Introduction
Inflammatory bowel disease (IBD) refers to a group
of conditions characterised by chronic inflammation of the bowel,
including Crohn's disease (CD) and ulcerative colitis. A number of
factors are associated with the development of IBD, including
genetic, environmental, microbial and immune response factors
(1,2). It has been reported that microRNAs
(miRNAs) serve important roles in IBD (3–7). For
instance, several studies have demonstrated an association between
miR-21 and IBD in patients and animal models (8,9). In
addition, Brain et al (10)
provided a profile of how nucleotide-binding oligomerization
domain-containing protein 2 (NOD2) induced miR-29 to limit
interleukin-23 release in human dendritic cells (DCs). NOD2
mutations have also been reported to be associated with CD
(11). Brest et al
(12) identified that a variant in
immunity-related GTPase family M protein (IRGM) gene altered a
binding site for miR-196 and caused the deregulation of
IRGM-dependent xenophagy in CD. In a murine model of IBD, miR-146
regulated NOD2-derived gut inflammation and served an important
role in amplifying inflammatory responses (13). Other functions of miR-146 were also
confirmed to be associated with IBD, including the regulation of
the Toll-like receptor (TLR) signalling pathways, T cell
differentiation and regulator T (Treg) cell function (7,14).
Circular RNA (circRNA) is a type of RNA that can
form a covalently closed loop. The 5′ cap or 3′ poly(A) tail are
not found in circRNA, and this characteristic results in the
difference of circRNAs from other linear RNAs. In the 1970s,
circRNAs were detected in viruses and extracted from certain
eukaryotic cells; however, their functions were not fully
understood at that time (15,16).
In 2010, Poliseno et al (17) demonstrated that expressed
pseudogenes were able to regulate the expression of coding genes.
Salmena et al (18) further
hypothesised that circRNAs are competing endogenous RNAs (ceRNAs).
This hypothesis suggested that miRNAs do not interact with
messenger RNAs (mRNAs) unidirectionally, but they interact with the
pool of mRNAs, transcribed pseudogenes and long noncoding RNAs
(18). In 2013, Memczak et
al (19) described circRNAs as
a large class of RNAs with regulatory potency, with thousands of
circRNAs potentially existing stably in eukaryotic cells. These
authors further revealed that the human circRNA CDR1as was able to
be densely bound by miRNA (19).
circRNAs reportedly function as miRNA sponges, while they have not
been found to function as coding proteins to date. Currently, the
most likely role of circRNAs is considered to be as ceRNAs.
Although miRNAs are known to serve important roles
in the pathogenesis of IBD, the roles of circRNAs in the
development of IBD remain unclear. The present study investigated
the expression levels of circRNAs, considered as miRNA sponges, in
the colonic tissues of CD patients. In addition, one of the
potential circRNA-miRNA-gene networks was predicted by further
examining a circRNA candidate.
Materials and methods
Patients and samples
Between April 2015 and March 2017, 13 patients with
active CD (8 males and 5 females, mean age 30.69±6.25, from 18 to
38, without any other known diseases) who were treated at the Renji
Hospital (Shanghai, China) and 13 healthy controls (7 males and 6
females, mean age 32.77±5.89, from 24 to 43, without any other
known diseases), who were physicians at this hospital, were
enrolled into the present study. The diagnosis of CD was
established based on the medical history, endoscopy, biopsy,
imaging and serological studies. The diagnosis of CD was confirmed
by two different gastroenterologists. All patients were classified
into the L3 (ileocolon) type of CD according to the Montreal
Classification, and exhibited a Harvey-Bradshaw index score of ≥5
(20,21). All colonic samples were collected
under colonoscopy prior to treatment. Samples were stored in
RNAlater stabilization reagent (Qiagen, Valencia, CA, USA)
following the protocol provided by the manufacturer. Samples from 3
random patients with active CD and 3 random healthy controls were
initially selected for circRNA microarray analysis. The remaining
samples from 10 CD patients and 10 healthy controls were used for
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) verification. All patients and healthy donors were well
informed of the study details, and provided written informed
consent. The experiments were approved by the Ethics Committee of
Renji Hospital, School of Medicine, Shanghai Jiao Tong University
(Shanghai, China).
RNA isolation
Total RNA was extracted from the colonic samples
using TRIzol™ reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) following the protocol provided by the
manufacturer. Next, RNA was purified with the RNeasy Mini kit
(Qiagen) according to the manufacturer's guidelines. A NanoDrop
ND-1000 spectrophotometer (Thermo Fisher Scientific, Inc.) was used
to measure the concentration of RNA based on the
OD260/OD280 ratio.
circRNA microarray analysis
Arraystar Human Circular RNA Array (Arraystar, Inc.,
Rockville, MD, USA) analysis was respectively performed in 3
samples from patients and controls to determine the expression
profile of circRNAs. The sample preparation and microarray
hybridization were performed based on the standard protocols
provided by Arraystar. Briefly, total RNA from each sample was
amplified and transcribed into fluorescent complementary RNA
utilizing random primers, according to the Arraystar Super RNA
Labelling protocol (Arraystar, Inc.). The labelled circRNAs were
hybridised onto the Arraystar Human circRNA Array (6×7K; Arraystar,
Inc.). Subsequent to washing the slides, the arrays were scanned by
the Axon GenePix 4000B microarray scanner (Molecular Devices, LLC,
Sunnyvale, CA, USA). The scanned images were then imported into the
Axon GenePix Pro 6.0 software (Molecular Devices, LLC) for grid
alignment and data extraction. The R package (https://www.r-project.org/) was used for quantile
normalization and subsequent expression matrix processing.
