as the results indicated
strong positive staining of CUL4A in the liver cancer tissues,
while staining of CUL4A in the paracancerous tissues was weak, and
quantification of staining demonstrated significantly higher CUL4A
expression in liver cancer tissues compared with paracancerous
tissue (Fig. 1C). Therefore, these
results indicated that human liver cancer tissues exhibited higher
expression of CUL4A compared with normal tissue.
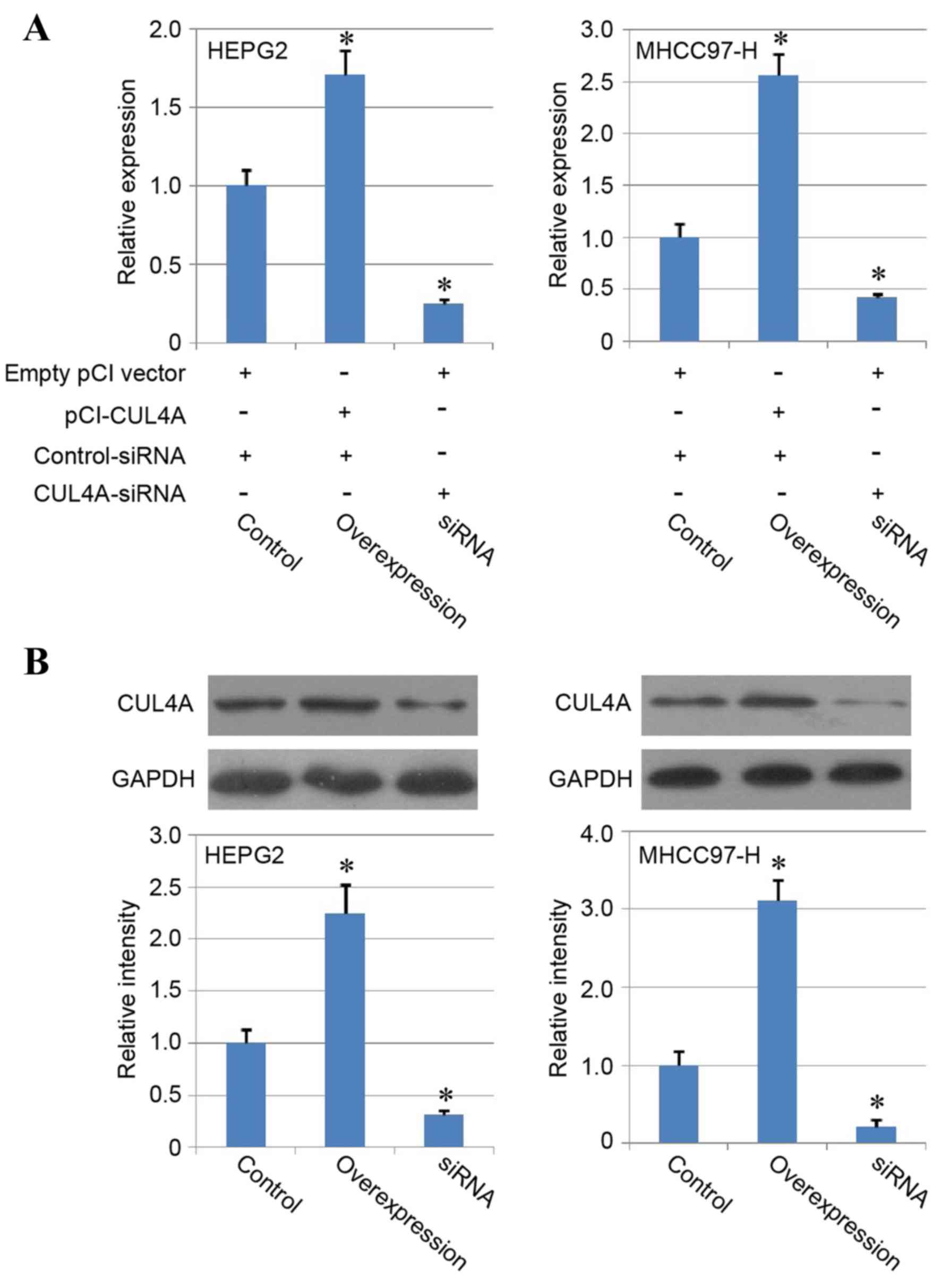
CUL4A overexpression and knockdown in
liver cancer cell lines
To further investigate the role of CUL4A in the
biological function of human liver cancer cells, HEPG2 and MHCC97-H
cancer cell lines were employed in the present study.
