Introduction
Mechanical stimulation is an important environmental
factor that influences bone metabolism (1). The molecular signals driving the
regenerative process in distraction osteogenesis involves a complex
system of cellular behaviours triggered by mechanical strain
(1). Previously, attention has
been focused on the role of mechanical forces in the
differentiation of stem cells, including the differentiation of
mesenchymal stem cells (MSCs) into osteoblasts (2,3).
However, whether mechanical stimuli can modulate the osteogenic
differentiation of human amniotic epithelial cells (hAECs) remains
unknown.
hAECs can be derived from the full-term placenta and
express embryonic stem cell (ESC) markers, including stage-specific
embryonic antigen-4 (SSEA-4) and octamer-binding protein 4 (Oct-4)
(4,5). According to Miki et al
(5), the amnion is derived from
the totipotent epiblast and is considered an alternative source of
stem cells due to the expression of SSEA-4. Furthermore, as Toda
et al (6) reported that
various advantages make the amnion a potentially useful and
noncontroversial source of cells for transplantation and
regenerative medicine. The considerable advantages described in
detail were as follows: hAECs can differentiate into all three germ
layers; they demonstrate low immunogenicity and anti-inflammatory
functions; and they prompt no ethical concerns due to not requiring
the sacrifice of human embryos for their isolation, therefore
avoiding the current controversies associated with the use of human
ESCs (5,6).
hAECs have the potential to differentiate into all
three germ layers: Endoderm (liver, pancreas), mesoderm
(cardiomyocyte) and ectoderm (neural cells) in vitro
(7,8). According to Kakishita et al
(9), AECs can differentiate into
mature neural cells that synthesize and release acetylcholine,
norepinephrine and dopamine. Wlodarski et al (10) demonstrated that WISH cells (an
amniotic epithelial cell line) grafted intramuscularly into rats or
into Syrian hamsters survived 14 days and induced both cartilage
and bone formation. It has also been reported that hAECs can
differentiate into osteoblasts following stimulation with a pulsed
electromagnetic field (11).
Therefore, stimulation of hAECs in this manner has been proposed as
a potential source of stem cells for cell therapy.
Previously, stem cell-based therapies have been
utilized for their ability to regenerate osteoblasts and have been
demonstrated to be efficacious in experimental contexts and human
disease. In addition, dexamethasone, β-glycerol phosphate and
ascorbic acid have been identified as osteogenic differentiation
inducers in hAECs (12).
Furthermore, reports of hAECs differentiating into osteoblasts upon
induction by mechanical stretching are rare. In the present study,
the hypothesis that mechanical stretching serves important roles in
the process of osteogenic differentiation of hAECs was
confirmed.
Materials and methods
Isolation and culture of hAECs
Placentas were donations from six patients (from
February 15th 2015 to August 4th 2017; mean age, 32±4.5) of
Yongchuan Hospital of Chongqing Medical University (Chongqing,
China). All patients gave written informed consent. The present
study was performed according to the Declaration of Helsinki and
approved by the Medical Ethics Committee of the Yongchuan Hospital
of Chongqing Medical University.
As described by Miki et al (13), the amnion layer was mechanically
peeled from human placenta and digested with 0.05% trypsin + 0.02%
EDTA for 30 min at 37°C (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). Following filtration with 200-mesh filter and
centrifugation at 200 × g for 5 min at 4°C, the cells were cultured
in low glucose Dulbecco's modified Eagle's medium (DMEM; Gibco;
Thermo Fisher Scientific, Inc.) at a density of 1×105
cells/cm2 and placed in a 37°C and 5% CO2
humidified incubation chamber. The medium was supplemented with 5%
foetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 2
mmol/l-glutamine, 1% non-essential amino acids and 55 µmol/l
β-mercaptoethanol. New culture medium was supplied after 24 h of
subculture. Experiments were performed once the cells were fused
into a single layer and reached 80–90% confluence.
Identification of hAECs
The hAECs were identified immunocytochemically by
the specific epithelial cell marker cytokeratin-19 (CK19) and the
ESC marker Oct-4, the cell surface marker cluster of
differentiation 44 (CD44), and the stem cell surface marker SSEA-4
(all Abcam, Cambridge, UK).
hAECs (3rd passage) were seeded onto cover slips in
6-well plates. At 80–90% confluence, they were incubated with
primary antibodies (1:300) at 4°C and secondary antibodies (Goat
anti-Mouse/rabbit IgG (H+L) secondary antibody; Alex Fluor 488/594;
1:200; Jackson) at 25°C following a standard immunocytochemistry
protocol including fixing with 4% polyoxymethylene at 4°C for 30
min, permeabilizing with 1% Triton-X100 for 10 min at 25°C and
blocking with 5% bovine serum albumin (BSA; Sigma-Aldrich,
Darmstadt, Germany, USA) for 30 min at 25°C. Eventually,
4′,6-diamidino-2-phenylindole (DAPI; 1:1,000; Invitrogen; Thermo
Fisher Scientific, Inc.) was used to mark the nucleus for 10 min at
25°C. The stained cells were imaged on a confocal inverted
microscope (Olympus FluoView FV 1,000; Olympus Corporation, Tokyo,
Japan).
