Introduction
Osteoporosis is becoming a serious public health
issue as aging of the global population accelerates. Bone
marrow-derived mesenchymal stem cells (BMSCs) are multipotent
progenitor cells with the capacity of differentiating into
osteoblasts, adipocytes, chondrocytes, myoblasts and neurocytes
(1,2). The differentiation of osteoblasts and
adipocytes from BMSCs, as well as the balance between osteoclastic
bone resorption and osteoblastic bone formation, contribute to the
maintenance of bone volume (3).
With increasing age, the imbalance between osteogenesis and
adipogenesis of BMSCs results in a decrease in bone mineral density
leading to osteoporosis (4).
Therefore, the balance between bone marrow osteogenesis and
adipogenesis may represent another therapeutic target for the
prevention or treatment of osteoporosis. However, the mechanisms
regulating the osteogenesis of BMSCs require further
exploration.
miRNAs are a series of 20–24 nt non-coding RNAs that
act as epigenetic negative regulators of their target mRNAs by
directly binding to the 3′-untranslated region (3′UTR)
post-transcriptionally (5,6). Recently, miRNAs have been shown to
play a key role in bone homeostasis and to regulate a variety of
cellular functions, including differentiation, proliferation and
migration (7–9). For example, miR-133 directly targets
Runx2 and regulates BMP-2-mediated osteogenic differentiation of
C2C12 cells (10) and
β-glycerophosphate (β-GP)-induced vascular smooth muscle cell
(VSMC) osteoblastic differentiation (7). In addition, miR-125b has been shown
to suppress osteogenic differentiation of BMSCs via targeting Sp7
(11). miR-124 also plays an
important role in the differentiation of BMSCs, including myogenic
(12), neuronal (13), and osteogenic differentiation
(14). However, the mechanisms
involved in miR-124-mediated osteogenic differentiation of BMSCs
require elucidation.
In the present study, the miR-124 regulation of
osteogenic differentiation of BMSCs was investigated and it was
revealed that miR-124 regulated the osteogenic differentiation of
BMSCs by targeting Sp7.
Materials and methods
Culture of BMSCs and cell
transfection
Human BMSCs were isolated from bone marrow samples
as described in our previous study (15). The specimens were from three
subjects [aged 24 (female), 26 (male) and 30 years (male)]
undergoing routine therapeutic surgery at the Department of
Orthopaedic Surgery from December 2016 to March 2017, The Second
Xiang-Ya Hospital, Central South University (Changsha, China). All
of the subjects provided informed consent. Approval for the present
study was acquired from the Medical Ethics Committee of The Second
Xiang-Ya Hospital, Central South University. The bone marrow
samples were harvested using standard Ficoll-Hypaque density
sedimentation methods. BMSCs were selected based on adhesion and
proliferation on a tissue culture plastic substrate. Cells from
passages 3–5 were used in the experiments.
For cell transfection, miR-124 mimics, miR-124
inhibitor, and the negative controls, or Sp7 small interfering RNA
(siRNA) oligos (RiboBio, Guangzhou, China) were transfected into
BMSCs using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) according to the manufacturer's
instructions. After 6 h of co-culture, the cells were cultured in
Dulbecco's modified Eagle's medium (DMEM) supplemented with 10%
foetal bovine serum (FBS; both Gibco; Thermo Fisher Scientific,
Inc.) and cultured for a further 24–48 h.
Osteogenic differentiation
To induce osteogenic differentiation, BMSCs were
cultured in osteogenic medium (basal medium + 50 µg/ml
phospho-ascorbate + 10 mM β-GP) for 3–7 days. To assess osteogenic
differentiation, an ALP activity assay (16) was performed using the
spectrophotometric measurement of p-nitrophenol release at
37°C.
Alizarin Red S staining
Alizarin Red S staining was performed as previously
described (17). Briefly, BMSCs
cultured in osteogenic medium for 14 days were fixed in 4%
paraformaldehyde for 30 min at room temperature and stained with 2%
(pH 4.2) Alizarin Red S solution for 1–3 min at 37°C. Next, cells
were washed with phosphate-buffered saline (PBS) three times to
eliminate non-specific staining. The stained matrixes were assessed
and photographed using a digital microscope. To quantify calcium
levels, cells were washed with PBS and decalcified with 0.6 N HCl
for 24 h. Calcium content was determined by measuring the
concentration of calcium in the HCl supernatant by atomic
absorption spectroscopy. Following decalcification, the cells were
washed with PBS three times and solubilized with 0.1 N NaOH/0.1%
SDS. The protein content was measured with a BCA protein assay
(Beyotime Biotechnology, Shanghai, China). The calcium content of
the cell layer was normalized to the protein content.
Western blotting
To investigate the protein levels of Runx2 and Sp7,
western blotting was performed as previously described (16,18).