Overexpression of CUL4A was induced with pCI vector transfection
(overexpression group) and knockdown of CUL4A in liver cancer cells
was performed with using siRNA (siRNA group). Following
transfection, the mRNA and protein expression of CUL4A was
determined with RT-qPCR and western blotting, respectively, to
evaluate the overexpression and siRNA efficiency, and the results
confirmed significant upregulation of CUL4A in the overexpression
group and downregulation of CUL4A expression in the siRNA group
compared with the control group (P<0.05; Fig. 2A and B), indicating the enhancing
effect of the pCI-CUL4A vector and inhibiting function of the siRNA
on CUL4A expression in liver cancer cell lines.
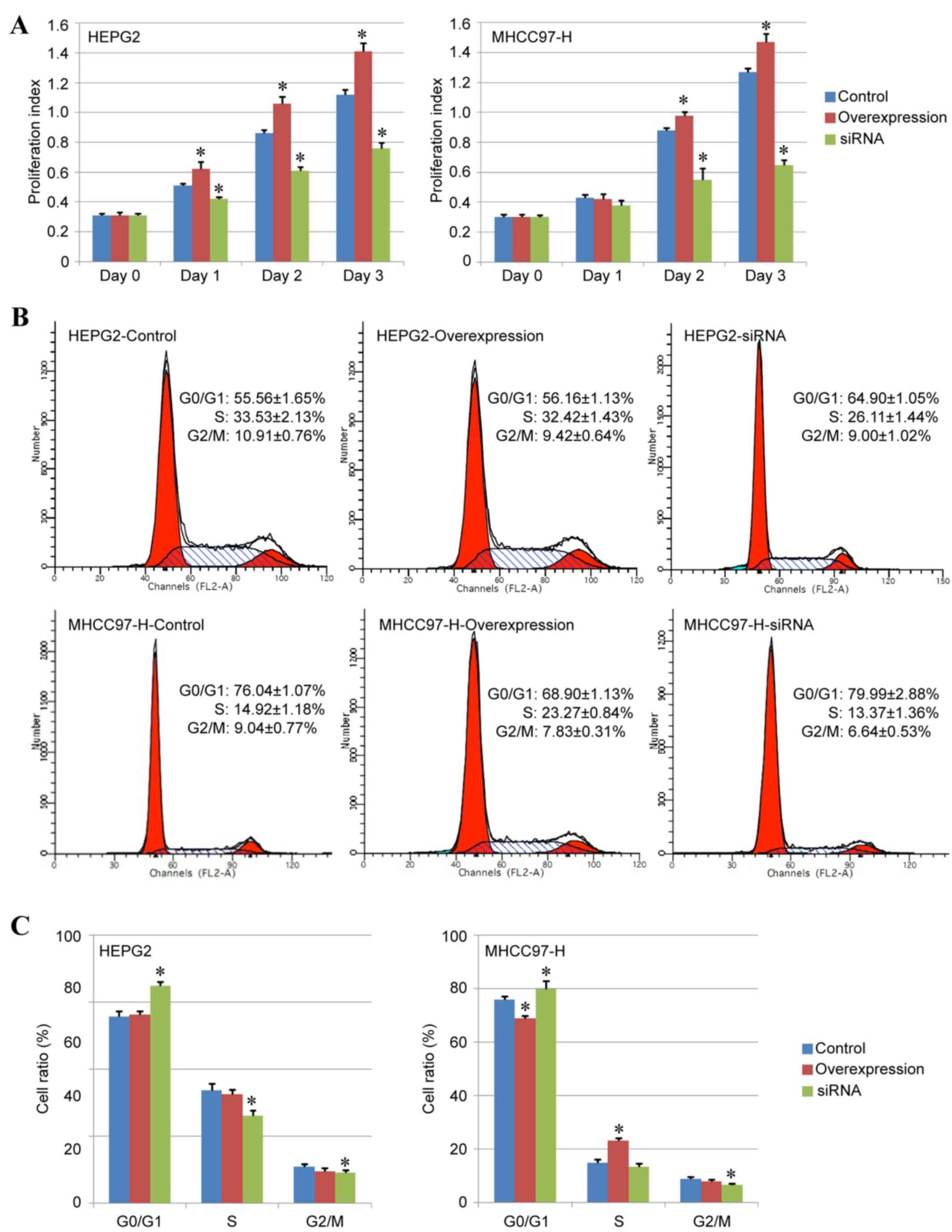
Effect of CUL4A on liver cancer cell
proliferation
The present study analyzed the differences in cell
proliferation ability among the control, overexpression and siRNA
groups. The CCK-8 detection assay indicated that the proliferation