Mechanical stretch stimulation
In the mechanical stretch group, the 3rd to 5th
passage hAECs were seeded reached 60–70% confluency on silicone
membranes coated with collagen type I, transferred onto a uniaxial
stretch chamber (size, 4.0×7.0 cm), the maximum uniaxial stretched
length is 7.35 cm, using a machine similar to that in Hata et
al (14) and stretched for 2,
6, 12 and 24 h with 5% elongation at a frequency of 0.5 Hz
(15,16). The positive control group was
treated with osteogenic induction medium (OM; DMEM supplemented
with 100 nmol/l dexamethasone, 50 mg/l ascorbic acid and 5 mmol/l
β-glycerophosphate) for 21 days. The stretch-biochemical combined
stimulation group was treated with stretching with 5% elongation at
0.5 Hz for 2, 6, 12 and 24 h following 21 days of OM stimulation.
The cells were collected using TRIzol™ (Thermo Fisher Scientific,
Inc.) or radioimmunoprecipitation assay (RIPA) lysis buffer
(Beyotime Institute of Biotechnology, Haimen, China) at each time
point. Each experiment was conducted in five replicates.
RNA interference
4 µg of runt-related transcription factor 2
(Runx2)/core binding factor α 1 (Cbfa1)-specific short hairpin RNA
(shRNA) plasmid and 4 µg of scrambled shRNA control plasmid was
constructed, followed by cationic lipid (Lipofectamine™ 2000
Invitrogen; Thermo Fisher Scientific, Inc.) transfection for 48 h
at 37°C. OM induction and stretch was performed following
transfection for 24 h. The sequences of Runx2/Cbfa1-shRNA and
scrambled shRNA, designed based on NCBI GeneBank data, are
demonstrated in Table I.
 | Table I.Sequences of Runx2-shRNA and scrambled
control. |
Table I.
Sequences of Runx2-shRNA and scrambled
control.
| Name | Sequence |
|---|
| Runx2-shRNA | F:
5′-GATCCCCAGACTTACTGTATGTATAGTTCAAGAGACTATACATACAGTAAGTCTTTTTTGGAAA-3′ |
|
| R:
5′-AGCTTTTCCAAAAAAGACTTACTGTATGTATAGTCTCTTGAACTATACATACAGTAAGTCTGGG-3′ |
| Control shRNA | F:
5′-GATCCCCTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTTGGAAA-3′ |
|
| R:
5′-AGCTTTTCCAAAAATTCTCCGAACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGAGAAGGG-3′ |
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The hAECs in each group were collected with TRIzol
and the corresponding RNA samples were obtained following
extraction and purification. For extraction, 1 ml TRIzol was used
to collect total RNA and 0.2 ml chloroform was added to each sample
to extract RNA. Following incubation at 4°C for 5 min, samples were
centrifuged at 12,000 × g for 15 min at 4°C. Then, the upper
aqueous fraction was collected by the addition of 0.5 ml ethanol.
Followed centrifugation at 12,000 × g for 15 min at 4°C, 75%
ethanol was used to purify the extracted RNA. Following air-drying,
RNA was eluted in 20 µl RNase-free water. RNA concentrations were
quantified by spectrophotometry (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) and cDNA was synthesized using a RT kit
(FSQ-101; Toyobo Life Science, Osaka, Japan) at 37°C for 5 min and
then 95°C for 5 min. To detect the accumulation of PCR products,
EvaGreen (Bio-Rad Laboratories, Inc.) with the CFX-96™ (Bio-Rad
Laboratories, Inc.) Real-Time PCR Detection System was used
followed by 40 cycles of: 95°C for 5 min, 95°C for 10 sec, 59°C for
1 min. The sequences of qPCR primers and program are presented in
Table II. The β-actin gene was
used as an internal reference (17).
 | Table II.Primer sequences for reverse
transcription-quantitative polymerase chain reaction. |
Table II.
Primer sequences for reverse
transcription-quantitative polymerase chain reaction.
| Gene | GenBank ID | Primer sequence
(5′-3′) |
|---|
| ALP | NM_000478 | F:
5′GGAACTCCTGACCCTTGACC 3′ |
|
|
| R:
5′TCCTGTTCAGCTCGTACTGC 3′ |
| OC | NM_001199662 | F:
5′AGGGCAGCGAGGTAGTGAA 3′ |
|
|
| R:
5′TCCTGAAAGCCGATGTGGT 3′ |
| Runx2/Cbfa1 | NM_001015051 | F:
5′TTACTTACACCCCGCCAGTC 3′ |
|
|
| R:
5′TATGGAGTGCTGCTGGTCTG 3′ |
| β-catenin | NM_020248 | F:
5′CCTATGCAGGGGTGGTCAAC 3′ |
|
|
| R:
5′CGACCTGGAAAACGCCATCA 3′ |
| Cyclin D1 | NM_053056.2 | F:
5′TTTGTTGTGTGTGCAGGGAG 3′ |
|
|
| R:
5′TTTCTTCTTGACTGGCACGC 3’ |
| β-actin | NM_001099771 | F:
5′ACTATCGGCAATGAGCGGTTC 3′ |
|
|
| R:
5′ATGCCACAGGATTCCATACCC 3′ |
Western blot analysis
The proteins from each group of hAECs were
separately collected in RIPA lysis buffer (Santa Cruz
Biotechnology, Inc., Dallas, TX, USA). Following centrifugation at
4°C and 16,000 × g for 10 min, the supernatant proteins were
collected. The concentration was determined by the bicinchoninic
acid method, followed by loading 20 µg total proteins onto the
SDS-PAGE gels (4% separation gel, 5% spacer gel) and transferred to
0.45 µm Immobilon polyvinylidene difluoride membranes (EMD
Millipore, Billerica, MA, USA). The membranes were blocked with 5%
BSA in Tris Buffered Saline with Tween® 20 (TBST) at
25°C for 1 h, incubated with the indicated primary antibodies
[anti-β-catenin (1:1,000; cat. no. ab16051), anti-Runx2 (1:1,000;
ab23981), anti-Cyclin D1 (1:1,000; cat. no. ab226977),
anti-alkaline phosphatase (1:1,000; cat. no. ab229126) and
anti-osteocalcin (1:1,000; cat. no. ab93876; Abcam)] at 4°C
overnight and then incubated with the corresponding secondary
antibodies (Goat anti-mouse/rat secondary antibodies; 1:3,000; cat.