Total protein was extracted from BMSCs using
radioimmunoprecipitation assay lysis buffer (Beijing Solarbio
Science & Technology Co., Ltd., Beijing, China) and the
concentration of protein was determined a Bicinchoninic Acid
protein kit. A total of 30 µg protein was loaded per lane in 10%
electrophoresis gel and separated with sodium dodecyl sulphate-PAGE
and transferred to polyvinylidene fluoride membranes (Millipore,
Billerica, MA, USA). Membranes were blocked with 5% non-fat milk in
Tris-buffered saline for at least 1 h at room temperature. The
membranes were incubated with primary antibodies, including
anti-Runx2 (cat. no. ab76956; 1:1,000), anti-Sp7 (cat. no.
ab209484, 1:1,000; both Abcam, Cambridge, UK), and anti-β-actin
(cat. no. AM1021B, 1:3,000; Abgent, Inc., San Diego, CA, USA) at
4°C overnight, followed by incubation with the horseradish
peroxidase-conjugated goat anti-rabbit (cat. no. sc-2004, 1:5,000)
or horseradish peroxidase-conjugated goat anti-mouse (cat. no.
sc-2005, 1:5,000; both Santa Cruz Biotechnology, Santa Cruz, CA,
USA) secondary antibodies at 37°C for 1 h. The immunoreactive bands
were visualised using the enhanced chemiluminescence Plus Western
blot detection kit (Amersham Biosciences UK Ltd., Little Chalfont,
Buckinghamshire, UK) and densitometric quantification of band
intensity from three independent experiments was carried out with
Image-Pro Plus 6.0 software (Media Cybernetics, Inc., Rockville,
MD, USA). The relative expression levels of target proteins were
normalized to the intensity of the β-actin band.
RT-qPCR analysis
Total small RNA was extracted using the mirVana
miRNA Isolation Kit (Ambion, Austin, TX, USA). The cDNA was
synthesised via an All-in-One™ miRNA first-strand cDNA synthesis
kit (AMRT-0060; GeneCopoeia, Inc., Rockville, MD, USA). Briefly, a
25 µl reverse transcription reaction was carried out for 60 min at
37°C, 5 min at 85°C, and then held at 4°C. To analyse the
expression of miR-124, RT-qPCR was performed with All-in-One™ qPCR
Mix (QP003; GeneCopoeia, Inc.) using the LightCycler® 96
System (Roche Diagnostics, Indianapolis, IN, USA). For qPCR
analysis, 20 µl reactions were incubated in a 96-well optical plate
at 95°C for 10 min, followed by 40 cycles of 95°C for 10 sec, 62°C
for 20 sec, and 72°C for 20 sec. U6 snRNA was used as the
reference. The miR-124 (HmiRQP0074) and U6 (HmiRQP9001) primers
were purchased from GeneCopoeia. The relative standard curve method
(2−∆∆Cq) was used to determine the relative mRNA and
miRNA expression (19). Results
are expressed as fold changes relative to the relevant control.
qPCR was performed in triplicate and the results are presented as
the mean ± standard error of samples.
Luciferase reporter assay
A segment of the Sp7 3-untranslated region (3′UTR)
containing the predicted miR-124 binding sites was amplified by
PCR. The PCR products were purified and inserted into the
XbaI-FseI site downstream of the stop codon in the
pGL3 control luciferase reporter vector (Promega Corporation,
Madison, WI, USA), resulting in WT-pGL3-Sp7. To construct
MUT-pGL3-Sp7, the Quick Change Site-Directed Mutagenesis kit
(Stratagene; Agilent Technologies, Inc., Santa Clara, CA, USA) was
used to induce point mutations in the 3′UTR of WT-pGL3-Sp7. The
primer sequences for the PCR are shown in Table I. BMSCs were cotransfected with the
luciferase reporter carrying WT-pGL3-Sp7 or MUT-pGL3-Sp7 and
miR-124 mimics or scramble control oligos. After 48 h, luciferase
activity was detected with the luciferase assay system (Promega
Corporation).
 | Table I.Nucleotide sequences of primers for
WT and mutant reporter plasmids. |
Table I.
Nucleotide sequences of primers for
WT and mutant reporter plasmids.
| Gene | Primer sequence (5′
to 3′) |
|---|
| WT-Sp7 | F:
5′-GCTCTAGAGCCCATCTCCACCTCCAGT-3′ |
|
| R:
5′-GGCCGGCCATCCCTTCCATTCCACAA-3′ |
| MUT-Sp7 | F:
5′-AAGCCGCTTCGTGTCTATCCTGTAACGTTGGA-3 |
|
| R:
5′-TTCGGCGAAGCACAGATAGGACATTGCAACCT-3 |
Statistical analysis
The results are presented as means ± standard
deviation. Analysis was performed with SPSS software (version 17.0;
SPSS Inc., Chicago, IL, USA). For comparisons of two groups, the
Student's t-test was used, and comparisons between values of more
than two groups were evaluated by one-way analysis of variance
(ANOVA) together with a Tukey's post hoc test. A level of P<0.05
was considered statistically significant. Representative figures
are shown in the study.