index of the overexpression group was higher compared with the
control group, while the index of the siRNA group was significantly
lower compared with the control group, indicating that the
expression of CUL4A may be essential for liver cancer cell
proliferation and downregulation of CUL4A expression may inhibit
the proliferation ability of liver cancer cells (Fig. 3A).
In addition, the effects of CUL4A overexpression and
CUL4A-siRNA on the cell cycle of HEPG2 and MHCC97-H cancer cells
were analyzed by fluorescence-activated cell sorting. The results
demonstrated that compared with the control group, CUL4A
overexpression reduced the percentage of
G0/G1 phase and increased the percentage of S
phase MHCC97-H cells (P<0.05; Fig.
3B and C), but exhibited no notable effects in HEPG2 cells
(Fig. 3B and C). Conversely,
treatment with CUL4A-siRNA increased the percentage of
G0/G1 phase cells and decreased the
percentage of S phase and G2/M phase cells compared with the
control group (P<0.05; Fig. 3B and
C), therefore inhibiting cell proliferation ability in both
liver cancer cell lines.
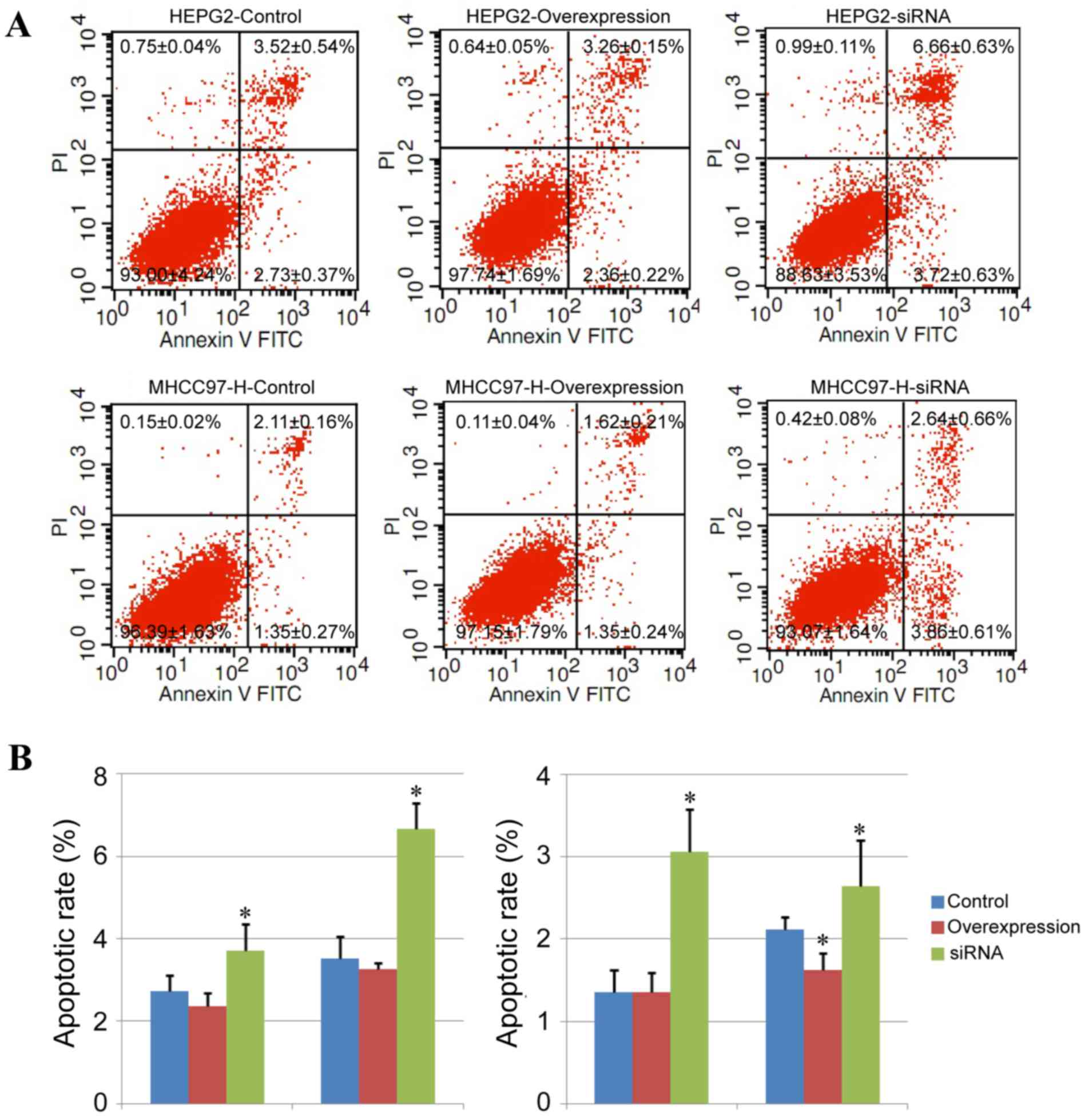
Effect of CUL4A on liver cancer cell
apoptosis
The degree of cell apoptosis in different groups was
further analyzed with Annexin V-FITC/PI double staining in the
present study. In the HEPG2 and MHCC97-H cell lines, the
downregulation of CUL4A expression led to an increased percentage
of early-stage apoptotic cells (Annexin V-FITC-positive and
PI-negative cells; P<0.05) and late-stage apoptotic cells
(Annexin V-FITC-positive and PI-positive cells; P<0.05) compared
with the control group (Fig. 4A and
B). Additionally, CUL4A overexpression only decreased the
percentage of late-stage apoptotic cells in MHCC97-H cells weakly
(P<0.05), with no notable alterations induced by CUL4A
overexpression in HEPG2 cells (Fig. 4A
and B).
