Introduction
Epithelial ovarian cancer (EOC) is the fifth leading
cause of cancer-associated mortality among women worldwide
(1). During the past decades, the
early detection rate and prognosis of EOC have been substantially
improved (2). However, the
prognosis for the advanced stages of the disease has not improved
significantly in more than two decades (3). Further investigations on the
mechanisms of EOC progression are required to aid the development
of novel diagnostic markers and therapeutic targets.
Only 2% of the human genome accounts for
protein-coding regions, yet >70% of the human genome is
transcribed into RNAs that do not encode proteins; these RNAs are
called non-coding RNAs (4).
Non-coding RNAs are generally divided into two classes according to
their size. Long non-coding RNAs (lncRNAs) are defined as a class
of non-coding RNAs with a length >200 base pairs. In recent
years, numerous lncRNAs have been discovered, and the pivotal roles
of lncRNAs in almost every aspect of cell biology have been
gradually revealed, such as epigenetic regulation via molecular
scaffolding, regulation of mRNA processing, molecular decoying and
lncRNA-derived peptides (5).
Cancer susceptibility candidate 2 (CASC2), an lncRNA
located at chromosome 10q26, was originally identified as a
downregulated gene in endometrial cancer (6). Previous studies suggested that CASC2
may act as a tumor suppressor gene, with epigenetic and genetic
alterations contributing to gene inactivation (6–13).
Downregulation of CASC2 may provide a growth advantage in EOC cells
(7). Recent studies identified
that CASC2 is associated with tumor development and prognosis in
various cancer types, including renal cell carcinoma, osteosarcoma,
bladder cancer, pancreatic cancer, endometrial cancer,
hepatocellular carcinoma, glioma, non-small cell lung cancer and
gastric cancer (8–13). However, little is known regarding
the role of CASC2 in EOC.
The present study aimed to investigate the
expression, clinical significance and functional role of CASC2 in
EOC. The expression levels of CASC2 in EOC cells and tissues were
measured, and the association between CASC2 expression and the
clincopathological characteristics of patients with EOC were
statistically analyzed. Furthermore, CASC2 was overexpressed and
silenced in EOC cells, and the functional alterations in the EOC
cells were evaluated.
Patients and methods
Patients and tissue samples
A total of 126 female patients (aged 56.9±13.2 years
old, ranging between 35–82 years old) with EOC who underwent
surgical resection between August 2010 and August 2011 at Shanghai
Jiao Tong University Affiliated Sixth People's Hospital (Shanghai,
China) were enrolled in the present study, and written informed
consent was obtained from each patient. The collected EOC tissues
and paired tumor adjacent tissues were frozen in liquid nitrogen
immediately following surgical resection and were stored at −80°C
until use. The exclusion criteria were patients who had undergone
presurgical anticancer therapy and were diagnosed with two or more
malignances. The present study was approved by the Ethics Committee
of the Shanghai Jiao Tong University Affiliated Sixth People's
Hospital.
Cell lines and cell culture
The EOC cell lines (SKOV3, OV90, A2780, ES2 and
IGROV-1) and an immortalized ovarian epithelial cell line (Moody)
(14) were purchased from the Cell
Bank of Type Culture Collection of the Chinese Academy of Science
(Beijing, China). Cells were cultured in RPMI-1640 medium
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA;
SKOV3) or Dulbecco's modified Eagle's medium (Invitrogen; Thermo
Fisher Scientific, Inc.; OV90, A2780, ES2 and IGROV-1) supplemented
with 10% fetal bovine serum (FBS; Life Technologies; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin and 100 mg/ml streptomycin
(Life Technologies; Thermo Fisher Scientific, Inc.) in a humidified
atmosphere containing 5% CO2 at 37°C.
Gene ectopic expression, interference
and cell transfection
Ectopic expression of CASC2 was achieved by
subcloning the CASC2 sequence into the pcDNA3.1 vector (Invitrogen;
Thermo Fisher Scientific, Inc.; pcDNA3.1-CASC2; 1.0 µg/µl), with an
empty pCDNA3.1 vector serving as an empty control. Small
interfering (si)RNAs [siCASC2-1 and siCASC2-2 (sequences
unavailable); 5 nM] and non-targeting siRNA [siRNA negative control
(siNC)] were all purchased from Genewiz, Inc. (Suzhou, China). When
EOC cells were 60–70% confluent, transfection was performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. After 5
h of incubation, the transfection medium was replaced with the
appropriate culture medium without antibiotics, followed by
incubation for 48 h. The cells were subsequently used in
experiments. The expression levels of CASC2 following ectopic
expression and interference were confirmed by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR).
RNA extraction and RT-qPCR
Total RNA was extracted from EOC tissues and cell
lines with TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. cDNA
was reverse transcribed from total RNA (2.5 µg) using the
Superscript III kit (Life Technologies; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. Reverse
transcription was performed according to the following temperature
protocol: Incubation for 5 min at 25°C, followed by 60 min at 42°C
and then 70°C for 5 min. The relative expression of selected genes
were quantified and analyzed by real-time PCR using the iQ
SYBRGreen PCR Supermix kit (Bio-Rad Laboratories, Inc., Hercules,
CA, USA). The qPCR thermocycling conditions used were as follows:
Initial denaturation at 95°C for 5 min; followed by 40 cycles of
denaturation at 95°C for 30 sec, annealing at 50°C for 30 sec and
extension at 72°C for 30 sec. The primers were as follows: CASC2
sense, 5′-GCACATTGGACGGTGTTTCC-3′; CASC2 antisense,
5′-CCCAGTCCTTCACAGGTCAC-3′; and GAPDH sense,
5′-AGAAGGCTGGGGCTCATTTG-3′; GAPDH antisense,
5′-AGGGGCCATCCACAGTCTTC-3′. The expression levels of the target
genes were calculated using the 2−ΔΔCq method based on
the cycle threshold values of the genes compared with those of
GAPDH (15). All samples were
typically analyzed in triplicate in at least three independent
runs.