Results
Expression of miR-124 is decreased
during osteogenic differentiation of BMSCs
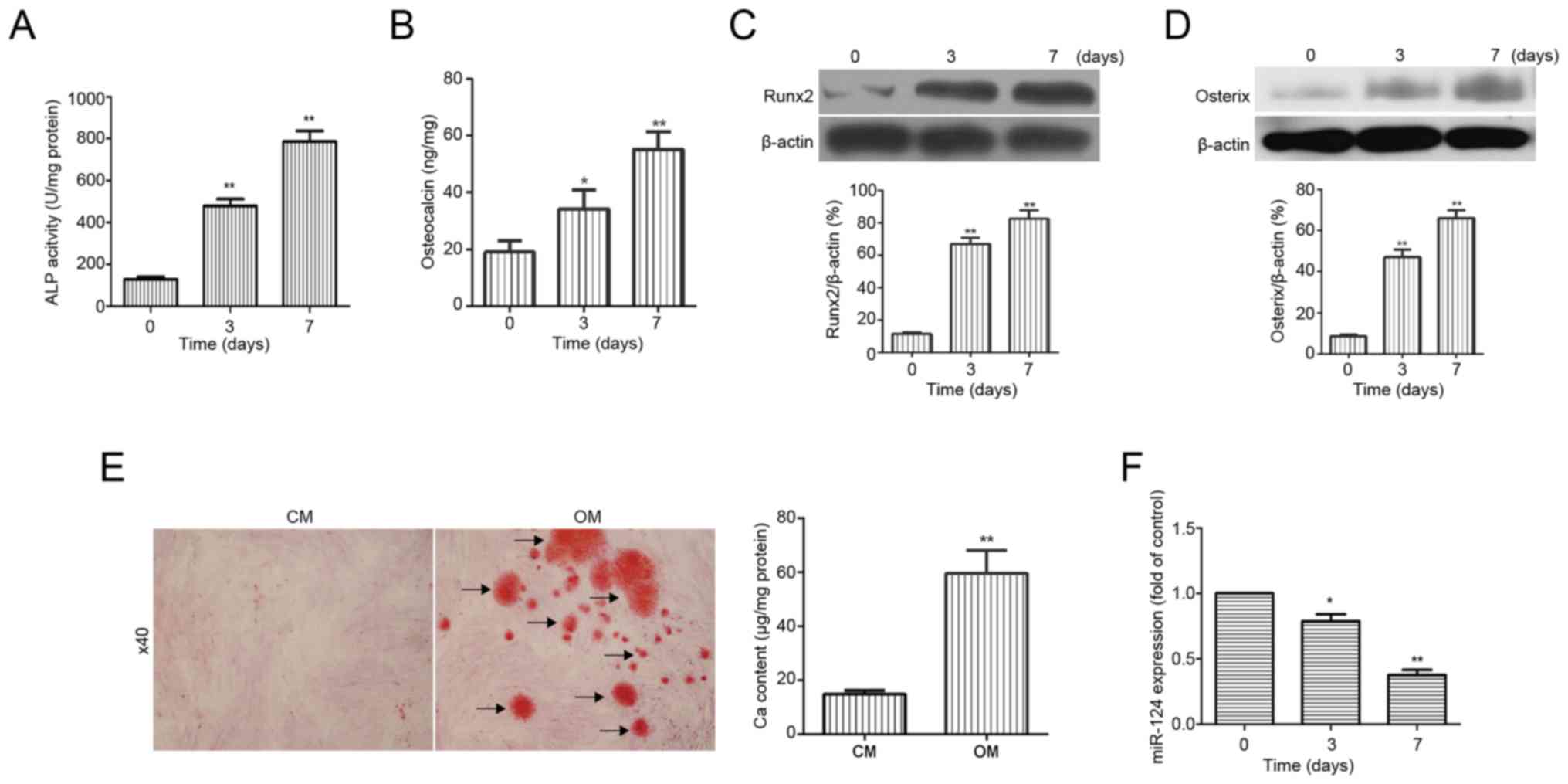
To examine the potential osteogenic differentiation
ability of BMSCs, human BMSCs were cultured in osteogenic medium.
The osteoblastic differentiation markers ALP activity, OC
secretion, and osterix and Runx2 expression (20,21)
were assessed. As expected, ALP activity and OC secretion, as well
as the protein expression of osterix and Runx2, were significantly
increased (Fig. 1A-D). In
addition, Alizarin Red S staining verified that mineralization
nodules were formed in BMSCs cultured in osteogenic medium for 14
days (Fig. 1E). However, RT-qPCR
showed that the expression of miR-124 was considerably decreased in
a time-dependent manner during osteogenic differentiation of BMSCs
(Fig. 1F). These results showed
that BMSCs cultured in osteogenic medium could differentiate into
osteoblasts and that miR-124 may be involved in osteogenic
differentiation of BMSCs.
miR-124 inhibits osteogenic
differentiation of BMSCs
To investigate whether miR-124 exerts a regulatory
effect on osteogenic differentiation of BMSCs, a gain- and
loss-of-function approach was used to overexpress or inhibit the
expression of miR-124, respectively. RT-qPCR verified that miR-124
mimics induced miR-124 expression by more than 70-fold whereas the
miR-124 inhibitor significantly decreased the expression of miR-124
(Fig. 2A). In addition, ALP
activity and OC and Runx2 protein expression were significantly
decreased in miR-124-overexpressing cells while inhibition of
miR-124 expression led to opposite results (Fig. 2B-D). Taken together, our results
demonstrated that miR-124 could regulate osteogenic differentiation
of BMSCs.
