Introduction
Metal-dependent protein phosphatase (PPM), formerly
referred to as a member of the protein phosphatase 2C (PP2C)
family, is one of two major serine/threonine protein phosphatase
families in eukaryotes (1). Ppm1a
and Ppm1b, closely related members of the PPM family with an amino
acid identity of 75%, catalyze the dephosphorylation of a variety
of phosphorylated proteins that are involved in intracellular
signaling and cellular responses but show distinct cellular
localization (2–4).
Ppm1a catalyzes the dephosphorylation of the p38
mitogen-activated protein kinase (MAPK), MAPK kinase (MKK) 4, MKK6
in the stress-activated protein kinase (SAPK) cascade and Smad2/3
in the bone morhogenic protein (BMP) signaling pathway (5–7),
whereas Ppm1b dephosphorylates transforming growth factor
(TGF)-β-activated kinase 1 (TAK1), a member of the stress-activated
protein kinase (SAPK) cascade (8,9).
Ppm1a and Ppm1b also dephosphorylate IκB kinase, a protein kinase
that is involved in the nuclear factor kappa κB (NFκB) pathway in
cellular systems, and appears to down-regulate or terminate
cytokine-induced NFκB activation (10,11).
A proteomic analysis has also shown that TNFα enhances the
interaction of 14-3-3ε with TAK1 and Ppm1b, suggesting that it is
involved in cross talk with the MAPK signaling pathway (12). It therefore appears that Ppm1a and
Ppm1b have both unique and redundant functions.
AMP-activated protein kinase (AMPK) is activated in
response to increased AMP concentrations and induces the activation
of metabolic pathways that generate ATP, while also repressing ATP
consumption, and hence can be regarded as a sensor for cellular
energy status (13,14). Chida et al (15) reported that Ppm1a and Ppm1b
dephosphorylate the alpha-subunit of AMPK in their N-myristoylated
forms. In addition, Ppm1b activates the
peroxisome-proliferator-activated receptor gamma (PPARγ) via the
dephosphorylation of Ser112 (16). Because PPARγ is a ligand-activated
transcriptional factor of the nuclear receptor superfamily that
regulates genes that are involved in differentiation, metabolism,
and immunity (17), it is possible
that Ppm1b might also be involved in the regulation of these
metabolic processes. The association of Ppm1b with glioma amplified
sequence 41 (GAS41), a member of a protein family that is
characterized by the presence of an N-terminal YEATS domain,
catalyzes the dephosphorylation of Ser366 of p53,
leading to a decrease in p53 levels (18). Because the hyperactivation of p53
causes accelerated senescence, the depletion of Ppm1b causes
premature senescence, caused by the sustained activation of p53
(19). Among the five distinct
Ppm1b isoforms that are produced by the mouse (20–23),
three are predominantly expressed in pachytene spermatocytes and in
more highly differentiated germ cells, suggesting that they play a
role in the spermatogenic process. The levels of Ppm1b are
increased during the course of the first wave of spermatogenesis in
the neonatal male mouse (24).
Ppm1b-deficient mice with the C57BL/6 background
have been established by a gene-knockout technique (4). Fertilized Ppm1b−/− oocytes
die without cleavage, but Ppm1b+/− mice develop normally
and show no vast phenotypic abnormalities in the first observation.
Since these observations, a Ppm1b gene-trap mouse line
(Ppm1b+/d), which contains a retroviral gene trap in the
second intron of the Ppm1b locus, was developed and employed in
studies concerning the Rip3-mediated regulation of necroptosis
(25). Homozygous gene-trap mice
(Ppm1bd/d) are viable and are born with a normal
Mendelian distribution. However, these mice express lower levels of
the wild-type Ppm1b, which appears to support the viability of the
mice.
Because the expression of Ppm1b in germ cells
implies that the gene might play a role in reproduction, we
attempted to clarify this issue. We changed the genetic background
of Ppm1b+/− mice from the C57BL/6 strain to the ICR
strain because the latter strain is more fertile and produces more
pups. However, no Ppm1b−/− mice were born again, an
observation that prompted us to examine the fertilizing ability of
Ppm1b+/− mice from the viewpoints of histology and
protein levels. The findings indicate that the Ppm1b+/−
ICR female mice showed impaired ovulation in response to hormonal
stimuli, which is consistent with lower levels of the Ppm1b protein
in the ovaries of these mice.
