Introduction
At present, obesity poses a major health problem
worldwide. Obesity is associated with mortality and multiple
comorbidities, including cancer and various cardiometabolic
disorders (1,2). Modulation of adipogenesis may be used
in the treatment of obesity (3).
Additionally, elucidating the molecular mechanisms underlying
adipogenesis may have important applications, in particular for the
development of novel treatments for obesity and other related
metabolic disorders.
Mesenchymal stem cells from various types of
tissues, including bone marrow, are able to differentiate into
adipocytes, similar to the pluripotent stem cells found in fat
tissue (4). Bone marrow human
mesenchymal stem cells (hMSCs) exhibit self-renewal capabilities
and are multipotent (5). A
previous in vitro study suggested that hMSCs may
differentiate, not only into adipocytes (6), but also into other cell types,
including chondrocytes (7) and
osteoblasts (8). Therefore, hMSC
cultures may represent an optimal model for analyzing the molecular
mechanisms that regulate adipogenesis in humans (9).
The leukemia inhibitory factor (LIF) receptor (LIFR)
consists of α and β subunits (10,11).
LIFR can be activated by LIF, which induces heterodimerization of
the two subunits of the receptor, thus activating protein
phosphorylation and downstream signaling pathways involved in
various biological processes (12–14).
LIF has been reported to exhibit various additional functions in
adipocytes (15,16). However, to the best of our
knowledge, the number of studies investigating LIFR in the context
of adipogenic differentiation remains limited. In the present
study, the expression levels of LIFR were revealed to progressively
increase during adipogenic differentiation of hMSCs, suggesting
that LIFR may be involved in this process.
In order to investigate the regulatory role of LIFR
in adipogenesis, lentivirus-mediated LIFR knockdown was performed
in hMSCs, and the expression levels of LIF and LIFR were analyzed
during adipogenic differentiation. Silencing LIFR expression
significantly suppressed adipocyte differentiation. Furthermore,
the expression levels of LIF and LIFR exhibited opposite trends.
Collectively, the present results suggested that LIFR may promote
adipogenic differentiation, whereas LIF may negatively regulate
this process.
Materials and methods
Cell culture and adipocyte
differentiation
Human bone marrow mesenchymal stem cells (hMSCs;
HUXMA-01001) were purchased from Cyagen Biosciences, Inc. Cells
were evaluated for specific surface protein expression using flow
cytometry. Flow cytometry was performed using FACSCalibur (BD
Biosciences). The following fluorescent-conjugated monoclonal
antibodies were used in this study: Rat allophycocyanin (APC)
anti-human/mouse CD44 (1:50; cat. no. 17-0441-82; clone IM7;
eBioscience; Thermo Fisher Scientific, Inc.), APC mouse anti-human
CD29 (1:50; cat. no. 303008; clone TS2/16, BioLegend, Inc.), FITC
mouse anti-human CD105 (1:50; cat. no. 561443; Endoglin; clone 266;
BD Biosciences), APC mouse anti-human CD45 (1:50; cat. no. 555485;
clone H130; BD Biosciences) and APC mouse antihuman CD14 (1:50;
cat. no. 17-0149-42; clone 61D3; eBioscience; Thermo Fisher
Scientific, Inc.). The cells were washed with PBS with 3% fetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) after
detachment with 0.05% trypsin-EDTA (Gibco; Thermo Fisher
Scientific, Inc.). A total 5×105 cells were re-suspended
in 50 µl of PBS with 3% FBS, blocked with 10 µl of FcR Blocking
Reagent (cat. no. 130-059-901; Miltenyi Biotec GmbH) for 10 min at
room temperature, and incubated with each type of the antibodies as
listed above, for 30 min at 4°C in the dark. After incubation, the
cells were washed twice in PBS with 3% FBS, and re-suspended in 500
µl of PBS with 3% FBS for flow cytometry. The fluorescence
intensity of the cells was evaluated using a FACSAria instrument,
and data were analyzed with the FlowJov10.0.7 software (FlowJo
LLC). hMSCs were confirmed to be positive for CD44, CD29 and CD105
(>70%) and negative for CD45 and CD14 (<5%).
Cells were grown to a cell density of
5×104 cells/cm2 in OriCell hMSCs growth
medium (cat. no. HUXMA-9001c; Cyagen Biosciences, Inc.),
supplemented with 10% FBS, 100 IU/ml penicillin (Gibco; Thermo
Fisher Scientific, Inc.), 100 µg/ml streptomycin (Gibco; Thermo
Fisher Scientific, Inc.) and 0.4% glutamine (Gibco; Thermo Fisher
Scientific, Inc.). Cells were maintained at 37°C in an atmosphere
containing 5% CO2 and 95% humidity. Cells were passaged
using 0.25% trypsin-EDTA (Gibco; Thermo Fisher Scientific, Inc.)
every 3–4 days. hMSCs were used at the fifth passage.
Confluent hMSCs were incubated for 2 additional days
and adipogenesis was induced using Dulbecco's modified Eagle's
medium containing 10% FBS, 10 µg/ml insulin, 0.5 mM
3-isobutyl-1-methylxanthine and 0.5 mM dexamethasone. All reagents
were obtained from Gibco (Thermo Fisher Scientific, Inc.).
Adipogenic differentiation was induced for 9 days and the medium
was replaced every 3 days.