Differentially expressed circRNAs between the two groups that
presented a statistically significant variance were confirmed by
filtering with a volcano plot. For the selection of circRNA
candidates, differentially expressed circRNAs were defined as
having a fold-change of >2 and P<0.01. Hierarchical
clustering was then performed to identify distinguishable patterns
of circRNA expression among the samples. Since miR-146 has already
been demonstrated to be involved in the pathogenesis of CD
(7), one of the circRNAs that
putatively targeted both miR-146a and miR-146b was selected as the
candidate.
RT-qPCR
To validate the circRNA expression profile detected
using the Arraystar Human circRNA Array, the expression level of
hsa-circRNA-102685 (circRNA-102685) was further validated by
RT-qPCR in the remaining samples from 10 CD patients and 10 healthy
controls. Briefly, total RNA was incubated for 1 h at 37°C with 0
units (serving as the mock treatment) or 20 units of RNase R
(Epicentre; Illumina, Inc., San Diego, CA, USA) (22). RNA was subsequently purified with
the RNeasy Mini kit (Qiagen), and reverse transcribed to synthesise
cDNA with ReverTra Ace qPCR RT Kit (Toyobo, Osaka, Japan). The
process of reverse transcription was performed according with the
manufacturer's instructions: Incubation of RNA solution at 65°C for
5 min, followed by incubation at 37°C for 15 min, and lastly
heating at 98°C for 5 min. Next, the AFD9600 Real-time PCR
Detection system (Hangzhou AGS MedTech Co., Ltd., Hangzhou, China)
and the SYBR Green qPCR SuperMix (Invitrogen; Thermo Fisher
Scientific, Inc.) were applied to perform qPCR assay. Primers were
designed using Primer Premier 6.0 (Premier Biosoft International,
Palo Alto, CA, USA), and were as follows: circRNA-102685,
5′-ACATTCTGCCAGTGCGTCAAC-3′ (forward) and
5′-GAATCTTCAGGACAACGTGGAGAG-3′ (reverse); GAPDH,
5′-ACTTTGGTATCGTGGAAGGACTCAT-3′ (forward) and
5′-GTTTTTCTAGACGGCAGGTCAGG-3′ (reverse). GAPDH served as an
internal standard control. The PCR conditions included a 95°C
denaturation for 5 min, followed by 40 cycles at 95°C for 5 sec and
61°C for 30 sec. Three independent samples were tested for each CD
patient and control individual, and all reactions were performed in
triplicate. The quantification cycle (Cq) was applied to define the
expression of circRNA-102685, and the 2−ΔΔCq method was
used to calculate the relative expression levels (23).
Bioinformatics analysis and biological
function prediction
Arraystar's homemade miRNA target prediction
software based on TargetScan (24)
and miRanda (25) was applied for
the prediction of circRNA-miRNA interactions. The top 5 potential
target miRNAs were selected from the analysis. IBD-associated
miRNAs were selected based on the review published by Kalla et
al (7), while TargetScan
(http://www.targetscan.org) was used to
predict the putative target genes of miRNAs. A visualised
circRNA-miRNA-gene network was subsequently generated using the
Cytoscape software platform (https://cytoscape.org/). Gene Ontology (GO) enrichment
analysis was conducted for annotating the functions of the target
genes, while Kyoto Encyclopaedia of Genes and Genomes (KEGG)
pathway enrichment analysis was also performed to determine the
involved pathways (26–28).
Statistical analysis
Student's t-test was used for continuous variables,
and data are expressed as the mean ± standard deviation. P<0.05
was considered to indicate a statistically significant difference.
Statistical analyses were performed using R package (version 3.4.4)
and GraphPad Prism software (version 5.0; GraphPad Software, Inc.,
La Jolla, CA, USA).