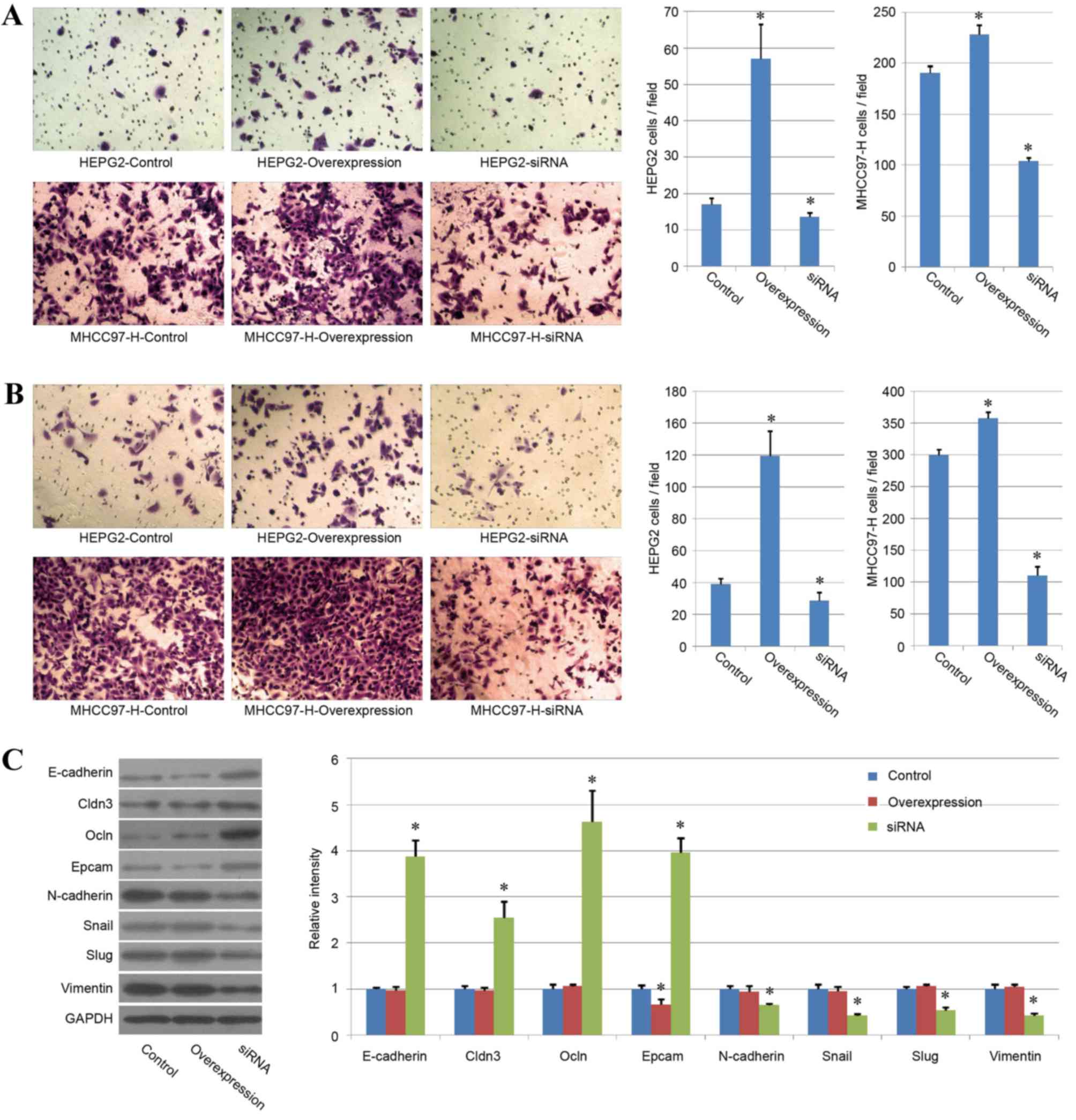
CUL4A promotes liver cancer cell
migration and invasion
The present study also analyzed the variations in
cell migration and invasion ability among the different treatment
groups. The same tendencies were observed for the results of both
evaluations. CUL4A overexpression increased cell migration and
invasion ability in the two liver cancer cell lines and the
CUL4A-siRNA group exhibited a significant reduction in cell
migration and invasion ability, compared with the control group
(P<0.05; Fig. 5A and B). These
results indicated a key role of CUL4A in liver cancer migration and
invasion.