Materials and methods
Animals
Ppm1b-deficient mice were originally generated using
embryonic stem cells with a C57BL/6 and b129Sv mixed background
using a gene-targeting technique (4). We mated Ppm1b+/− male mice
with female congenic ICR mice and backcrossed the litters more than
10 times to produce ICR mice. The animal room climate was
maintained under specific pathogen-free conditions at a constant
temperature of 20–22°C with a 12-h alternating light-dark cycle,
with food and water available ad libitum. The genotype of
the mice was determined by polymerase chain reaction (PCR) analysis
using specific primers for Ppm1b and knockout allele, as reported
previously (4). Animal experiments
were performed in accordance with the Declaration of Helsinki under
a protocol approved by the Animal Research Committee of Yamagata
University.
Assessing reproductive ability of
female mice
The fertilizing ability of the Ppm1b+/+
and Ppm1b+/− female mice was examined under conventional
breeding conditions. Individual Ppm1b+/+ and
Ppm1b+/− female mouse at 11 weeks old were cohabitated
with a sexually mature Ppm1b+/+ male mouse and kept
until childbirth.
Counting sperm numbers
The number of sperm cells in the cauda epididymis
was determined as described in a previous study (26). The cauda epididymis was dissected
from the mice, transferred to phosphate buffered saline (PBS), and
minced into small pieces. After incubation for 15 min, the released
sperm were suspended in the incubation solution to obtain a
homogeneous mixture. Sperm numbers were counted under a light
microscope.
Immunoblot analyses
The mice were anesthetized by diethyl ether and then
sacrificed by thoracotomy and exsanguination. Blood was mostly
collected from the heart in the presence of
ethylenediaminetetraacetic acid. Animals without reflex but with
beating heart were regarded as anaesthetized, and those without
heart beat were regarded as dead. Then, testes and ovary were
dissected from the dead mice. After dissection, one testis and one
ovary from an individual male and female mouse, respectively, were
frozen in liquid nitrogen and stored at −80°C until used. After
thawing on ice, the ovaries and testes were homogenized with a
glass-teflon homogenizer in lysis buffer, which contained 25 mM
Tris-HCl, pH 7.5, 150 mM NaCl, 1% (w/v) Nonidet P-40, 1% (w/v)
sodium deoxycholate, and 0.1% (w/v) SDS, supplemented with a
protease inhibitor cocktail (P8340, Sigma-Aldrich). After
centrifugation at 17,400 × g for 15 min at 4°C, the protein content
in the supernatant was determined using a BCA protein assay reagent
(Thermo Fisher Scientific). The proteins (20 µg/lane) were
separated on 12 or 15% SDS-polyacrylamide gels and blotted onto
polyvinylidene difluoride (PVDF) membranes (GE Healthcare). The
blots were blocked with 5% skim milk in Tris-buffered saline
containing 0.1% Tween-20 (TBST), and were then incubated overnight
with the primary antibodies diluted in TBST containing 1% skim
milk. The primary antibodies (diluted 1:1,000) used were; Ppm1b
(AF4396, R&D Systems), glyceraldehyde-3-phosphate dehydrogenase
(GAPDH) (sc-25778, Santa Cruz Biotechnology), and β-actin
(sc-69879, Santa Cruz Biotechnology). After incubation with
horseradish peroxidase-conjugated 2nd antibodies (Santa Cruz
Biotechnology), the bands were detected using the Immobilon western
chemiluminescent HRP substrate (Millipore) on an image analyzer
(ImageQuant LAS500, GE Healthcare). After reprobing, the same
membranes were reacted with either anti-GAPDH antibody for
testicular proteins or anti-β-actin antibody for ovarian proteins
because reactivity of the antibodies to non-specific proteins in
testes and ovaries was different.
Counting follicles in the whole
ovaries
The numbers of follicles in whole ovaries were
counted according to a previously described method (27). One ovary from an individual female
mouse was used for the analyses. Ovaries were collected from
females at 11-weeks of age, fixed in 15% buffered formalin,
embedded in paraffin, and 7 µm thick sections were prepared. To
determine follicle numbers, every 4th serial section was stained
with hematoxylin and eosin (H&E). Photographs of the sections
were obtained using a BZ-X700 microscope (Keyence) and the number
of follicles was counted.
Measurement of plasma progesterone by
an enzyme immunoassay
Blood was collected from the tail vein of the mice
at the estrous stage in the presence of an excess of
ethylenediaminetetraacetic acid. After centrifugation at 800 × g
for 5 min, plasma progesterone concentrations in the mice were
measured using a Progesterone enzyme-linked immunosorbent assay
(ELISA) kit (ADI-900-011, Enzo Life Sciences) according to the
manufacturer's instructions.
Oocyte collection after
superovulation
Hormone-stimulated ovulation was induced in mice, as
described previously (28).