Results
circRNA expression profiling in colon
tissues of CD
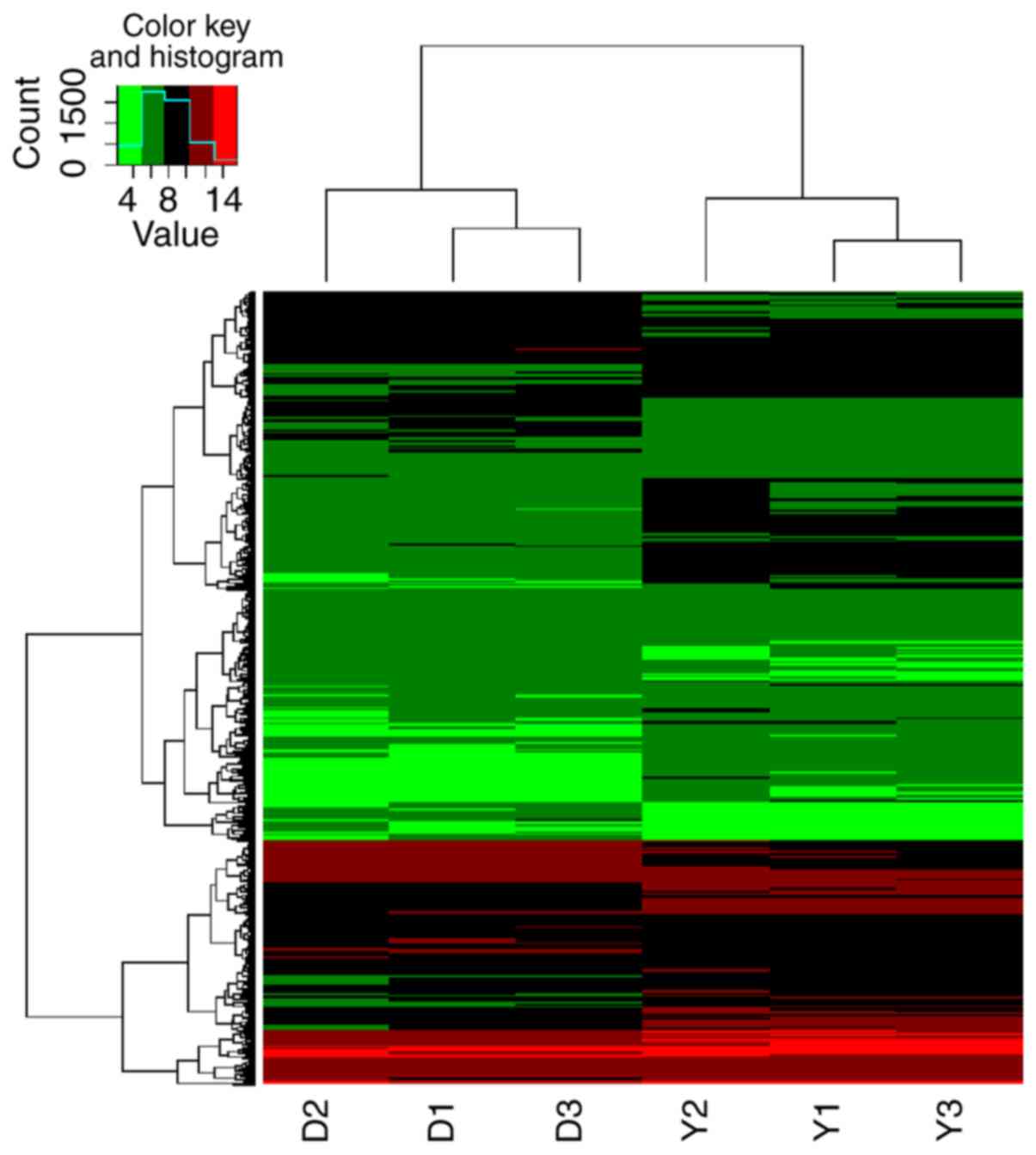
Overall, 218 circRNAs were identified to be
differentially expressed in colon tissues between CD patients and
controls. A total of 163 circRNAs were upregulated in the CD group,
and these putatively targeted 435 miRNAs. In addition, 55 circRNAs
were downregulated in the CD group and putatively targeted 207
miRNAs. Hierarchical clustering and patterns of circRNA expression
are displayed in Fig. 1. Of these
putative miR-146-targeting circRNAs, 4 were upregulated and 3 were
downregulated (Table I).
circRNA-102685 was the only upregulated circRNA that putatively
targeted both miR-146a and miR-146b, and was thus selected for
further investigation.
 | Table I.Differentially expressed circRNAs
that putatively target microRNA-146 in Crohn's disease. |
Table I.
Differentially expressed circRNAs
that putatively target microRNA-146 in Crohn's disease.
| circRNA | Fold change | P-value |
|---|
| Upregulated |
|
|
|
hsa_circRNA_102774 | 8.564309 | <0.001 |
|
hsa_circRNA_101753 | 7.143933 | 0.006 |
|
hsa_circRNA_102685 | 3.020609 | 0.004 |
|
hsa_circRNA_102261 | 4.532291 | <0.001 |
| Downregulated |
|
|
|
