Introduction
Diabetes mellitus is one of the principal risk
factors for the development of vascular complications. Several
studies have highlighted the involvement of platelets, as a
component of the coagulation system, in the development of
vasculopathies, which is a multifactorial process (1–3).
Individuals with type 2 diabetes mellitus may also be characterized
by a high incidence of blood platelet dysfunction (4,5).
As the energy produced by mitochondria is required
for fundamental processes of platelet functioning, platelet
mitochondria may be regarded as a factor for the development of
disorders of platelet adhesion and aggregation and vasculopathies
in diabetes mellitus. However, the potential involvement of
platelet mitochondria in mechanisms underlying these dysfunctions
remains unclear. In platelets, which are anucleated cells, the
mitochondria are the primary source of adenosine 5′-triphosphate
(ATP) production from acetyl-coenzyme A oxidation and oxidative
phosphorylation (6,7). Adenine nucleotides are required for
platelet activation and aggregation, and for platelet bioenergetic
maintenance (1,6,8). It
has been established that ATP itself or its breakdown to adenosine
5′-diphosphate (ADP) may activate platelets (8). In addition, intracellular ADP and ATP
levels have an essential role in blood platelet activation though
the regulation of intracellular calcium (9).
A number of reports have indicated that
hyperglycemia may increase reactive oxygen species production and
subsequent oxidative stress (10–12).
In addition, hyperglycemia-induced oxidative stress is a principal
contributor to mitochondrial damage in cells (13). Although the majority of
mitochondrial proteins are encoded by the nuclear genome, numerous
important proteins involved in oxidative phosphorylation are
encoded by mitochondrial DNA (mtDNA) such as NADH dehydrogenase 1,
cytochrome b, cytochrome c oxidase I and ATP synthase 6. Increased
oxidative stress reduces ability to repair and histone protection,
which affects the mitochondria and damages mtDNA. Once the
proportion of mutated mtDNA rises above a threshold, mitochondrial
membrane potential (ΔΨm), mitochondrial ATP synthesis and the
cellular ATP/ADP ratio decrease. If ATP demands are not met, this
may increase the risk of a number of pathologies, including cell
death and organ failure, are known to be associated with disease or
aging (14).
At least 20 different types of deletions in mtDNA
have been documented to accumulate in aging human tissues (15). Among these deletions, a common
marker of mtDNA damage associated with oxidative stress is the
4,977-base pair (bp) common deletion (mtDNA4977)
(16). The deleted mtDNA for this
mutation contains all or part of the genes encoding four
polypeptides for complex I, one for complex IV, two for complex V
involved in oxidative phosphorylation, and five tRNA genes. It has
been demonstrated that the mtDNA4977 deletion is
associated with metabolic machinery defects that are acquired
during aging. Furthermore, there may be a correlation between an
accumulation of mtDNA4977 deletions and reduced ATP
production in a number of tissues (14,17).
The mtDNA4977 deletion accumulates faster
in tissues with greater metabolic activity and minimal cell
turnover, including those in the brain and heart (18). Elevated levels of these deletions
have also been detected in peripheral leucocytes, which have a high
turnover activity, from patients with a variety of diseases
associated with oxidative stress, including type 2 diabetes
mellitus (18). The frequency of
mtDNA4977 deletions may be elevated in platelets, as
well as whole blood cells, in response to increased oxidative
damage in diabetes mellitus. This may result in bioenergetic
impairment and platelet functional alteration. To the best of the
authors' knowledge, the frequency and proportion of deleted mtDNA
has not yet been detected in the platelets of patients with type 2
diabetes mellitus.
Taking these previous findings into account, the aim
of the present study was to assess the mitochondrial common
deletion in the platelets of patients with type 2 diabetes
mellitus, in the context of mitochondrial function and platelet
reactivity. To make this assessment, the mtDNA4977
deletion frequency, mtDNA copy number, ΔΨm, adenine nucleotide
level and P-selectin (CD62p) expression was measured in the
peripheral blood platelets of patients with type 2 diabetes, with
and without complications. The obtained findings were compared with
data from age- and body mass index (BMI)-matched healthy control
subjects. Data presented indicate that the mtDNA4977
deletion occurred in platelets more frequently in patients with
diabetes and that there is a requirement for further studies to
explore its clinical significance.
Materials and methods
Study population and collection of
specimens
The present study was conducted on outpatients
receiving anti-diabetic treatment at the Istanbul Training and
Research Hospital (Istanbul, Turkey) between June 2015 and February
2016. This study included 66 patients (31 men, 35 women) with
diabetes and 23 (13 men, 10 women) age- and BMI-matched healthy
subjects. Patients with type 2 diabetes mellitus were divided into
three groups based on plasma glycated hemoglobin (HbA1c) levels,
and the presence or absence of chronic diabetic complications:
Group I, good glycemic control (HbA1c <7) without complications;
Group II, poor glycemic control (HbA1c ≥7) without complications;
and Group III, poor glycemic control (HbA1c ≥7) with complications.
The patients were evaluated for micro- and macrovascular
complications via clinical and laboratory tests. Patients with
diabetic complications had at least one of the following:
Neuropathy, peripheral vascular disease, cerebrovascular disease,
nephropathy or retinopathy.
The inclusion criteria for patients and healthy
volunteers were: Age range, 45–70 years; BMI, ≥22 kg/m2;
and a history of type 2 diabetes lasting ≥5 years. Exclusion
criteria for the patients were: A medical history including
hematological disorders that affected platelet count or function,
malignant disease, hypertension, chronic inflammatory disease,
infectious disease or autoimmune disease. For healthy control
subjects, the exclusion criteria were: A known history of chronic,
inflammatory or malignant disease, and the use of antithrombotic
drugs. All patients provided written informed consent. The present
study was approved by the Istanbul Training and Research Hospital
Ethics Committee (approval no. 339/2013; Istanbul, Turkey), and was
performed according to the criteria set out by the Declaration of
Helsinki.