no. ZB2305/ZB2306; Zsbio; OriGene Technologies, Inc., Beijing,
China) at 25°C for 1 h. Images were obtained with an enhanced
chemiluminescence kit (Thermo Fisher Scientific, Inc.) on film and
the images were captured, and analysed with a gel imaging system
(2.0 version of quantity one 1-D, Bio-Rad Laboratories, Inc.).
Statistical analysis
The data are presented as the mean ± standard
deviation. Single-tailed Student's t-test of unpaired data or
one-way analysis of variance, followed by the least significance
difference performed in SPSS 13.0 software (SPSS, Inc., Chicago,
IL, USA) was used for comparisons between groups, n=3. P<0.05
was considered to indicate a statistically significant
difference.
Results
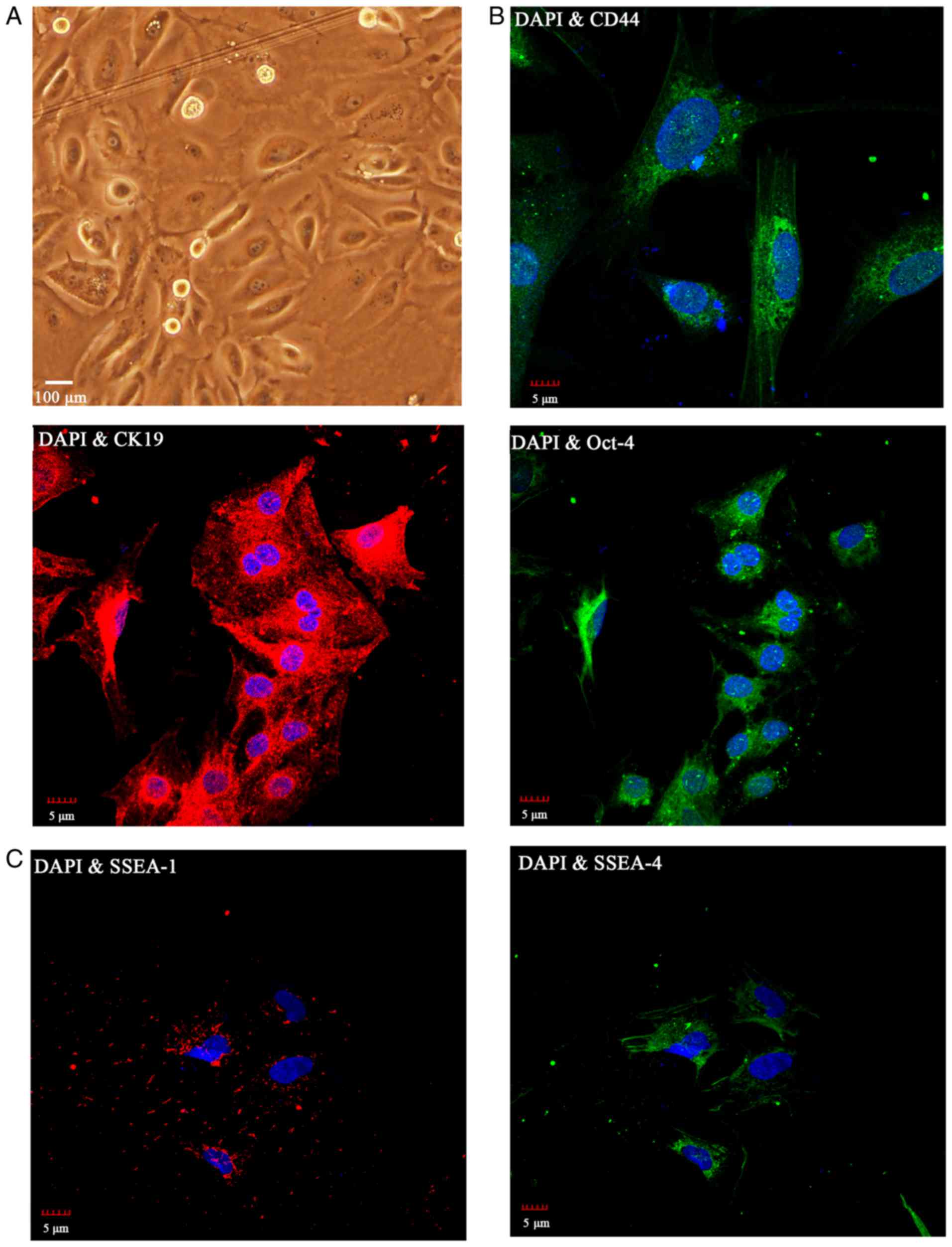
Morphology and characteristics of
isolated hAECs
hAECs were successfully isolated from full-term
amnion without disease. Following 3 days in culture, the hAECs
demonstrated the typical morphology of cobblestone-shape
epithelial-like cells (Fig.
1A).
Expression of the epithelial cell marker CK19, and
the ESC markers CD44 and Oct-4 was detected in the primary cultures
of hAECs by immunocytochemistry (Fig.