Lentiviral infection and screening of
hMSCs
A lentiviral vector used to induce LIFR knockdown
was purchased from Shanghai GeneChem Co., Ltd. A short hairpin RNA
(shRNA) targeting human LIFR was designed (target sequence,
5′-TCCCATTGTTGCACCAAAT-3′). In addition, a negative control (NC)
shRNA was used (sequence, 5′-TTCTCCGAACGTGTCACGT-3′). Chemically
synthesized DNA oligonucleotides (Shanghai GeneChem Co., Ltd.) were
cloned into the pGV248-green fluorescent protein (GFP) vector
lentivirus (Shanghai GeneChem Co., Ltd.). hMSCs were infected with
LIFR shRNA vector or with NC. The lentivirus titer was determined
through serial dilutions. hMSCs were seeded into 6-well plates at a
density of 5×104 cells/cm2, and cultured to
20–30% confluence. Cells were infected with 1×108
transducing units/ml lentivirus (volume, 10 µl; multiplicity of
infection, 5) in complete medium with 5 µg/ml polybrene (Shanghai
GeneChem Co., Ltd.). Subsequently, the infected cells were
incubated at 37°C for 10 h, after which, the medium was replaced
with fresh medium and cells were incubated at 37°C for an
additional 72 h. Infected cells were screened after culturing for
48 h in medium containing 0.5 µg/ml puromycin. The medium was
replaced after 1–2 days, and screening continued for 6 days.
Fluorescence microscopy was performed on living cells in order to
evaluate the efficiency of lentiviral infection. Additionally,
prior to adipogenic differentiation, LIFR expression was assessed
using quantitative PCR (qPCR) to examine infection efficiency after
infection 72 h.
During adipogenic differentiation of hMSCs, the
development of intracellular lipid droplets was assessed at various
time points (3, 6 and 9 days) using Oil Red O. Subsequently, cells
were harvested for analysis of the mRNA and protein expression
levels of LIF, LIFR, peroxisome proliferator-activated receptor γ
(PPARγ), CCAAT enhancer binding protein α (C/EBPα) and aP2.
Oil Red O staining and lipid
measurement
After washing with PBS, fixation was performed with
4% formalin at room temperature for 30 min. Samples were then
washed twice with PBS and incubated for 30 min with 60% saturated
Oil Red O (Sigma-Aldrich; Merck KGaA) at room temperature. After
two further washes with PBS, the cells were visualized using light
microscopy (IX73; Olympus Corporation). Subsequently, stained cells
were treated with isopropanol and a microplate reader (Bio-Rad
Laboratories, Inc.) was used to measure the optical density at 490
nm, in order to quantify the intracellular levels of lipid
droplets.
Reverse transcription (RT)-qPCR
RNA was extracted with TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. First-strand cDNA was obtained using the
Reverse Transcription System and Oligo (dT), according to the
manufacturer's protocol (Thermo Fisher Scientific, Inc.). qPCR was
performed using a SYBR Premix Ex Taq kit (Toyobo Life Science) with
a 7300 real-time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The PCR thermocycling conditions consisted of an
initial denaturation at 95°C for 1 min, followed by 40 cycles at
95°C for 15 sec and at 60°C for 34 sec. Relative expression levels
were quantified using the 2−ΔΔCq method (17). The expression levels of the target
genes were normalized to the expression levels of β-actin. The
sequences of the primers used are listed in Table I.
 | Table I.Primer sequences used for reverse
transcription-quantitative PCR analysis. |
Table I.
Primer sequences used for reverse
transcription-quantitative PCR analysis.
| Gene symbol | Primer sequence
(5′-3′) | Amplicon length
(bp) |
|---|
| PPARγ | F:
GGGATGTCTCATAATGCCATCAG | 97 |
|
| R:
GCCCTCGCCTTTGCTTTG |
|
| C/EBPα | F:
CCAAGAAGTCGGTGGACAAGAAC | 122 |
|
| R:
CACCTTCTGCTGCGTCTCCA |
|
| aP2 | F:
GGATGATAAACTGGTGGTGGAATG | 123 |
|
| R:
CAGAATGTTGTAGAGTTCAATGCGA |
|
| LIFR | F:
AGCCTCAAGCAAAACCAGAA | 144 |
|
| R:
TTGGCCTGAGGTCTGTAACC |
|
| LIF | F:
CTGTTGGTTCTGCACTGGAA | 154 |
|
| R:
CCCCTGGGCTGTGTAATAGA |
|
| β-actin | F:
GCGAGAAGATGACCCAGATCATGT | 160 |
|
| R:
TACCCCTCGTAGATGGGCACA |
|
Western blotting
Cells were lysed using RIPA buffer [50 mM Tris (pH
7.4), 1% Triton X-100, 150 mM NaCl, 0.1% SDS, 1% sodium
deoxycholate, 1 mmol/l sodium orthovanadate, 50 mmol/l sodium
fluoride, 1 mM EDTA and 2 µg/ml leupeptin]. The amount of protein
was measured using a Micro bicinchoninic acid Protein assay kit
following the protocol provided by the manufacturer (Thermo Fisher
Scientific, Inc.). Cell samples were diluted 10, 20 and 40 times in
NaCl 0.9% and after 2 h incubation at 37°C, absorbance was measured
using the Bio-Rad iMark microplate reader (Bio-Rad Laboratories,
Inc.). The extracted proteins were boiled for 5 min in 5× SDS
sample buffer. Electrophoresis was performed on 15 µg of protein
loaded onto 10% SDS-PAGE, followed by transfer to PVDF membranes
(EMD Millipore). Membranes were blocked for 90 min with 5% skim
milk for 120 min at room temperature, followed by incubation at 4°C
overnight with primary antibodies. The primary antibodies used
were: Mouse anti-LIFR (1:1,000; cat. no. ab89792), rabbit
anti-PPARγ (1:1,000; cat. no. ab191407), rabbit anti-C/EBPα
(1:1,000; cat. no. ab40764), rabbit anti-aP2 (1:1,000; cat. no.
ab92501) and mouse anti-β-actin (1:2,000; cat. no. ab173838; all
Abcam). Subsequently, membranes were incubated for 60 min with the
appropriate secondary antibody at room temperature: Anti-mouse
horseradish peroxidase (HRP)-conjugated IgG (1:5,000; cat. no.
7076P2; Cell Signaling Technology, Inc.) or anti-rabbit
HRP-conjugated IgG (1:5,000; cat. no. 7074P2; Cell Signaling
Technology, Inc.). Finally, enhanced chemiluminescence (cat. no.