Platelet preparation
Platelets from peripheral blood were prepared by
differential centrifugation, according to the following method:
Peripheral blood (10 ml) was collected in a vacutainer tube
containing containing 3.8% sodium citrate and was processed within
30 min of collection. Samples were centrifuged at 160 × g for 10
min at 22°C, and the platelet-rich plasma (PRP) was carefully
aspirated, leaving behind the erythrocytes, prior to centrifugation
again at 160 × g for 5 min at 22°C to remove the leukocytes. The
PRP was diluted with PBS (1:1) and carefully layered onto the
Histopaque-1077 (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)
(2:1) for leukocyte depletion. Following centrifugation at 350 × g
for 20 min at 22°C, the opaque interface and the upper layer were
transferred into a clean conical centrifuge tube and diluted with
PBS. Subsequently, the platelet suspension was filtered with a 6-µm
pluriStrainer (pluriSelect Life Science, Leipzig, Germany), which
was put onto a separate sterile centrifuge tube, using a connector
ring to remove traces of contaminating leukocytes. Following
centrifugation for 10 min at 1,500 × g at 22°C, the platelet pellet
was washed with PBS. The platelet counts of prepared samples were
analyzed with a Cell-Dyn Coulter counter (Abbott Pharmacuetical Co.
Ltd., Lake Bluff, IL, USA).
mtDNA extraction
mtDNA was isolated from washed platelets with the
BioVision mitochondrial DNA isolation kit (cat. no. K280-50;
BioVision, Inc., Milpitas, CA, USA). The concentration of DNA was
determined with a NanoDrop™ 2000 spectrophotometer (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and the samples were stored at
−80°C until further experimentation.
Analysis of the amount of mtDNA and
the mtDNA4977 bp deletion by polymerase chain reaction
(PCR)
The levels of mtDNA4977 deletion and
mitochondrial reference fragment in mtDNA isolated from platelet
samples were determined by fluorescence-based quantitative (q)PCR
as described previously (19–21),
with certain modifications. The fragment with no deletions and the
conservative region of mtDNA was taken as reference for the total
mtDNA. The following primer sets were used: Normal mtDNA
(mtDNAn; internal control) forward,
5′-AACATACCCATGGCCAAC-3′ and reverse, 5′-TCAGCGAAGGGTTGTAGTAGC-3′
(249 bp); and mtDNA4977 deletion forward,
5′-TATGGCCCACCATAATTACCC-3′ and reverse,
5′-AAGCGAGGTTGACCTGTTAGG-3′ (270 bp). Purified PCR products were
cloned into the pGEM-T Easy vector (Promega Corporation, Madison,
WI, USA), according to the manufacturer's protocol, and
dose-dependent plasmid-constructed (mtDNAn and
mtDNA4977) standards were used in each run of PCR for
qPCR optimization and sensitivity analysis. The plasmids were
further confirmed by Sanger sequencing method using the ABI Prism
3130XL Genetic Analyzer and the BigDye Terminator v3.1 Ready
Reaction Kit (Applied Biosystems; Thermo Fisher Scientific, Inc.).
DNA samples were subjected to qPCR with the CFX Connect™ Real-Time
PCR Detection system (Bio-Rad Laboratories, Inc., Hercules, CA,
USA) beginning with an initial denaturation step at 95°C for 10
min, followed by for 40 cycles of 94°C for 15 sec and 60°C for 1
min, using the iTaq Universal SYBR Green Supermix (Bio-Rad
Laboratories, Inc.). PCR amplifications were performed in a 20-µl
reaction volume, containing 20 ng DNA template and a 10 nmol/l
concentration of each primer. Average quantitation cycle (Cq)
values were calculated to determine the mtDNA content and deletion
load in samples (22). Each
measurement was repeated two or three times and normalized against
a serial dilution of the corresponding plasmid clones at a known
concentration. The quantity of each target gene in the samples was
subsequently calculated according to the corresponding standard
curve. The formula published by the Genomics and Sequencing Center
of the University of Rhode Island (Kingston, RI, USA; cels.uri.edu/gsc/cndna.html) was used to
calculate the mtDNA copy number. The formula used was: Number of
copies=(amount of DNA ×6.022×1023)/(length of amplicon
×1×109 ×650). The level of mtDNA4977 deletion
was expressed as a percentage ratio of the deleted mtDNA copy
number with respect to the total mtDNA copy number.
ATP/ADP assay
Frozen platelet cell samples were rapidly thawed and
the cell debris was pelleted by centrifugation (10,000 × g for 3
min at room temperature). The supernatant was diluted in 3%
perchloric acid. Samples were subsequently neutralized with 75 µl
ice-cold 2M KOH, 2 mM Na2 EDTA, 50 mM
3-(N-morpholino)propanesulfonic acid, which was incubated on ice
for 10 min. The precipitate was pelleted by centrifugation (10,000
× g for 1 min at room temperature) and the ADP/ATP levels of the
supernatant were determined using the EnzyLight ADP/ATP Ratio Assay
kit (BioAssay Systems, Hayward, CA, USA), according to the
manufacturer's protocol, by comparison with the appropriate
standards. Protein concentrations in samples were determined using
the Bio-Rad DC™ Protein Assay kit (Bio-Rad Laboratories, Inc.) and
ADP/ATP content was expressed as nmol/mg protein.
ΔΨm assay
ΔΨm was assessed using a JC-10 fluorometric assay
kit (AAT Bioquest, Inc., Sunnyvale, CA, USA) according to the
manufacturer's protocol. JC-10 dye loading solution (20 µM) was
added to each well and cells were incubated for 30 min at 37°C. Red
fluorescent JC-10 aggregates were detected at 540 nm excitation and
590 nm emission. Green fluorescence due to the ΔΨm collapse was
detected at 490 nm excitation and 525 nm emission. The ratio of
fluorescence intensities on emission at 525/590 was used to
determine mitochondrial membrane depolarization. Data were recorded
in relative fluorescence units and normalized to the total protein
amount using the Bradford assay (Sigma-Aldrich; Merck KGaA). The
JC-10 aggregate/monomer ratio was proportional to ΔΨm.
