Introduction
Colorectal cancer (CRC) is a common malignancy that
ranks as the second leading cause of cancer-associated mortality in
men and women in the USA (1).
Despite improvements in CRC therapy, CRC remains a major public
health problem, and it is estimated that there are 1,000,000
individuals suffering from CRC worldwide, with the mortality rate
is as high as ~50% in certain developed countries (2). The tumor stage is the most important
prognostic indicator for CRC. However, the tumors are often
diagnosed at an intermediate or late stage, and the pathological
staging cannot accurately predict recurrence (3). An immunochemical test has been used
in CRC screening, which is considered more useful than colonoscopy
in the Chinese population, and is less invasive and more accurate
than colonoscopy (4,5). The progression of CRC is a complex
multigene, multistep, multistage process involving certain specific
molecular alterations. A number of genes and pathways have been
revealed to be involved the occurrence and development of CRC. For
example, mutations of tissue inhibitor of metalloproteinases 2 and
metalloproteinases are associated with the tumorigenesis and
certain biological behaviors of CRC (6). The activation of Wnt/β-catenin
signaling occurs in the majority of cases of CRC, and activation of
this pathway is an early event in CRC tumorigenesis (7). However, the molecular mechanisms
associated with CRC require further investigation, and it is
important to identify novel biomarkers that may guide the diagnosis
and therapy of CRC.
MicroRNAs (miRNAs) post-transcriptionally regulate
the expression of numerous genes. miRNAs can silence gene
expression by binding to the 3′untranslated regions (3′-UTRs) of a
target mRNA, resulting in gene degradation or translation
termination (8). Increasing
evidence indicates that miRNAs are crucial in several types of
cancer. miRNAs can regulate cell growth, cell death, cell
proliferation and differentiation, in addition to tumorigenesis.
Several signaling pathways and genes are reported as regulatory
targets of miRNAs in cancer, including B-cell lymphoma 2 apoptosis
regulator and sirtuin 1 genes in breast cancer (9), and KRas and Notch pathways in
pancreatic cancer (10).
Furthermore, miRNAs may also be useful as cancer biomarkers and
treatment targets. However, the specific regulatory mechanism of
miRNAs in CRC remains to be fully elucidated.
In the present study, bioinformatics methods were
used to identify differentially expressed genes (DEGs) and
differentially expressed miRNAs (DEMs) in CRC tissue samples
compared with non-cancerous samples. The construction of the
differential co-regulation network and miRNA-gene regulatory
network may assist in improving current understanding of the
regulatory mechanisms of miRNAs in CRC.
Materials and methods
Microarray data
The mRNA expression and miRNA profiles of the
GSE32323 (11) and GSE53592
datasets were respectively downloaded from the Gene Expression
Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) database. GSE53592
consisted of data from six samples, three CRC tissue samples and
three surrounding control tissue. The miRNA expression profile was
detected using the GPL8786 [miRNA-1_0] Affymetrix miRNA Array
platform (Affymetrix, Inc., Santa Clara, CA, USA; http://www.affymetrix.com/analysis/index.affx). The
mRNA dataset GSE32323 contained 44 samples, including 17 pairs of
cancer and non-cancerous tissue samples from patients with CRC,
five pairs treated with 5-aza-2′-deoxycitidine and untreated cell
line samples. Gene expression profiles were measured using the
GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array
(Affymetrix, Inc.; http://www.affymetrix.com) platform.
Identification of DEMs and DEGs
For the mRNA dataset, the raw data were background
corrected and normalized using the affy package version 1.58.0
(https://bioconductor.org/packages/release/bioc/html/affy.html)
in R version 2.10.0 (12).
If more than one probe corresponded to only one gene, the average
expression value of these probes was considered as the expression
value of the gene. The DEMs and DEGs in the CRC tissue samples
compared with surrounding control tissue samples (DEGs-CC) and the
DEGs in cell samples treated with 5-aza-2′-deoxycitidine compared
with untreated samples (DEGs-TC) were identified using the limma
package (http://bioconductor.org/packages/release/bioc/html/limma.html)
of R. The DEMs were identified according to the following
criteria: false discovery rate (FDR) corrected P-value of P<0.05
and |log2(fold change)| >1. The screening criteria
for the DEGs was |log2(fold change)| >1 and Benjamini
and Hochberg corrected P-value of P<0.05. Hierarchical
clustering analysis of CRC tissue samples and non-cancerous samples
based on the expression value of these DEGs was performed. The DEGs
in cell samples treated with 5-aza-2′-deoxycitidine compared with
untreated samples were identified via the limma package with the
criteria of P<0.05 and |log2(fold change)|
>0.5.
Functional and pathway enrichment
analysis
The Database for Annotation, Visualization and
Integrated Discovery (DAVID; http://david.ncifcrf.gov/) is a widely used web-based
tool for genomic functional annotations. To understand the
biological functions of the DEGs, Gene Ontology (GO) terms and
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analyses were performed based on DAVID. The GO terms and KEGG
pathways that contained more than two genes were selected.
P<0.05 was used as the threshold to select the enriched GO terms
and KEGG pathways.
Construction of the differential
co-regulation network
DCGL is an R package for screening
differentially co-expressed genes (DCGs) and differentially
co-expressed links (DCLs). It analyzes the expression correlation
based on the exact co-expression changes to distinguish the
significant co-expression changes and relatively minor ones. In the
present study, differential co-regulation pairs were identified via
DCGL version 2.1.2 in the CRC tissue samples compared with
non-cancerous tissue samples, and the
5-aza-2′-deoxycitidine-treated cell line samples compared with the
untreated cell line samples, and a differential co-regulation
network was constructed based on the data.