Briefly, 4- or 11-week-old Ppm1b+/+ and
Ppm1b+/− female ICR mice were injected intraperitoneally
(i.p.) with 5 IU of pregnant mare serum gonadotropin (PMSG)
(Asuka-seiyaku). At 48 h after the administration of PMSG, 5 IU of
human chorionic gonadotropin (hCG) (Asuka-seiyaku) was administered
i.p. At 14 h after hCG administration, oocytes in the ampullary
sites of both fallopian tubes were collected and counted. For
repeated-superovulation, PMSG/hCG injections were repeated at
interval of 7 days.
Statistical analyses
The results are expressed as the mean ± standard
error of the mean (SEM). Statistical analysis was performed using
the Student t-test. Data were analyzed by two-way analysis of
variance followed by Tukey's multiple comparisons test when
comparisons were made in datasets containing more than two groups.
All data were analyzed using the GraphPad Prism 6 software.
P-values <0.05 were considered to indicate statistically
significant differences.
Results
Establishing Ppm1b+/− ICR
mice
Ppm1b-deficient mice were originally established
under a C57BL/6 background (4),
which is commonly used but produces limited numbers of pups
compared to the ICR background. We therefore changed the genetic
background of the mice from C57BL/6 to ICR mice by mating the
original Ppm1b+/− C57BL/6 mice and backcrossing the
litters to the ICR mice more than 10 times. However, the breeding
of Ppm1b+/− male and Ppm1b+/− female ICR mice
under conventional conditions resulted in the birth of only