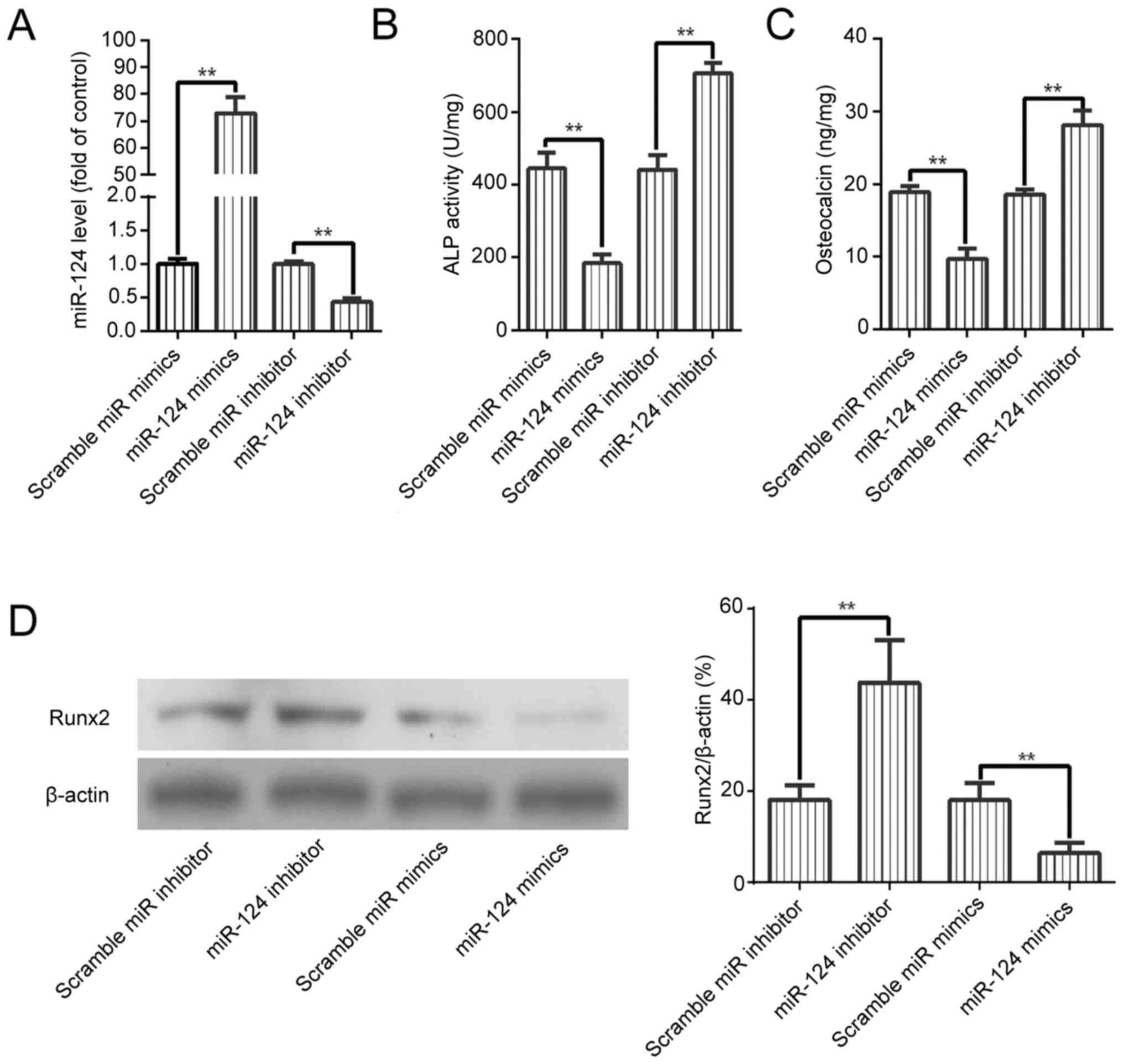 | Figure 2.miR-124 regulates osteogenic
differentiation of BMSCs. (A) BMSCs were transfected with scramble
miR mimics, miR-124 mimics, scramble miR inhibitor, or miR-124
inhibitor. The expression of miR-124 was determined by RT-qPCR.
(B-D) BMSCs were transfected and cultured in OM. ALP activity, OC
secretion, and the level of Runx2 were assessed. Data are expressed
as mean ± SD, n=3. **P<0.01. BMSCs, bone marrow-derived
mesenchymal stem cells; ALP, alkaline phosphatase; OC, osteocalcin;
CM, control medium; OM, osteogenic medium. |
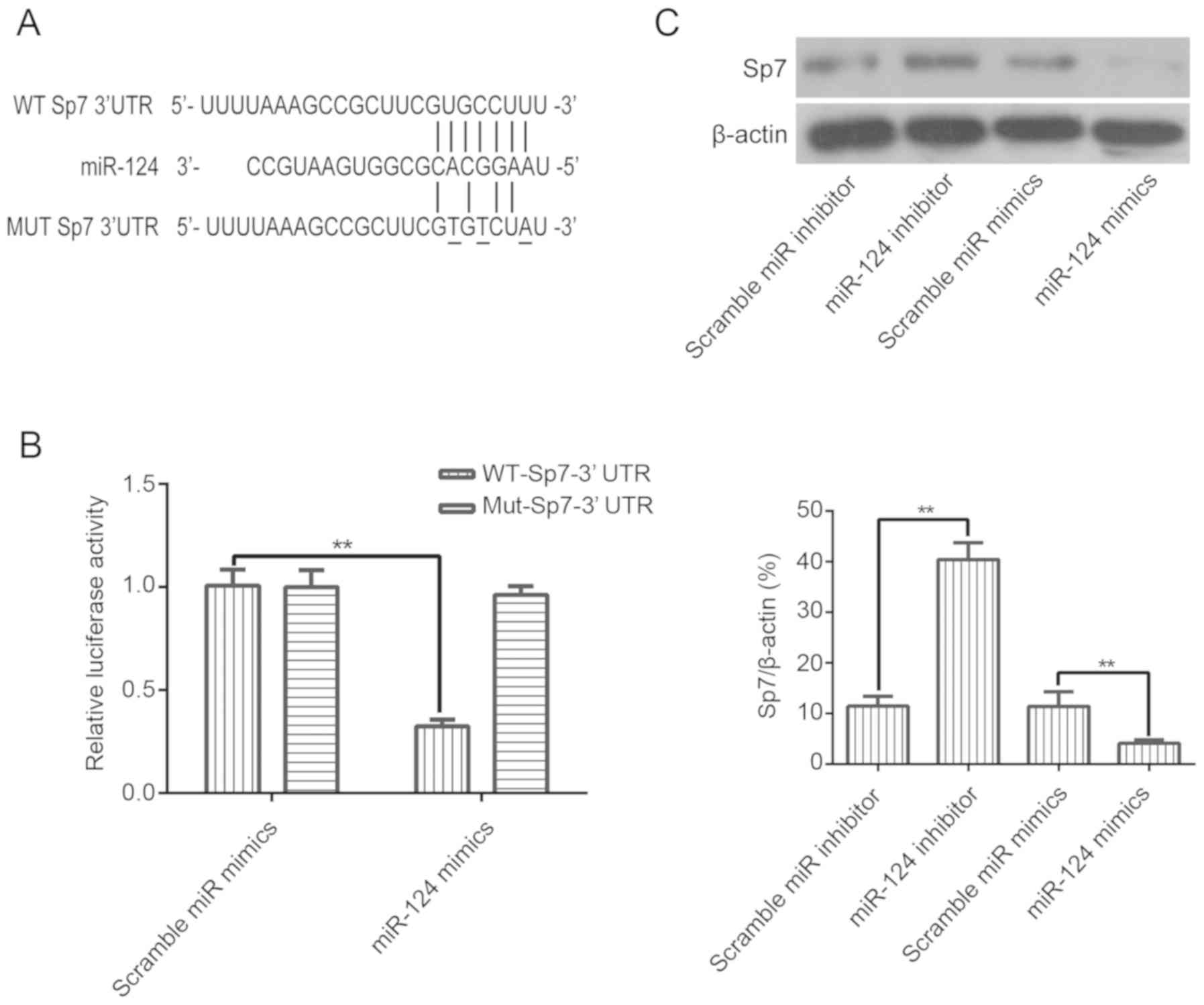
Sp7 is the target gene of miR-124
To investigate the mechanism underlying miR-124
regulation of osteogenic differentiation of BMSCs, we searched for
potential targets of miR-124 using miRNA target prediction
algorithms and identified Sp7 as a putative target gene of miR-124
(Fig. 3A). To confirm this target,
a luciferase reporter assay was performed to evaluate whether
miR-124 could directly target the 3′UTR of Sp7. Luciferase
reporters containing the wild-type (WT) or mutant (MUT) 3′UTR
uncoding sequences for the predicted miRNA binding sites of Sp7
were introduced with miR-124 mimics into BMSCs. The results showed
that overexpression of miR-124 decreased the luciferase activity of
the WT-Sp7 3′UTR reporter genes while mutation of Sp7 binding sites
abolished this repression. These results suggested that Sp7 is a
direct target of miR-124 (Fig.
3B). Moreover, overexpression of miR-124 in BMSCs resulted in
decreased expression of Sp7, whereas inhibition of miR-124
expression upregulated the expression of Sp7 (Fig. 3C). These results provide evidence
that miR-124 directly targets Sp7.
Sp7 is involved in the miR-124
regulation of osteogenic differentiation of BMSCs
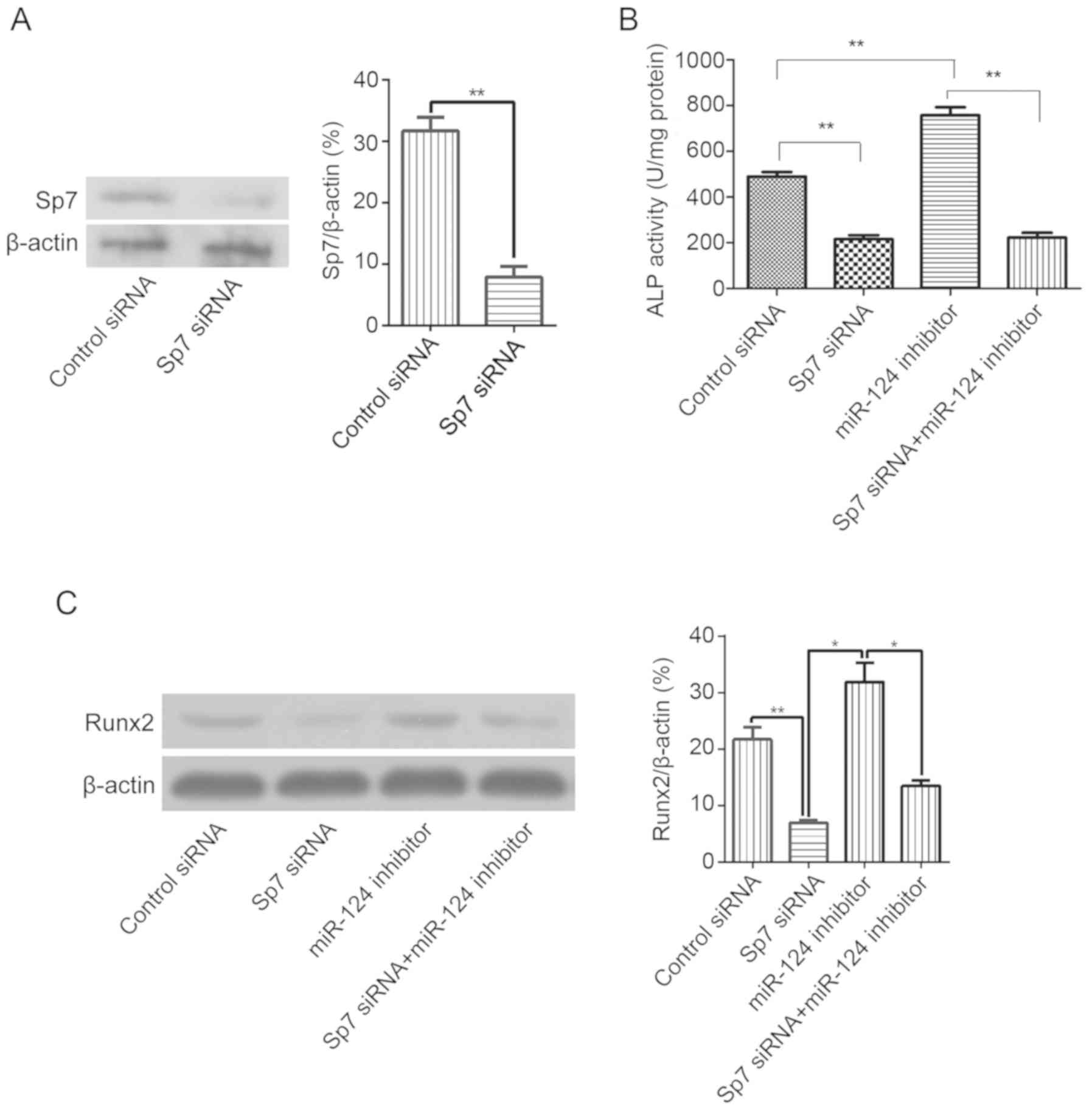
To further confirm that miR-124 relies on Sp7 to
regulate osteogenic differentiation of BMSCs, the effect of Sp7
siRNA treatment on osteoblastic markers involved in the osteogenic