Construction of the miRNA-gene
regulatory network
The targets of the DEMs were identified based on the
TargetScan version 6.2 (http://www.targetscan.org/) database. The overlaps
between the DEGs and the targets of DEMs were selected. The
miRNA-gene regulatory network was established and visualized using
Cytoscape software version 3.1.0 (http://www.cytoscape.org/).
Results
DEGs and DEMs
A total of 145 DEMs were identified in the CRC
samples compared with surrounding control tissue samples. The top
20 DEMs according to the P-value are listed in Table I. Additionally, there were 1,284
DEGs-CC, including 638 downregulated DEGs and 646 upregulated DEGs.
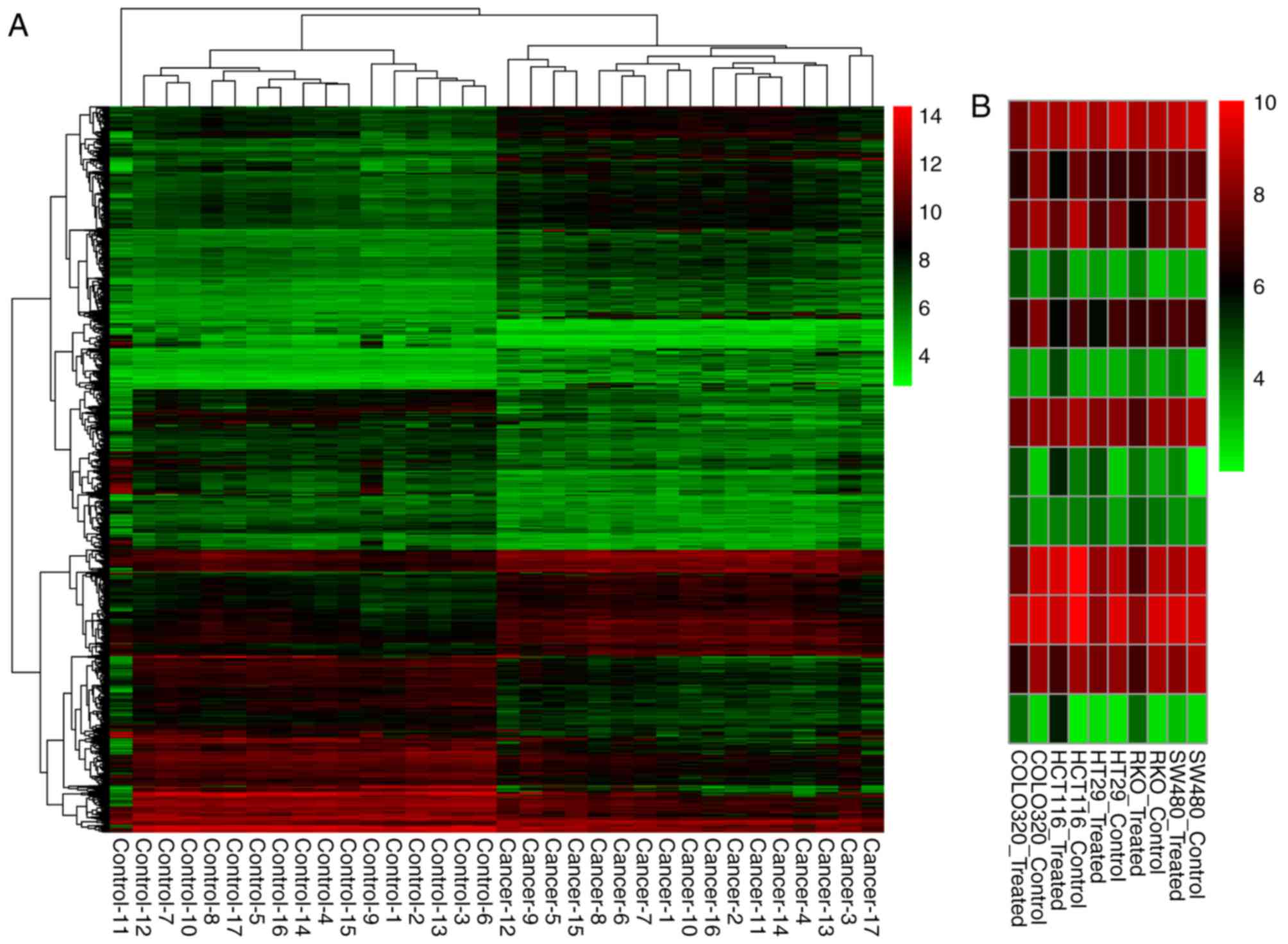
Cluster analysis of the CRC tissue samples and non-cancerous tissue
samples based on the DEGs-CC expression values is shown in Fig. 1A. Furthermore, there were 101
DEGs-TC, including 42 downregulated DEGs and 59 upregulated DEGs.
The top 20 DEGs in DEGs-CC and DEGs-TC are listed in Table IIA and Table IIB, respectively. There were 13
overlaps between the DEGs-CC and DEGs-TC. The expression value
heatmap of these overlaps in the 5-aza-2′-deoxycitidine-treated
cell line samples and untreated cell line samples is shown in
Fig. 1B.
 | Table I.Top 20 differentially expressed miRs
in colorectal cancer tissue samples compared with surrounding
control tissue samples. |
Table I.