1B). The stem cell surface marker SSEA-4 was highly expressed
in the hAECs, as indicated by the confocal images; whereas, SSEA-1
was negative (Fig. 1C). Together,
the results suggested that the hAECs that were obtained were
CK19-positive and expressed stem cell markers, CD44, Oct-4 and
SSEA-4, which is consistent with previous reports (4,5).
Based on the DAPI and marker staining that were performed in
Fig. 1, the majority of the cells
that were isolated were indeed hAECs.
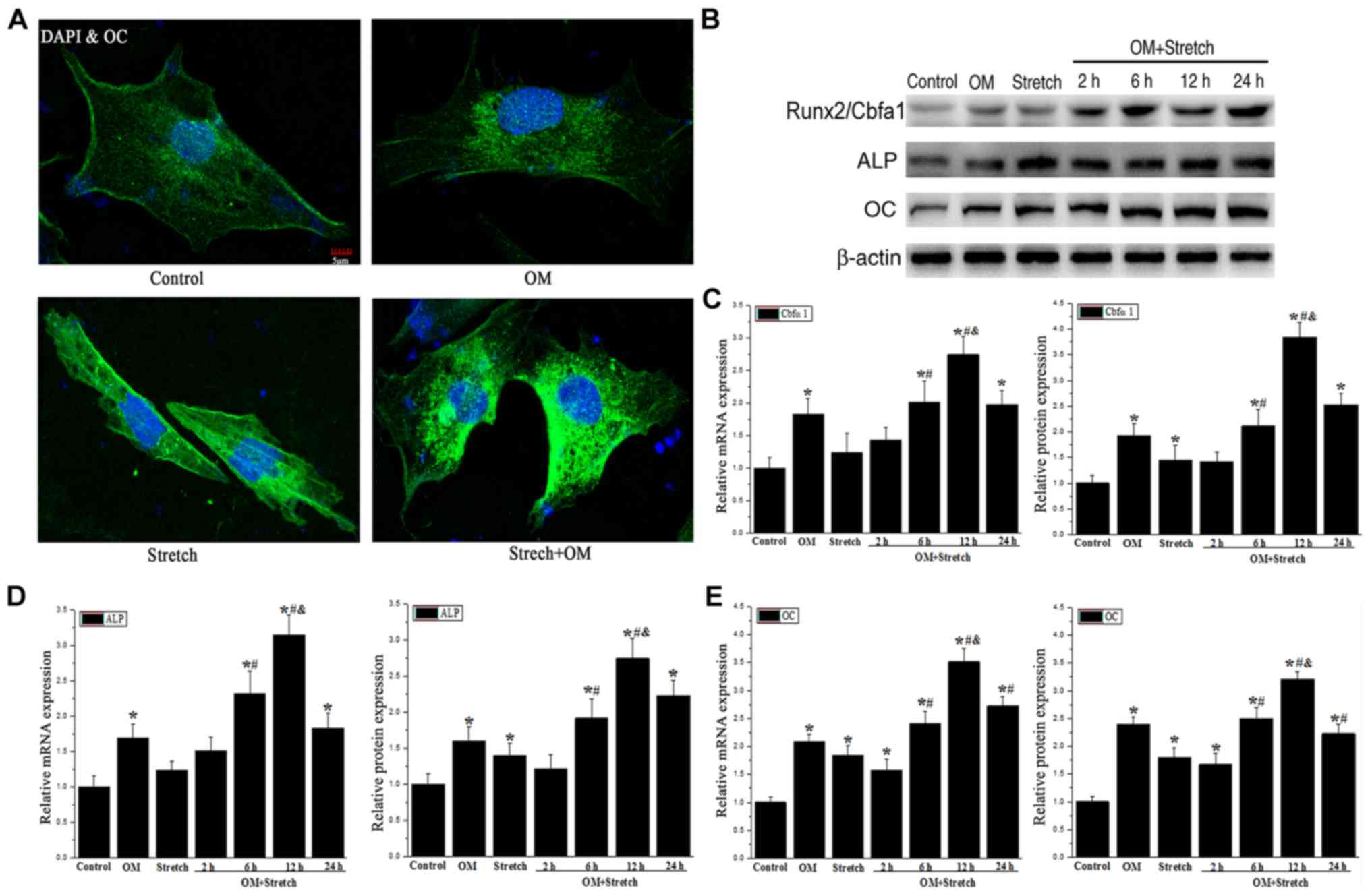
Stretch regulates the osteogenic
differentiation of AECs
To determine whether the mechanical stimulation
affected the osteogenic differentiation of the hAECs, the protein
level of osteocalcin was detected by immunocytochemistry. The data
demonstrated that the osteocalcin (OC) level was induced at 12 h
post-treatment by OM and OM + stretch compared with the control
(Fig. 2A). This indicated that
hAECs can be induced to differentiate into osteogenic-like
cells.
To investigate the whether that mechanical stretch
induces hAECs to differentiate into osteoblasts, RT-qPCR and
western blotting were performed to determine the expression levels
of osteoblast markers, including Runx2/Cbfa1, alkaline phosphatase
(ALP) and OC. The results suggested that the expression levels of
Runx2/Cbfa1, ALP and OC demonstrated were significantly increased
in the OM-induced group, the positive control group, and there was
an increasing trend in the stretch group but with no significant
differences compared with the control group (Fig. 2B-E).
Compared with the stretch or OM group alone, the
mRNA and protein expression levels of Runx2/Cbfa1, ALP and OC
increased following 6 h of treatment with OM combined with stretch
(OM + stretch) and peaked at 12 h but decreased at 24 h (P<0.05;
Fig. 2B-E). In brief, the above
results suggested that the combination of OM and stretch had a
synergistic effect on the osteogenic differentiation of hAECs.