Ppm1b+/+ and Ppm1b+/− ICR mice (Table I), an observation that is
consistent with the selective death of the Ppmlb−/−
embryos reported for mice with the C57BL/6 background (4). Accordingly we analyzed only
Ppm1b+/− ICR mice in comparison with Ppm1b+/+
ICR mice throughout the study.
 | Table I.Comparison of female fertility
between Ppm1b+/+ and Ppm1b+/− ICR mice. |
Table I.
Comparison of female fertility
between Ppm1b+/+ and Ppm1b+/− ICR mice.
|
|
|
| Number of
neonates |
|---|
|
|
|
|
|
|---|
| Ppm1b genotype Male
× female | Number of mating
pairs | Litter size | +/+ | +/- |
|---|
| +/+ × +/+ | 6 | 14.17±2.32 | 14.17±2.32 | 0 |
| +/+ × +/- | 14 | 14.21±2.75 | 7.50±2.03 |
6.71±1.94 |
| +/- × +/- | 7 | 13.57±2.57 | 3.71±1.25 | 10.00±2.08 |
Characteristics of Ppm1b+/−
male ICR mice
We first examined sexually mature male mice because
the involvement of Ppm1b in the spermatogenic process was
suspected, based on the unique expression of isoforms in
spermatogenic cells (20–23). Generally male mice are sexually
matured after 8 weeks, so that we chose mice at 11 weeks old when
they were fully matured and most reproductive. Ppm1b+/−
male ICR mice were viable, with normal growth and showed no obvious
abnormalities. The body weights of the Ppm1b+/− male ICR
mice and Ppm1b+/+ male ICR mice at 11-weeks of age were
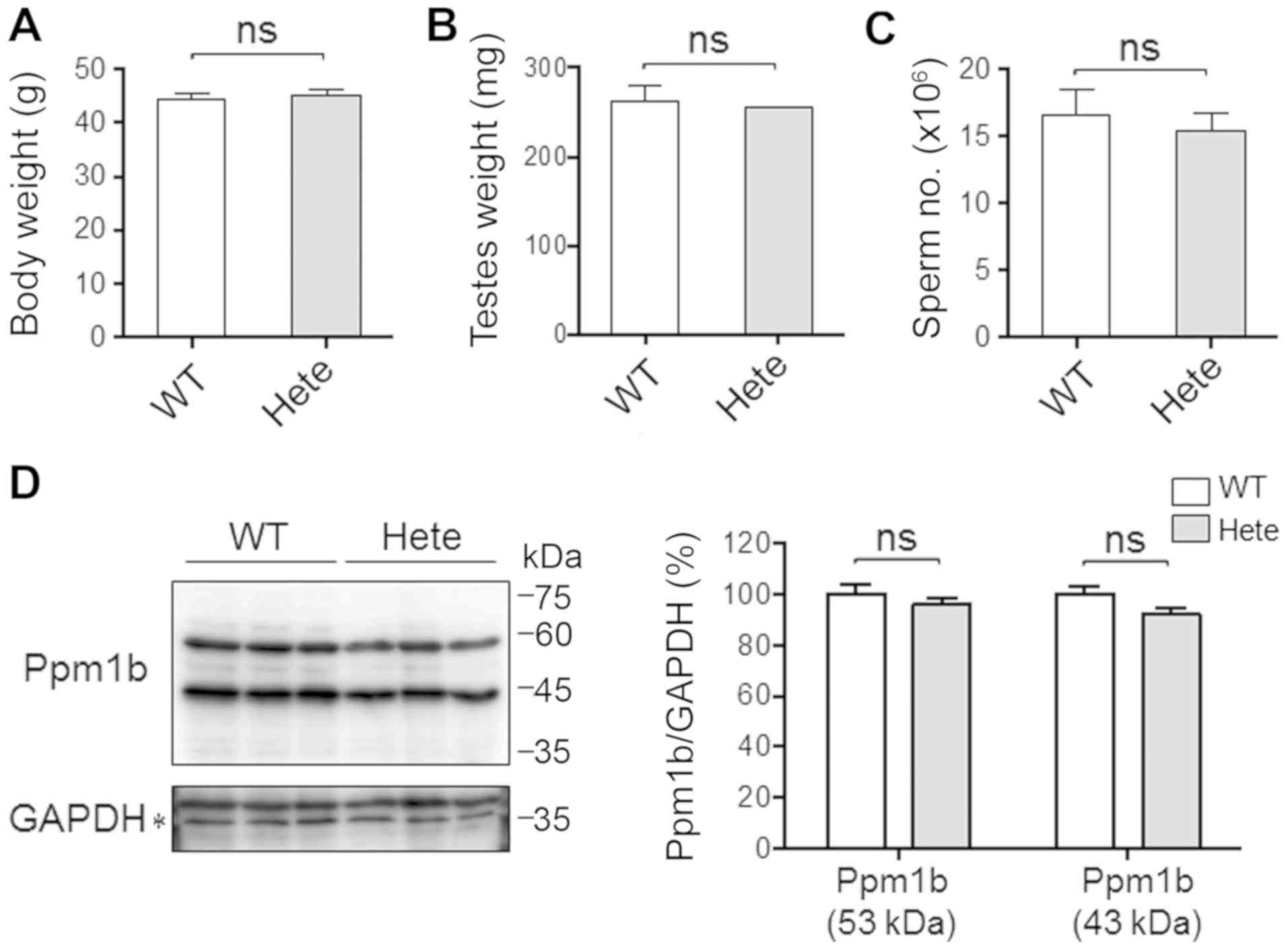
identical (Fig. 1A). No
significant difference in testes weight (Fig. 1B) or sperm numbers (Fig. 1C) was detected between
Ppm1b+/− and Ppm1b+/+ male ICR mice, which is