Top 20 differentially expressed miRs
in colorectal cancer tissue samples compared with surrounding
control tissue samples.
| miR ID | P-value | LogFC |
|---|
| miR-195-5p |
3.62×10−05 | −4.04 |
| miR-302c-5p |
1.52×10−04 | 3.22 |
| miR-4328 |
2.71×10−04 | −3.75 |
| miR-28-3p |
2.80×10−04 | −4.90 |
| miR-186-5p |
3.11×10−04 | −4.28 |
| miR-320a |
3.18×10−04 | −2.85 |
| miR-30b-5p |
3.99×10−04 | −3.68 |
| miR-101-3p |
5.02×10−04 | −2.89 |
| miR-30c-5p |
5.30×10−04 | −2.11 |
| miR-140-3p |
6.33×10−04 | −2.42 |
| miR-145-5p |
7.54×10−04 | −3.34 |
| miR-143-3p |
8.34×10−04 | −5.10 |
| miR-378e |
8.40×10−04 | −4.35 |
| miR-708-5p |
9.01×10−04 | 4.26 |
| miR-125b-5p |
9.56×10−04 | −3.67 |
| miRPlus-C1066 |
1.08×10−03 | 3.12 |
| miR-320b |
1.11×10−03 | −2.08 |
| miR-3158-5p |
1.29×10−03 | 2.26 |
| miR-1973 |
1.31×10−03 | −2.18 |
| miR-4748 |
1.49×10−03 | 3.47 |
 | Table II.Top 20 DEGs-CC and DEGs-TC. |
Table II.
Top 20 DEGs-CC and DEGs-TC.
| A, Top 20
DEGs-CC |
|---|
|
|---|
| Gene | Adjusted
P-value | LogFC |
|---|
| ENC1 |
2.70×10−12 | 2.548145 |
| CLDN1 |
2.70×10−12 | 2.735442 |
| MMP28 |
1.87×10−11 | −2.33266 |
| HILPDA |
1.87×10−11 | 2.375653 |
| FOXQ1 |
1.87×10−11 | 5.546198 |
| ASCL2 |
1.87×10−11 | 2.24839 |
| SCARA5 |
2.88×10−11 | −2.0155 |
| ABCA8 |
2.88×10−11 | −1.263 |
| CHGA |
6.66×10−11 | −1.98874 |
| C10orf54 |
7.64×10−11 | −1.4338 |
| MYC |
8.02×10−11 | 2.925688 |
| ST6GALNAC6 |
9.43×10−11 | −2.36517 |
| DIEXF |
9.43×10−11 | 1.409167 |
| SMPD1 |
1.03×10−10 | −1.11622 |
| PLEKHA8P1 |
1.64×10−10 | 1.146974 |
| ABI3BP |
1.72×10−10 | −1.3326 |
| AJUBA |
2.03×10−10 | 1.757746 |
| SLC11A2 |
2.47×10−10 | 1.208803 |
| MTERF3 |
2.66×10−10 | 1.755627 |
| SPIB |
2.92×10−10 | −1.91248 |
|
| B, Top 20
DEGs-TC |
|
| Gene | P-value | LogFC |
|
| GAGE3 |
5.09×10−15 | 4.320948 |
| CXorf67 | 0.000186 | 0.866842 |
| MAEL | 0.000313 | 2.889106 |
| ACRC | 0.00077 | 4.065277 |
| TKTL1 | 0.000883 | 4.047408 |
| COX7B2 | 0.001184 | 1.485046 |
| PDIA2 | 0.001524 | 0.680444 |
| KIF11 | 0.002012 | −1.16205 |
| PAGE2B | 0.002021 | 2.791034 |
| CDC45 | 0.002089 | −0.93697 |
| AOC3 | 0.003546 | 1.582814 |
| DCAF4L1 | 0.003678 | 2.629287 |
| USHBP1 | 0.003725 | 0.529246 |
| NAA11 | 0.004706 | 0.730605 |
| SSX1 | 0.004742 | 1.379837 |
| ENTPD3-AS1 | 0.004955 | −0.53635 |
| DAZL | 0.005373 | 3.35696 |
| EID3 | 0.006215 | 0.708792 |
| OVAAL | 0.006445 | 1.746998 |
| TDRD12 | 0.006801 | 2.10315 |
Enriched GO terms and pathways