Transwell assay and Matrigel
assay
For the Transwell assay and the Matrigel assay,
cells (1×105) were suspended in 200 µl serum-free medium
and were seeded into the upper chamber of Matrigel-coated (Matrigel
assay) or uncoated (Transwell assay) Boyden chambers (8 µM pore
size; BD Biosciences, Franklin Lakes, NJ, USA). Medium containing
20% FBS was added to the lower chamber. After 24 h incubation,
cells remaining in the upper chamber were removed with cotton
swabs, and cells that had invaded through the membrane were fixed
with ethanol for 30 min at room temperature, and subsequently
stained with Giemsa for another 30 min at room temperature. The
cells that migrated or invaded were counted in five independent
fields using an Olympus CKX53 inverted light microscope
(magnification, ×10; Olympus Corporation, Tokyo, Japan), and images
were obtained (magnification, ×4). The results presented are
representative of three independent experiments with technical
duplicates.
Cell Counting kit-8 (CCK-8) assay
To analyze the effect of CASC2 on cell
proliferation, a CCK-8 assay was performed using a CCK-8 kit (Roche
Applied Science, Penzberg, Germany). In total, 5,000 cells were
seeded into each well of 96-well plates after 48 h of transfection.
CCK-8 was used to quantify the absorbance measurements at 450 nm at
the indicated time points (0, 24, 48, 72 and 96 h). The absorbance
values were normalized to those of cells transfected with empty
vector or siNC. The results represent the average of three
replicates under the same conditions.
Colony formation assay
For the colony formation assay, cells were incubated
in 6-well plates (1,000 cells/well) in triplicates, and
subsequently cultured in a humidified incubator with 5%
CO2 for 10–14 days. The medium was changed every 3 days.
Subsequent to incubation, the cell colonies were incubated in
methanol for 10 min at room temperature, stained with 1% crystal
violet for 10 min at room temperature. All colonies in the 6-well
plate were manually counted using a light microscope
(magnification, ×10). The results presented are representative of
three independent experiments.
Statistical analysis
The data were analyzed with SPSS 19.0 (IBM Corp.,
Armonk, NY, USA) and are presented as the mean ± standard
deviation. To statistically evaluate the association between CASC2
expression and the clinical features of patients with EOC, the
patients with EOC (n=126) were dichotomized with the mean level of
CASC2 expression serving as the cutoff value. Associations between
clinicopathological features and CASC2 expression levels were
assessed using the χ2 test. Student's t-test was used to
analyze the difference between two groups. One-way analysis of
variance and Dunnett's t-test was used for multiple comparisons.
Survival analysis was conducted using the Kaplan-Meier method. The
log-rank test was applied to compare the survival characteristics
between groups. Univariate and multivariate Cox's proportional
hazards regression models were applied to analyze the survival
data. P<0.05 was considered to indicate a statistically
significant difference.
Results
CASC2 is downregulated and correlates
with tumor progression in EOC
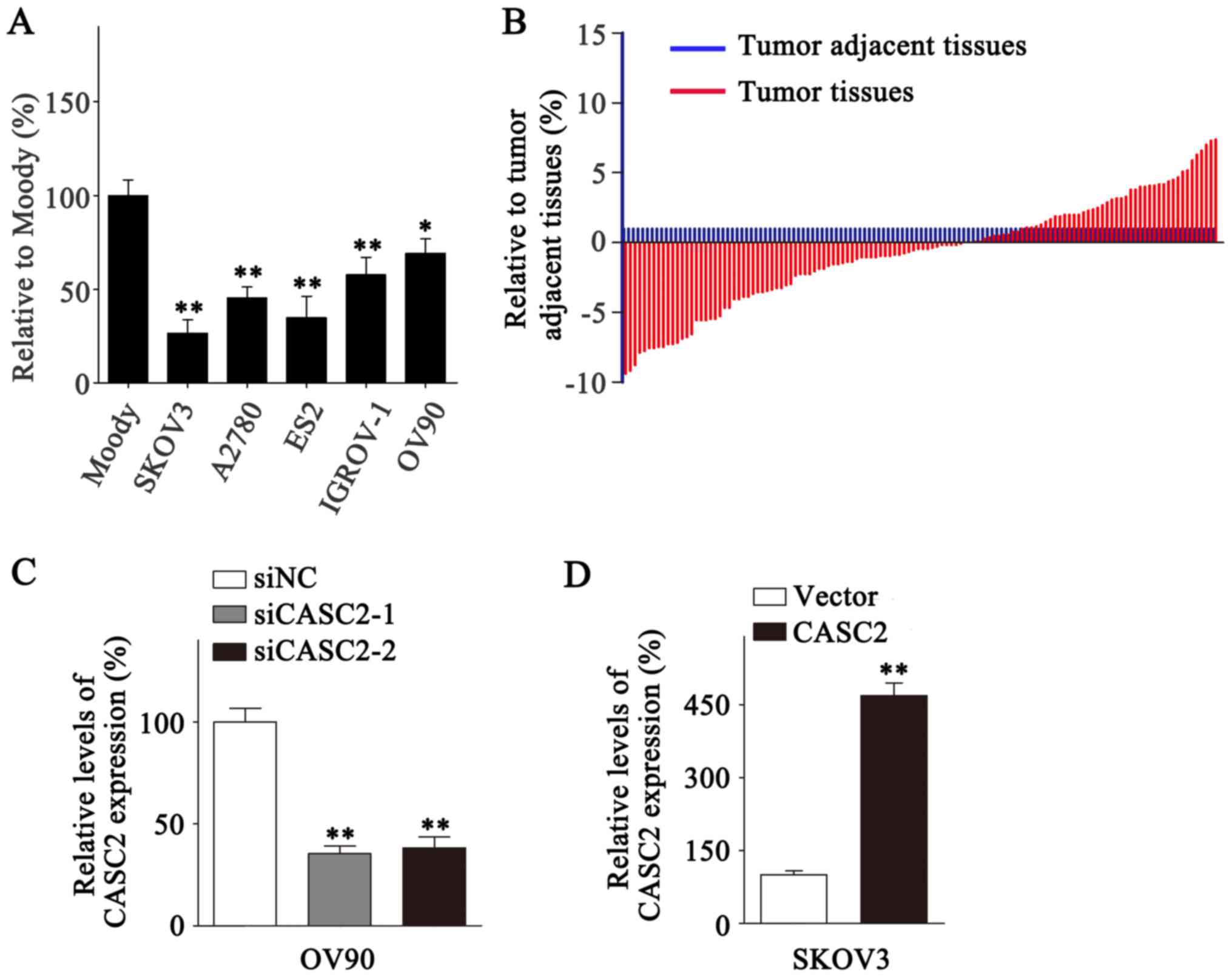
The expression of CASC2 was evaluated in five EOC
cell lines and one immortalized ovarian epithelial cell line
(Moody) with an RT-qPCR assay. CASC2 exhibited a significantly
higher expression level in Moody cells compared with the five EOC
cell lines (Fig. 1A; P<0.05).