Flow cytometry for platelet CD62p
expression
Platelet CD62p expression levels, as a marker of
platelet activation, were measured under the basal and activated
(with 1 µM ADP; Chrono-Log Corporation, Havertown, PA, USA; for 5
min at room temperature) conditions by flow cytometry. Blood was
collected into collection tubes containing 3.8% sodium citrate. PRP
was obtained from tube following 10 min of centrifugation at 160 ×
g at 22°C, and 50 µl PRP was diluted with 450 µl PBS (pH 7.4). A
volume of 100 µl of the mixture was incubated with fluorescein
isothiocyanate (FITC)-conjugated CD62p antibodies (1:100; cat. no.
555523; BD Biosciences, San Jose, CA, USA) and phycoethryin
(PE)-conjugated platelet glycoprotein IIb of IIb/IIIa complex
antibodies (1:100; cat. no. 55467; BD Biosciences, San Jose, CA,
USA) for 20 min at room temperature in the dark. As control
experiments, platelets were incubated with PE-conjugated
non-specific mouse immunoglobulin G1 (1:100; cat. no. 406608;
BioLegend, Inc., San Diego, CA, USA). Following gentle mixing,
samples were washed once in 2 ml PBS and centrifuged at 1,500 × g
for 5 min at 22°C. The supernatant was discarded and samples were
fixed with 1% paraformaldehyde (500 µl). The fixed samples were
analyzed at room temperature for up to 4 h, and data acquisition
was done by flow cytometry. Platelet populations were sorted by
side-scatter (SSC) and forward scatter (FSC), and the FL1 channel
detected the FITC fluorescence intensity. Data from the
fluorescence-activated cell sorting process was further analyzed
using WinMDI version 2.8. software (Scripps Research Institute,
USA). Data were expressed as the percentage of positive events.
Statistical analysis
Differences between groups with normally distributed
data were tested using one-way analysis of variance (ANOVA)
followed by Tukey's post-hoc test. Kruskal-Wallis with Dunn's
post-hoc test was used for non-parametric data analyses. Each
experiment was repeated ≥2 times, and data were expressed as the
mean ± standard deviation. P≤0.05 was considered to indicate a
statistically significant difference. Spearman's correlation
coefficients were estimated to determine the correlation between
variables. Analyses were performed using the GraphPad Prism 5.0
statistical package (GraphPad Software, Inc., La Jolla, CA, USA).
Cv values were calculated as: % Cv=(standard deviation of
measurement/measurement average) ×100.
Results
Clinical and laboratory
characteristics among the studied groups with type 2 diabetes
Clinical characteristics and laboratory data of
patients with type 2 diabetes mellitus and healthy subjects are
presented in Table I. Age, sex,
BMI, diabetic duration, fasting plasma glucose and HbA1c values
were used as a clinical characteristic parameters for patients and
healthy subjects. No significant differences were observed in the
age and BMI values among the patient and control groups
(P>0.05). Fasting plasma glucose levels in type 2 diabetic
patients in groups II and III were significantly increased compared
with those in group I (HbA1c <7; P<0.001). In addition, the
fasting plasma glucose levels of all patient groups were
significantly increased compared with the control group
(P<0.001). HbA1c values in type 2 diabetic patients in groups II
and III were significantly higher compared with group I
(P<0.001). All patient groups had significantly higher HbA1c
values, compared with those in the control group (P<0.001).
 | Table I.Clinical characteristics and
laboratory data of patients with diabetes and healthy subjects. |
Table I.
Clinical characteristics and
laboratory data of patients with diabetes and healthy subjects.
| Clinical
characteristics | Group I | Group II | Group III | Healthy
subjects |
|---|
| Subjects, n | 20 | 20 | 26 | 23 |
| Age, years | 59.14±7.83 | 58.61±7.49 | 60.27±7.29 | 58.38±6.82 |
| Sex,
male/female | 9/11 | 8/12 | 14/12 | 13/10 |
| BMI,
kg/m2 | 25.11±2.80 | 25.85±2.27 | 26.15±2.55 | 24.95±2.65 |
| Diabetic duration,
years |
7.51±1.62a |
9.45±2.24c |
11.31±3.81d | N/A |
| Fasting plasma
glucose, mg/dl | 118±10b,e | 144±25c,e | 162±35d,e | 85±7 |
| HbA1c, % |
5.96±0.74b,e |
7.88±0.70c,e |
9.27±1.20d,e | 5.07±0.59 |
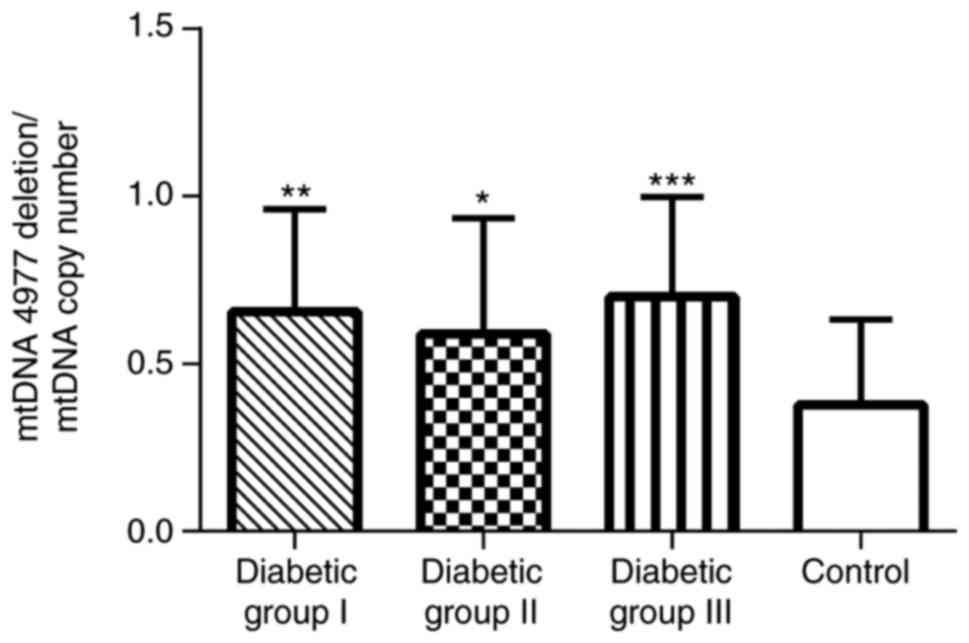
mtDNA4977 bp deletion and
mtDNA copy number in the blood platelets of patients with type 2
diabetes
A qPCR assay was performed in order to measure the
frequency of mtDNA4977 deletion. This mutation was
detected in all examined platelet samples, with the exception of
two diabetic patients and one non-diabetic subject. Although the
mtDNA4977 carriers were identified at a similar
frequency in patients with type 2 diabetes mellitus (96.9%) and
controls (95.6%), the level of mtDNA4977 deletion was
higher in each of the three patient groups, compared with the
control group (Fig. 1), as
demonstrated using the Kruskal-Wallis test and Dunn's multiple
comparison test. However, the levels did not differ significantly
among the three subgroups of the patients.