hsa_circRNA_101598 | 3.31592 | 0.008 |
|
hsa_circRNA_101863 | 2.424199 | 0.003 |
|
hsa_circRNA_100189 | 2.337042 | 0.002 |
circRNA candidate selection and
validation with RT-qPCR
According to the aforementioned results,
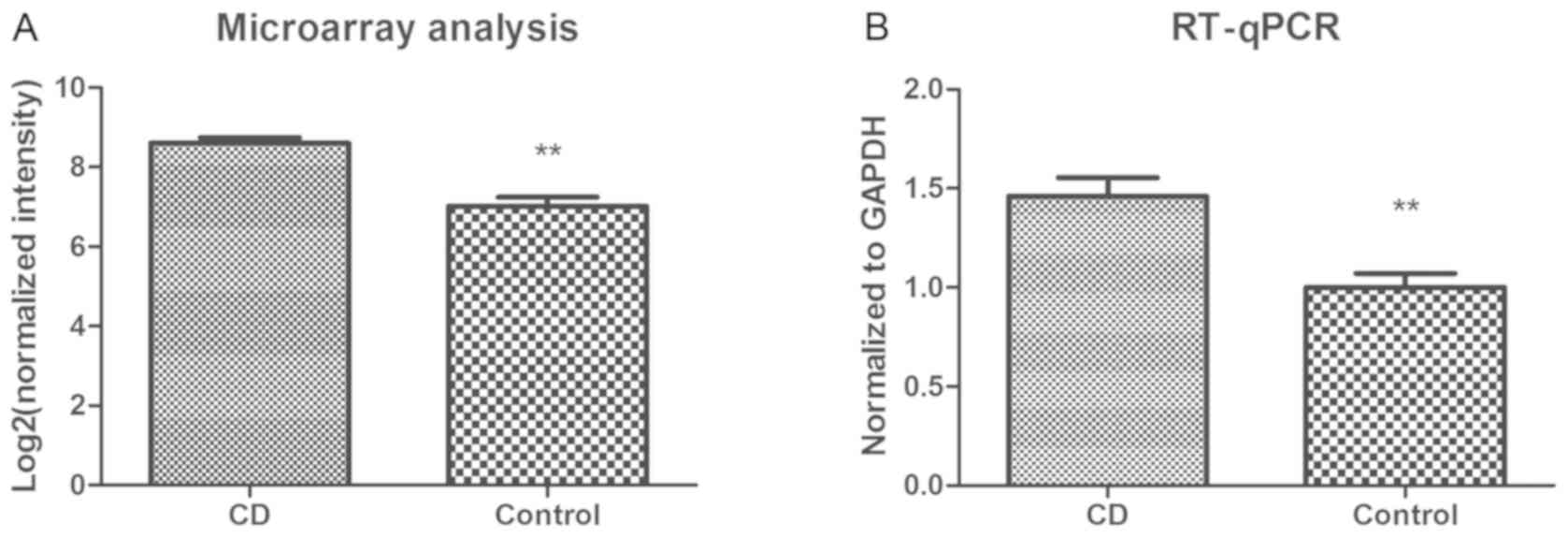
circRNA-102685 was selected as a candidate for further
verification. In the circRNA microarray analysis, the genomic locus
of circRNA-102685 was detected to be on chromosome 2, and the
predicted best transcript was uc002rpd.3. Furthermore,
circRNA-102685 was upregulated by 3.02-fold in the CD group as
compared with the control group (P<0.01). According to the
results of RT-qPCR validation, circRNA-102685 was upregulated by
1.46-fold in the CD group as compared with the control group
(P<0.01; Fig. 2).
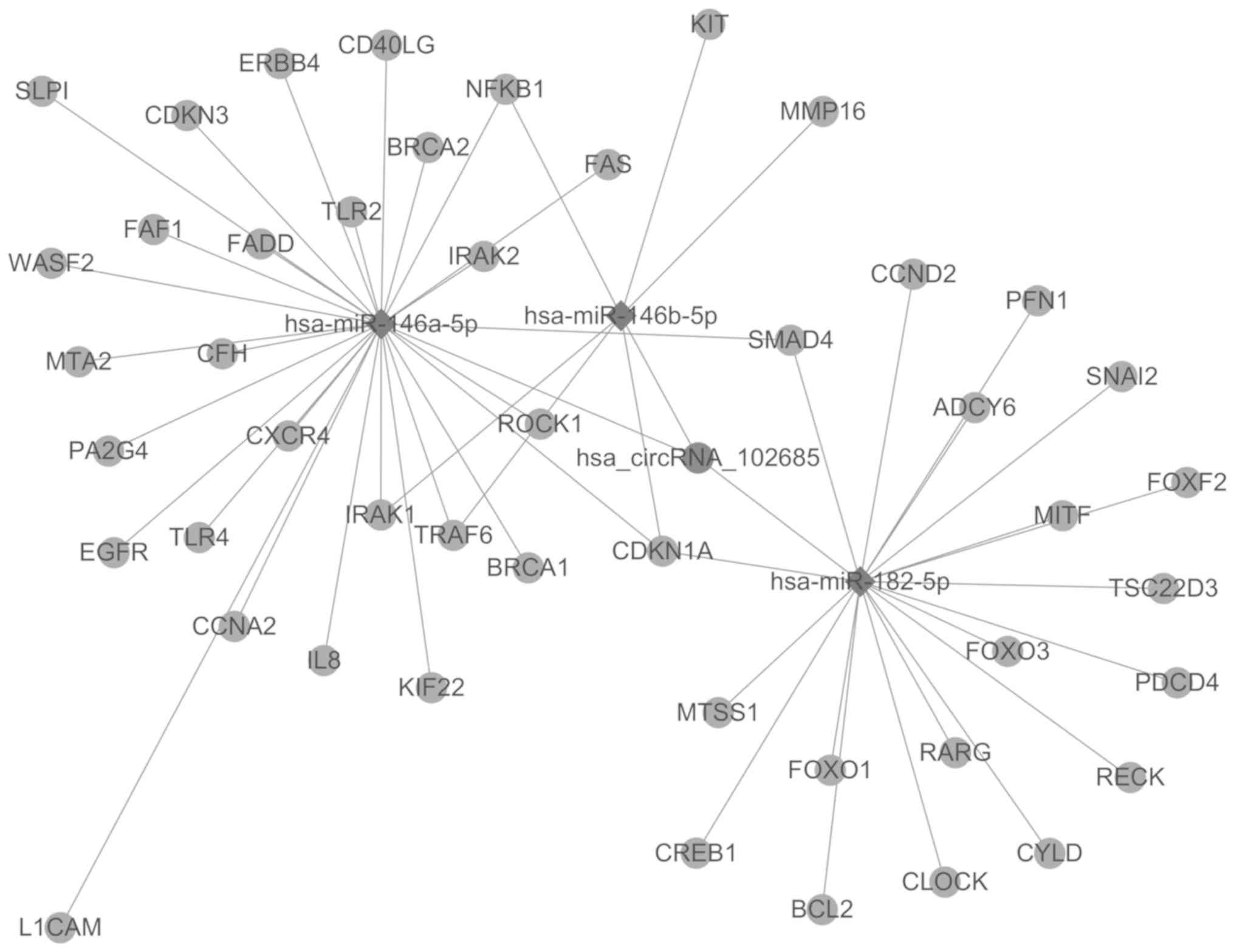
Candidate circRNA-miRNA-gene network
prediction
As circRNAs are potential upstream molecular sponges
for their target miRNAs, the TargetScan and miRanda tools were used
to predict the circRNA-miRNA-gene network. The top 3 putative miRNA
targets of circRNA-102685 were hsa-miR-146b-5p, hsa-miR-182-5p and
hsa-miR-146a-5p. In total, 47 target genes of these 3 miRNAs were
identified by target gene prediction using TargetScan database.