Further detection of CASC2 expression levels in the 126 EOC tissues
and paired tumor adjacent tissues revealed that CASC2 was
downregulated in the EOC tissues compared with the tumor adjacent
tissues [1 vs. (−0.5±3.92); (Fig.
1B)]. The results demonstrated that CASC2 downregulation was
associated with histological subtype (P<0.001), lymph node
metastasis (P=0.038), histological grade (P<0.001) and tumor
size (P=0.001; Table I).
Therefore, it may be postulated that CASC2 may be a tumor
suppressor in EOC and is able to repress tumor progression.
 | Table I.Correlation between long non-coding
RNA CASC2 expression and epithelial ovarian cancer
clinicopathological characteristics. |
Table I.
Correlation between long non-coding
RNA CASC2 expression and epithelial ovarian cancer
clinicopathological characteristics.
| Parameters | Number of patients,
n=126 | CASC2 high/low,
50/76 | P-value |
|---|
| Age |
|
| 0.945 |
| <60
years | 60 | 24/36 |
|
| ≥60
years | 66 | 26/40 |
|
| Histology
subtype |
|
| <0.001 |
|
Serous | 52 | 46/6 |
|
|
Others | 74 | 30/44 |
|
| CA125 |
|
| 0.768 |
| <35
U/ml | 31 | 13/18 |
|
| ≥35
U/ml | 95 | 37/58 |
|
| Lymph node
metastasis |
|
| 0.038 |
|
Yes | 77 | 25/52 |
|
| No | 49 | 25/24 |
|
| Residual tumor
size |
|
| 0.218 |
| <1
cm | 80 | 35/45 |
|
| ≥1
cm | 46 | 15/31 |
|
| Histological
grade |
|
| <0.001 |
|
G1+G2 | 68 | 37/31 |
|
| G3 | 58 | 13/45 |
|
| Tumor size |
|
| 0.001 |
| <2
cm | 62 | 34/28 |
|
| ≥2
cm | 64 | 16/48 |
|
| FIGO stage |
|
| 0.519 |
|
I+II | 59 | 25/34 |
|
|
III+IV | 67 | 25/43 |
|
CASC2 inhibits the migration, invasion
and proliferation of EOC cells
Cancer metastasis and proliferation are two primary
factors leading to tumor progression. Therefore, the functional
role of CASC2 in EOC metastasis and cell proliferation was
investigated in in vitro studies. To better elucidate the
function of CASC2 in EOC cells, siRNAs were used to interfere with
the expression of CASC2 in OV90 cells due to their relatively high
CASC2 expression level among the six EOC cell lines (Fig. 1A and 1C), and ectopic overexpression was
conducted in SKOV3 cells due to their relatively low CASC2
expression level among the six EOC cell lines (Fig. 1A and 1D). CASC2 expression was significantly
downregulated and upregulated in OV90 and SKOV3 cells, respectively
following transfection (Fig. 1C and
D; P<0.01).
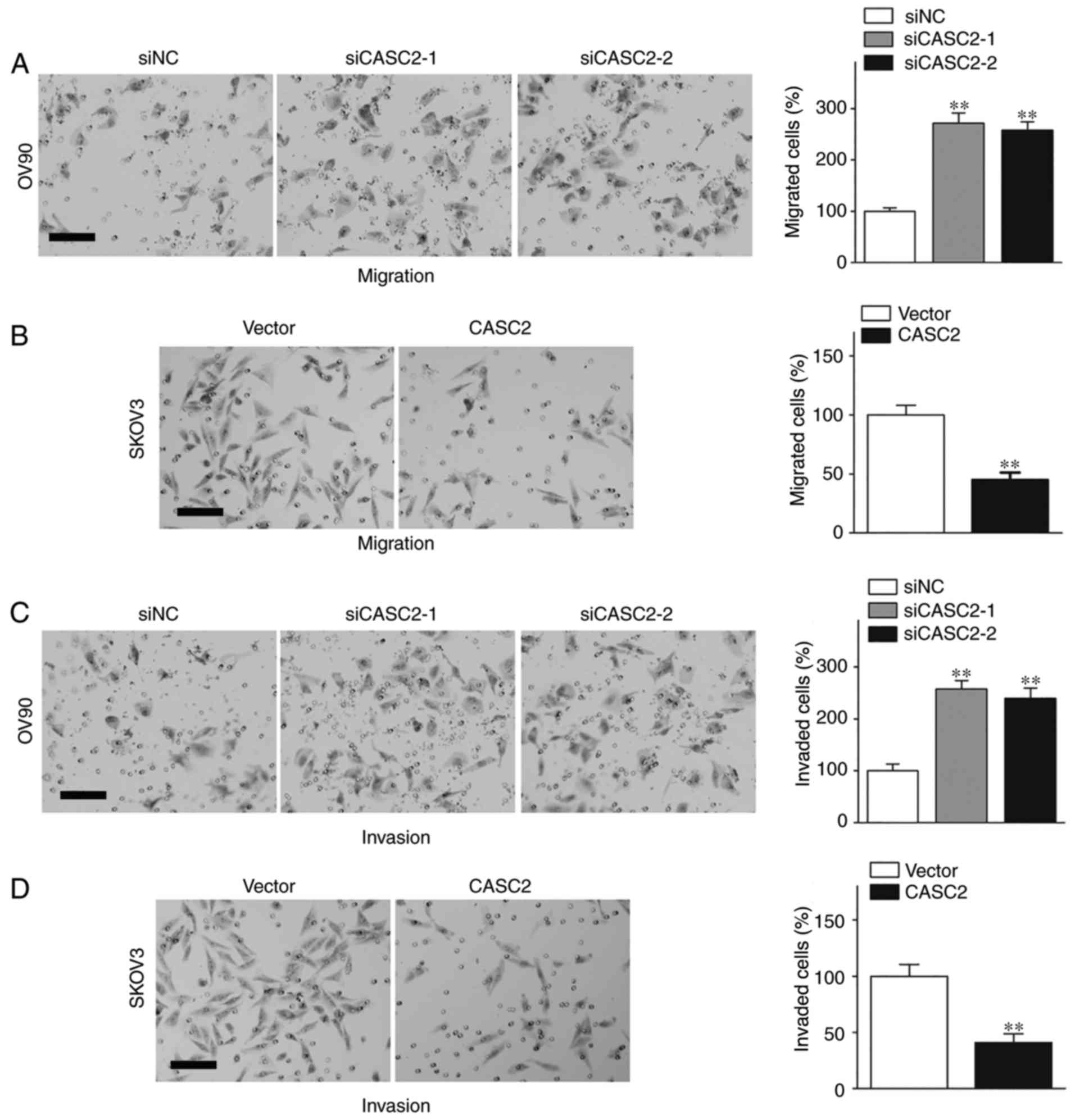
The effects of CASC2 interference and overexpression
on the motility of EOC cells were determined with the Transwell
assay and the Matrigel assay. The results of the migration assay
demonstrated that OV90 cells with silenced CASC2 exhibited
significantly increased migration ability in the Transwell assay
(Fig. 2A; P<0.01); whereas,
CASC2 upregulation significantly suppressed the migration of SKOV3
cells (Fig. 2B; P<0.01).