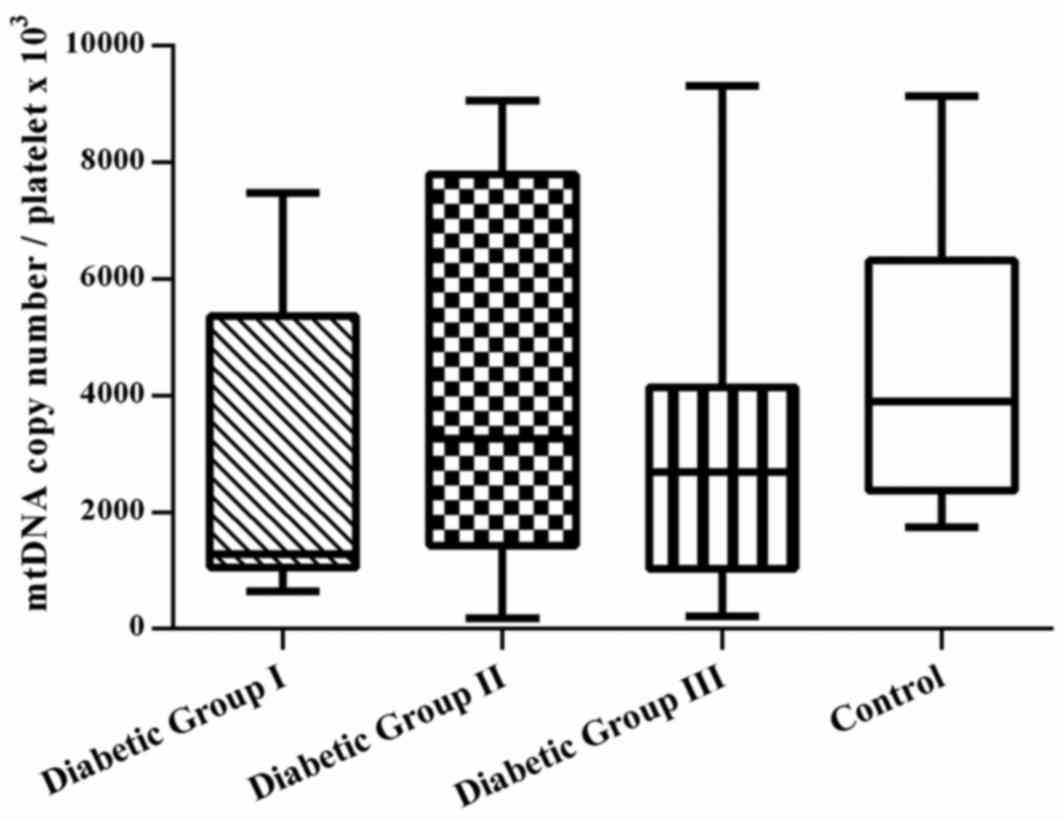
Notably, the coefficient of variation (CV) value for
mtDNA4977 deletion/mtDNA copy number was higher in the
healthy control group, compared with the patient groups (group I,
48%; group II, 59%; group III, 43%; and control group, 75%).
Differences in mitochondrial DNA copy number were not observed in
platelets among the groups. However, the range of CV values
observed in the patient groups was larger (76–102%; Fig. 2).
ATP/ADP levels in platelets
As presented in Table
II, no significant differences in platelet ATP and ADP content
were observed among the groups (P>0.05). In addition, relative
ATP/ADP content was similar among the groups. However, the CV
values of the ATP levels were ~90% for group III.
 | Table II.Adenine nucleotide content in resting
platelets of patients with type 2 diabetes and healthy
subjects. |
Table II.
Adenine nucleotide content in resting
platelets of patients with type 2 diabetes and healthy
subjects.
| Measure | Group I n=20 | Group II n=20 | Group III n=26 | Healthy subjects
n=23 |
|---|
| ATP (nmol/mg
platelet protein) | 22.13±12.92 | 24.70±14.90 | 19.95±18.05 | 26.98±11.78 |
| ADP (nmol/mg
platelet protein) | 14.46±8.04 | 15.33±8.52 | 13.17±5.97 | 17.04±7.89 |
|
ATP/ADPa | 1.56±0.37 | 1.61±0.37 | 1.53±0.42 | 1.61±0.32 |
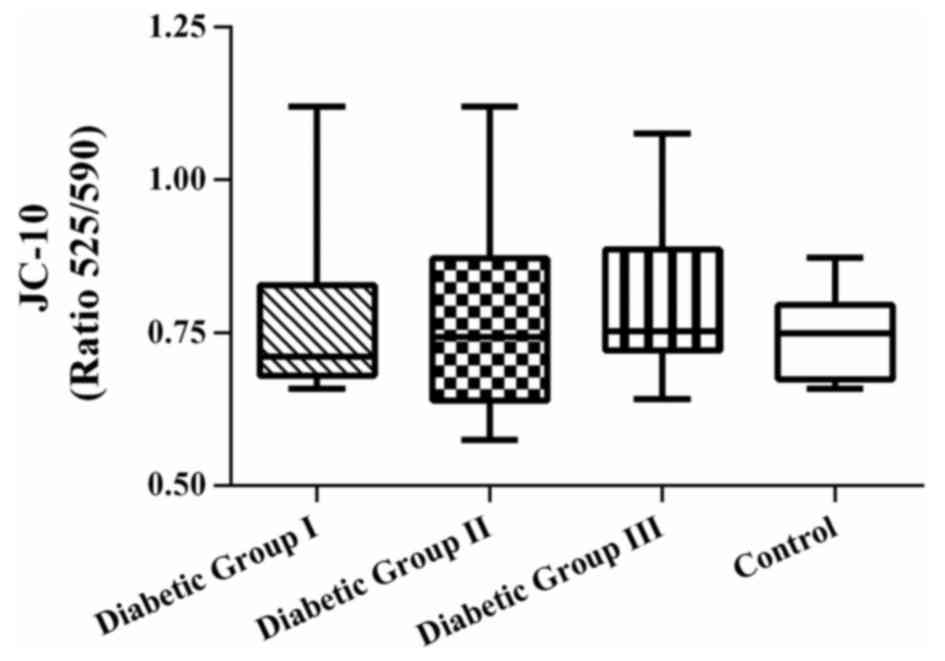
JC-10 assay
ΔΨm was measured as a marker of mitochondrial
bioenergetic efficiency, in addition to ATP. JC-10 is a sensitive
cationic and lipophilic fluorescent probe that was used to monitor
ΔΨm alterations in platelets. The results revealed that there were
no significant differences in the patient groups, compared with the
control group (P>0.05; Fig.