consistent with results reported for mice with the C57BL/6
background (4). The antibody
against Ppm1b indicated the presence of two bands corresponding to
43 and 53 kDa in size in all testes, irrespective of the mouse
genotype. The 53-kDa protein is the form that is expressed
ubiquitously, and the 43-kDa protein is the form that is expressed
specifically in the testis and liver (20–23).
While the expression of two out of five Ppm1 isoforms could be
confirmed, the levels of both the 43 and 53-kDa Ppm1b proteins in
the testes were not significantly different between
Ppm1b+/+ and Ppm1b+/− mice at 11-weeks of age
(Fig. 1D). It therefore appears
that the Ppm1b+/− male ICR mice are normal in terms of
spermatogenesis.
Ovarian expression of Ppm1b in
mice
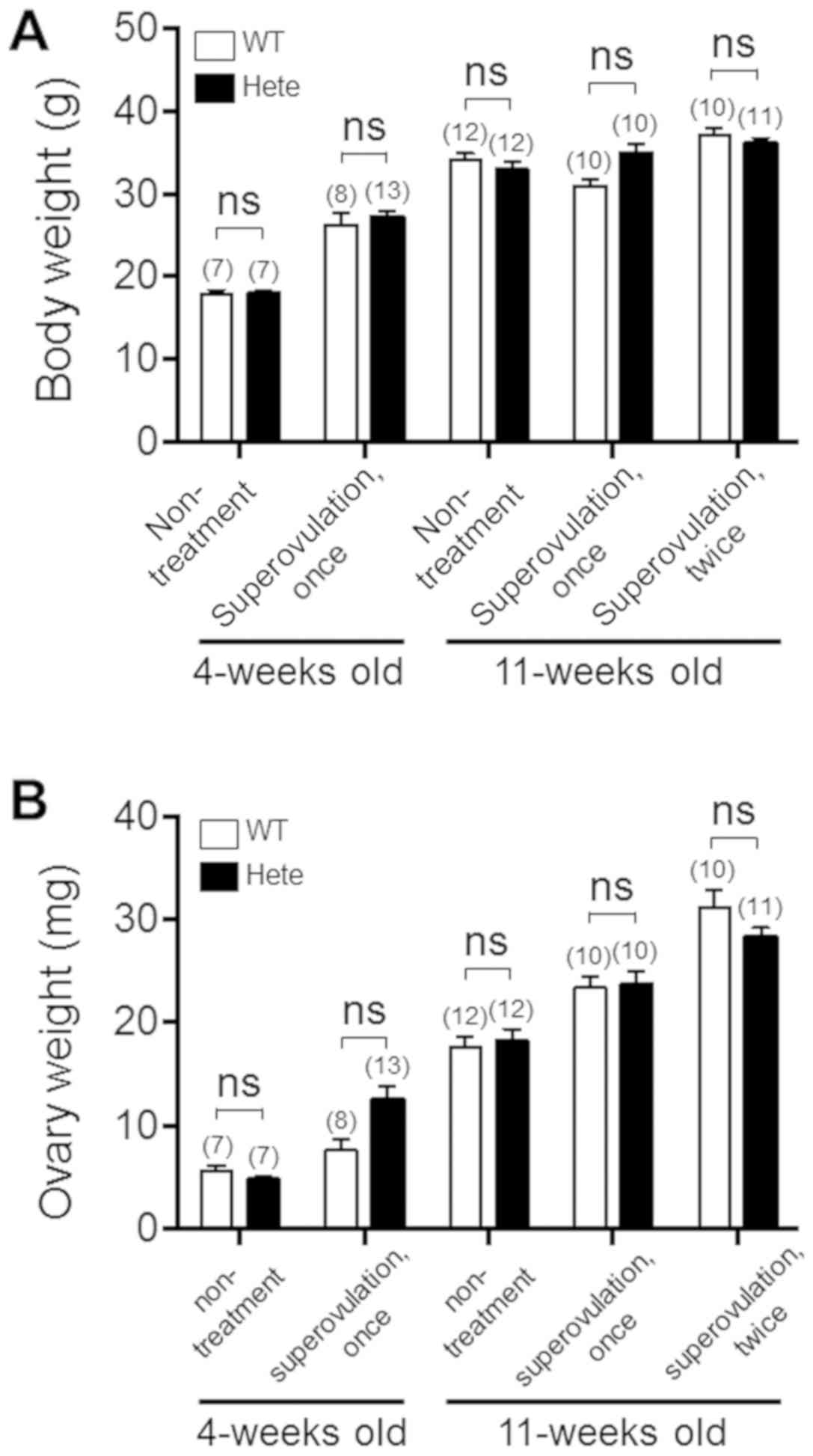
Ppm1b+/− female ICR mice also developed
normally and had normal body and ovary weights compared with the
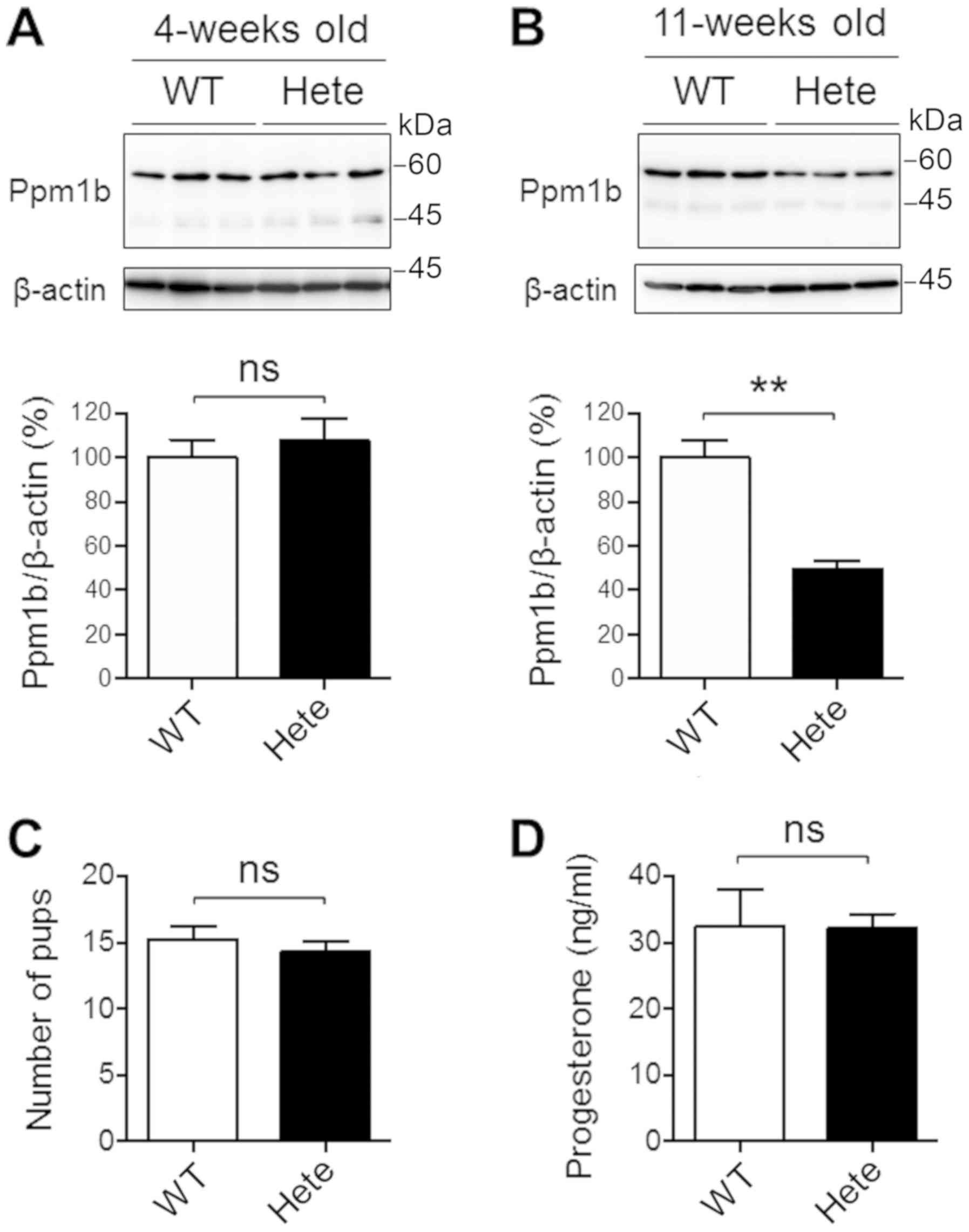
age-matched Ppm1b+/+ female ICR mice (Fig. 2). We then performed immunoblot
analyses of proteins isolated from the ovaries. An antibody against
Ppm1b indicated the presence of a band corresponding to a size of
53 kDa from all ovaries. The levels of the Ppm1b protein in ovaries
were not significantly different between Ppm1b+/+ and
Ppm1b+/− mice at 4-weeks of age (Fig. 3A). However, it was down-regulated
to about 50% in the Ppm1b+/− mice compared to the
Ppm1b+/+ mice at 11-weeks of age (Fig. 3B). Thus, the expression of the
Ppm1b gene appeared to be altered during the sexual maturation of