Accordingly, CASC2 deficiency significantly promoted invasion in
OV90 cells (Fig. 2C; P<0.01),
and ectopic overexpression of CASC2 significantly decreased
invasion in SKOV3 cells (Fig. 2D;
P<0.01). Therefore, CASC2 is able to inhibit the metastasis of
EOC cells.
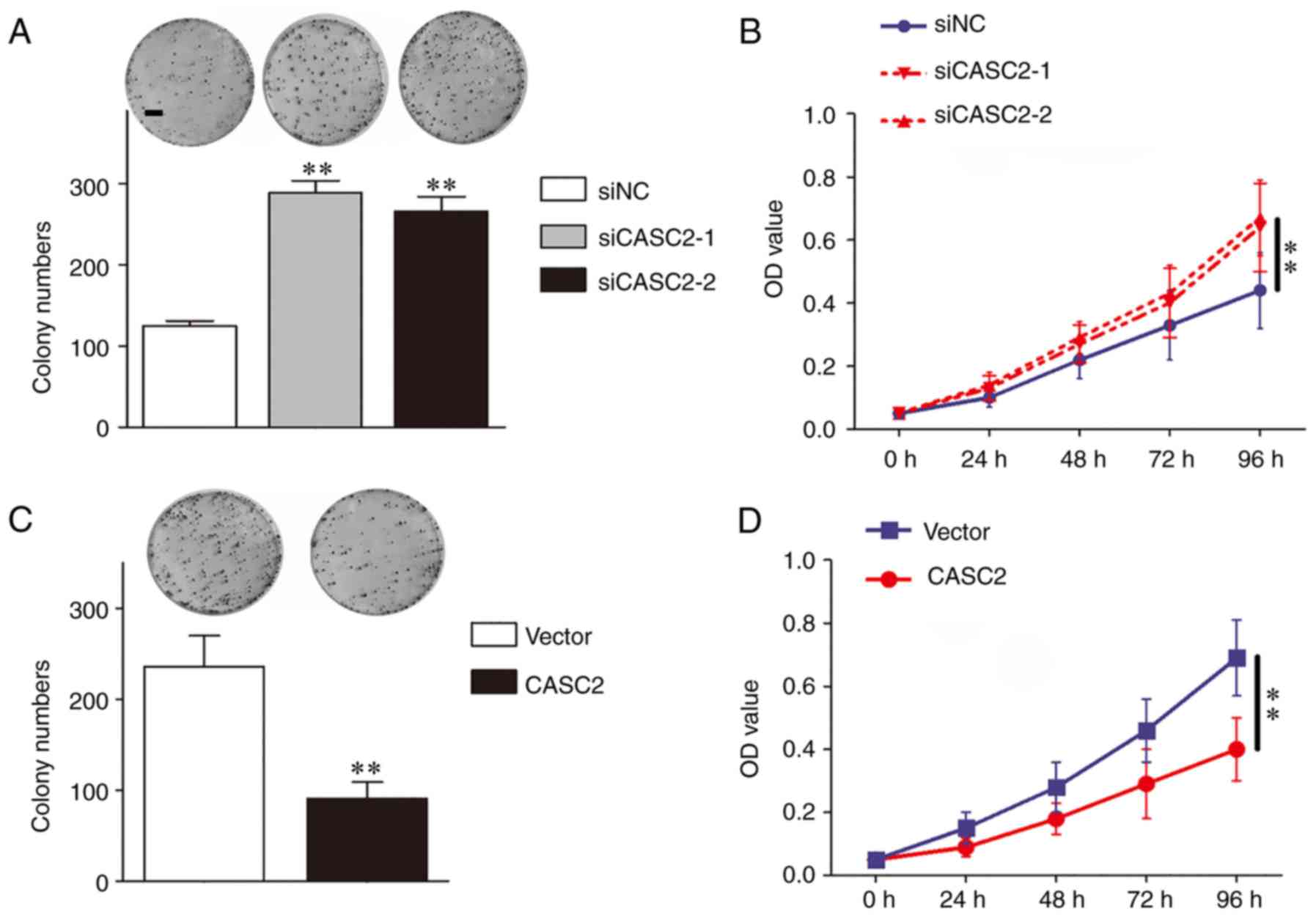
The CCK-8 assay and the colony formation assay were
used to investigate the role of CASC2 in EOC cell proliferation.
The results identified that CASC2 silencing increased the colony
numbers and the optical density values of OV90 cells in the colony
formation assay and CCK-8 assay, respectively (Fig. 3A and B). However, CASC2
upregulation reduced the proliferative ability of SKOV3 cells
(Fig. 3C and 3D). Overall, CASC2 suppressed the
proliferation of EOC cells.