P0018; BeyoECL Plus; Beyotime Institute of Biotechnology) was
performed to evaluate the intensity of the protein bands.
Quantification of LIF
concentration
LIF protein concentration was assessed in the cell
culture supernatant using an ELISA kit (cat. no. SX01091; Shanghai
Senxiong Biotech Industry Co., Ltd.), according to the
manufacturer's protocol. Absorbance was measured at 450 nm
following background correction. LIF concentration was calculated
using a standard curve.
Statistical analysis
All experiments were performed in triplicate. Data
are presented as the mean ± standard deviation. SPSS (version 16.0;
SPSS, Inc.) was used to perform statistical analyses. Statistical
analysis was performed using one-way analysis of variance (ANOVA)
and Tukey's post-hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
LIFR expression during adipogenic
differentiation
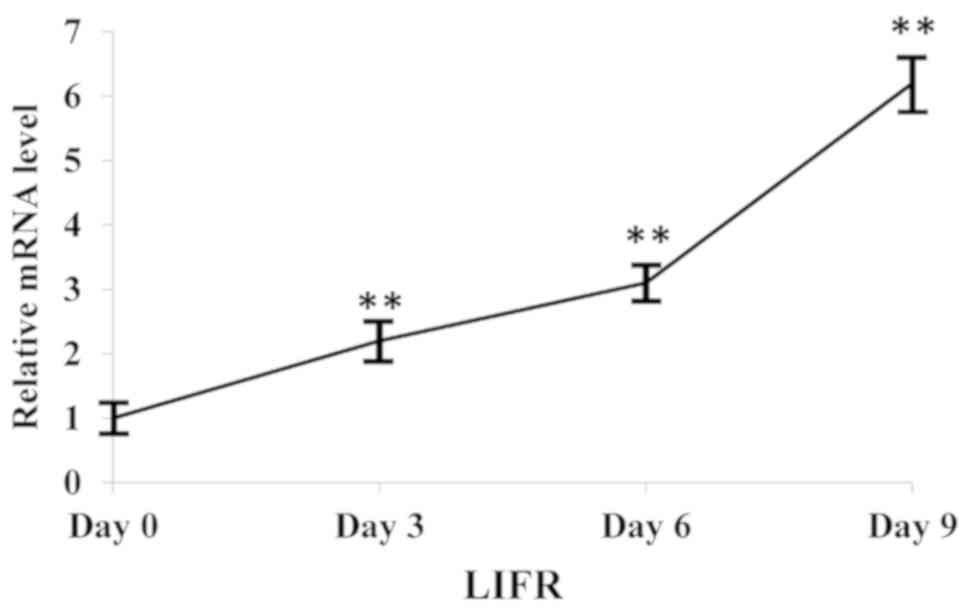
The expression levels of LIFR progressively
increased during adipogenic differentiation (Fig. 1), suggesting that LIFR may be
involved in the adipogenic differentiation of hMSCs.
Identification of stably infected
hMSCs
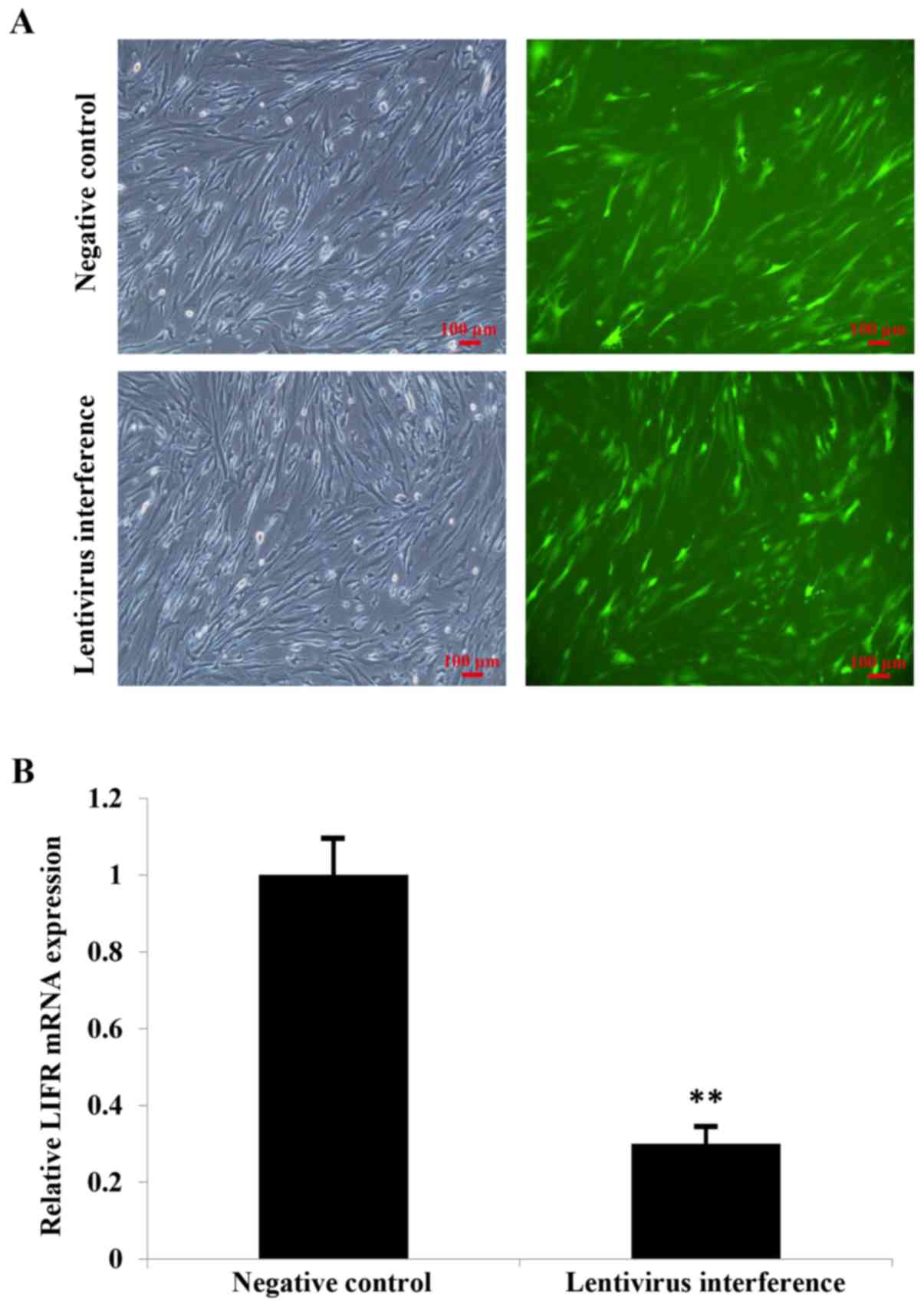
hMSCs with stable infection were identified by
treating cells with 0.5 µg/ml puromycin for 6 days. Inverted
fluorescence microscopy was used to identify GFP-positive cells in
both the NC group and in cells with knockdown of LIFR, indicating
stable infection (Fig. 2A).