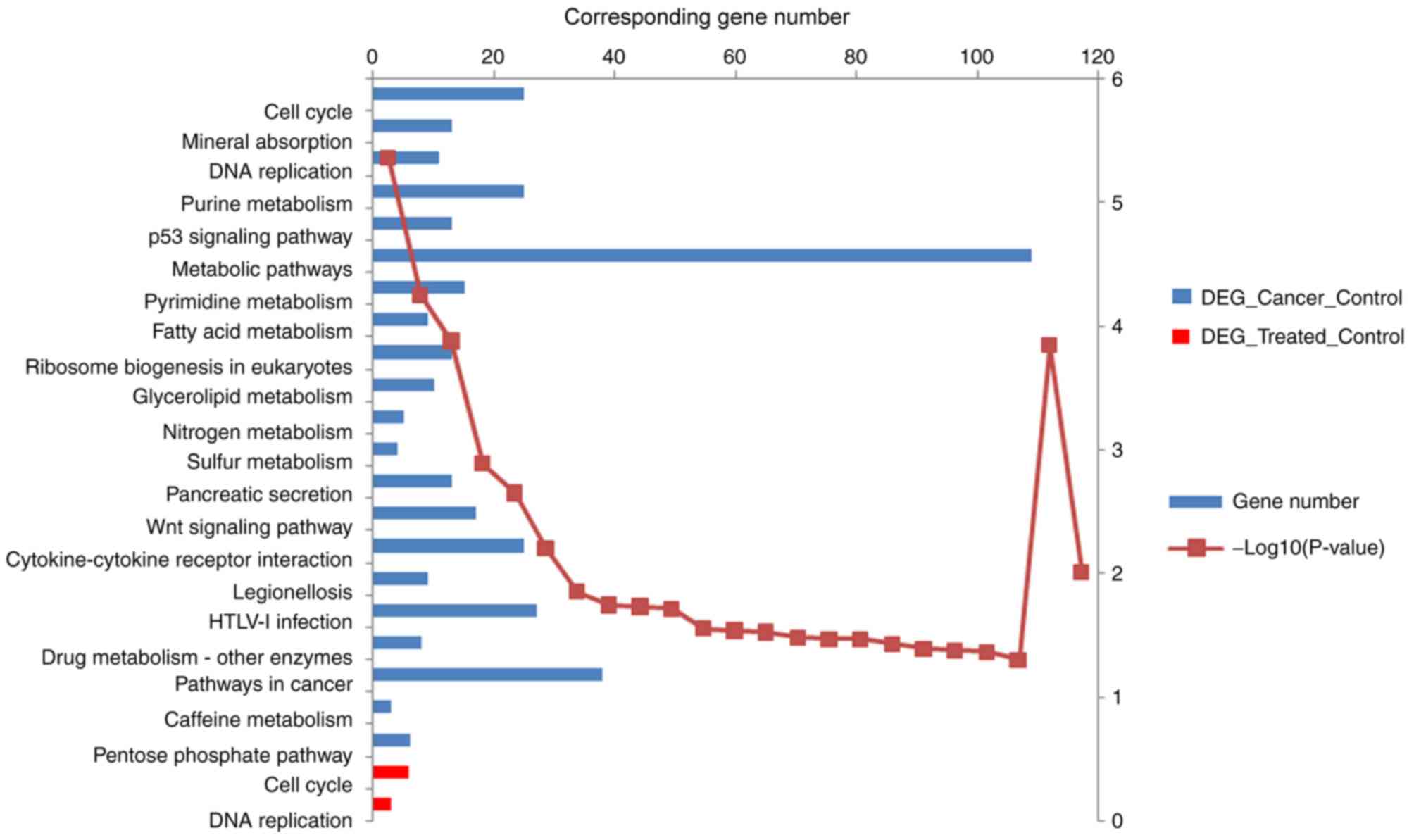
The DEGs-CC were enriched in 196 GO terms and 23
KEGG pathways, and the DEGs-TC were enriched in 46 GO terms and two
KEGG pathways. The top 10 GO terms of DEGs-CC and DEGs-TC are
listed in Table III according to
P-value. Fig. 2 shows the enriched
KEGG pathways of the DEGs-CC and DEGs-TC.
 | Table III.Top 10 GO terms of DEGs-CC and
DEGs-TC. |
Table III.
Top 10 GO terms of DEGs-CC and
DEGs-TC.
| GO ID | GO term | Genes (n) | P-value |
|---|
| DEGs-CC |
|
GO:0007067 | Mitotic nuclear
division | 46 |
6.21×10−10 |
|
GO:0051301 | Cell division | 56 |
2.07×10−09 |
|
GO:0000082 | G1/S transition of
mitotic cell cycle | 22 |
2.93×10−06 |
|
GO:0007062 | Sister chromatid
cohesion | 21 |
1.27×10−05 |
|
GO:0006260 | DNA
replication | 26 |
3.27×10−05 |
|
GO:0007052 | Mitotic spindle
organization | 10 |
9.93×10−05 |
|
GO:0045926 | Negative regulation
of growth | 8 |
1.41×10−04 |
|
GO:0008283 | Cell
proliferation | 44 |
2.02×10−04 |
|
GO:0042127 | Regulation of cell
proliferation | 27 |
2.39×10−04 |
|
GO:0006271 | DNA strand
elongation involved in DNA replication | 7 |
2.55×10−04 |
| DEGs-TC |
|
GO:0051301 | Cell division | 12 |
3.57×10−07 |
|
GO:0000082 | G1/S transition of
mitotic cell cycle | 7 |
5.74×10−06 |
|
GO:0000731 | DNA synthesis
involved in DNA repair | 5 |
1.64×10−05 |
|
GO:0006260 | DNA
replication | 7 |
6.20×10−05 |
|
GO:0007062 | Sister chromatid
cohesion | 6 |
9.07×10−05 |
|
GO:0000732 | Strand
displacement | 4 |
1.99×10−04 |
|
GO:0007059 | Chromosome
segregation | 5 |
2.29×10−04 |
|
GO:0007067 | Mitotic nuclear
division | 7 |
7.82×10−04 |
|
GO:0007283 |
Spermatogenesis | 8 | 0.001517 |
|
GO:0007076 | Mitotic chromosome
condensation | 3 | 0.001938 |
Differential co-regulation
network
There were two types of association between the
co-expression gene pairs and the regulatory factors. One is that
the two co-expressed genes had a common transcription factor, and
the co-expression association is not known or predicted by the
transcription factor, gene regulation pairs (TF-bridged-DCL pairs).
The other is that the co-expression pair is a known or predicted
transcription factor, a gene regulation pair in itself
(TF2target-DCL pairs). In the present study, 5,306 TF-bridged-DCL
pairs and four TF2target-DCL pairs were obtained. The top 20 nodes
according to degree are listed in Table IV.
 | Table IV.Top 20 nodes in the differential
co-regulation network. |
Table IV.