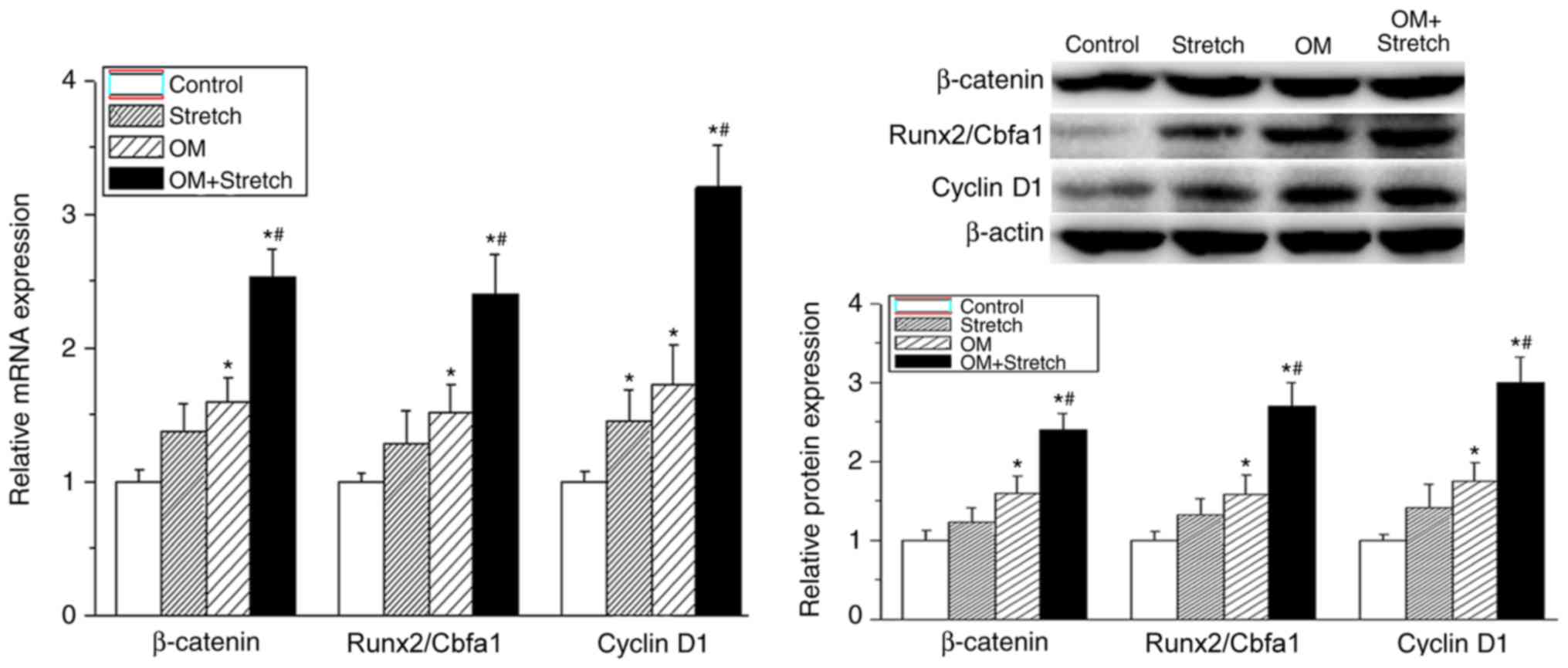
Mechanical and biochemical stimulation
modulates the osteogenic differentiation of hAECs via the
Wnt/β-catenin signalling pathway
The Wnt/β-catenin signalling pathway has been known
to affect the bone formation and homeostasis. A previous report
indicated that the Wnt/β-catenin signalling pathway enhanced
osteogenic differentiation from human periodontal ligament
fibroblasts (18). In the present
study, the mRNA and protein expression levels of β-catenin and
cyclin D1 were assessed following different treatments to confirm
whether Wnt/β-catenin signalling also has an important role in the
osteogenic differentiation of hAECs. As shown in Fig. 3, the mRNA and protein expression
levels of β-catenin, Runx2/Cbfa1 and cyclin D1 were significantly
increased in the OM group (P<0.05). Furthermore, the combined
application of stretching (the cells were stretched for 12 h) with
OM led to significant upregulation of β-catenin, Runx2/Cbfa1 and
cyclin D1 expression compared with the other treatments
(P<0.05). This suggested that the Wnt/β-catenin signalling
pathway may mediate the osteogenic differentiation of AECs.