The efficiency of lentiviral infection was confirmed
by RT-qPCR analysis. The expression levels of LIFR were reduced
~3-fold following LIFR knockdown compared with cells in the NC
group (Fig. 2B).
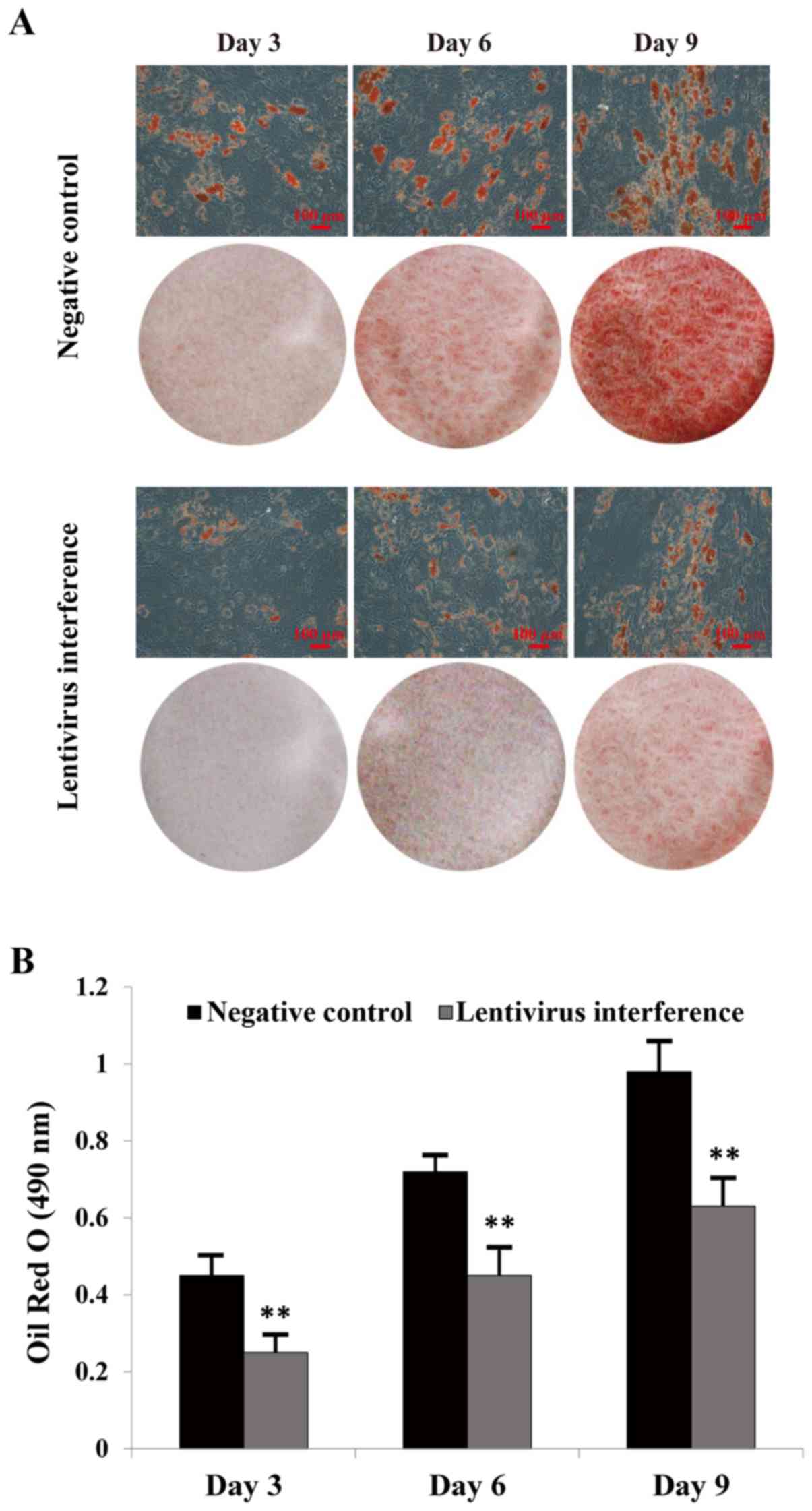
Oil Red O staining
Adipogenesis was investigated in hMSCs at various
time points (3, 6 and 9) following incubation in adipogenic medium.
The majority of cells exhibited cytoplasmic lipid vesicles, as
assessed by Oil Red O staining (Fig.
3A). LIFR knockdown inhibited adipogenic differentiation.
Furthermore, the intracellular lipid content and the number of
lipid droplets were lower in the LIFR-knockdown group compared with
in the NC group (Fig. 3B).
mRNA expression levels of LIF, LIFR
and adipogenic markers during adipogenic differentiation
The expression levels of LIF, LIFR, PPARγ, C/EBPα
and aP2 were measured by RT-qPCR in LIFR-knockdown and NC groups
(Fig. 4). LIFR downregulation
significantly suppressed adipocyte differentiation, alongside
inhibition of PPARγ, C/EBPα and aP2 expression (Fig. 4A-C). Furthermore, the mRNA
expression levels of LIF were significantly decreased following
induction of adipogenic differentiation in the negative control and
LIFR knockdown groups, respectively (Fig. 4E). Conversely, the mRNA expression
levels of LIFR exhibited an opposite trend (Fig. 4D).