3).
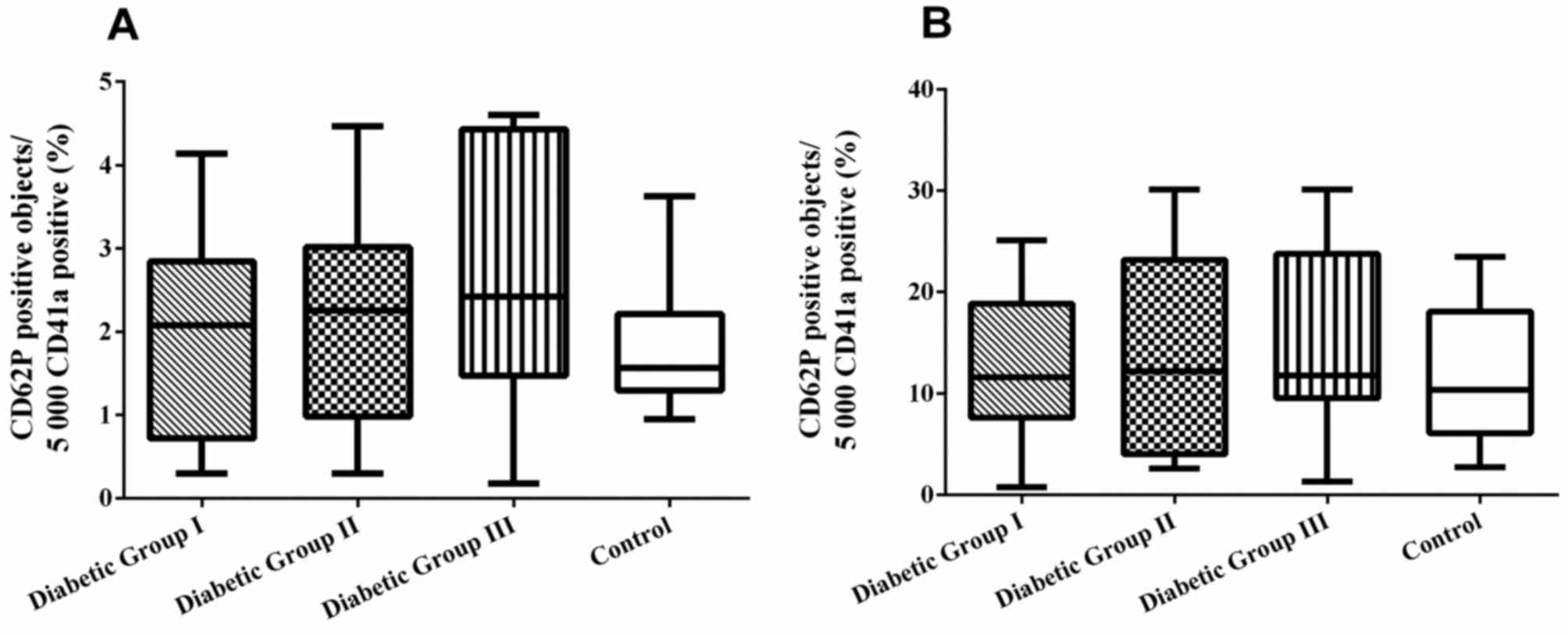
Expression of CD62p
The level of basal platelet activation was measured
by detecting the expression of surface platelet marker, CD62p. The
level of blood platelet responsiveness to ADP (1 µM) was also
measured. The percentage of CD62p+ platelets was not
significantly different among the four groups (P>0.05; Fig. 4A). Furthermore, no significant
differences in the percentage of CD62p expression on ADP-activated
platelets was observed between the patient groups and healthy
control subjects (ANOVA; P>0.05; Fig. 4B).
Associations between
mtDNA4977 deletion and other studied parameters
To better characterize the consequences of increased
mtDNA4977 deletion in platelets, the potential
associations between this parameter and other bioenergetics
parameters were studied, including platelet activation marker
expression and mtDNA copy number. Simple correlation analyses
demonstrated that the deletion level was positively correlated with
mtDNA copy number in patients with complications (Table III). However, no significant
correlations were detected between the accumulation of
mtDNA4977 and with the other studied parameters in
patient groups.
 | Table III.Correlations between
mtDNA4977 deletion and the other detected parameters in
each diabetes subgroup. |
Table III.
Correlations between
mtDNA4977 deletion and the other detected parameters in
each diabetes subgroup.
| Parameter | Group I | Group II | Group III |
|---|
| ATP vs.
mtDNA4977 deletion | P=0.662,
r=−0.214 | P=0.632,
r=−0.176 | P=0.528,
r=0.203 |
| mtDNA copy number
vs. mtDNA4977 deletion | P=0.925,
r=0.024 | P=0.714,
r=0.087 | P=0.274,
r=0.223 |
| ΔΨm vs.
mtDNA4977 deletion | P=0.200,
r=−0.571 | P=0.296,
r=0.370 | P=0.362,
r=0.275 |
| P-selectin with ADP
stimulation vs. mtDNA4977 deletion | P=0.662,
r=−0.264 | p=0.470,
r=0.261 | P=0.793,
r=−0.082 |
Discussion
There is experimental evidence that
hyperglycemia-induced oxidative stress may markedly decrease the
mutation threshold required for mitochondrial dysfunction (23). Additionally, a number of reports
have suggested that an increased mtDNA mutation rate in various
cell types, including the skeletal muscle, kidney and pancreas, is
associated with diabetes mellitus (24–26).