differentiation of BMSCs was determined. As shown in Fig. 4A, Sp7 was successfully knocked down
by Sp7-targeting siRNA. Transfection with Sp7 siRNA significantly
reduced ALP activity and the protein levels of Runx2, whereas
transfection with the miR-124 inhibitor significantly increased ALP
activity and Runx2 expression. Nevertheless, Sp7 siRNA completely
blocked the ALP activity induced by miR-124 inhibitors (Fig. 4B and C). These data confirmed that
miR-124-dependent inhibition of ALP activity was dependent on
Sp7.
Discussion
In the present study, it was demonstrated that
miR-124 was downregulated during osteogenic differentiation of bone
marrow-derived mesenchymal stem cells (BMSCs) and involved in
regulating the osteogenic differentiation of BMSCs. In addition,
our results confirmed that Sp7 is a direct target of miR-124 and is
also involved in regulating the osteogenic differentiation of
BMSCs. Taken together, our results showed that miR-124 negatively
regulates osteogenic differentiation of BMSCs by targeting Sp7.
MSCs are commonly isolated from adult bone marrow
and have the potential to differentiate into adipose tissue, bone,
cartilage, tendon, and muscle, and thus hold great potential for
therapeutic applications (1,22,23).
Recent studies have shown that certain miRNAs may play a key role
in osteogenic differentiation of BMSCs (24,25).
However, the molecular mechanisms governing osteogenic
differentiation of BMSCs involved in the pathogenesis of bone
homeostasis remain unclear. In the present study, human BMSCs were
isolated and cultured from three individual donors. Osteogenic
medium was used to induce osteogenic differentiation of BMSCs,
which was confirmed by an increase in ALP activity, OC secretion,
Runx2 and Sp7 protein expression, and the formation of mineralized
nodules.
miRNAs, important post-transcriptional regulators in
gene expression (26), play a
pivotal role in the functions of BMSCs. Recently, many miRNAs have
been reported to modulate the phenotypes and differentiation of
BMSCs (27–29). A previous study demonstrated that