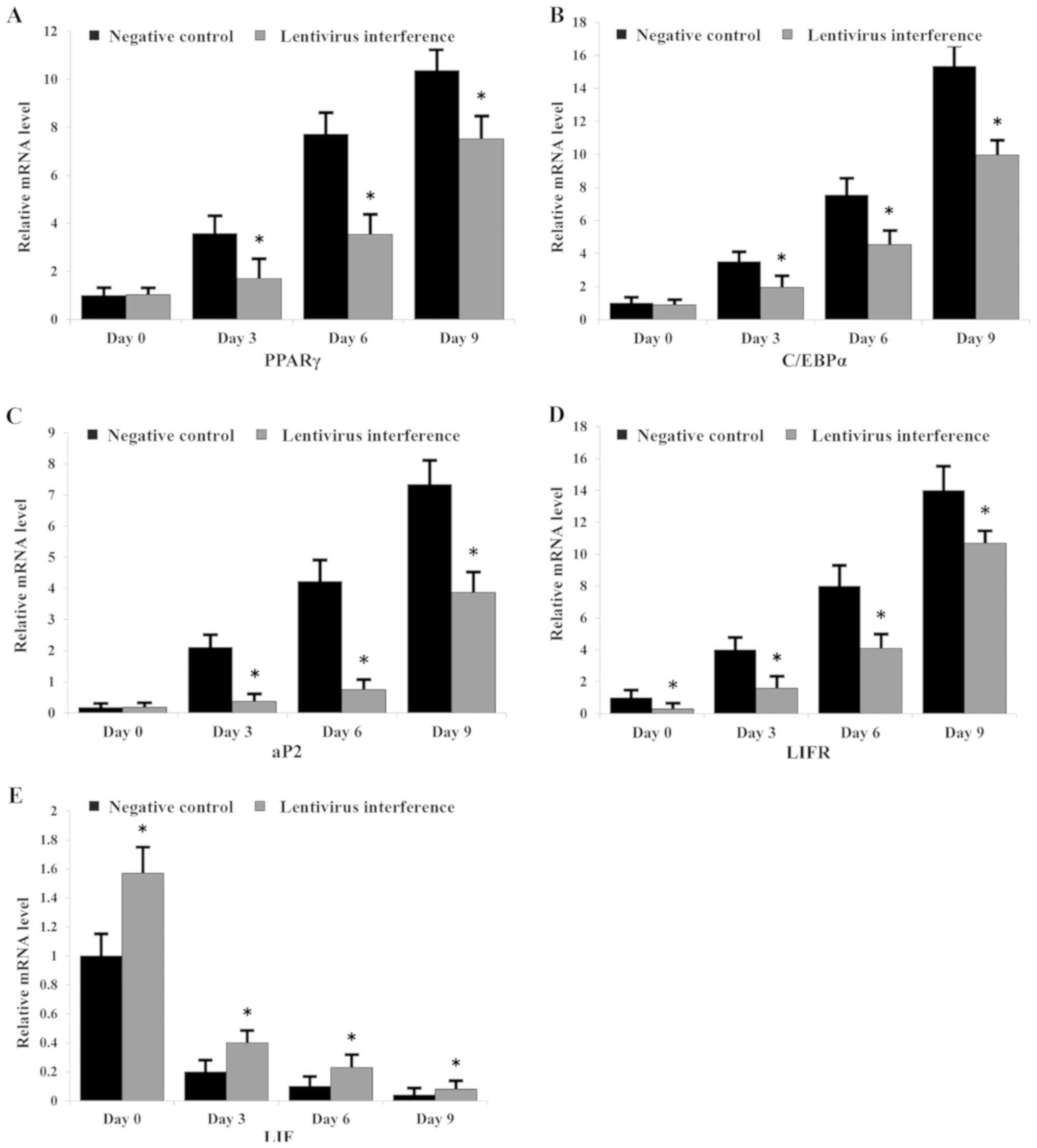 | Figure 4.mRNA expression levels of LIF, LIFR
and adipogenic markers during adipogenic differentiation.
Expression levels of (A) PPARγ, (B) C/EBPα, (C) aP2, (D) LIFR and
(E) LIF in each group of cells, as assessed by reverse
transcription-quantitative PCR. *P<0.05 vs. negative control.