Next, the circRNA-102685-miRNA-gene network was constructed, and is
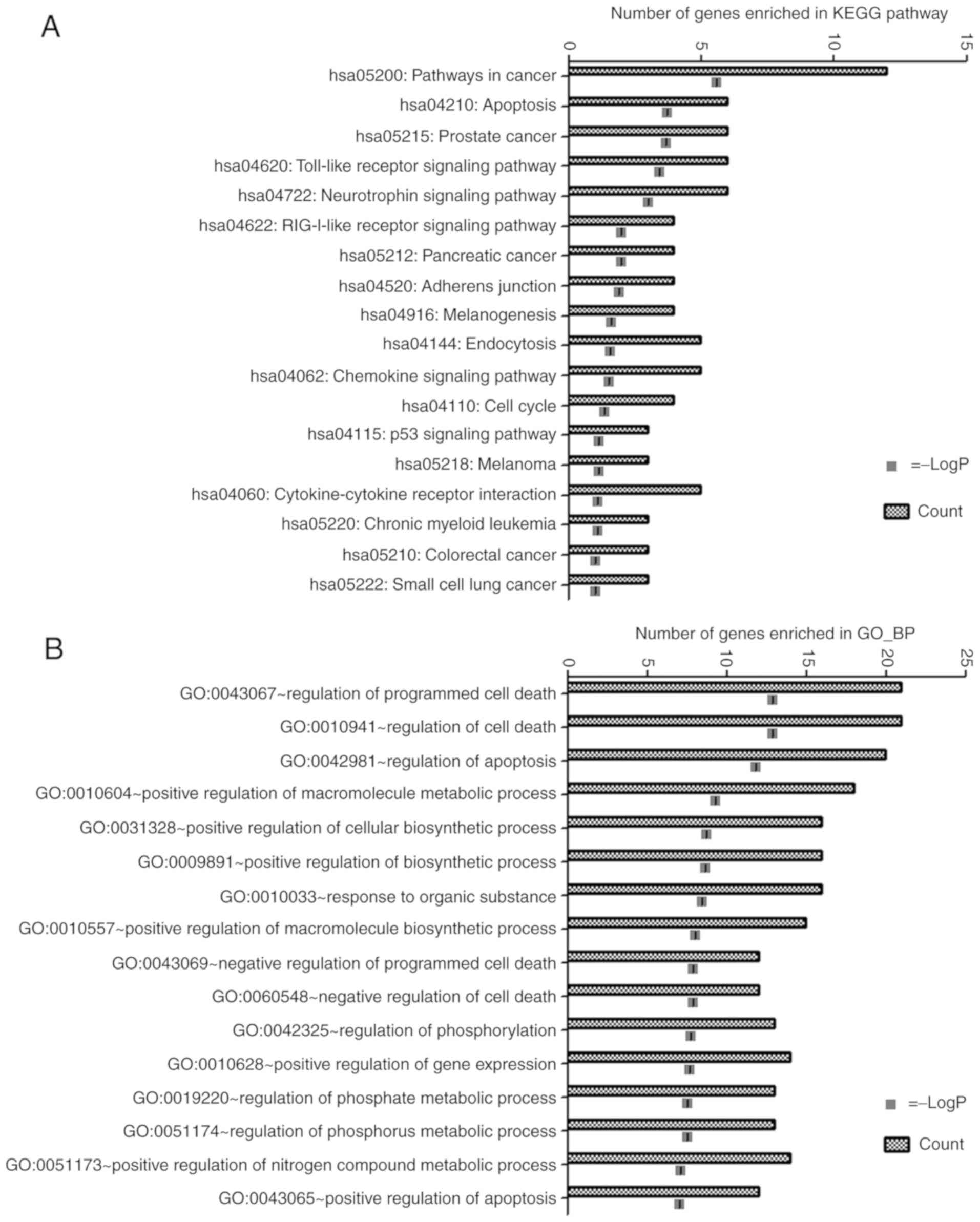
displayed in Fig. 3. Subsequent
prediction of biological pathways provided a profile of the
potential functions of these target genes. The target genes were
strongly associated with KEGG pathways that were associated with
cancer (including pathways in cancer, prostate cancer, pancreatic
cancer, melanoma, chronic myeloid leukaemia, colorectal cancer and
small cell lung cancer), cell growth and death (including
apoptosis, cell cycle, and p53 signalling pathways), immune system
(including TLR, RIG-I-like receptor and chemokine signalling
pathways), nervous system (namely neurotrophin signalling pathway),
cellular community-eukaryotes (adherens junction), endocrine system
(melanogenesis), transport and catabolism (endocytosis), and
signalling molecules and interaction (cytokine-cytokine receptor
interaction). The significant KEGG pathways are displayed in
Fig. 4A. Furthermore, GO analysis
revealed that these target genes were mainly associated with cell
death, apoptosis and phosphorylation (Fig. 4B).