miR-144-3p negatively regulated osteogenic differentiation and
proliferation of MSCs by specifically targeting Smad4 (27). A study by Jeong et al
suggested that miR-194 could determine whether MSCs would
differentiate into osteoblasts or adipocytes by targeting COUP-TFII
(28). Yang et al showed
that TNF-α inhibited osteoblastic differentiation of MSCs by
suppressing the functional axis of miR-21 and Spry1 (29). These results indicated that miRNAs
play a vital role in regulating the osteogenic differentiation of
MSCs.
Recently, miR-124 has received much attention from
researchers for its key role in the different differentiation
pathways of MSCs. For example, Qadir et al showed that
miR-124 acts as a negative regulator of myogenic differentiation of
MSCs by targeting Dlx5 (12) and
another study showed that miR-124 regulated cardiomyocyte
differentiation of BMSCs by targeting STAT3 mRNA (30). In addition, Zou et al
demonstrated that miR-124 plays an important role in the
differentiation of BMSCs into neurons (13). In the present study, it was also
identified that miR-124 was downregulated during osteogenic
differentiation of BMSCs and involved in regulating osteogenic
differentiation of BMSCs. That is, ALP activity, OC secretion and
Runx2 protein expression were significantly decreased in
miR-124-overexpressing BMSCs while inhibition of the expression of
miR-124 led to opposite results. These results are consistent with
one previous study, which showed that miR-124 is also negatively
correlated with osteogenic differentiation of MSCs by targeting
Dlx3, Dlx5, and Dlx2 (14).
However, the role of miR-124 in the osteogenic differentiation of
BMSCs in vivo needs further study.
Sp7, a member of the Sp family of
zinc-finger-containing transcription factors, is a bone-specific
transcription factor that plays a crucial role in osteoblast
mineralization (31,32). Previous studies have shown that
BMSCs are multipotent and exhibit different levels of Sp7
expression depending on the differentiation commitment (33,34).