aP2, fatty acid binding protein 4; C/EBPα, CCAAT enhancer binding
protein α; LIF, leukemia inhibitory factor; LIFR, LIF receptor;
PPARγ, peroxisome proliferator-activated receptor γ. |
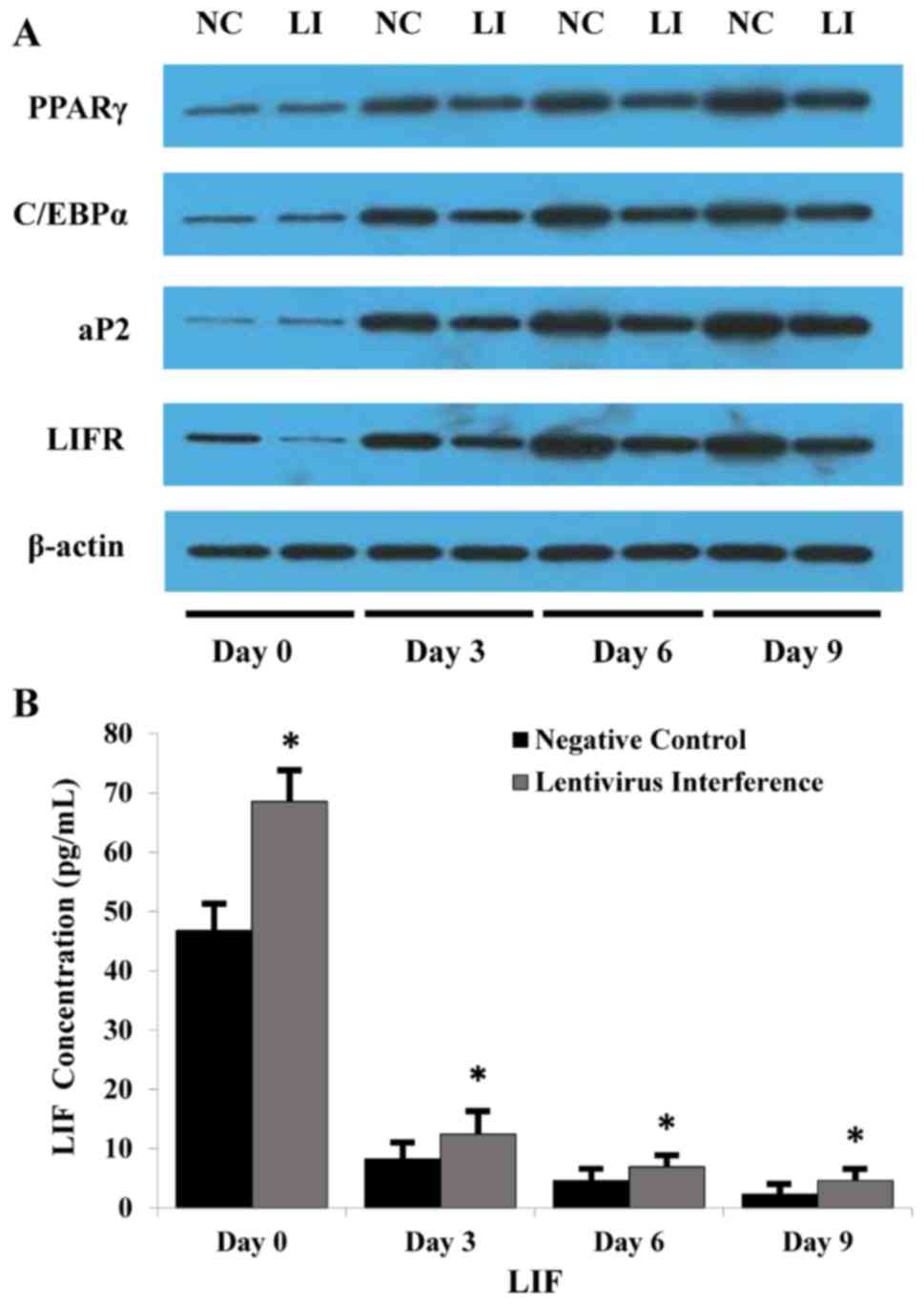
Protein expression levels of LIF, LIFR
and markers during adipogenesis
LIFR, PPARγ, C/EBPα and aP2 mRNA expression was
elevated in each group, as aforementioned. Consistently, western
blotting revealed that the protein expression levels of PPARγ,
C/EBPα and aP2 appeared to be decreased during adipogenesis
following LIFR knockdown (Fig.
5A). Additionally, ELISA was performed to measure the
concentration of LIF in the medium, and LIF was decreased during
adipogenesis in the negative control and LIFR knockdown groups
(Fig. 5B).
Discussion
Adipogenic differentiation serves an important role
in the development of obesity. The adipogenic differentiation of
hMSCs through preadipocytes may represent an important source of
adipose tissue during obesity (3,4,18).
In the present study, LIFR expression was increased during the
adipogenic differentiation of hMSCs, suggesting that this protein
may be involved in the regulation of adipogenesis. To investigate
this hypothesis, stable lentivirus-mediated infection was performed
to efficiently downregulate the expression of LIFR in hMSCs.
Subsequently, adipogenic differentiation of hMSCs was examined at
different time points. LIFR downregulation significantly inhibited
adipogenesis and the accumulation of intracellular lipids. The
present findings indicated that LIFR may promote adipogenesis in
these cells.
A previous study confirmed that microRNA
(miR)-377-3p targeted LIFR and regulates adipogenic differentiation
(19); however, the function of
LIFR in adipogenesis had not been investigated. Therefore, the
present study aimed to investigate the role of LIFR in adipogenic
differentiation at various time points. In addition, the mRNA and
protein expression levels of various molecular markers of
adipogenesis, including PPARγ, C/EBPα and aP2, were examined. LIFR
downregulation in hMSCs caused a significant reduction in the mRNA
and protein expression levels of these markers during
differentiation. PPARγ and C/EBPα are transcription factors that
promote adipogenesis, which may act synergistically (20,21).
In addition, aP2 modulates lipid storage and metabolism, acting
downstream of PPARγ and C/EBPα signaling (22,23).
The present results suggested that LIFR downregulation may be
associated with a decrease in the expression levels of these three
molecules, thus impairing adipogenic differentiation. Previous
studies have demonstrated that these adipogenic markers are
important transcription factors involved in adipogenic regulation,
and in the pathophysiology of obesity and several
endocrine-metabolic disorders (24–26).
The present results indicated that LIFR may also be associated with
these diseases.
In investigating LIFR function, it is important to
examine LIF. LIF is a glycoprotein secreted by hMSCs (27) that is able to interact with LIFR,
activating its downstream signaling pathway. Although previous
studies have investigated the impact of LIF on adipogenic
differentiation, the previous results were inconsistent or
contradictory. For example, a previous study reported that LIF
inhibits differentiation of 3T3-L1 adipocytes in vitro
(28); however, in Ob1771 cells,
LIF was revealed to promote differentiation (16). To the best of our knowledge,
pre-adipocyte cell lines, including 3T3-L1 and Ob1771, are
immortalized and may not fully mimic human primary cells due to
their murine origin. Since primary hMSCs extracted from the bone
marrow may more closely mirror the physiological context of
adipogenic differentiation in humans, these cells were selected and
investigated in the present study.