Top 20 nodes in the differential
co-regulation network.
| Gene | Degree |
|---|
| SGK1 | 842 |
| SOX9 | 615 |
| SLC25A23 | 557 |
| SLC20A1 | 479 |
| EDN3 | 424 |
| PRKAR2B | 255 |
| PAX5 | 250 |
| CDCA7 | 246 |
| WT1 | 231 |
| GPX2 | 227 |
| TPD52L1 | 203 |
| KLF9 | 181 |
| DAND5 | 178 |
| PSG1 | 178 |
| SP1 | 178 |
| C3orf70 | 177 |
| PIPOX | 162 |
| CREB1 | 161 |
| E2F | 152 |
| E2F1 | 152 |
miRNA-gene regulatory network
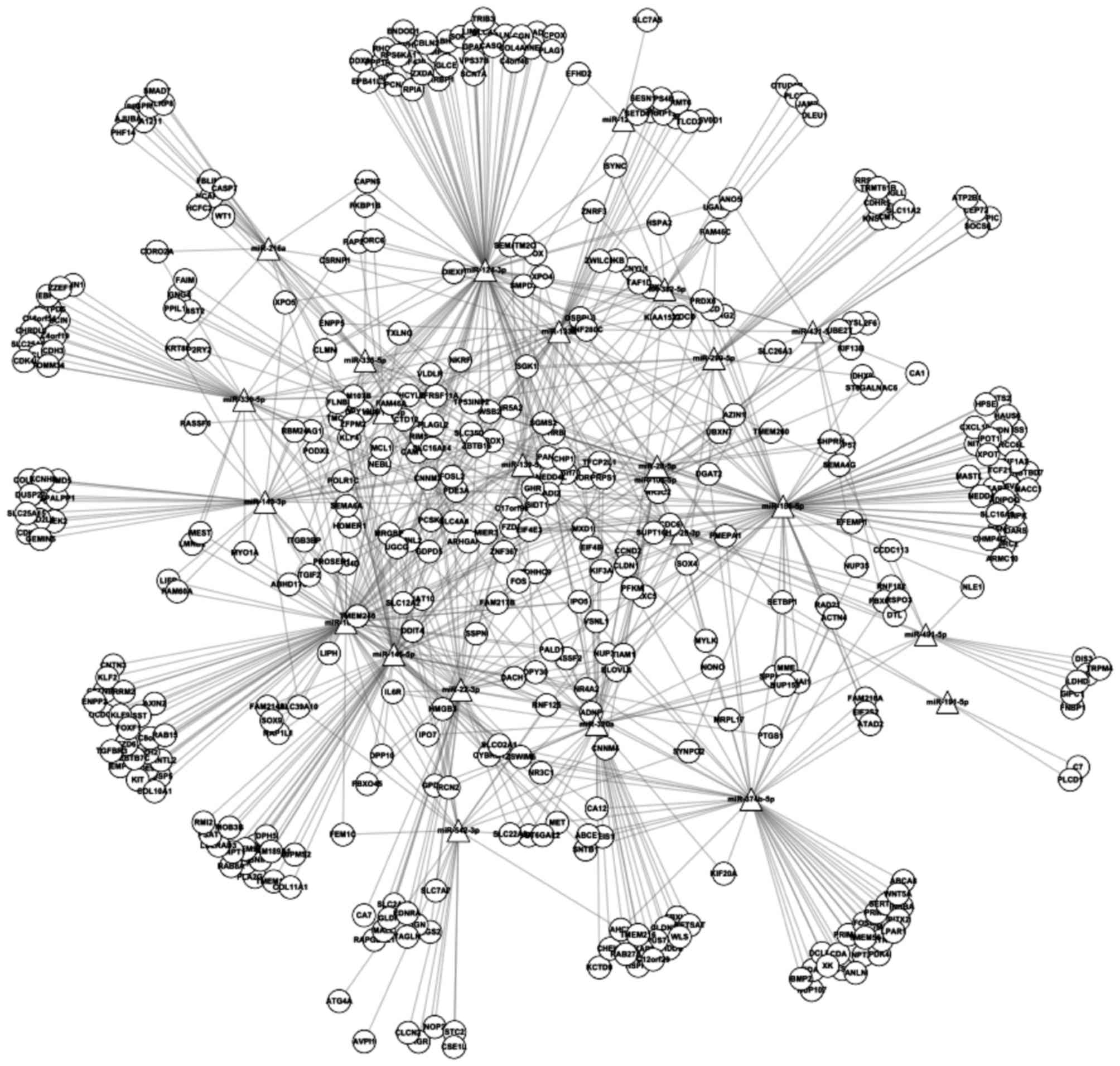
A total of 795 regulatory pairs between the DEMs and
the DEGs were identified. Based on these pairs, the miRNA-gene
regulatory network was constructed, which contained 451 nodes
(Fig. 3). The top 20 nodes in the
miRNA-gene network according to the degree are listed in Table V.
 | Table V.Top 20 nodes in the miR-gene
regulatory network. |
Table V.
Top 20 nodes in the miR-gene
regulatory network.
| Node | Degree |
|---|
| miR-124-3p | 94 |
| miR-101-3p | 75 |
| miR-186-5p | 74 |
| miR-145-5p | 65 |
| miR-320a | 51 |
| miR-374b-5p | 51 |
| miR-22-3p | 43 |
| miR-133b | 37 |
| miR-143-3p | 36 |
| miR-330-5p | 33 |
| miR-139-5p | 30 |
| miR-140-3p | 25 |
| miR-299-5p | 24 |
| miR-542-3p | 24 |
| miR-216a | 18 |
| miR-28-3p | 17 |
| miR-335-5p | 17 |
| miR-28-5p | 16 |
| miR-382-5p | 16 |
| miR-10a-5p | 15 |
Discussion
The top two enriched GO terms of the DEGs-CC were
‘mitotic nuclear division’ and ‘cell division’, and were ‘cell
division’ and ‘G1/S transition of mitotic cell cycle’ for the
DEGs-TC. These processes are critical in cancer development.
Mitotic nuclear division is a key step in cell division, which is
an essential process in tumorigenesis (13). Cell division increases the risk of
various genetic errors. DNA adducts or other forms of
single-stranded DNA damage are usually converse to gaps or
mutations in the process of cell division (14). Tumor development is caused by the
activation or altered expression of proto-oncogenes to oncogenes
and the inactivation of tumor suppressor genes (15). Oncogenes can be activated by
amplification translocation, or mutation. The loss or inactivation
of tumor suppressor genes can promote cell division (16). Additionally, targeting the protein
cell division cycle 7 kinase is a novel method for treating cancer.