Discussion
CD is a life-long disease involving chronic
gastrointestinal inflammation; however, its causes are currently
not fully understood. As the functions of miRNAs have been reported
to be associated with CD (7), the
current study aimed to examine the profile of circRNAs in this
disease, which are another type of RNAs. circRNAs can function as
miRNA sponges, and may work together with miRNAs to regulate the
expression of target genes (29).
In addition, circRNAs are very stable in eukaryotic cells, with a
half-life that can reach 48 h, while the half-life of miRNAs is
only 10 h (30). This
characteristic makes circRNAs stable molecular markers in
diseases.
circRNAs have been reported to be potential
molecular markers in different types of cancer (31). However, a number of studies have
demonstrated that circRNAs may also be involved in certain
non-cancer diseases. For instance, Wang et al (32) observed that a heart-associated
circRNA targeting miR-223 was able to protect the heart, and serve
as a potential therapeutic target in the treatment of cardiac
hypertrophy and heart failure. Furthermore, the circRNA CDR1as was
reported to be associated with neurodegenerative conditions
(33). In chronic autoimmune
disease, circRNAs are differentially expressed. The study by Liu
et al (34) indicated that
circ-CER may be associated with degenerative changes in the joint
cartilage, which can be a potential target in osteoarthritis.
As a type of chronic autoimmune disease, CD was
investigated in the present study, and alterations in the
expression of circRNAs in CD patients were examined. The results
demonstrated that >200 circRNAs were differentially expressed
between CD patients and healthy controls. These circRNAs may
function as sponges of miRNAs and serve different roles in the
pathogenesis of CD.
Among the differentially expressed circRNAs
identified in the current study, 7 candidates putatively targeted
miR-146. Previously, miR-146 has been reported to be involved in
the TLR signalling pathway, and thus serves a role in the
pathogenesis of CD (35). Taganov
et al (35) identified that
the expression of both miR-146a and miR-146b in monocytes was
induced by lipopolysaccharide, flagellin, peptidoglycan and
exposure to TLR ligands. In animal experiments, Nata et al
(36) demonstrated that miR-146b
improved intestinal inflammation by activating nuclear factor-κB.
Furthermore, in a small sample study, Fasseu et al (37) identified that miR-146 was
upregulated in CD tissues compared with the controls. A review by
Xu and Zhang (5) summarised that
miR-146 may affect the differentiation, maturation and functions of
several different immune cells, including DCs, helper T cells and
Treg cells. The present study identified that circRNA-102685 was
upregulated by performing a circRNA array analysis, and that it
potentially targeted both miR-146a and miR-146b. Thus,
circRNA-102685 was selected as a candidate circRNA for further
validation by RT-qPCR. The RT-qPCR results demonstrated that
circRNA-102685 was upregulated in colon tissues obtained from CD
patients as compared with the healthy controls. Therefore,
circRNA-102685 may serve a role in regulating target gene
expression via miR-146 in the pathogenesis of CD.
Further investigation focused on the potential
circRNA-miRNA-gene network. In the current study, it was predicted
that circRNA-102685 potentially functioned as a sponge of miR-146.
In the predicted circRNA-miRNA-gene network, a number of important
genes were identified that are known to be associated with the
pathogenesis of CD, such as IRAK1, IRAK2, BCL2, NFκB1, CREB1 and
TRAF6 (38–42). Furthermore, the prediction of KEGG
pathways presented several top pathways associated with
circRNA-102685, including apoptosis, TLR and p53 signalling
pathways. These pathways have been demonstrated to be associated
with IBD (43–46). Other pathways, such as the
RIG-I-like receptor signalling pathway, chemokine signalling
pathway and cytokine-cytokine receptor interaction pathway, are
potentially associated with IBD (47–49).
The association between cancer and IBD has already been
demonstrated (50), and the
prediction conducted in the current study identified the
involvement of certain cancer-associated pathways. GO enrichment
analysis presented similar results. However, the role of
circRNA-102685 in these pathways requires further research.
There are several limitations in the current study.
As the size and number of biopsy tissues were limited in patients
with active CD, it was not possible to verify the expression of
other circRNAs. In addition, only 13 CD patients and 13 controls
were enrolled in the study. More studies on the role of circRNAs in
IBD should be performed in the future.