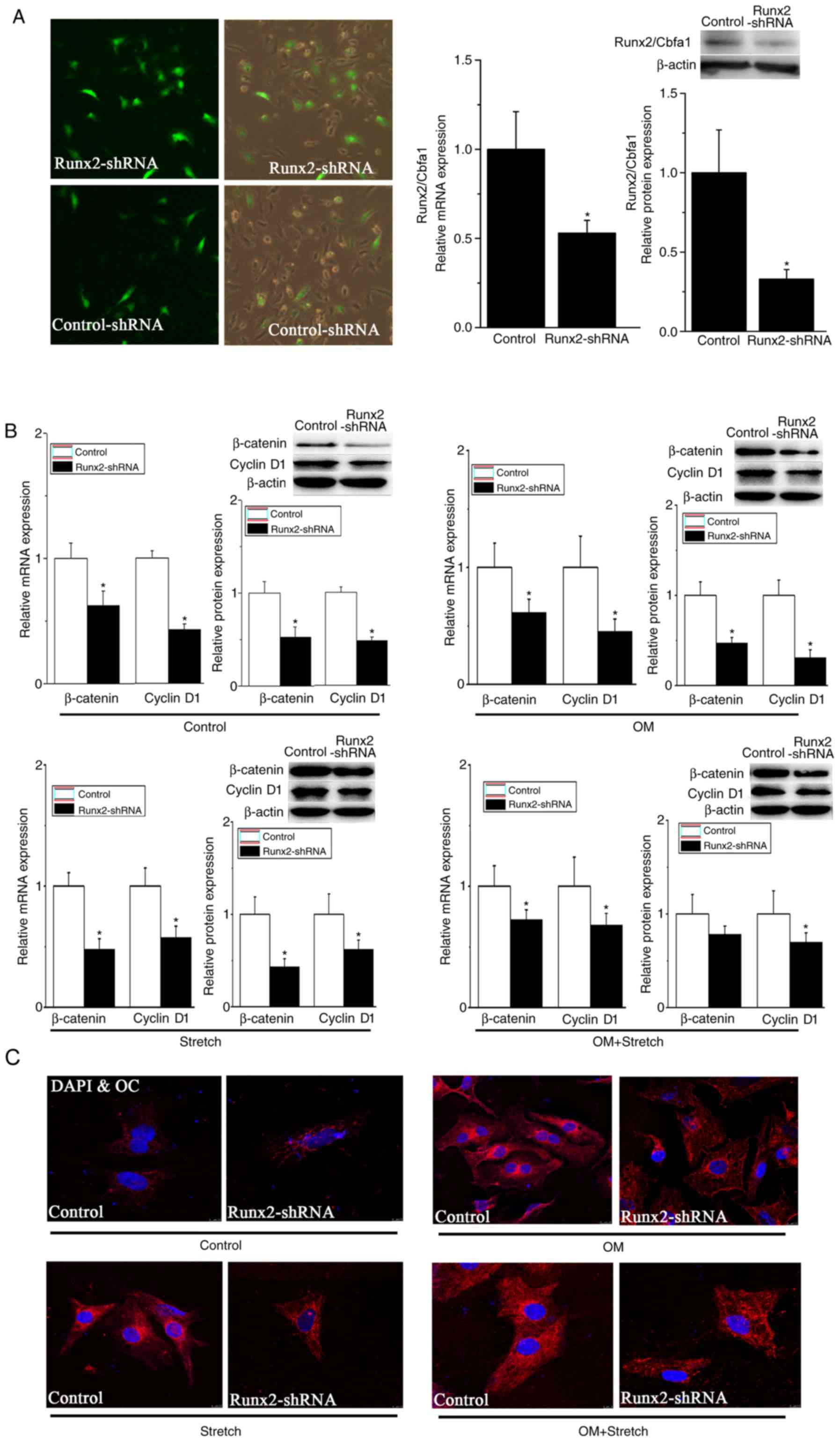
Based on these results, a Runx2-targeting shRNA was
constructed and transfected into hAECs prior to OM or stretch
stimulation to further investigate the mechanism of the
Wnt/β-catenin signalling pathway in osteoblastic differentiation of
hAECs. As exhibited in Fig. 4A,
the mRNA and protein levels of Runx2/Cbfa1 were decreased by ~50
and 70%, respectively, following Runx2-shRNA transfection. The gene
and protein levels of β-catenin and cyclin D1, as detected by
RT-qPCR and western blotting, were significantly decreased
following transfection of Runx2-shRNA in the basal and
OM/stretch-stimulated groups (P<0.05; Fig. 4B).
To examine whether Runx2-shRNA transfection could
inhibit the differentiation process, confocal microscopy was used
to assess the expression of OC in hAECs. The immunocytochemistry
results in Fig. 4C demonstrated
that the OC expression was decreased in the transfected group
compared with the control group, in the basal and stimulated
groups, demonstrating that the osteogenic differentiation of AECs
was blocked by cutting off the Wnt/β-catenin signalling pathway.
These results demonstrated that combined OM and stretch had an
important role in the modulation of osteogenic differentiation of
AECs via the Wnt/β-catenin signalling pathway.
Discussion
The present study revealed certain significant
results: i) Among all the treatment groups, the combined
application of mechanical stretch and biochemical osteogenic
stimulation of the hAECs has the most intense effects on gene or
protein expression; and ii) the Wnt/β-catenin signalling pathway
had an important role in the osteogenic differentiation of the
hAECs induced by stretch and/or biochemical stimulation. These
results suggest that mechanical stretch, particularly when combined
with biochemical stimulation, had an important role in the
modulation of osteogenic differentiation of hAECs. Wnt/β-catenin
signalling leads to the differentiation of the hAECs into
osteogenic lineage following mechanical stretch stimulation of
osteogenic transcription factors.
In present study, the hAECs derived from full-term
placenta seem to retain the capability to differentiate and to
produce cell types from all three germ layers. Furthermore, hAECs
are considered an ideal source of stem cells because of their
ability to produce growth factors and catecholamines, and to
differentiate into specific lineages and there are few concerns
about their limitations (5,6).
These characteristics make the hAECs good candidates for use in the
present study.