The G1/S transition in the mitotic cell cycle is a key step in
which DNA replication is initiated. Deregulation of the cell cycle
facilitates the aberrant cell proliferation that is characteristic
of cancer (17). Cyclin D1 and
cyclin-dependent kinase 4 are important regulators of the G1/S
transition (18). The
overexpression of cyclin D1 is reported to be involved in the
development and progression of certain types of cancer, including
esophageal and breast cancer (19–21).
As a major regulator of the G1/S transition, cyclin D1 is an
oncogenic driver in human cancer and may also serve as a
therapeutic target (19). The top
two enriched pathways for DEGs-CC were ‘cell cycle’ and ‘mineral
absorption’, whereas the top two enriched pathways for DEGs-TC were
‘cell cycle’ and ‘DNA replication’. The association between the
cell cycle and cancer development is well established. In addition,
the cell cycle phase can be used as a prognostic marker and
therapeutic target in various types of cancer (22). DNA replication, a fundamental
cellular process, is closely linked to cell proliferation.
Inhibiting DNA replication-initiation proteins can induce the
apoptosis of cancer cells (23).
In CRC, DNA replication can serve as a prognostic biomarker in
young patients with CRC. However, the association between mineral
absorption and cancer development remains to be fully
elucidated.
The top five genes in the differential co-regulation
network were serum/glucocorticoid regulated kinase 1 (SGK1),
SRY-box 9 (SOX9), solute carrier family 25 member 23
(SLC25A23), solute carrier family 20 member 1
(SLC20A1) and endothelin 3 (EDN3). SGK1
encodes a serine/threonine protein kinase, which is also associated
with cell survival pathways (24).
SGK1 is also involved in the stimulation of motility,
adhesiveness and invasiveness, and thus contributes to tumor
metastasis (25). It has been
reported that inhibition of the function of SGK1
significantly decreased breast cancer cell adhesion, suggesting a
role for SGK1 in the aggressive phenotype of cancer cells
(25). In CRC, SGK1 was
important for colorectal tumor growth and may be an attractive
pharmacological target for cytostatic therapy. SOX9 is an
important downstream factor induced by β-catenin (26). SOX9 is a high mobility group
box transcription factor involved in embryonic development and
required for differentiation, lineage commitment and
epithelial-mesenchymal transition (27). High expression of SOX9 is
associated with poor prognosis in various types of human cancer,
including breast cancer, prostate cancer, melanomas and CRC
(28–30). In CRC, a previous study reported
that high expression of SOX9 was an independent poor risk
factor in the prognosis of CRC, which may be used to predict
clinical outcomes for patients with CRC (31). Another study demonstrated that
SOX9 is important in the development, progression and
metastasis of CRC, and that the combined detection of SOX9
and caudal type homeobox 2 may be useful as a marker to evaluate
the degree of malignancy and the prognosis of CRC (32). Solute carrier family 25 member 23,
solute carrier family 20 member 1, and endothelin 3 are associated
with cancer development (33–35).
However, their specific roles in CRC require further
investigation.
The top five nodes in the miRNA-gene regulatory
network were miR-124-3p, miR-101-3p, miR-186-5p, miR-145-5p
and miR-320a. miR-124-3p is a brain-enriched miRNA
involved in the regulation of gastrulation and neural development.
It functions as a tumor suppressor by targeting important genes,
including Rac family small GTPase 1 (Rac1), sphingosine kinase 1,
Rho-associated coiled-coil containing protein kinase 2 and enhancer
of zeste 2 polycomb repressive complex 2 subunit. A previous study
reported that the levels of miR-124-3p were downregulated in
breast cancer tissues, and that miR-124-3p suppresses the
expression of Cbl protooncogene, E3 ubiquitin protein ligase and
negatively regulates the proliferation and invasion of breast
cancer cells (36). In
astrocytoma, miR-124-3p has been shown to regulate cell
proliferation, invasion, apoptosis and bioenergetics by targeting
Pim-1 proto-oncogene, serine/threonine kinase (37). However, the regulatory the
mechanism of miR-124-3p in CRC remains to be fully
elucidated. miR-320a is considered to be a metastatic
suppressor, and high expression is associated with improved
outcomes in patients with CRC. Therefore, miR-320a may be an
important suppressor of the development and metastasis of CRC.
Several genes and pathways have been reported to be targets of
miR-320a. For example, a previous study demonstrated that
miR-320a suppresses the progression of CRC by targeting Rac1
(38). Another report showed that
miR-320 inhibits Wnt/β-catenin signaling by targeting the
3′-UTR of β-catenin (39).
miR-145-5p is also a tumor suppressor miRNA, and it was
previously demonstrated that miR-145 was downregulated in
certain types of cancer, including CRC, ovarian cancer and B-cell
tumors (40–42). The downregulation of
miR-145-5p has been correlated with poor prognosis in
patients with CRC, and angiogenesis was inhibited in CRC by
transfecting cancer cells with miR-145-5p (43). miR-101-3p and
miR-186-5p also regulated processes in human cancer
(44,45), although their roles in CRC remain
to be fully elucidated.
In conclusion, bioinformatics analysis was used to
identify important miRNAs and genes associated with the oncogenesis
of CRC. miR-124-3p, miR-145-5p and miR-320a may be
critical in regulating processes in CRC. SGK1 and
SOX9 may be key genes that affect tumor progression of CRC.
However, further investigations are required to further confirm
this conclusion.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this article.