In conclusion, the present study revealed that
circRNAs were differentially expressed in colon tissues of CD
patients in comparison with the healthy controls. circRNA
expression alterations may serve an important role in the
pathogenesis of CD. The expression changes of hsa-circRNA-102685
may be involved in the process of CD. Certain associated pathways
that may be involved in the process of CD were identified, and
these may serve as novel targets in the diagnosis and treatment of
this disease.
Acknowledgements
The authors would like to thank Dr Si Bo Zhu (Fudan
University) for his help in the bioinformatics analysis.
Funding
This research was supported by the National Natural
Science Foundation of China (grant nos. 81500424 and 81670497).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
YQQ and JS designed the study. YQQ worked on the
bioinformatics analysis and drafted the manuscript. CWC collected
the samples and performed the verification work. QZ and ZHR
co-worked on the analysis and critically reviewed the
interpretation of data. All authors read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
All patients and healthy donors were well informed
of the study details, and provided written informed consent. The
experiments were approved by the Ethics Committee of Renji
Hospital, School of Medicine, Shanghai Jiao Tong University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lee SH, Kwon JE and Cho ML: Immunological
pathogenesis of inflammatory bowel disease. Intest Res. 16:26–42.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
de Mattos BR, Garcia MP, Nogueira JB,
Paiatto LN, Albuquerque CG, Souza CL, Fernandes LG, Tamashiro WM
and Simioni PU: Inflammatory bowel disease: An overview of immune
mechanisms and biological treatments. Mediators Inflamm.
2015:4930122015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dalal SR and Kwon JH: The role of MicroRNA
in inflammatory bowel disease. Gastroenterol Hepatol (NY).
6:714–722. 2010.
|
|
4
|
Chapman CG and Pekow J: The emerging role
of miRNAs in inflammatory bowel disease: A review. Therap Adv
Gastroenterol. 8:4–22. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xu XM and Zhang HJ: miRNAs as new
molecular insights into inflammatory bowel disease: Crucial
regulators in autoimmunity and inflammation. World J Gastroenterol.
22:2206–2218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schaefer JS, Attumi T, Opekun AR, Abraham
B, Hou J, Shelby H, Graham DY, Streckfus C and Klein JR: MicroRNA
signatures differentiate Crohn's disease from ulcerative colitis.
BMC Immunol. 16:52015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kalla R, Ventham NT, Kennedy NA, Quintana
JF, Nimmo ER, Buck AH and Satsangi J: MicroRNAs: New players in
IBD. Gut. 64:504–517. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Adams AT, Kennedy NA, Hansen R, Ventham
NT, O'Leary KR, Drummond HE, Noble CL, El-Omar E, Russell RK,
Wilson DC, et al: Two-stage genome-wide methylation profiling in
childhood-onset Crohn's disease implicates epigenetic alterations
at the VMP1/MIR21 and HLA loci. Inflamm Bowel Dis. 20:1784–1793.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu F, Dong F, Arendovich N, Zhang J, Huang
Y and Kwon JH: Divergent influence of microRNA-21 deletion on
murine colitis phenotypes. Inflamm Bowel Dis. 20:1972–1985. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brain O, Owens BM, Pichulik T, Allan P,
Khatamzas E, Leslie A, Steevels T, Sharma S, Mayer A, Catuneanu AM,
et al: The intracellular sensor NOD2 induces microRNA-29 expression
in human dendritic cells to limit IL-23 release. Immunity.
39:521–536. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wehkamp J, Harder J, Weichenthal M, Schwab
M, Schäffeler E, Schlee M, Herrlinger KR, Stallmach A, Noack F,
Fritz P, et al: NOD2 (CARD15) mutations in Crohn's disease are
associated with diminished mucosal alpha-defensin expression. Gut.
53:1658–1664. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brest P, Lapaquette P, Souidi M, Lebrigand
K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier JF,
Hébuterne X, et al: A synonymous variant in IRGM alters a binding
site for miR-196 and causes deregulation of IRGM-dependent
xenophagy in Crohn's disease. Nat Genet. 43:242–245. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ghorpade DS, Sinha AY, Holla S, Singh V
and Balaji KN: NOD2-nitric oxide-responsive microRNA-146a activates
Sonic hedgehog signaling to orchestrate inflammatory responses in
murine model of inflammatory bowel disease. J Biol Chem.
288:33037–33048. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He X, Jing Z and Cheng G: MicroRNAs: New
regulators of Toll-like receptor signalling pathways. Biomed Res
Int. 2014:9451692014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hsu MT and Coca-Prados M: Electron
microscopic evidence for the circular form of RNA in the cytoplasm
of eukaryotic cells. Nature. 280:339–340. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Poliseno L, Salmena L, Zhang J, Carver B,
Haveman WJ and Pandolfi PP: A coding-independent function of gene
and pseudogene mRNAs regulates tumour biology. Nature.
465:1033–1038. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Harvey RF and Bradshaw JM: A simple index
of Crohn's-disease activity. Lancet. 1:5141980. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Satsangi J, Silverberg MS, Vermeire S and
Colombel JF: The Montreal classification of inflammatory bowel
disease: Controversies, consensus, and implications. Gut.