Furthermore, it has been reported that mitochondrial dysfunction
occurs with increased accumulation of various mtDNA mutations,
including mtDNA4977, in the peripheral blood mononuclear
cells of patients with diabetes (18). Over the lifetime of an organism,
nuclear and mitochondrial mutations tend to accumulate in adult
stem cells, including bone marrow stem cells (27). Therefore, mtDNA4977
deletion, a common mutation of the mitochondrial genome, is
expected to be at a high frequency in platelets, which derived from
megakaryocytes, with a shorter lifespan as well as in blood stem
cells.
As hyperglycemia may be a risk factor for increased
mitochondrial mutation accumulation in stem cells, it was
hypothesized that the mtDNA4977 deletion may have
accumulated in the platelets of patients with diabetes. In the
present study, it was demonstrated that mtDNA4977 was
accumulated in peripheral blood platelets in the majority of cases.
Furthermore, the accumulation of the mtDNA4977 deletion
was more pronounced in the platelets of patients with type 2
diabetes compared with healthy subjects. To the best of the
authors' knowledge, this is the first study to suggest a potential
link between the mtDNA4977 deletion and diabetes,
specifically in platelets.
The present data suggested that there was 50–75%
interindividual variability in the levels of mtDNA4977 deletion in
patients with diabetes and healthy subjects. The findings provided
further evidence that the mtDNA4977 deletion may be
common in the population, as demonstrated in previous studies of
numerous cell types, including human leukocytes (18,19,24).
Numerous factors may contribute to the variability in the level of
mtDNA deletion in platelets. According to simple correlation
analysis, age, which was among the candidate causative factors for
this interindividual variation, was not responsible for the
mtDNA4977 deletion level in the present study (data not
shown). This result was consistent with a similar observation in
the platelets of non-medicated patients with early Parkinson's
disease, age-matched healthy controls and young subjects by Sandy
et al (28). In contrast to
our data, Biagini et al (29) did not detect this deletion in
platelets obtained from both young and old individuals. On the
other hand, these studies may have a potential contamination of
platelet samples with other cellular components of blood. In
addition, it is noteworthy that the amount the mtDNA4977
bp deletion in the study of Biagini et al (29) has been detected by
semi-quantitative PCR. Evidence from human and animal studies
demonstrates that abnormal circulating platelet reactivity
typically occurs in pathological states associated with diabetes
mellitus (30–32). Studies have proposed a number of
mechanisms concerning this phenomenon, including abnormalities in
mitochondrial respiration and altered adenine nucleotide metabolism
(6,8,33,34).
Although ADP is the principal promoter of platelet aggregation,
previous reports indicate that ATP contributes to platelet
activation (35,36). Additionally, it has been revealed
that abnormal platelet reactivity occurs in patients with diabetes
mellitus (8).
The high level of mtDNA4977 deletion in
platelets raises the possibility of reduced ATP production in
platelets, as a previous study indicated that ATP synthesis
declines in cells as mtDNA4977 deletion frequency
increases (14). However, the
exact effects of mtDNA deletion remain unclear. Given the lack of
specific literature on this topic, it may be important to evaluate
whether the increase in mtDNA4977 deletions observed in
the platelets of patients with diabetes consequently affects
platelet mitochondrial function and reactivity.
In addition, it was hypothesized that a higher
frequency of mtDNA4977 deletions in the platelets of
patients with diabetes was associated with abnormal platelet
mitochondrial function. In the present study, no platelet
mitochondrial dysfunction differences were observed between
patients with type 2 diabetes and healthy subjects, as platelet
adenine nucleotide content and ΔΨm were similar between the two
groups. Furthermore, the results of basal platelet activation and
platelet responsiveness to ADP by platelet P-selectin expression
demonstrated that the accumulation of mtDNA4977 deletion
in platelets of diabetic patients was not accompanied by abnormal
platelet reactivity. Furthermore, correlations between the
mtDNA4977 deletion and the studied parameters were not
identified, suggesting that the increased platelet
mtDNA4977 deletion may not have had a major role in
mitochondrial and platelet function. Inconsistent results have been
reported in the literature concerning blood platelet mitochondrial
activity, in which increased (32), reduced (37) and unaltered (38) mitochondrial function has been
demonstrated in diabetes. There is evidence that platelet function
abnormalities are multifactorial, although it is not clear whether
platelet abnormalities are intrinsic to the platelet or are a
consequence of circulating factors that affect platelet function.
Undoubtedly, these discrepancies may be due to numerous reasons,
including differences in participant selection, disease stage,
treatment type and measuring techniques (39,40).
For example, a report demonstrated that there were no abnormalities
in platelet function in patients with moderately controlled type 2
diabetes (41). Due to these and
other potential variables, comparisons among studies are difficult.
Mitochondria are dynamic organelles, and mtDNA copy number
generally varies depending on the energy demand of the cell or
tissue. Alterations have been observed in a variety of pathologies
and in the presence of various environmental stressors, such as
diabetes, hypoxia and oxidative stress (42,43).
Additionally, a number of previous studies on different human
tissues suggested that higher mtDNA content may be involved in a
protective mechanism against damage induced by such stressors to
regulate energy balance (44).
Considering the lack of association between increased
mtDNA4977 deletion and ATP/ADP levels in platelets from
the diabetic patients compared with healthy subjects, mtDNA copy
number analysis was also performed to evaluate whether mtDNA
content had a compensatory role in response to diabetes. Although
mtDNA content in platelets was lower in the good glycemic control
group compared with the other groups, the results revealed that the
differences were not statistically significant. As Tang et
al (37) reported that the
copy number is significantly higher in various cell and cell lines,
including fibroblasts, due to mtDNA4977 deletion
accumulation, the present study evaluated the correlation between
mtDNA4977 deletion frequency and mtDNA copy number in
platelets, using Spearman's correlation coefficient. However, no
correlations were observed between the mtDNA4977
deletion frequency and mtDNA copy number in each diabetic group,
suggesting that increased mitochondrial deletion in platelets had
not induced mtDNA copy number alterations, as a compensatory effect
in response to mtDNA deletion.