Authors' contributions
JH analyzed the microarray data. XY and JL performed
functional and pathway enrichment analyses, and constructed the
networks. DK designed the experiments and drafted the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wu JT, Kakar S, Nelson RL, Mihalov ML,
Hayward B, Gilbert PB and Ghosh L: Prognostic significance of DCC
and p27Kip1 in colorectal cancer. Appl Immunohistochem Mol Morphol.
13:45–54. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Draht MX, Riedl RR, Niessen H, Carvalho B,
Meijer GA, Herman JG, van Engeland M, Melotte V and Smits KM:
Promoter CpG island methylation markers in colorectal cancer: The
road ahead. Epigenomics. 4:179–194. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marisa L, de Reyniès A, Duval A, Selves J,
Gaub MP, Vescovo L, Etienne-Grimaldi MC, Schiappa R, Guenot D,
Ayadi M, et al: Gene expression classification of colon cancer into
molecular subtypes: Characterization, validation, and prognostic
value. PLoS Med. 10:e10014532013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wong BC, Wong WM, Cheung KL, Tong TS,
Rozen P, Young GP, Chu KW, Ho J, Law WL, Tung HM, et al: A
sensitive guaiac faecal occult blood test is less useful than an
immunochemical test for colorectal cancer screening in a Chinese
population. Aliment Pharmacol Ther. 18:941–946. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Quintero E, Castells A, Bujanda L,
Cubiella J, Salas D, Lanas Á, Andreu M, Carballo F, Morillas JD,
Hernández C, et al: Colonoscopy versus fecal immunochemical testing
in colorectal-cancer screening. N Engl J Med. 366:697–706. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Park KS, Kim SJ, Kim KH and Kim JC:
Clinical characteristics of TIMP2, MMP2, and MMP9 gene
polymorphisms in colorectal cancer. J Gastroenterol Hepatol.
26:391–397. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Aguilera O, Fraga MF, Ballestar E, Paz MF,
Herranz M, Espada J, García JM, Muñoz A, Esteller M and
González-Sancho JM: Epigenetic inactivation of the Wnt antagonist
DICKKOPF-1 (DKK-1) gene in human colorectal cancer. Oncogene.
25:4116–4121. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pauli A, Rinn JL and Schier AF: Non-coding
RNAs as regulators of embryogenesis. Nat Rev Genet. 12:136–149.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li L, Yuan L, Luo J, Gao J, Guo J and Xie
X: MiR-34a inhibits proliferation and migration of breast cancer
through down-regulation of Bcl-2 and SIRT1. Clin Exp Med.
13:109–117. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vorvis C, Koutsioumpa M and Iliopoulos D:
Developments in miRNA gene signaling pathways in pancreatic cancer.
Future Oncol. 12:1135–1150. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Khamas A, Ishikawa T, Shimokawa K, Mogushi
K, Iida S, Ishiguro M, Mizushima H, Tanaka H, Uetake H and Sugihara
K: Screening for epigenetically masked genes in colorectal cancer
Using 5-Aza-2′-deoxycytidine, microarray and gene expression
profile. Cancer Genomics Proteomics. 9:67–75. 2012.PubMed/NCBI
|
|
12
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ames BN and Gold LS: Too many rodent
carcinogens: Mitogenesis increases mutagenesis. Science.
249:970–971. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ramel C: Short-term testing-are we looking
at wrong endpoints? Mut Res. 205:13–24. 1988. View Article : Google Scholar
|
|
15
|
Stanbridge EJ: Identifying tumor
suppressor genes in human colorectal cancer. Science. 247:12–13.
1990. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Preston-Martin S, Pike MC, Ross RK, Jones
PA and Henderson BE: Increased cell division as a cause of human
cancer. Cancer Res. 50:7415–7421. 1990.PubMed/NCBI
|
|
17
|
Williams GH and Stoeber K: The cell cycle
and cancer. J Pathol. 226:352–364. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Martín-Romero FJ, Santiago-Josefat B,
Correa-Bordes J, Gutierrez-Merino C and Fernandez-Salguero P:
Potassium-induced apoptosis in rat cerebellar granule cells
involves cell-cycle blockade at the G1/S transition. J Mol
Neurosci. 15:155–165. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Alao JP: The regulation of cyclin D1
degradation: Roles in cancer development and the potential for
therapeutic invention. Mol Cancer. 6:242007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bartkova J, Lukas J, Müller H, Lützhøt D,
Strauss M and Bartek J: Cyclin D1 protein expression and function
in human breast cancer. Int J Cancer. 57:353–361. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang W, Zhang YJ, Kahn SM, Hollstein MC,
Santella RM, Lu SH, Harris CC, Montesano R and Weinstein IB:
Altered expression of the cyclin D1 and retinoblastoma genes in
human esophageal cancer. Proc Natl Acad Sci USA. 90:9026–9030.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Diaz-Moralli S, Tarrado-Castellarnau M,
Miranda A and Cascante M: Targeting cell cycle regulation in cancer
therapy. Pharmacol Ther. 138:255–271. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Feng D, Tu Z, Wu W and Liang C: Inhibiting
the expression of DNA replication-initiation proteins induces
apoptosis in human cancer cells. Cancer Res. 63:7356–7364.
2003.PubMed/NCBI
|
|
24
|
Brunet A, Park J, Tran H, Hu LS, Hemmings
BA and Greenberg ME: Protein kinase SGK mediates survival signals
by phosphorylating the forkhead transcription factor FKHRL1
(FOXO3a). Mol Cell Biol. 21:952–965. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tangir J, Bonafé N, Gilmore-Hebert M,
Henegariu O and Chambers SK: SGK1, a potential regulator of c-fms
related breast cancer aggressiveness. Clin Exp Metastasis.