Previous studies have demonstrated that stem cell
differentiation is affected by various factors, including the
following: Static stretching increases cell proliferation and
activates synergistic osteoblastic differentiation of mesenchymal
cells; transforming growth factor-β1 induced by microenvironments
adjacent to distracted areas in vivo is thought to be
involved in the regulation of bone synthesis and turnover (19); the renin-angiotensin system has
been reported to mediate vascular endothelial growth factor
synthesis in MSCs (20);
biochemical stimulation has been reported to mediate the
differentiation of hAECs into osteoblasts (12); cyclic stretching has been reported
to promote osteogenesis-associated gene expression in
osteoblast-like cells through a cofilin-associated mechanism
mechanical stimuli (21); and
mechanical loading is known to be an important factor in regulating
bone growth and development (22).
Thus, in the present study, it was hypothesized that hAECs
differentiate into osteoblasts following stimulation by mechanical
stretching.
The results of the present study indicate that
treating cultured AECs with mechanical stretching altered cell
morphology and modestly increased the gene and protein expression
levels of Runx2/Cbfa1, ALP and OC in the hAECs. However, the
combined application of stretch with OM led to further induction of
Runx2/Cbfa1, ALP and OC gene and protein expression compared with
either treatment alone, which demonstrated that combining stretch
and biochemical osteogenic stimulation was synergistic for the
osteogenic differentiation of the hAECs.
The expression of β-catenin in vitro and
in vivo may contribute to the enhanced osteogenic
differentiation of MSCs and bone formation in vivo (19,20,23,24).
Under physiological conditions, cytoplasmic β-catenin is maintained
at low levels by a continuous process of phosphorylation-dependent
ubiquitination and degradation in the absence of Wnt ligand. Upon
Wnt stimulation, Wnt binds with a membrane receptor called
frizzled, leading to the dimerization of membrane and cytoplasmic
proteins. As a result, the kinases that phosphorylate and
destabilize β-catenin are inhibited, and β-catenin then accumulates
in the nucleus, where it associates with transcription factors such
as T-cell-specific transcription factor/lymphoid enhancing factor
to activate the expression of Wnt-responsive genes, including Runx2
and cyclin D (24–27). In the present study, the expression
levels of β-catenin and cyclin D1 were upregulated following
osteogenic differentiation of hAECs induced by mechanical
stretching. Furthermore, combined stretching and biochemical
stimulation induced the expression of β-catenin and cyclin D1.
Runx2/Cbfa1, the runt domain-containing
transcription factor, is a bone-specific product of the Cbfa1 gene
that is essential for bone formation. It is one of the earliest and
most specific markers during osteogenesis, and induces
osteoblast-specific gene expression in vitro (28–30).
The expression of Runx2/Cbfa1 in response to different stimuli was
examined, and the data demonstrated that the gene and protein
expression was induced by the combination of mechanical and
biochemical stimulation. However, the upregulation of Runx2/Cbfa1
was decreased by shRNA targeting Runx2/Cbfa1 into hAECs, as was the
expression of β-catenin and cyclin D1. Notably, the OC protein
level, as examined by confocal microscopy, was decreased following
shRNA knockdown of Runx2/Cbfa1. Thus, the present observations
demonstrated that mechanical stretching, and the combination of
stretching and biochemical stimuli may induce the osteogenic
differentiation of hAECs via activation of Wnt/β-catenin
signalling.
In conclusion, the results of the present study
demonstrated that the combined use of mechanical stretching and
biochemical stimulation was synergistic for the osteogenic
differentiation of hAECs, and that this differentiation may be
partially mediated via the Wnt/β-catenin signalling pathway.
Modulation of this may be a novel approach in bone regenerative
medicine.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Yongchuan
Science and Technology Commission (grant no. YCSTC,
2015nc5006).
Availability of data and materials
The analysed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
FL and HL designed experiments; KM, MZ and FY
carried out experiments; JM analysed results. FL, HL and KM wrote
the manuscript.
Ethics approval and consent to
participate
All patients gave written informed consent. The
present study was performed according to the Declaration of
Helsinki and approved by the Medical Ethics Committee of the
Yongchuan Hospital of Chongqing Medical University.
Patient consent for publication
Patients gave their consent for the present data to
be published.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Duncan RL and Turner CH:
Mechanotransduction and the functional response of bone to
mechanical strain. Calcif Tissue Int. 57:344–358. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Qi MC, Zou SJ, Han LC, Zhou HX and Hu J:
Expression of bone-related genes in bone marrow MSCs after cyclic
mechanical strain: Implications for distraction osteogenesis. Int J
Oral Sci. 1:143–150. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Maul TM, Chew DW, Nieponice A and Vorp DA:
Mechanical stimuli differentially control stem cell behavior:
Morphology, proliferation, and differentiation. Biomech Model
Mechanobiol. 10:939–953. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ilancheran S, Moodley Y and Manuelpillai
U: Human fetal membranes: A source of stem cells for tissue
regeneration and repair? Placenta. 30:2–10. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Miki T, Mitamura K, Ross MA, Stolz DB and
Strom SC: Identification of stem cell marker-positive cells by
immunofluorescence in term human amnion. J Reprod Immunol.
75:91–96. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Toda A, Okabe M, Yoshida T and Nikaido T:
The potential of amniotic membrane/amnion-derived cells for
regeneration of various tissues. J Pharmacol Sci. 105:215–228.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hodge A, Lourensz D, Vaghjiani V, Nguyen
H, Tchongue J, Wang B, Murthi P, Sievert W and Manuelpillai U:
Soluble factors derived from human amniotic epithelial cells
suppress collagen production in human hepatic stellate cells.