Low CASC2 expression is an independent
risk factor for poor prognosis
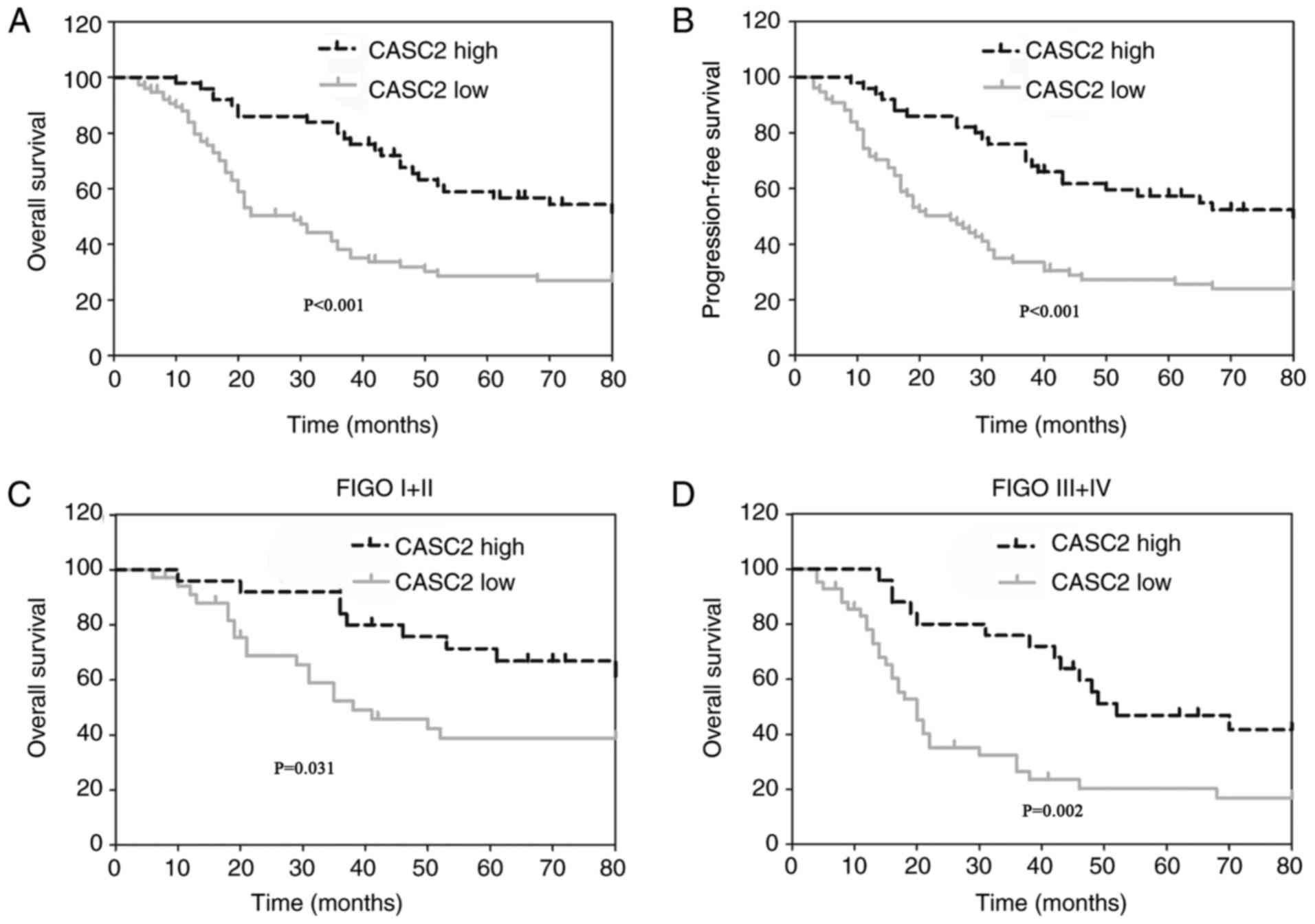
To verify the prognostic role of CASC2 in EOC,
statistical analysis was performed to evaluate the association
between CASC2 expression level and EOC prognosis. Notably, patients
with EOC with low CASC2 expression demonstrated a markedly poorer
overall survival rate (Fig. 4A)
and progression-free survival rate (Fig. 4B) compared with patients with high
CASC2 expression. In addition, the subtype analysis identified that
low CASC2 expression predicted shorter overall survival time in
patients with EOC with Fédération Internationale de Gynécologie et
d'Obstétrique (FIGO) (16) I/II
(Fig. 4C) and FIGO III/IV
(Fig. 4D).
Furthermore, the risk factors for overall survival
in EOC were analyzed by univariate analysis and multivariate
analysis. Lymph node metastasis (HR=1.754; 95% CI=1.075–2.860;
P=0.024), advanced FIGO stage (HR=1.977; 95% CI=1.234–3.169;
P=0.005) and low CASC2 expression (HR=0.403; 95% CI=0.245–0.663;
P<0.001) were revealed to be risk factors for poor overall
survival (Table II). The
subsequent multivariate analysis identified advanced FIGO stage
(HR=1.989; 95% CI=1.237–3.197; P=0.005) and low CASC2 expression
(HR=0.417; 95% CI=0.251–0.693; P=0.001) as independent risk factors
of EOC overall survival (Table
II). In addition, the risk factors for progression-free
survival were analyzed. As expected, lymph node metastasis
(HR=2.021; 95% CI=1.238–3.300; P=0.005), advanced FIGO stage
(HR=1.799; 95% CI=1.139–2.842; P=0.012) and low CASC2 expression
(HR=0.405; 95% CI=0.249–0.659; P<0.001) were identified as risk
factors for poor progression-free survival (Table III). The subsequent multivariate
analysis identified the following three factors as independent risk
factors for progression-free survival in EOC, lymph node metastasis
(HR=1.680; 95% CI=1.019–2.768; P=0.042), advanced FIGO stage
(HR=1.810; 95% CI=1.142–2.870; P=0.012), low CASC2 expression
(HR=0.426; 95% CI=0.260–0.699; P=0.001; Table III). Taken together, these
results suggest that CASC2 may be a promising biomarker for the
prediction of prognosis in patients with EOC.
 | Table II.Univariate and multivariate analysis
of clinicopathological features for overall survival of patients
with epithelial ovarian cancer. |
Table II.
Univariate and multivariate analysis
of clinicopathological features for overall survival of patients
with epithelial ovarian cancer.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Parameters | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age ≥60 years vs.