Previous studies have also shown that miR-125b regulates the
osteogenic differentiation of both BMSCs (11) and VSMCs (35) by targeting Sp7. Yang et al
demonstrated that miR-93 regulates osteoblast mineralization
through a novel miR-93/Sp7 regulatory feedback loop (32). In the present study, bioinformatic
analysis predicted that Sp7 is a potential target of miR-124 and it
was demonstrated that Sp7 is the direct target of miR-124. On the
one hand, miR-124 negatively regulates the expression of Sp7. On
the other hand, overexpression of miR-124 significantly suppressed
the luciferase activity of the WT-Sp7 3′UTR reporter but not that
of the MUT-Sp7 3′UTR reporter. Moreover, knockdown of the
expression of Sp7 using siRNA blocked the osteogenic
differentiation of BMSCs induced by the miR-124 inhibitor,
suggesting that the effect of miR-124 in regulating osteogenic
differentiation is dependent on Sp7. Collectively, the data
supported that miR-124 attenuates osteogenic differentiation of
BMSCs by directly targeting Sp7.
In conclusion, the present study identified for the
first time that miR-124 is a significant regulator of osteogenic
differentiation of BMSCs by targeting Sp7. This finding provides
insight into the mechanisms of osteogenic differentiation of BMSCs
and suggests that miR-124 may be an important potential therapeutic
target for a variety of bone diseases including osteoporosis.
Acknowledgements
Not applicable.
Funding
The present study was supported by funding from the
National Natural Science Foundation of China (nos. 81770881 and
81870623), the Fundamental Research Funds for the Central
Universities of Central South University (nos. 2018zzts918 and
2018zzts048) and the National Basic Research Program of China (973
Program) (no. 2014CB942903).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
LQY and FL conceived and designed the experiments.
JZT and XL performed the experiments, analyzed the data and
prepared all the figures. JYZ, FX, RRC, FW and XBL provided
technical support, made substantial contributions to data analysis
and revised the manuscript critically for important intellectual
content. JZT and XL wrote the manuscript. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
All of the subjects provided informed consent.
Approval for the present study was acquired from the Medical Ethics
Committee of The Second Xiang-Ya Hospital, Central South
University.
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
References
|
1
|
Pittenger MF, Mackay AM, Beck SC, Jaiswal
RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S and
Marshak DR: Multilineage potential of adult human mesenchymal stem
cells. Science. 284:143–147. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tuli R, Tuli S, Nandi S, Wang ML,
Alexander PG, Haleem-Smith H, Hozack WJ, Manner PA, Danielson KG
and Tuan RS: Characterization of multipotential mesenchymal
progenitor cells derived from human trabecular bone. Stem Cells.
21:681–693. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rachner TD, Khosla S and Hofbauer LC:
Osteoporosis: Now and the future. Lancet. 377:1276–1287. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nuttall ME and Gimble JM: Controlling the
balance between osteoblastogenesis and adipogenesis and the
consequent therapeutic implications. Curr Opin Pharmacol.
4:290–294. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cui RR, Li SJ, Liu LJ, Yi L, Liang QH, Zhu
X, Liu GY, Liu Y, Wu SS, Liao XB, et al: MicroRNA-204 regulates
vascular smooth muscle cell calcification in vitro and in vivo.
Cardiovasc Res. 96:320–329. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lin X, Xu F, Cui RR, Xiong D, Zhong JY,
Zhu T, Li F, Wu F, Xie XB, Mao MZ, et al: The arterial
calcification is regulated via a miR-204/DNMT3a regulatory circuit
both in vitro and in female mice. Endocrinology. 159:2905–2916.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liao XB, Zhang ZY, Yuan K, Liu Y, Feng X,
Cui RR, Hu YR, Yuan ZS, Gu L, Li SJ, et al: MiR-133a modulates
osteogenic differentiation of vascular smooth muscle cells.
Endocrinology. 154:3344–3352. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu Y, Xu F, Pei HX, Zhu X, Lin X, Song
CY, Liang QH, Liao EY and Yuan LQ: Vaspin regulates the osteogenic
differentiation of MC3T3-E1 through the PI3K-Akt/miR-34c loop. Sci
Rep. 6:255782016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu Y, Zhang XL, Chen L, Lin X, Xiong D,
Xu F, Yuan LQ and Liao EY: Epigenetic mechanisms of bone
regeneration and homeostasis. Prog Biophys Mol Biol. 122:85–92.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Z, Hassan MQ, Volinia S, van Wijnen AJ,
Stein JL, Croce CM, Lian JB and Stein GS: A microRNA signature for
a BMP2-induced osteoblast lineage commitment program. Proc Natl
Acad Sci USA. 105:13906–13911. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen S, Yang L, Jie Q, Lin YS, Meng GL,
Fan JZ, Zhang JK, Fan J, Luo ZJ and Liu J: MicroRNA125b suppresses
the proliferation and osteogenic differentiation of human bone
marrowderived mesenchymal stem cells. Mol Med Rep. 9:1820–1826.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qadir AS, Woo KM, Ryoo HM, Yi T, Song SU
and Baek JH: MiR-124 inhibits myogenic differentiation of
mesenchymal stem cells via targeting Dlx5. J Cell Biochem.
115:1572–1581. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zou D, Chen Y, Han Y, Lv C and Tu G:
Overexpression of microRNA-124 promotes the neuronal
differentiation of bone marrow-derived mesenchymal stem cells.