As EMT has been regarded as the key process in
cancer cell migration and invasion (24), the expression of certain key genes
associated with EMT was analyzed by western blotting to confirm the
effect of CUL4A in HEPG2 cells. The present study demonstrated that
the protein expression of epithelial genes (E-cadherin, Cldn3, Ocln
and Epcam) was upregulated in the CUL4A-siRNA group and the
expression of mesenchymal genes (N-cadherin, Snail, Slug and
vimentin) was reduced in the CUL4A siRNA group (P<0.05; Fig. 5C), compared with the control group,
indicating that CUL4A may affect liver cancer cell migration and
invasion by regulating EMT. However, the overexpression of CUL4A
exhibited few effects on the expression of most mesenchymal or
epithelial genes.
Discussion
Previous studies have indicated that CUL4A serves an
important function in the progression of various cancer types
(10,12). However, to the best of our
knowledge, previous studies have not extensively investigated the
role of CUL4A in human liver cancer. Recently, an inverse
correlation was reported between the expression of CUL4A and
patient survival, and a positive correlation with lymphatic and
venous invasion (13). In
addition, the expression of CUL4A in liver cancer tissues was
associated with patient HBeAg status, and knockdown of CUL4A
ameliorated the motility of liver cancer cell lines by regulating
the expression of EMT-associated molecules (13). Recently, another group identified
that a novel lncRNA, uc.134, may repress liver cancer progression
by inhibiting the CUL4A-mediated ubiquitination of the LATS1
protein, indicating that the application of uc.134 lncRNA may offer
a promising treatment approach for liver cancer and that CUL4A may
have an important role in liver cancer progression (14). In the present study, CUL4A was
observed to exhibit increased expression in human liver cancer
tissues compared with in paracancerous tissues. In addition, the
inhibition of CUL4A using siRNA led to the reduction of cell
proliferation, cell migration and invasion, and enhanced the
percentage of cell apoptosis, indicating the key function of CUL4A
in liver cancer progression.
However, a detailed understanding of the molecule
mechanism of CUL4A function in human liver cancer remains unclear.
For example, further investigation is required to determine why the
expression level of CUL4A may be upregulated in liver cancer.
Numerous complex genetic and epigenetic alterations were previously
reported in hepatocytes during liver cancer progression, and those
alterations led to transformation of normal hepatocytes and
resulted in hepatocarcinogenesis (6). Thus, other factors may contribute to
the upregulation of CUL4A in liver cancer progression. HBV
infection has been considered as the most important cause of liver
cancer worldwide. Recently, one study indicated that the majority
of liver cancer cases were HBsAg-positive and that HBV infection
directly upregulated the expression of CUL4A in liver cancer cells,
indicating the regulatory role of HBV on the expression of CUL4A
(13). However, the exact
mechanisms underlying the function of HBV in CUL4A regulation
require in vitro and in vivo investigation.
In present study, the results demonstrated that
CUL4A overexpression increased the proliferation of human liver
cancer cell lines, while the downregulation of CUL4A suppressed
cell proliferation and enhanced cell apoptosis. However, different
cell lines revealed different results in the present study. For
example, CUL4A overexpression increased the percentage of S phase
and reduced the percentage of G0/G1 cells,
and decreased the percentage of late-stage apoptotic cells in
MHCC97-H cells, but demonstrated no notable effects in HEPG2 cells.
These varying effects require further investigation. Thus, the
basal expression levels of CUL4A in various cell lines may be
different and cancer cells with high expression levels of CUL4A may
exhibit a certain degree of tolerance to CUL4A overexpression.
However, investigation of additional cell lines is required to
confirm this hypothesis, as well as the mechanism underlying this
phenomenon.
CUL4A overexpression did not result in significant
alteration of mesenchymal or epithelial gene expression. However,
CUL4A overexpression significantly increased cell migration and
invasion ability in the two liver cancer cell lines. This may be
due to the high expression of mesenchymal genes and low expression
of epithelial genes that is typically observed in liver cancer cell
lines (25–27). Therefore, CUL4A overexpression may
have been unable to increase mesenchymal or decrease epithelial
gene expression further. However, CUL4A overexpression still
increased cell viability by promoting cell proliferation and
inhibiting cell apoptosis. Cell migration and invasion ability was
also promoted, likely through other signaling pathways. This
hypothesis requires further investigate to elucidate the mechanisms
of CUL4A overexpression on migration, invasion and viability.