the ovaries, suggesting the existence of a haploinsufficiency of
the gene at the sexually mature stage.
Characteristics of Ppm1b+/−
female ICR mice
Because Ppm1b was down-regulated in
Ppm1b+/− ovaries at 11-weeks of age, we hypothesized
that ovary structure and/or function may have been altered. We
performed a histological analysis of ovaries from mice at 11-weeks
of age. H&E-stained ovarian sections from the
Ppmlb+/− ICR mice appeared to be similar to those from
the Ppm1b+/+ ICR mice (data not shown). The same numbers
of offspring were produced when Ppm1b+/+ and
Ppm1b+/− female ICR mice were mated with
Ppm1b+/+ male ICR mice (Fig. 3C), which suggests that
Ppm1b+/− female ICR mice have a normal fertility. In
addition, no differences in plasma progesterone levels were
detected at the estrous stage of the mice (Fig. 3D).
The number of ovulated oocytes by
superovulation is reduced in Ppmlb+/− ICR mice
Because the process of ovulation is similar to
inflammation that accompanies oxidative stress with elevated
reactive oxygen and nitrogen species (29), the induction of ovulation may
expose a latent impairment caused by a Ppm1b haploinsufficiency. To
address this issue, we evaluated ovarian function under conditions
of superovulation by administering PMSG followed by hCG to
stimulate ovulation (a well-accepted protocol) (28) in female mice at 4- and 11-weeks of
age. After superovulation, the weights of ovaries from the
Ppmlb+/− ICR mice were found to be similar to those from
the Ppm1b+/+ ICR mice (Fig.