Neural Regen Res. 9:1241–1248. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qadir AS, Um S, Lee H, Baek K, Seo BM, Lee
G, Kim GS, Woo KM, Ryoo HM and Baek JH: miR-124 negatively
regulates osteogenic differentiation and in vivo bone formation of
mesenchymal stem cells. J Cell Biochem. 116:730–742. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liang QH, Jiang Y, Zhu X, Cui RR, Liu GY,
Liu Y, Wu SS, Liao XB, Xie H, Zhou HD, et al: Ghrelin attenuates
the osteoblastic differentiation of vascular smooth muscle cells
through the ERK pathway. PLoS One. 7:e331262012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang W, Guo H, Jing H, Li Y, Wang X,
Zhang H, Jiang L and Ren F: Lactoferrin stimulates osteoblast
differentiation through PKA and p38 pathways independent of
lactoferrin's receptor LRP1. J Bone Miner Res. 29:1232–1243. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yuan L, Chan GC, Fung KL and Chim CS:
RANKL expression in myeloma cells is regulated by a network
involving RANKL promoter methylation, DNMT1, microRNA and TNFα in
the microenvironment. Biochim Biophys Acta. 1843:1834–1838. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu GY, Liang QH, Cui RR, Liu Y, Wu SS,
Shan PF, Yuan LQ and Liao EY: Leptin promotes the osteoblastic
differentiation of vascular smooth muscle cells from female mice by
increasing RANKL expression. Endocrinology. 155:558–567. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Peng YQ, Xiong D, Lin X, Cui RR, Xu F,
Zhong JY, Zhu T, Wu F, Mao MZ, Liao XB and Yuan LQ: Oestrogen
inhibits arterial calcification by promoting autophagy. Sci Rep.
7:35492017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu SS, Liang QH, Liu Y, Cui RR, Yuan LQ
and Liao EY: Omentin-1 stimulates human osteoblast proliferation
through PI3K/Akt signal pathway. Int J Endocrinol. 2013:3689702013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qian J, Hu Y, Zhao L, Xia J, Li C, Shi L
and Xu F: Protective role of adipose-derived stem cells in
staphylococcus aureus-induced lung injury is mediated by RegIIIγ
secretion. Stem Cells. 34:1947–1956. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu F, Hu Y, Zhou J and Wang X: Mesenchymal
stem cells in acute lung injury: Are they ready for translational
medicine? J Cell Mol Med. 17:927–935. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Eskildsen T, Taipaleenmäki H, Stenvang J,
Abdallah BM, Ditzel N, Nossent AY, Bak M, Kauppinen S and Kassem M:
MicroRNA-138 regulates osteogenic differentiation of human stromal
(mesenchymal) stem cells in vivo. Proc Natl Acad Sci USA.
108:6139–6144. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kang H and Hata A: The role of microRNAs
in cell fate determination of mesenchymal stem cells: Balancing
adipogenesis and osteogenesis. BMB Rep. 48:319–323. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ying H, Kang Y, Zhang H, Zhao D, Xia J, Lu
Z, Wang H, Xu F and Shi L: MiR-127 modulates macrophage
polarization and promotes lung inflammation and injury by
activating the JNK pathway. J Immunol. 194:1239–1251. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang C, Geng J, Wei X, Zhang R and Jiang
S: MiR-144-3p regulates osteogenic differentiation and
proliferation of murine mesenchymal stem cells by specifically
targeting Smad4. FEBS Lett. 590:795–807. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jeong BC, Kang IH, Hwang YC, Kim SH and
Koh JT: MicroRNA-194 reciprocally stimulates osteogenesis and
inhibits adipogenesis via regulating COUP-TFII expression. Cell
Death Dis. 5:e15322014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang N, Wang G, Hu C, Shi Y, Liao L, Shi
S, Cai Y, Cheng S, Wang X, Liu Y, et al: Tumor necrosis factor α
suppresses the mesenchymal stem cell osteogenesis promoter miR-21
in estrogen deficiency-induced osteoporosis. J Bone Miner Res.
28:559–573. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cai B, Li J, Wang J, Luo X, Ai J, Liu Y,
Wang N, Liang H, Zhang M, Chen N, et al: microRNA-124 regulates
cardiomyocyte differentiation of bone marrow-derived mesenchymal
stem cells via targeting STAT3 signaling. Stem Cells. 30:1746–1755.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nakashima K, Zhou X, Kunkel G, Zhang Z,
Deng JM, Behringer RR and de Crombrugghe B: The novel zinc
finger-containing transcription factor osterix is required for
osteoblast differentiation and bone formation. Cell. 108:17–29.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang L, Cheng P, Chen C, He HB, Xie GQ,
Zhou HD, Xie H, Wu XP and Luo XH: miR-93/Sp7 function loop mediates
osteoblast mineralization. J Bone Miner Res. 27:1598–1606. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tu Q, Valverde P and Chen J: Osterix
enhances proliferation and osteogenic potential of bone marrow
stromal cells. Biochem Biophys Res Commun. 341:1257–1265. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou X, Zhang Z, Feng JQ, Dusevich VM,
Sinha K, Zhang H, Darnay BG and de Crombrugghe B: Multiple
functions of Osterix are required for bone growth and homeostasis
in postnatal mice. Proc Natl Acad Sci USA. 107:12919–12924. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Goettsch C, Rauner M, Pacyna N, Hempel U,
Bornstein SR and Hofbauer LC: miR-125b regulates calcification of
vascular smooth muscle cells. Am J Pathol. 179:1594–1600. 2011.
View Article : Google Scholar : PubMed/NCBI
|