The present study confirmed the association between
the expression of CUL4A and human liver cancer, and indicated that
CUL4A may represent a novel target in the treatment of human liver
cancer. Further analysis for each pathway associated with CUL4A
expression and an enhanced understanding of the regulatory
mechanism of those genes in various cancer cells may contribute to
the development of novel drugs or gene therapy methods for the
treatment of patients with liver cancer and potentially other types
of cancer.
In conclusion, the present study indicated that the
mRNA and protein expression levels of CUL4A were markedly higher in
human liver cancer tissues compared with human paracancerous
tissues. The overexpression of CUL4A in human liver cancer cells
increased the cell proliferation, cell migration and invasion, and
reduced the percentage of cell apoptosis, while CUL4A knockdown
exhibited opposing effects. The results of the present indicated
the key function of CUL4A expression in liver cancer, as well as
the potential of CUL4A in the diagnosis and treatment of human
liver cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Science
Research Foundation of Yunnan Provincial Department of Education
(grant no. 2013C239) and the Postdoctoral Support Research Project
of Kunming Human Resources and Social Security Bureau.
Availability of data and materials
The analyzed datasets generated during the study are
available from the corresponding author on reasonable request.
Author's contributions
LL conceived, designed and supervised the
experiments. GC, XZ, ZT, DW, DL, PZ and JC performed the
experiments. The data were analyzed by GC, JC, FW and QL. GC and LL
contributed the reagents, materials and analysis tools. GC and LL
wrote the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Committee on
the Ethics of Animal Experiments and Human Subject Research of the
First People's Hospital of Kunming. All volunteers involved in the
present study provided written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Altekruse SF, Henley SJ, Cucinelli JE and
McGlynn KA: Changing hepatocellular carcinoma incidence and liver
cancer mortality rates in the United States. Am J Gastroenterol.
109:542–553. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tang A, Hallouch O, Chernyak V, Kamaya A
and Sirlin CB: Epidemiology of hepatocellular carcinoma: Target
population for surveillance and diagnosis. Abdom Radiol (NY).
43:13–25. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xu IM, Lai RK, Lin SH, Tse AP, Chiu DK,
Koh HY, Law CT, Wong CM, Cai Z, Wong CC and Ng IO: Transketolase
counteracts oxidative stress to drive cancer development. Proc Natl
Acad Sci USA. 113:E725–E734. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nishida N and Goel A: Genetic and
epigenetic signatures in human hepatocellular carcinoma: A
systematic review. Curr Genomics. 12:130–137. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu M, Jiang L and Guan XY: The genetic
and epigenetic alterations in human hepatocellular carcinoma: A
recent update. Protein Cell. 5:673–691. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nishida N and Kudo M: Clinical
significance of epigenetic alterations in human hepatocellular
carcinoma and its association with genetic mutations. Dig Dis.
34:708–713. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yasui K, Arii S, Zhao C, Imoto I, Ueda M,
Nagai H, Emi M and Inazawa J: TFDP1, CUL4A, and CDC16 identified as
targets for amplification at 13q34 in hepatocellular carcinomas.