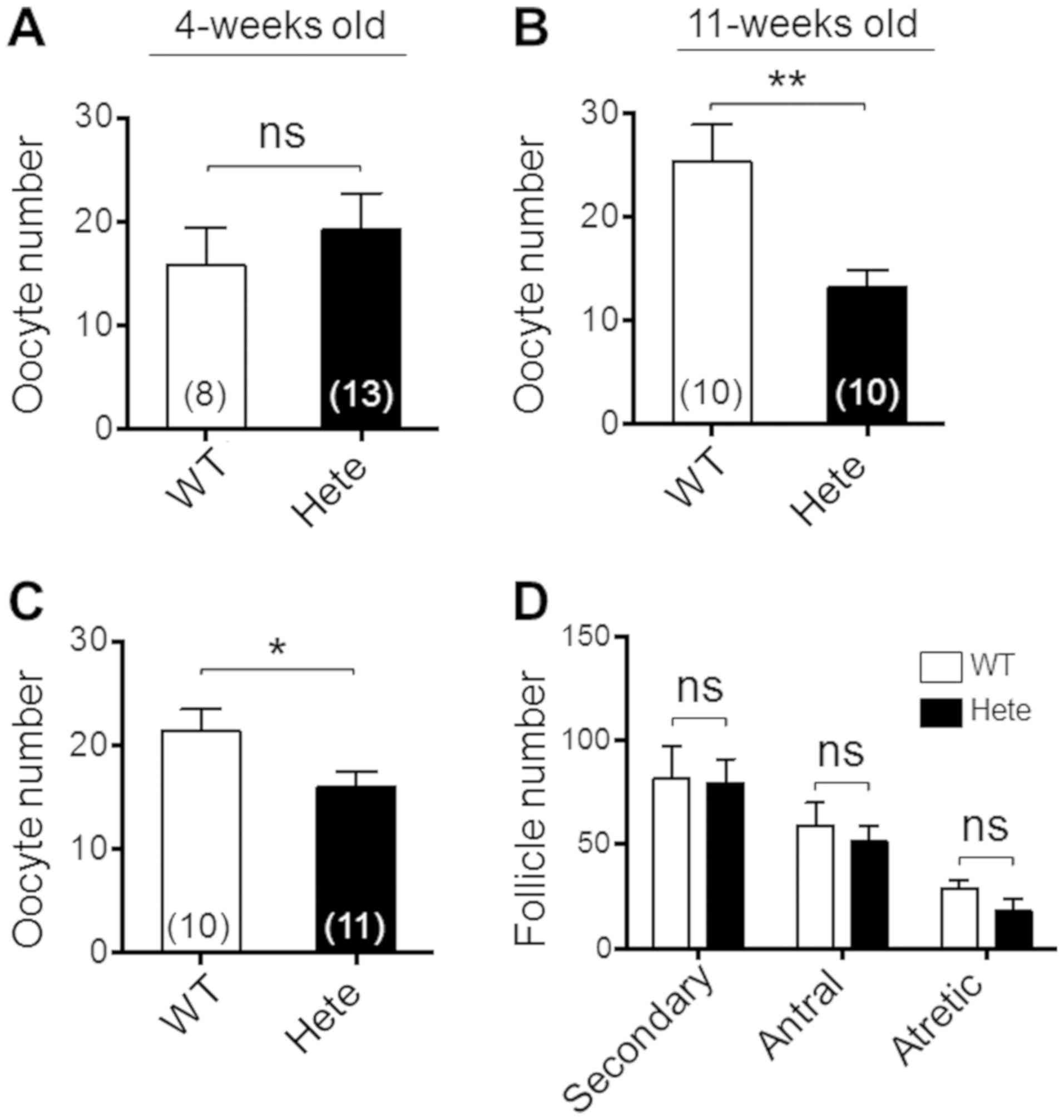
2B) and no difference was found in the number of ovulated
oocytes at 4-weeks of age (Fig.
4A). However, at 11-weeks of age the number of ovulated oocytes
was smaller in the Ppmlb+/− ICR mice compared to the
Ppm1b+/+ ICR mice (Fig.
4B), although no difference was observed in ovary weight
(Fig. 2B). Interestingly, the
number of ovulated oocytes caused by the hormone treatment remained
less in Ppmlb+/− ICR mice at the second superovulation
(Fig. 4C). Although the
macroscopic appearance of the ovaries was normal (data not shown),
the decreased ovulation could have been caused by impaired
folliculogenesis in the Ppm1b+/− ICR mice. We attempted
to validate this possibility by counting follicles in whole ovaries
at the developed stages. The results indicated that the number of
follicles at stages later than the secondary follicle in whole
ovaries were similar between these genotypic mice (Fig. 4D). Thus, the hormone treatment
stimulated the ovulation process, but not folliculogenesis, an
observation that appears to be due to a defect resulting from a
haploinsufficiency of the Ppm1b gene.