<60 years | 1.056 | 0.944–2.403 | 0.086 |
|
|
|
| Histology subtype
serous vs. others | 1.414 | 0.893–2.239 | 0.139 |
|
|
|
| CA125 <35 U/ml
vs. ≥35 U/ml | 1.768 | 0.987–3.168 | 0.055 |
|
|
|
| Lymph node
metastasis yes vs. no | 1.754 | 1.075–2.860 | 0.024 | 1.431 | 0.866–2.358 | 0.160 |
| Residual tumor size
<1 cm vs. ≥1 cm | 1.113 | 0.698–1.775 | 0.653 |
|
|
|
| Histological grade
G1+G2 vs. G3 | 1.052 | 0.665–1.664 | 0.827 |
|
|
|
| Tumor size ≥2 cm
vs. <2 cm | 1.061 | 0.672–1.674 | 0.799 |
|
|
|
| FIGO stage (III+IV)
vs. (I +II) | 1.977 | 1.234–3.169 | 0.005 | 1.989 | 1.237–3.197 | 0.005 |
| CSCA2 low vs.
high | 0.403 | 0.245–0.663 | <0.001 | 0.417 | 0.251–0.693 | 0.001 |
 | Table III.Univariate and multivariate analysis
of clinicopathological features for progression-free survival of
patients with epithelial ovarian cancer. |
Table III.
Univariate and multivariate analysis
of clinicopathological features for progression-free survival of
patients with epithelial ovarian cancer.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Parameters | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age ≥60 years vs.
<60 years | 1.460 | 0.925–2.303 | 0.104 |
|
|
|
| Histology subtype
serous vs. others | 1.485 | 0.947–2.328 | 0.085 |
|
|
|
| CA125 <35 U/ml
vs. ≥35 U/ml | 1.583 | 0.911–2.749 | 0.103 |
|
|
|
| Lymph node
metastasis yes vs. no | 2.021 | 1.238–3.300 | 0.005 | 1.680 | 1.019–2.768 | 0.042 |
| Residual tumor size
<1 cm vs. ≥1 cm | 1.173 | 0.744–1.850 | 0.492 |
|
|
|
| Histological grade,
G1+G2 vs. G3 | 1.155 | 0.734–1.818 | 0.533 |
|
|
|
| Tumor size ≥2 cm
vs. <2 cm | 1.127 | 0.720–1.764 | 0.600 |
|
|
|
| FIGO stage (III+IV)
vs. (I +II) | 1.799 | 1.139–2.842 | 0.012 | 1.810 | 1.142–2.870 | 0.012 |
| CASC2 low vs.
high | 0.405 | 0.249–0.659 | <0.001 | 0.426 | 0.260–0.699 | 0.001 |
Discussion
Due to the various functional roles of lncRNAs in
cancer biology, an increasing number of studies have focused on the
role of lncRNAs in different types of cancer. Through analysis of
the differential expression of lncRNAs in normal tissues and cancer
tissues, lncRNAs have been observed to be involved in the initial
of carcinogenesis (17,18). A larger number of lncRNAs have been
discovered as a result of the extensive use of next-generation
sequencing technologies, and additional functional roles of lncRNAs
have been detected in carcinogenesis and cancer progression
(17,18). Through analysis of the expression
profiles of lncRNAs in EOC, specific differentially expressed
lncRNAs have been identified (19). Further comprehensive analysis of
the expression profiles of lncRNAs in EOC revealed that an
eight-lncRNA signature may be a measure for predicting the
chemotherapeutic response and identifying patients with platinum
resistance, who may benefit from other more effective therapies
(20). At present, a number of
lncRNAs have been suggested to be associated with EOC progression,
chemoresistance and prognosis, including deleted in lymphocytic
leukemia 1 (21), H19, imprinted
maternally expressed transcript (22), HOX transcript antisense RNA
(23–26), nuclear paraspeckle assembly
transcript 1 (27,28) and metastasis associated lung
adenocarcinoma transcript 1 (29).
Mechanistically, lncRNAs have been identified to regulate EOC
progression by mediating cancer cell proliferation, metastasis,
inflammasome formation, and the epithelial to mesenchymal
transition (23,30,31).
Numerous lncRNAs are involved in regulating EOC biology through
various ways. In addition, certain lncRNAs, including AB073614
(32), colon cancer associated
transcript 2 (33) and
neuroblastoma associated transcript 1 (34), have been demonstrated to be
potential prognostic markers in patients with EOC. However, the
significance of CASC2 in EOC has not been elucidated yet.
The lncRNA CASC2 was discovered in 2004 in patients
with endometrial carcinoma as a potential tumor suppressor
(6). CASC2 transcripts may be
classified into three subgroups; CASC2a, CASC2b and CASC2c
(5). Evidence suggested that the
expression levels of CASC2b and CASC2c mRNA were similar in normal
and neoplastic endometrial tissues; whereas, CASC2a was identified
to be downregulated in neoplastic samples compared with the normal
counterparts (5). Since then,
further studies in other types of neoplasia have been conducted,
and the mechanisms and the interactions of CASC2 in cancer have
been better elucidated (35); in
addition, CASC2 has become synonymous with CASC2a. Aberrant
expression of CASC2 was detected in renal cell carcinoma (12), osteosarcoma (36), pancreatic cancer (13), endometrial cancer (6), hepatocellular carcinoma (37), glioma (10,38,39),
non-small cell lung cancer (8) and
gastric cancer (11).
Functionally, different previous studies have verified that CASC2
may be involved in cancer tumorigenesis, autophagy, proliferation,
invasion, metastasis and apoptosis (35). Mechanistically, CASC2 has been
demonstrated to function by interacting with microRNAs (38,40–43),
the phosphatase and tensin homology pathway (13,44)
and the Wnt/β-catenin signaling pathway (45). Furthermore, survival analyses
identified that low CASC2 expression predicts poor prognosis in
thyroid cancer (46), glioma
(10), astrocytoma (40) and non-small cell lung cancer
(8). Taken together, these results
suggest that CASC2 may act as a tumor suppressor in a wide range of
cancer types through different signaling pathways.
The present study investigated the significance of
CASC2 in EOC by determining its baseline expression level. In
accordance with the results of previous studies (35), CASC2 demonstrated significantly
decreased expression levels in EOC cells and tissues. In addition,
the clinical significance analysis of the association CASC2
expression and the clinicopathological features of patients with
EOC identified that low CASC2 expression was associated with the
serous histological subtype, lymph node metastasis, poor
histological grade and large tumor size, all of which contribute to
EOC progression. Indeed, the prognostic evaluation revealed that
patients with low CASC2 expression exhibited a markedly poorer
overall survival rate and progression-free survival rate. In
addition, further analysis revealed that patients with FIGO stage
I/II or III/IV demonstrated a poorer overall survival rate if they
additionally had low CASC2 expression. Furthermore, low CASC2
expression was confirmed to be an independent risk factor for poor
prognosis in EOC. These results suggested that CASC2 has a primary
role in inhibiting EOC progression in patients. To verify the
function of CASC2 in EOC cells, CASC2 was overexpressed or
silenced, and the results of the functional studies confirmed that
CASC2 may inhibit the proliferation and metastasis of EOC cells.