Hepatology. 35:1476–1484. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hori T, Osaka F, Chiba T, Miyamoto C,
Okabayashi K, Shimbara N, Kato S and Tanaka K: Covalent
modification of all members of human cullin family proteins by
NEDD8. Oncogene. 18:6829–6834. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sharma P and Nag A: CUL4A ubiquitin
ligase: A promising drug target for cancer and other human
diseases. Open Biol. 4:1302172014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dohna M, Reincke M, Mincheva A, Allolio B,
Solinas-Toldo S and Lichter P: Adrenocortical carcinoma is
characterized by a high frequency of chromosomal gains and
high-level amplifications. Genes Chromosomes Cancer. 28:145–152.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu Y, Wang Y, Ma G, Wang Q and Wei G:
CUL4A is overexpressed in human pituitary adenomas and regulates
pituitary tumor cell proliferation. J Neurooncol. 116:625–632.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pan Y, Wang B, Yang X, Bai F, Xu Q, Li X,
Gao L, Ma C and Liang X: CUL4A facilitates hepatocarcinogenesis by
promoting cell cycle progression and epithelial-mesenchymal
transition. Sci Rep. 5:170062015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ni W, Zhang Y, Zhan Z, Ye F, Liang Y,
Huang J, Chen K, Chen L and Ding Y: A novel lncRNA uc.134 represses
hepatocellular carcinoma progression by inhibiting CUL4A-mediated
ubiquitination of LATS1. J Hematol Oncol. 10:912017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
López-Terrada D, Cheung SW, Finegold MJ
and Knowles BB: Hep G2 is a hepatoblastoma-derived cell line. Hum
Pathol. 40:1512–1515. 2009. View Article : Google Scholar
|
|
16
|
Liu P, Feng Y, Dong D, Liu X, Chen Y, Wang
Y and Zhou Y: Enhanced renoprotective efect of IGF-1 modifed human
umbilical cord-derived mesenchymal stem cells on gentamicin-induced
acute kidney injury. Sci Rep. 6:202872016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu P, Cai J, Dong D, Chen Y, Liu X, Wang
Y and Zhou Y: Effects of SOX2 on proliferation, migration and
adhesion of human dental pulp stem cells. PloS One.
10:e01413462015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tao S, Liu P, Luo G, de la Vega Rojo M,
Chen H, Wu T, Tillotson J, Chapman E and Zhang DD: p97 Negatively
regulates NRF2 by extracting ubiquitylated NRF2 from the KEAP1-CUL3
E3 complex. Mol Cell Biol. 37:e00660–e00616. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cai J, Zhang Y, Liu P, Chen S, Wu X, Sun
Y, Li A, Huang K, Luo R, Wang L, et al: Generation of tooth-like
structures from integration-free human urine induced pluripotent
stem cells. Cell Reg (Lond). 2:62013.
|
|
21
|
Liu P, Feng Y, Dong C, Yang D, Li B, Chen
X, Zhang Z, Wang Y, Zhou Y and Zhao L: Administration of BMSCs with
muscone in rats with gentamicin-induced AKI improves their
therapeutic efficacy. PloS One. 9:e971232014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu P, Feng Y, Dong C, Liu D, Wu X, Wu H,
Lv P and Zhou Y: Study on therapeutic action of bone marrow derived
mesenchymal stem cell combined with vitamin E against acute kidney
injury in rats. Life Sci. 92:829–837. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao L, Feng Y, Chen X, Yuan J, Liu X,
Chen Y, Zhao Y, Liu P and Li Y: Effects of IGF-1 on neural
differentiation of human umbilical cord derived mesenchymal stem
cells. Life Sci. 151:93–101. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Heerboth S, Housman G, Leary M, Longacre
M, Byler S, Lapinska K, Willbanks A and Sarkar S: EMT and tumor
metastasis. Clin Transl Med. 4:62015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jayachandran A, Dhungel B and Steel JC:
Epithelial-to-mesenchymal plasticity of cancer stem cells:
Therapeutic targets in hepatocellular carcinoma. J Hematol Oncol.
9:742016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Panebianco C, Saracino C and Pazienza V:
Epithelial-mesenchymal transition: Molecular pathways of hepatitis
viruses-induced hepatocellular carcinoma progression. Tumour Biol.
35:7307–7315. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ogunwobi OO and Liu C: Therapeutic and
prognostic importance of epithelial-mesenchymal transition in liver
cancers: Insights from experimental models. Crit Rev Oncol Hematol.
83:319–328. 2012. View Article : Google Scholar : PubMed/NCBI
|