55:749–753. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li L, Guo J, Chen Y, Chang C and Xu C:
Comprehensive CircRNA expression profile and selection of key
CircRNAs during priming phase of rat liver regeneration. BMC
Genomics. 18:802017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila. Genome Biol.
5:R12003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pasquinelli AE: MicroRNAs and their
targets: Recognition, regulation and an emerging reciprocal
relationship. Nat Rev Genet. 13:271–282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
The Gene Ontology Consortium: Expansion of
the gene ontology knowledgebase and resources. Nucleic Acids Res.
45:D331–D338. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P
and Wu M: CircRNA: Functions and properties of a novel potential
biomarker for cancer. Mol Cancer. 16:942017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang K, Long B, Liu F, Wang JX, Liu CY,
Zhao B, Zhou LY, Sun T, Wang M, Yu T, et al: A circular RNA
protects the heart from pathological hypertrophy and heart failure
by targeting miR-223. Eur Heart J. 37:2602–2611. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lukiw WJ: Circular RNA (circRNA) in
Alzheimer's disease (AD). Front Genet. 4:3072013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu Q, Zhang X, Hu X, Dai L, Fu X, Zhang J
and Ao Y: Circular RNA related to the chondrocyte ECM regulates
MMP13 expression by functioning as a MiR-136 ‘Sponge’ in human
cartilage degradation. Sci Rep. 6:225722016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Taganov KD, Boldin MP, Chang KJ and
Baltimore D: NF-kappaB-dependent induction of microRNA miR-146, an
inhibitor targeted to signaling proteins of innate immune
responses. Proc Natl Acad Sci USA. 103:12481–12486. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nata T, Fujiya M, Ueno N, Moriichi K,
Konishi H, Tanabe H, Ohtake T, Ikuta K and Kohgo Y: MicroRNA-146b
improves intestinal injury in mouse colitis by activating nuclear
factor-kappaB and improving epithelial barrier function. J Gene
Med. 15:249–260. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fasseu M, Tréton X, Guichard C, Pedruzzi
E, Cazals-Hatem D, Richard C, Aparicio T, Daniel F, Soulé JC,
Moreau R, et al: Identification of restricted subsets of mature
microRNA abnormally expressed in inactive colonic mucosa of
patients with inflammatory bowel disease. PLoS One. 5:e131602010.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhernakova A, Festen EM, Franke L, Trynka
G, van Diemen CC, Monsuur AJ, Bevova M, Nijmeijer RM, van 't Slot
R, Heijmans R, et al: Genetic analysis of innate immunity in
Crohn's disease and ulcerative colitis identifies two
susceptibility loci harboring CARD9 and IL18RAP. Am J Hum Genet.
82:1202–1210. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Catarzi S, Marcucci T, Papucci L, Favilli
F, Donnini M, Tonelli F, Vincenzini MT and Iantomasi T: Apoptosis
and Bax, Bcl-2, Mcl-1 expression in neutrophils of Crohn's disease
patients. Inflamm Bowel Dis. 14:819–825. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sartor RB: Mechanisms of disease:
Pathogenesis of Crohn's disease and ulcerative colitis. Nat Clin
Pract Gastroenterol Hepatol. 3:390–407. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Diegelmann J, Czamara D, Le Bras E,
Zimmermann E, Olszak T, Bedynek A, Göke B, Franke A, Glas J and
Brand S: Intestinal DMBT1 expression is modulated by Crohn's
disease-associated IL23R variants and by a DMBT1 variant which
influences binding of the transcription factors CREB1 and ATF-2.
PLoS One. 8:e777732013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shen J, Qiao Y, Ran Z and Wang T:
Different activation of TRAF4 and TRAF6 in inflammatory bowel
disease. Mediators Inflamm. 2013:6479362013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Nunes T, Bernardazzi C and de Souza HS:
Cell death and inflammatory bowel diseases: Apoptosis, necrosis,
and autophagy in the intestinal epithelium. Biomed Res Int.
2014:2184932014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mudter J and Neurath MF: Apoptosis of T
cells and the control of inflammatory bowel disease: Therapeutic
implications. Gut. 56:293–303. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cario E: Toll-like receptors in
inflammatory bowel diseases: A decade later. Inflamm Bowel Dis.
16:1583–1597. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Goretsky T, Dirisina R, Sinh P, Mittal N,
Managlia E, Williams DB, Posca D, Ryu H, Katzman RB and Barrett TA:
p53 mediates TNF-induced epithelial cell apoptosis in IBD. Am J
Pathol. 181:1306–1315. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ireton RC and Gale M Jr: RIG-I like
receptors in antiviral immunity and therapeutic applications.
Viruses. 3:906–919. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Singh UP, Singh NP, Murphy EA, Price RL,
Fayad R, Nagarkatti M and Nagarkatti PS: Chemokine and cytokine
levels in inflammatory bowel disease patients. Cytokine. 77:44–49.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xavier RJ and Podolsky DK: Unravelling the
pathogenesis of inflammatory bowel disease. Nature. 448:427–434.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Terzić J, Grivennikov S, Karin E and Karin
M: Inflammation and colon cancer. Gastroenterology.
138:2101–2114.e5. 2010. View Article : Google Scholar : PubMed/NCBI
|