A number of previous studies have investigated the
role of either LIF or LIFR (15,16,27,28).
Similarly, our previous study investigated the function of LIFR as
a miR-377 target gene involved in adipogenic differentiation
(19). To the best of our
knowledge, the present study is the first to investigate the
relationship between the expression levels of LIF and LIFR during
adipogenic differentiation. Previous studies have reported that the
expression patterns of LIF and LIFR are similar in certain tissues
and cells (29,30). In the present study, the expression
levels of LIF and LIFR exhibited opposite trends during adipogenic
differentiation of hMSCs. After initiation of adipogenesis, the
expression levels of LIF were decreased, whereas the expression
levels of LIFR were increased. LIFR knockdown was also associated
with an increased expression of LIF compared with in the NC group,
thus suggesting that LIF may be a negative regulator of adipogenic
differentiation. LIF is an inhibitor of cell differentiation in
mouse embryonic stem cells (31,32).
Although LIF overexpression and/or knockout were not performed in
the present study, results from previous studies support our
hypothesis of the role of LIF in cell differentiation. The present
results suggested that LIFR and LIF may serve opposite roles during
adipogenic differentiation of hMSCs. Previous studies investigated
the use of hMSCs for the treatment of aplastic anemia (33,34).
Notably, the relationship between the expression levels of LIF and
LIFR, and hematopoietic and bone marrow differentiation is an
interesting aspect to address in future studies.
In addition to LIF, LIFR may interact with other
ligands, including oncostatin M and ciliary neurotrophic factor,
which may promote the activation of multiple downstream signaling
pathways involved in adipogenic differentiation (12,35,36),
including the mitogen-activated protein kinase and Janus
kinase/signal transducer and activator of transcription signaling
pathways (12–14,37).
Collectively, LIFR may be associated with various ligands and
signaling pathways involved in adipogenic differentiation.
In conclusion, the present study suggested that LIFR
may be a novel positive regulator of adipogenic differentiation.
Conversely, LIF may negatively regulate adipogenic differentiation.
The present findings suggested that LIFR-targeting treatments may
represent novel potential strategies to treat obesity and other
associated disorders.
Acknowledgements
Not applicable.
Funding
The present study was supported by The National
Natural Science Foundation of China (grant no. 81460221), the
Jiangxi Province Natural Science Foundation of China (grant nos.
20161BAB205197 and 20132BAB205012) and the Development Plan of
Young and Middle-aged Teachers in General Universities of Jiangxi
Province (grant no. 2012-132).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
TW, YY and WL designed the present study. RY, XX and
HY performed the experiments. JW analyzed the data. TW wrote the
paper.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kopelman PG: Obesity as a medical problem.
Nature. 404:635–643. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kahn BB and Flier JS: Obesity and insulin
resistance. J Clin Invest. 106:473–481. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Keller M, Hopp L, Liu X, Wohland T, Rohde
K, Cancello R, Klös M, Bacos K, Kern M, Eichelmann F, et al:
Genome-wide DNA promoter methylation and transcriptome analysis in
human adipose tissue unravels novel candidate genes for obesity.
Mol Metab. 6:86–100. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Otto TC and Lane MD: Adipose development:
From stem cell to adipocyte. Crit Rev Biochem Mol Biol. 40:229–242.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pittenger MF, Mackay AM, Beck SC, Jaiswal
RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S and
Marshak DR: Multilineage potential of adult human mesenchymal stem
cells. Science. 284:143–147. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Helder MN, Knippenberg M, Klein-Nulend J
and Wuisman PI: Stem cells from adipose tissue allow challenging
new concepts for regenerative medicine. Tissue Eng. 13:1799–1808.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Barry F, Boynton RE, Liu B and Murphy JM:
Chondrogenic differentiation of mesenchymal stem cells from bone
marrow: Differentiation-dependent gene expression of matrix
components. Exp Cell Res. 268:189–200. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Arinzeh TL: Mesenchymal stem cells for
bone repair: Preclinical studies and potential orthopedic
applications. Foot Ankle Clin. 10:651–665. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Subash-Babu P and Alshatwi AA: Aloe-emodin
inhibits adipocyte differentiation and maturation during in vitro
human mesenchymal stem cell adipogenesis. J Biochem Mol Toxicol.
26:291–300. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
del Valle I, Rudloff S, Carles A, Li Y,
Liszewska E, Vogt R and Kemler R: E-cadherin is required for the
proper activation of the Lifr/Gp130 signaling pathway in mouse
embryonic stem cells. Development. 140:1684–1692. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pan W, Cain C, Yu Y and Kastin AJ:
Receptor-mediated transport of LIF across blood-spinal cord barrier
is upregulated after spinal cord injury. J Neuroimmunol.
174:119–125. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Plun-Favreau H, Perret D, Diveu C, Froger
J, Chevalier S, Lelièvre E, Gascan H and Chabbert M: Leukemia
inhibitory factor (LIF), cardiotrophin-1, and oncostatin M share
structural binding determinants in the immunoglobulin-like domain
of LIF receptor. J Biol Chem. 278:27169–27179. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
White UA and Stephens JM: Neuropoietin
activates STAT3 Independent of LIFR activation in adipocytes.
Biochem Biophys Res Commun. 395:48–50. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Morton SD, Cadamuro M, Brivio S, Vismara
M, Stecca T, Massani M, Bassi N, Furlanetto A, Joplin RE, Floreani
A, et al: Leukemia inhibitory factor protects cholangiocarcinoma
cells from drug-induced apoptosis via a PI3K/AKT-dependent Mcl-1
activation. Oncotarget. 6:26052–26064. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ikeda S, Itoh S, Yamamoto Y, Matsushita K,
Naruse H and Hayashi M: Developmental stage-dependent effects of
leukemia inhibitory factor on adipocyte differentiation of murine
bone marrow stromal cells. Cell Biochem Biophys. 74:11–17. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Aubert J, Dessolin S, Belmonte N, Li M,
McKenzie FR, Staccini L, Villageois P, Barhanin B, Vernallis A,
Smith AG, et al: Leukemia inhibitory factor and its receptor
promote adipocyte differentiation via the mitogen-activated protein
kinase cascade. J Biol Chem. 274:24965–24972. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fernyhough ME, Hausman GJ, Guan LL, Okine
E, Moore SS and Dodson MV: Mature adipocytes may be a source of
stem cells for tissue engineering. Biochem Biophys Res Commun.