21:477–483. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yano F, Kugimiya F, Ohba S, Ikeda T,
Chikuda H, Ogasawara T, Ogata N, Takato T, Nakamura K, Kawaguchi H
and Chung UI: The canonical Wnt signaling pathway promotes
chondrocyte differentiation in a Sox9-dependent manner. Biochem
Biophys Res Commun. 333:1300–1308. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Barrionuevo F and Scherer G: SOX E genes:
SOX9 and SOX8 in mammalian testis development. Int J Biochem Cell
Biol. 42:433–436. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Darido C, Buchert M, Pannequin J, Bastide
P, Zalzali H, Mantamadiotis T, Bourgaux JF, Garambois V, Jay P,
Blache P, et al: Defective claudin-7 regulation by Tcf-4 and Sox-9
disrupts the polarity and increases the tumorigenicity of
colorectal cancer cells. Cancer Res. 68:4258–4268. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Passeron T, Valencia JC, Bertolotto C,
Hoashi T, Le Pape E, Takahashi K, Ballotti R and Hearing VJ: SOX9
is a key player in ultraviolet B-induced melanocyte differentiation
and pigmentation. Proc Natl Acad Sci USA. 104:13984–13989. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang H, Leav I, Ibaragi S, Wegner M, Hu
GF, Lu ML, Balk SP and Yuan X: SOX9 is expressed in human fetal
prostate epithelium and enhances prostate cancer invasion. Cancer
Res. 68:1625–1630. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lü B, Fang Y, Xu J, Wang L, Xu F, Xu E,
Huang Q and Lai M: Analysis of SOX9 expression in colorectal
cancer. Am J Clin Pathol. 130:897–904. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hu X, Yang W and Wu X: Expression of SOX9
and CDX2 in Colorectal Cancer. Chin J Surg Int Trad Western Med.
17:564–567. 2011.(In Chinese).
|
|
33
|
Schouten M, Fratantoni SA, Hubens CJ,
Piersma SR, Pham TV, Bielefeld P, Voskuyl RA, Lucassen PJ, Jimenez
CR and Fitzsimons CP: MicroRNA-124 and −137 cooperativity controls
caspase-3 activity through BCL2L13 in hippocampal neural stem
cells. Sci Rep. 5:124482015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sato K and Akimoto K: Expression levels of
KMT2C and SLC20A1 identified by information-theoretical analysis
are powerful prognostic biomarkers in estrogen receptor-positive
breast cancer. Clin Br Cancer. 17:e135–e142. 2017. View Article : Google Scholar
|
|
35
|
Wiesmann F, Veeck J, Galm O, Hartmann A,
Esteller M, Knüchel R and Dahl E: Frequent loss of endothelin-3
(EDN3) expression due to epigenetic inactivation in human breast
cancer. Breast Cancer Res. 11:R342009. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang Y, Chen L, Wu Z, Wang M, Jin F, Wang
N, Hu X, Liu Z, Zhang CY, Zen K, et al: miR-124-3p functions as a
tumor suppressor in breast cancer by targeting CBL. BMC Cancer.
16:8262016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Deng D, Wang L, Chen Y, Li B, Xue L, Shao
N, Wang Q, Xia X, Yang Y and Zhi F: MicroRNA-124-3p regulates cell
proliferation, invasion, apoptosis, and bioenergetics by targeting
PIM1 in astrocytoma. Cancer Sci. 107:899–907. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhao H, Dong T, Zhou H, Wang L, Huang A,
Feng B, Quan Y, Jin R, Zhang W, Sun J, et al: miR-320a suppresses
colorectal cancer progression by targeting Rac1. Carcinogenesis.
35:886–895. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hsieh IS, Chang KC, Tsai YT, Ke JY, Lu PJ,
Lee KH, Yeh SD, Hong TM and Chen YL: MicroRNA-320 suppresses the
stem cell-like characteristics of prostate cancer cells by
downregulating the Wnt/beta-catenin signaling pathway.
Carcinogenesis. 34:530–538. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li JM, Zhao RH, Li ST, Xie CX, Jiang HH,
Ding WJ, Du P, Chen W, Yang M and Cui L: Down-regulation of fecal
miR-143 and miR-145 as potential markers for colorectal cancer.
Saudi Med J. 33:24–29. 2012.PubMed/NCBI
|
|
41
|
Wu H, Xiao Z, Wang K, Liu W and Hao Q:
MiR-145 is downregulated in human ovarian cancer and modulates cell
growth and invasion by targeting p70S6K1 and MUC1. Biochem Biophys
Res Commun. 441:693–700. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xing AY, Wang B, Shi DB, Zhang XF, Gao C,
He XQ, Liu WJ and Gao P: Deregulated expression of miR-145 in
manifold human cancer cells. Exp Mol Pathol. 95:91–97. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Thuringer D, Jego G, Berthenet K, Hammann
A, Solary E and Garrido C: Gap junction-mediated transfer of
miR-145-5p from microvascular endothelial cells to colon cancer
cells inhibits angiogenesis. Oncotarget. 7:28160–28168. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li CY, Xiong DD, Huang CQ, He RQ, Liang
HW, Pan DH, Wang HL, Wang YW, Zhu HW and Chen G: Clinical value of
miR-101-3p and biological analysis of its prospective targets in
breast cancer: A study based on the cancer genome atlas (TCGA) and
bioinformatics. Med Sci Monit. 23:1857–1871. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li J, Xia L, Zhou Z, Zuo Z, Xu C, Song H
and Cai J: MiR-186-5p upregulation inhibits proliferation,
metastasis and epithelial-to-mesenchymal transition of colorectal
cancer cell by targeting ZEB1. Arch Biochem Biophys. 640:53–60.
2018. View Article : Google Scholar : PubMed/NCBI
|