Cytotherapy. 16:1132–1144. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Niknejad H, Peirovi H, Ahmadiani A,
Ghanavi J and Jorjani M: Differentiation factors that influence
neuronal markers expression in vitro from human amniotic epithelial
cells. Eur Cells Mater. 19:22–29. 2010. View Article : Google Scholar
|
|
9
|
Kakishita K, Elwan MA, Nakao N, Itakura T
and Sakuragawa N: Human amniotic epithelial cells produce dopamine
and survive after implantation into the striatum of a rat model of
Parkinson's disease: A potential source of donor for
transplantation therapy. Exp Neurol. 165:27–34. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wlodarski K, Moskalewski S, Skarzyńska S,
Póltorak A and Ostrowski K: Irradiation and the bone induction
properties of epithelial cells. Bull Acad Pol Sci Biol. 19:821–825.
1971.PubMed/NCBI
|
|
11
|
Wang Q, Wu W, Han X, Zheng A, Lei S, Wu J,
Chen H, He C, Luo F and Liu X: Osteogenic differentiation of
amniotic epithelial cells: Synergism of pulsed electromagnetic
field and biochemical stimuli. BMC Musculoskelet Disord.
15:2712014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Parolini O, Alviano F, Bagnara GP, Bilic
G, Bühring HJ, Evangelista M, Hennerbichler S, Liu B, Magatti M,
Mao N, et al: Concise review: Isolation and characterization of
cells from human term placenta: Outcome of the first international
workshop on placenta derived stem cells. Stem Cells. 26:300–311.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Miki T, Marongiu F, Dorko K, Ellis EC and
Strom SC: Isolation of amniotic epithelial stem cells. Curr Protoc
Stem Cell Biol Chapter. 1:Unit 1E.3. 2010. View Article : Google Scholar
|
|
14
|
Hata M, Naruse K, Ozawa S, Kobayashi Y,
Nakamura N, Kojima N, Omi M, Katanosaka Y, Nishikawa T, Naruse K,
et al: Mechanical stretch increases the proliferation while
inhibiting the osteogenic differentiation in dental pulp stem
cells. Tissue Eng Part A. 19:625–633. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang YQ, Li XT, Rabie AB, Fu MK and Zhang
D: Human periodontal ligament cells express osteoblastic phenotypes
under intermittent force loading in vitro. Front Biosci. 1:776–781.
2006. View Article : Google Scholar
|
|
16
|
Koike M, Shimokawa H, Kanno Z, Ohya K and
Soma K: Effects of mechanical strain on proliferation and
differentiation of bone marrow stromal cell line ST2. J Bone Miner
Metab. 23:219–225. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Heo JS, Lee SY and Lee JC: Wnt/β-catenin
signaling enhances osteoblastogenic differentiation from human
periodontal ligament fibroblasts. Mol Cells. 30:449–454. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim IS, Song YM and Hwang SJ: Osteogenic
responses of human mesenchymal stromal cells to static stretch. J
Dent Res. 89:1129–1134. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu C, Zhang JW, Hu L, Song YC, Zhou L,
Fan Y, Zhu HY, Wang Y and Li QP: Activation of the AT1R/HIF-1α/ACE
axis mediates angiotensin II-induced VEGF synthesis in mesenchymal
stem cells. Biomed Res Int. 2014:6273802014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao J, Fu S, Zeng Z, Li F, Niu Q, Jing D
and Feng X: Cyclic stretch promotes osteogenesis-related gene
expression in osteoblast-like cells through a cofilin-associated
mechanism. Mol Med Rep. 14:218–224. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu Y, Zhang P, Dai Q, Yang X, Fu R, Jiang
L and Fang B: Effect of mechanical stretch on the proliferation and
differentiation of BMSCs from ovariectomized rats. Mol Cell
Biochem. 382:273–282. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dao DY, Jonason JH, Zhang Y, Hsu W, Chen
D, Hilton MJ and O'Keefe RJ: Cartilage-specific β-CATENIN signaling
regulates chondrocyte maturation, generation of ossification
centers, and perichondrial bone formation during skeletal
development. J Bone Miner Res. 27:1680–1694. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jian H, Shen X, Liu I, Semenov M, He X and
Wang XF: Smad3-dependent nuclear translocation of beta-catenin is
required for TGFbeta1-induced proliferation of bone marrow-derived
adult human mesenchymal stem cells. Genes Dev. 20:666–674. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kestler HA and Kühl M: From individual Wnt
pathways towards a Wnt signaling network. Philos Trans R Soc Lond B
Biol Sci. 363:1333–1347. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Taurin S, Sandbo N, Qin Y, Browning D and
Dulin NO: Phosphorylation of beta-catenin by cyclic AMP-dependent
protein kinase. J Biol Chem. 281:9971–996. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Takemaru KI and Moon RT: The
transcriptional eoaetivator CBP interaets with beta-catenin to
activate gene expression. J Cell Biol. 149:249–254. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Komori T, Yagi H, Nomura S, Yamaguchi A,
Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, et al:
Targeted disruption of Cbfa1 results in a complete lack of bone
formation owing to maturational arrest of osteoblasts. Cell.
89:755–764. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xiao G, Jiang D, Ge C, Zhao Z, Lai Y,
Boules H, Phimphilai M, Yang X, Karsenty G and Franceschi RT:
Cooperative interactions between activating transcription factor 4
and Runx2/Cbfa1 stimulate osteoblast-specific osteocalcin gene
expression. J Biol Chem. 280:30689–30696. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mundlos S, Otto F, Mundlos C, Mulliken JB,
Aylsworth AS, Albright S, Lindhout D, Cole WG, Henn W, Knoll JH, et
al: Mutations involving the transcription factor CBFA1 cause
cleidocranial dysplasia. Cell. 89:773–779. 1997. View Article : Google Scholar : PubMed/NCBI
|