According to prior calculations, there would have to
be 17 participants in each study group in order to detect a
difference of 0.8 (80% power and 5% significance level). However,
the post-hoc power calculations based on the available data
indicated that the present study had a lower power for
mtDNA4977 deletion, mtDNA copy number and ATP levels due
to large interindividual variation. Therefore, it was concluded
that the analyses would gain credibility if they were performed
using data from a larger sample.
The findings of the present study demonstrated that
the mtDNA4977 deletion occurred in platelets more
frequently in patients with diabetes, compared with healthy donors,
although mtDNA4977 deletion levels were not associated
with glycemic control or the presence of diabetic complications.
Accumulation of mtDNA4977 deletion in the platelets of
diabetic patients did not appear to have a significant impact on
mitochondrial and/or platelet dysfunction when compared with
nondiabetic subjects. Furthermore, no differences in mtDNA copy
number between the diabetic groups and control group were noted,
suggesting copy number did not have a compensatory role in energy
balance regulation. Further studies are required in order to
understand the sources of inter-individual variability observed in
certain parameters.
Acknowledgements
The authors would like to thank Mr. Burhan Çağçağ
(Tissue Typing Laboratory, Cerrahpasa Faculty of Medicine, Istanbul
University, Turkey) for his technical assistance.
Funding
The present study was supported by The Turkish
Society of Hematology (grant no. 2015-4).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
AK, YH, İO, MN and NB were responsible for study
conception and design. YH and MN provided the patient specimens.
AK, NB, İO, FA, EY and TU performed data analysis. AK, YH, NB, MN,
İO, FA, EY and TU performed data interpretation. All authors
approved the final version of this manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Istanbul Training and Research Hospital (Grant no.
339/2013), and informed consent to participate in the study was
obtained from all patients involved.
Patient consent for publication
No identifying patient information is included in
the published manuscript. Participants provided their consent for
the publication of the data.
Competing interests
All the authors declare that they have no competing
interests.
References
|
1
|
Smyth SS, McEver RP, Weyrich AS, Morrell
CN, Hoffman MR, Arepally GM, French PA, Dauerman HL and Becker RC:
2009 Platelet Colloquium Participants: Platelet functions beyond
hemostasis. J Thromb Haemost. 7:1759–1766. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Salem MA, Adly AA, Ismail EA, Darwish YW
and Kamel HA: Platelets microparticles as a link between micro-and
macro-angiopathy in young patients with type 1 diabetes. Platelets.
26:682–688. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhao LS, Lin YY, Liu Y, Xu CY, Liu Y, Bai
WW, Tan XY, Li DZ and Xu JL: Doxazosin attenuates renal matrix
remodeling mediated by anti-α1-adrenergic receptor
antibody in a rat model of diabetes mellitus. Exp Ther Med.
14:2543–2553. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vinik AI, Erbas T, Park TS, Nolan R and
Pittenger GL: Platelet dysfunction in type 2 diabetes. Diabetes
Care. 24:1476–1485. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sobel BE and Schneider DJ: Platelet
function, coagulopathy, and impaired fibrinolysis in diabetes.
Cardiol Clin. 22:511–526. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Michno A, Skibowska A, Raszeja-Specht A,
Cwikowska J and Szutowicz A: The role of adenosine triphosphate
citrate lyase in the metabolism of acetyl coenzyme a and function
of blood platelets in diabetes mellitus. Metabolism. 53:66–72.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Skibowska A, Raszeja-Specht A and
Szutowicz A: Platelet function and acetyl-coenzyme A metabolism in
type 1 diabetes mellitus. Clin Chem Lab Med. 41:1136–1143. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Michno A, Bielarczyk H, Pawełczyk T,
Jankowska-Kulawy A, Klimaszewska J and Szutowicz A: Alterations of
adenine nucleotide metabolism and function of blood platelets in
patients with diabetes. Diabetes. 56:462–467. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Daniel JL, Dangelmaier C, Jin J, Kim YB
and Kunapuli SP: Role of intracellular signaling events in
ADP-induced platelet aggregation. Thromb Haemost. 82:1322–1326.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rolo AP and Palmeira CM: Diabetes and
mitochondrial function: Role of hyperglycemia and oxidative stress.
Toxicol Appl Pharmacol. 212:167–178. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Forbes JM, Coughlan MT and Cooper ME:
Oxidative stress as a major culprit in kidney disease in diabetes.
Diabetes. 57:1446–1454. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jay D, Hitomi H and Griendling KK:
Oxidative stress and diabetic cardiovascular complications. Free
Radic Biol Med. 40:183–192. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yamagishi SI, Edelstein D, Du XL and
Brownlee M: Hyperglycemia potentiates collagen-induced platelet
activation through mitochondrial superoxide overproduction.
Diabetes. 50:1491–1494. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Porteous WK, James AM, Sheard PW, Porteous
CM, Packer MA, Hyslop SJ, Melton JV, Pang CY, Wei YH and Murphy MP:
Bioenergetic consequences of accumulating the common 4977-bp
mitochondrial DNA deletion. Eur J Biochem. 257:192–201. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wei YH: Oxidative stress and mitochondrial
DNA mutations in human aging. Proc Soc Exp Biol Med. 217:53–63.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Krishnan KJ, Greaves LC, Reeve AK and
Turnbull DM: Mitochondrial DNA mutations and aging. Ann N Y Acad
Sci. 1100:227–240. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Drew B and Leeuwenburgh C: Ageing and
subcellular distribution of mitochondria: Role of mitochondrial DNA
deletions and energy production. Acta Physiol Scand. 182:333–341.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Botto N, Berti S, Manfredi S, Al-Jabri A,
Federici C, Clerico A, Ciofini E, Biagini A and Andreassi MG:
Detection of mtDNA with 4977 bp deletion in blood cells and
atherosclerotic lesions of patients with coronary artery disease.