However, the clinical specimens used in the present study were
collected in one medical center. Future studies investigating CASC2
should be performed in numerous different centers using a larger
sample size. The underlying mechanism by which CASC2 exerts its
function should be further explored.
In conclusion, CASC2 expression levels were
decreased in EOC cells and tissues, and a low CASC2 expression
level was associated with clinical progression in patients with
EOC. The results of the functional studies identified that CASC2
may inhibit the proliferation and metastasis of EOC cells.
Furthermore, patients with low CASC2 expression exhibited a poorer
overall survival rate and a shorter progression-free survival
period. Notably, low CASC2 expression was identified as an
independent risk factor for overall survival and progression-free
survival in patients with EOC. However, the detailed mechanisms of
the influence of CASC2 on EOC progression were not further verified
in the present study. The functional role of CASC2 as a tumor
suppressor was demonstrated in EOC, and CASC2 may be considered a
promising prognostic marker and therapeutic target in EOC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZX performed the statistical analysis of clinical
data, performed in vitro assays and wrote the manuscript. XZ
determined the expression levels of CASC2 in EOC cells and tissues,
and performed statistical analysis. YT designed the study,
performed the statistical analysis and wrote the manuscript. All
authors read and approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Shanghai Jiao Tong University Affiliated Sixth
People's Hospital. Written informed consent was obtained from each
patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Goff BA, Mandel L, Muntz HG and Melancon
CH: Ovarian carcinoma diagnosis. Cancer. 89:2068–2075. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bookman MA, Brady MF, McGuire WP, Harper
PG, Alberts DS, Friedlander M, Colombo N, Fowler JM, Argenta PA, De
Geest K, et al: Evaluation of new platinum-based treatment regimens
in advanced-stage ovarian cancer: A Phase III Trial of the
Gynecologic Cancer Intergroup. J Clin Oncol. 27:1419–1425. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Morales Rivea D, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–11672. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Baldinu P, Cossu A, Manca A, Satta MP,
Sini MC, Rozzo C, Dessole S, Cherchi P, Gianfrancesco F, Pintus A,
et al: Identification of a novel candidate gene, CASC2, in a region
of common allelic loss at chromosome 10q26 in human endometrial
cancer. Hum Mutat. 23:318–326. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Baldinu P, Cossu A, Manca A, Satta MP,
Sini MC, Palomba G, Dessole S, Cherchi P, Mara L, Tanda F and
Palmieri G: CASC2a gene is down-regulated in endometrial cancer.
Anticancer Res. 27:235–243. 2007.PubMed/NCBI
|
|
8
|
He X, Liu Z, Su J, Yang J, Yin D, Han L,
De W and Guo R: Low expression of long noncoding RNA CASC2
indicates a poor prognosis and regulates cell proliferation in
non-small cell lung cancer. Tumour Biol. 37:9503–9510. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gan Y, Han N, He X, Yu J, Zhang M, Zhou Y,
Liang H, Deng J, Zheng Y, Ge W, et al: Long non-coding RNA CASC2
regulates cell biological behaviour through the MAPK signalling
pathway in hepatocellular carcinoma. Tumor Biol.
39:10104283177062292017. View Article : Google Scholar
|
|
10
|
Wang R, Li Y, Zhu G, Tian B, Zeng W, Yang
Y and Li Z: Long noncoding RNA CASC2 predicts the prognosis of
glioma patients and functions as a suppressor for gliomas by
suppressing Wnt/β-catenin signaling pathway. Neuropsychiatr Dis
Treat. 13:1805–1813. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou J, Huang H, Tong S and Huo R:
Overexpression of long non-coding RNA cancer susceptibility 2
inhibits cell invasion and angiogenesis in gastric cancer. Mol Med
Rep. 16:5235–5240. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cao Y, Xu R, Xu X, Zhou Y, Cui L and He X:
Downregulation of lncRNA CASC2 by microRNA-21 increases the
proliferation and migration of renal cell carcinoma cells. Mol Med
Rep. 14:1019–1025. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu Y, Liang S, Zhou Y, Li S, Li Y and Liao
W: HNF1A/CASC2 regulates pancreatic cancer cell proliferation
through PTEN/Akt signaling. J Cell Biochem. 2017.(Epub ahead of
print). View Article : Google Scholar
|
|
14
|
Yang X, Wang J, Li WP, Jin ZJ and Liu XJ:
Desmocollin 3 mediates follicle stimulating hormone-induced ovarian
epithelial cancer cell proliferation by activating the EGFR/Akt
signaling pathway. Int J Clin Exp Patho. 8:6716–6723. 2015.
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Prat J: Staging classification for cancer
of the ovary, fallopian tube, and peritoneum. International journal
of gynaecology and obstetrics: The official organ of the
International Federation of Gynaecology and Obstetrics. 124:1–5.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bhan A, Soleimani M and Mandal SS: Long
Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jarroux J, Morillon A and Pinskaya M:
History, discovery, and classification of lncRNAs. Adv Exp Med
Biol. 1008:1–46. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shen L, Liu W, Cui J, Li J and Li C:
Analysis of long non-coding RNA expression profiles in ovarian
cancer. Oncol Lett. 14:1526–1530. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou M, Sun Y, Sun Y, Xu W, Zhang Z, Zhao
H, Zhong Z and Sun J: Comprehensive analysis of lncRNA expression
profiles reveals a novel lncRNA signature to discriminate
nonequivalent outcomes in patients with ovarian cancer. Oncotarget.
7:32433–32448. 2016.PubMed/NCBI
|
|
21
|
Wang LL, Sun KX, Wu DD, Xiu YL, Chen X,
Chen S, Zong ZH, Sang XB, Liu Y and Zhao Y: DLEU1 contributes to
ovarian carcinoma tumourigenesis and development by interacting
with miR-490-3p and altering CDK1 expression. J Cell Mol Med.