368:455–457. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li X, Yang Y, Yan R, Xu X, Gao L, Mei J,
Liu J, Wang X, Zhang J, Wu P, et al: miR-377-3p regulates
adipogenic differentiation of human bone marrow mesenchymal stem
cells by regulating LIFR. Mol Cell Biochem. 449:295–303. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Farmer SR: Transcriptional control of
adipocyte formation. Cell Metab. 4:263–273. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kudo M, Sugawara A, Uruno A, Takeuchi K
and Ito S: Transcription suppression of peroxisome
proliferator-activated receptor gamma2 gene expression by tumor
necrosis factor alpha via an inhibition of CCAAT/enhancer-binding
protein delta during the early stage ofadipocyte differentiation.
Endocrinology. 145:4948–4956. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rosen ED, Walkey CJ, Puigserver P and
Spiegelman BM: Transcriptional regulation of adipogenesis. Genes
Dev. 14:1293–1307. 2000.PubMed/NCBI
|
|
23
|
Zhang J, Huang Y, Shao H, Bi Q, Chen J and
Ye Z: Grape seed procyanidin B2 inhibits adipogenesis of 3T3-L1
cells by targeting peroxisome proliferator-activated receptor γ
with miR-483-5p involved mechanism. Biomed Pharmacother.
86:292–296. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Floyd ZE and Stephens JM: Controlling a
master switch of adipocyte development and insulin sensitivity:
Covalent modifications of PPARγ. Biochim Biophys Acta.
1822:1090–1095. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Furuhashi M, Tuncman G, Gorgun CZ,
Makowski L, Atsumi G, Vaillancourt E, Kono K, Babaev VR, Fazio S,
Linton MF, et al: Treatment of diabetes and atherosclerosis by
inhibiting fatty-acid-binding protein FABP4. Nature. 447:959–965.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hwang CS, Mandrup S, MacDougald OA, Geiman
DE and Lane MD: Transcriptional activation of the mouse obese (ob)
gene by CCAAT/enhancer binding protein alpha. Proc Natl Acad Sci
USA. 93:873–877. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Oskowitz AZ, Lu J, Penfornis P, Ylostalo
J, McBride J, Flemington EK, Prockop DJ and Pochampally R: Human
multipotent stromal cells from bone marrow and microRNA: Regulation
of differentiationand leukemia inhibitory factor expression. Proc
Natl Acad Sci USA. 105:18372–18377. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hogan JC and Stephens JM: Effects of
leukemia inhibitory factor on 3T3-L1 adipocytes. J Endocrinol.
185:485–496. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Margioula-Siarkou C, Prapas Y, Petousis S,
Milias S, Ravanos K, Kalogiannidis I, Mavromatidis G, Haitoglou C,
Prapas N and Rousso D: LIF and LIF-R expression in the endometrium
of fertile and infertile women: A prospective observational
case-control study. Mol Med Rep. 13:4721–4728. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li Y, Sun L, Zhao D, Ouyang J and Xiang M:
Aberrant expression of leukemia inhibitory factor receptor (LIFR)
and leukemia inhibitory factor (LIF) is associated with tubal
pregnancy occurrence. Turk J Med Sci. 45:214–220. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Saito M, Asai Y, Imai K, Hiratoko S and
Tanaka K: Connexin30.3 is expressed in mouse embryonic stem cells
and is responsive to leukemia inhibitory factor. Sci Rep.
7:424032017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cherepkova MY, Sineva GS and Pospelov VA:
Leukemia inhibitory factor (LIF) withdrawal activates mTOR
signaling pathway in mouse embryonic stem cells through the
MEK/ERK/TSC2 pathway. Cell Death Dis. 7:e20502016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cheng HC, Liu SW, Li W, Zhao XF, Zhao X,
Cheng M, Qiu L and Ma J: Arsenic trioxide regulates adipogenic and
osteogenic differentiation in bone marrow MSCs of aplastic anemia
patients through BMP4 gene. Acta Biochim Biophys Sin (Shanghai).
47:673–679. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lecourt S, Vanneaux V, Leblanc T, Leroux
G, Ternaux B, Benbunan M, Chomienne C, Baruchel A, Marolleau JP,
Gluckman E, et al: Bone marrow microenvironment in fanconi anemia:
A prospective functional study in a cohort of fanconi anemia
patients. Stem Cells Dev. 19:203–208. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Natesh K, Bhosale D, Desai A, Chandrika G,
Pujari R, Jagtap J, Chugh A, Ranade D and Shastry P: Oncostatin-M
differentially regulates mesenchymal and proneural signature genes
in gliomas via STAT3 signaling. Neoplasia. 17:225–237. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wagener EM, Aurich M, Aparicio-Siegmund S,
Floss DM, Garbers C, Breusing K, Rabe B, Schwanbeck R, Grötzinger
J, Rose-John S and Scheller J: The amino acid exchange R28E in
ciliaryneurotrophic factor (CNTF) abrogates interleukin-6
receptor-dependent but retains CNTF receptor-dependent signaling
via glycoprotein 130 (gp130)/leukemia inhibitory factor receptor
(LIFR). J Biol Chem. 289:18442–184450. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu GX, Zhu JC, Chen XY, Zhu AZ, Liu CC,
Lai Q and Chen ST: Inhibition of adipogenic differentiation of bone
marrow mesenchymal stem cells by erythropoietin via activating ERK
and P38 MAPK. Genet Mol Res. 14:6968–6977. 2015. View Article : Google Scholar : PubMed/NCBI
|