Mutat Res. 570:81–88. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee HC, Pang CY, Hsu HS and Wei YH:
Differential accumulations of 4,977 bp deletion in mitochondrial
DNA of various tissues in human ageing. Biochim Biophys Acta.
1226:37–43. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chuanzhong Y, Ming G, Fanglin Z, Haijiao
C, Zhen L, Shiping C and YongKang Z: Real-time quantitative reverse
transcription-PCR assay for renal cell carcinoma-associated antigen
G250. Clin Chim Acta. 318:33–40. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pushnova EA, Geier M and Zhu YS: An easy
and accurate agarose gel assay for quantitation of bacterial
plasmid copy numbers. Anal Biochem. 284:70–76. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delat C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fiorentino TV, Prioletta A, Zuo P and
Folli F: Hyperglycemia-induced oxidative stress and its role in
diabetes mellitus related cardiovascular diseases. Curr Pharm Des.
19:5695–5703. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liang P, Hughes V and Fukagawa NK:
Increased prevalence of mitochondrial DNA deletions in skeletal
muscle of older individuals with impaired glucose tolerance:
Possible marker of glycemic stress. Diabetes. 46:920–923. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kakimoto M, Inoguchi T, Sonta T, Yu HY,
Imamura M, Etoh T, Hashimoto T and Nawata H: Accumulation of
8-hydroxy-2′-deoxyguanosine and mitochondrial DNA deletion in
kidney of diabetic rats. Diabetes. 51:1588–1595. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lu H, Koshkin V, Allister EM,
Gyulkhandanyan AV and Wheeler MB: Molecular and metabolic evidence
for mitochondrial defects associated with beta-cell dysfunction in
a mouse model of type 2 diabetes. Diabetes. 59:448–459. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yao YG, Ellison FM, McCoy JP, Chen J and
Young NS: Age-dependent accumulation of mtDNA mutations in murine
hematopoietic stem cells is modulated by the nuclear genetic
background. Hum Mol Genet. 16:286–294. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sandy MS, Langston JW, Smith MT and Di
Monte DA: PCR analysis of platelet mtDNA: Lack of specific changes
in Parkinson's disease. Mov Disord. 8:74–82. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Biagini G, Pallotti F, Carraro S, Sgarbi
G, Pich MM, Lenaz G, Anzivino F, Gualandi G and Xin D:
Mitochondrial DNA in platelets from aged subjects. Mech Ageing Dev.
101:269–275. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tschoepe D, Roesen P, Esser J, Schwippert
B, Nieuwenhuis HK, Kehrel B and Gries FA: Large platelets circulate
in an activated state in diabetes mellitus. Semin Thromb Hemost.
17:433–438. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Watala C: Blood platelet reactivity and
its pharmacological modulation in (people with) diabetes mellitus.
Curr Pharm Des. 11:2331–2365. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Siewiera K, Kassassir H, Talar M, Wieteska
L and Watala C: Higher mitochondrial potential and elevated
mitochondrial respiration are associated with excessive activation
of blood platelets in diabetic rats. Life Sci. 148:293–304. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu F, Liu Y, Luo L, Lu Y, Yew DT, Xu J and
Guo K: Platelet mitochondrial dysfunction of DM rats and DM
patients. Int J Clin Exp Med. 8:6937–6946. 2015.PubMed/NCBI
|
|
34
|
Avila C, Huang RJ, Stevens MV, Aponte AM,
Tripodi D, Kim KY and Sack MN: Platelet mitochondrial dysfunction
is evident in type 2 diabetes in association with modifications of
mitochondrial anti-oxidant stress proteins. Exp Clin Endocrinol
Diabetes. 120:248–251. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rolf MG, Brearley CA and Mahaut-Smith MP:
Platelet shape change evoked by selective activation of P2X1
purinoceptors with alpha, beta-methylene ATP. Thromb Haemost.
85:303–308. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Oury C, Toth-Zsamboki E, Thijs C, Tytgat
J, Vermylen J and Hoylaerts MF: The ATP-gated P2X1 ion channel acts
as a positive regulator of platelet responses to collagen. Thromb
Haemost. 86:1264–1271. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tang Y, Schon EA, Wilichowski E,
Vazquez-Memije ME, Davidson E and King MP: Rearrangements of human
mitochondrial DNA (mtDNA): New insights into the regulation of
mtDNA copy number and gene expression. Mol Biol Cell. 11:1471–1485.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fink BD, Herlein JA, O'Malley Y and Sivitz
WI: Endothelial cell and platelet bioenergetics: Effect of glucose
and nutrient composition. PLoS One. 7:e394302012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bray PF: Platelet hyperreactivity:
Predictive and intrinsic properties. Hematol Oncol Clin North Am.
21:633–645. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
O'donnell CJ, Larson MG, Feng D,
Sutherland PA, Lindpaintner K, Myers RH, D'Agostino RA, Levy D and
Tofler GH: Framingham Heart Study: Genetic and environmental
contributions to platelet aggregation: The Framingham heart study.
Circulation. 103:3051–3056. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lemkes BA, Bähler L, Kamphuisen PW,
Stroobants AK, Van Den Dool EJ, Hoekstra JB, Nieuwland R, Gerdes VE
and Holleman F: The influence of aspirin dose and glycemic control
on platelet inhibition in patients with type 2 diabetes mellitus. J
Thromb Haemost. 10:639–646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Al-Kafaji G and Golbahar J: High
glucose-induced oxidative stress increases the copy number of
mitochondrial DNA in human mesangial cells. Biomed Res Int.
2013:7549462013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pastukh VM, Gorodnya OM, Gillespie MN and
Ruchko MV: Regulation of mitochondrial genome replication by
hypoxia: The role of DNA oxidation in D-loop region. Free Radic
Biol Med. 96:78–88. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Santos JM, Tewari S and Kowluru RA: A
compensatory mechanism protects retinal mitochondria from initial
insult in diabetic retinopathy. Free Radic Biol Med. 53:1729–1737.
2012. View Article : Google Scholar : PubMed/NCBI
|