21:3055–3065. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Medrzycki M, Zhang Y, Zhang W, Cao K, Pan
C, Lailler N, McDonald JF, Bouhassira EE and Fan Y: Histone h1.3
suppresses h19 noncoding RNA expression and cell growth of ovarian
cancer cells. Cancer Res. 74:6463–6473. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dong L and Hui L: HOTAIR promotes
proliferation, migration, and invasion of ovarian cancer SKOV3
cells through regulating PIK3R3. Med Sci Monit. 22:325–331. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Teschendorff AE, Lee SH, Jones A, Fiegl H,
Kalwa M, Wagner W, Chindera K, Evans I, Dubeau L, Orjalo A, et al:
HOTAIR and its surrogate DNA methylation signature indicate
carboplatin resistance in ovarian cancer. Genome Med. 7:1082015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Z, Cheng J, Wu Y, Qiu J, Sun Y and
Tong X: LncRNA HOTAIR controls the expression of Rab22a by sponging
miR-373 in ovarian cancer. Mol Med Rep. 14:2465–2472. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ozes AR, Miller DF, Ozes ON, Fang F, Liu
Y, Matei D, Huang T and Nephew KP: NF-κB-HOTAIR axis links DNA
damage response, chemoresistance and cellular senescence in ovarian
cancer. Oncogene. 35:5350–5361. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chai Y, Liu J, Zhang Z and Liu L:
HuR-regulated lncRNA NEAT1 stability in tumorigenesis and
progression of ovarian cancer. Cancer Med. 5:1588–1598. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
An J, Lv W and Zhang Y: LncRNA NEAT1
contributes to paclitaxel resistance of ovarian cancer cells by
regulating ZEB1 expression via miR-194. Onco Targets Ther.
10:5377–5390. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou Y, Xu X, Lv H, Wen Q, Li J, Tan L, Li
J and Sheng X: The long noncoding RNA MALAT-1 is highly expressed
in ovarian cancer and induces cell growth and migration. PLoS One.
11:e01552502016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mitra R, Chen X, Greenawalt EJ, Maulik U,
Jiang W, Zhao Z and Eischen CM: Decoding critical long non-coding
RNA in ovarian cancer epithelial-to-mesenchymal transition. Nat
Commun. 8:16042017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ren C, Li X, Wang T, Wang G, Zhao C, Liang
T, Zhu Y, Li M, Yang C, Zhao Y and Zhang GM: Functions and
mechanisms of long noncoding RNAs in ovarian cancer. Int J Gynecol
Cancer. 25:566–569. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cheng Z, Guo J, Chen L, Luo N, Yang W and
Qu X: A long noncoding RNA AB073614 promotes tumorigenesis and
predicts poor prognosis in ovarian cancer. Oncotarget.
6:25381–25389. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang S, Qing C, Huang Z and Zhu Y: The
long non-coding RNA CCAT2 is up-regulated in ovarian cancer and
associated with poor prognosis. Diagn Pathol. 11:492016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yan C, Jiang Y, Wan Y, Zhang L, Liu J,
Zhou S and Cheng W: Long noncoding RNA NBAT-1 suppresses
tumorigenesis and predicts favorable prognosis in ovarian cancer.
Onco Targets Ther. 10:1993–2002. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Palmieri G, Paliogiannis P, Sini MC, Manca
A, Palomba G, Doneddu V, Tanda F, Pascale MR and Cossu A: Long
non-coding RNA CASC2 in human cancer. Crit Rev Oncol Hematol.
111:31–38. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ba Z, Gu L, Hao S, Wang X, Cheng Z and Nie
G: Downregulation of lncRNA CASC2 facilitates osteosarcoma growth
and invasion through miR-181a. Cell Prolif. 51:2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zeng F, Le YG, Fan JC and Xin L: LncRNA
CASC2 inhibited the viability and induced the apoptosis of
hepatocellular carcinoma cells through regulating miR-24-3p. J Cell
Biochem. 119:6391–6397. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liao Y, Shen L, Zhao H, Liu Q, Fu J, Guo
Y, Peng R and Cheng L: LncRNA CASC2 interacts with miR-181a to
modulate glioma growth and resistance to TMZ through PTEN pathway.
J Cell Biochem. 118:1889–1899. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jiang C, Shen F, Du J, Fang X, Li X, Su J,
Wang X, Huang X and Liu Z: Upregulation of CASC2 sensitized glioma
to temozolomide cytotoxicity through autophagy inhibition by
sponging miR-193a-5p and regulating mTOR expression. Biomed
Pharmacother. 97:844–850. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu C, Sun Y, She X, Tu C, Cheng X, Wang
L, Yu Z, Li P, Liu Q, Yang H, et al: CASC2c as an unfavorable
prognosis factor interacts with miR-101 to mediate astrocytoma
tumorigenesis. Cell Death Dis. 8:e26392017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang W, He W, Gao J, Wang Y, Zang W, Dong
Z and Zhao G: Retraction notice to the long noncoding RNA CASC2
inhibits tumorigenesis through modulating the expression of PTEN by
targeting miR-18a-5p in esophageal carcinoma. Exp Cell Res.
361:30–38. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang Y, Liu Z, Yao B, Li Q, Wang L, Wang
C, Dou C, Xu M, Liu Q and Tu K: Long non-coding RNA CASC2
suppresses epithelial-mesenchymal transition of hepatocellular
carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol Cancer.
16:1232017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang P, Liu YH, Yao YL, Li Z, Li ZQ, Ma J
and Xue YX: Long non-coding RNA CASC2 suppresses malignancy in
human gliomas by miR-21. Cell Signal. 27:275–282. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Feng Y, Zou W, Hu C, Li G, Zhou S, He Y,
Ma F, Deng C and Sun L: Modulation of CASC2/miR-21/PTEN pathway
sensitizes cervical cancer to cisplatin. Arch Biochem Biophys.
623–624:20–30. 2017. View Article : Google Scholar
|
|
45
|
Pei Z, Du X, Song Y, Fan L, Li F, Gao Y,
Wu R, Chen Y, Li W, Zhou H, et al: Down-regulation of lncRNA CASC2
promotes cell proliferation and metastasis of bladder cancer by
activation of the Wnt/beta-catenin signaling pathway. Oncotarget.
8:18145–18153. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xiong X, Zhu H and Chen X: Low expression
of long noncoding RNA CASC2 indicates a poor prognosis and promotes
tumorigenesis in thyroid carcinoma. Biomed Pharmacother.
93:391–397. 2017. View Article : Google Scholar : PubMed/NCBI
|