Discussion
We found that the number of ovulated oocytes
resulting from stimulation by a hormone treatment were lower in
sexually mature Ppm1b+/− female ICR mice at 11-weeks of
age compared to Ppm1b+/+ ICR mice but were essentially
the same in immature mice at 4-weeks of age (Fig. 4). It therefore appears that a
haploinsufficiency of the Ppm1b gene exerted an influence over the
Ppm1b protein only in ovaries from sexually mature female mice at
11-weeks of age (Fig. 3). Thus,
Ppm1b expression may be regulated, at least in part, by female sex
hormones. However, there were no differences in the histological
appearance of the ovaries, litter sizes, or plasma progesterone
levels at the estrous stage, even in mice at 11-weeks of age
(Fig. 3). Although we did not
perform precise examination on ovarian function due to limited
amounts of samples, evaluation of basic ovarian function by
measuring hallmarks, such as anti-Müllerian hormone (AMH), might
help understanding roles of Ppm1b. There appears to be a
correlation between the number of ovulated oocytes by
superovulation and protein levels of Ppm1b in ovaries, although the
precise mechanism responsible for the Ppm1b gene expression is not
clear at this stage of our studies. Thus, the results obtained
herein suggest that Ppm1b plays roles in the ovulation process in
response to hormones.
Contrary to the impaired reproductive ability of
female mice, there were no differences in sperm numbers and other
phenotypes between Ppm1b+/+ and Ppm1b+/− male
ICR mice (Fig. 1). We detected two
Ppm1 protein bands that were different in size and which
corresponded to the Ppm1b isoforms that are specifically expressed
in differentiated male germ cells (20–23).
The levels of the Ppm1b isoforms were the same between
Ppm1b+/+ and Ppm1b+/− male ICR mice,
suggesting that a haploinsufficiency of the Ppm1b gene neither
affected the expression of these two isoforms nor the phenotypes in
the male reproductive system. The pool of primordial follicles is
maintained in a dormant state by a variety of inhibitory machinery
for follicular activation, including phosphatase and a tensin
homolog deleted on chromosome ten (PTEN) and Foxo3a, and the loss
of function of the inhibitory molecule leads to the premature
activation of the primordial follicle pool and follicle depletion
(30). The excessive activation of
the mammalian target of rapamycin complex 1 (mTORC1) actually
causes an accelerated activation and the depletion of primordial
follicles in mouse ovaries (31,32).
It has been reported that the pharmacological inhibition of mTORC1
activity by a rapamycin treatment consistently leads to the
suppression of primordial follicle activation (33). Based on these findings, we
hypothesized that a haploinsufficiency of the Ppm1b gene might have
caused the decline in the number of follicles and decreased numbers
of ovulated oocytes in the Ppm1b+/− ICR mice. However,
no difference was found in the number of developed follicles
between ovaries from Ppm1b+/− and Ppm1b+/+
mice (Fig. 4D). Thus, these
collective results suggest an opposite scenario; i.e. follicle
development proceeds at an excessive level in the
Ppm1b+/− mice, leading to the partial depletion of
matured follicles and a decrease in the number of ovulated oocytes.
This issue will need to be clarified in a future study.
Hormonal stimuli with PMSG and hCG induces the
activation of a variety of processes, including steroidogenesis,
and releases progesterone to maintain pregnancy (34). Steroidogenesis involves several
enzymatic processes, including oxygenase reactions that are
mediated by the P450 side-chain cleavage enzyme (P450scc) (35). Reactive oxygen species (ROS) are
produced as byproducts of these oxygenase-catalyzed reactions and
the action of molecules that are susceptible to oxidation are
impaired (36), as is typically
seen in the redox reactive molecule peroxiredoxin (37). In addition, hydrogen peroxide, a
stable and less toxic ROS, can regulate hormonal responses via its
ability to modulate redox-sensitive machinery such as epidermal
growth factor (EGF)-receptor signaling (38). Because tyrosine phosphatases
contain a redox-sensitive sulfhydryl residue at the catalytic
center and is prone to oxidative inactivation, hydrogen peroxide
that is produced by NADPH oxidases stimulate the signaling pathway,
as demonstrated in the case of EGF receptor signaling (39). However, Ppm1b is a metal-dependent
protein phosphatase that catalyzes the dephosphorylation of
phospho-serine/threonine in proteins (1) and is less sensitive to oxidative
modification than phosphotyrosine phosphatases. This suggests that
the direct inactivation of Ppm1b by ROS would be less likely.
Because many protein kinases catalyze the
phosphorylation of hydroxyl groups of serine/threonine residues in
signaling proteins, as typified in the MAPK cascade (3,5–7), a
dysfunction in Ppm1b may sustain the phosphorylation process active
state and enhance cellular proliferation after hormonal stimuli.
The interaction between mTOR and the MAPK signaling pathways and
their participation in the ovulation process is complex and is
currently not fully understood (40). The findings reported herein suggest
that the process is regulated by the dephosphorylation via Ppm1b
and is functional during ovulation in response to hormonal
stimulation. Experiments using conditional Ppm1b-null knockout mice
would provide a clear picture of this issue as well as the bona
fide roles of Ppm1b. In fact, for unknown reasons, we, as well
as Sasaki et al (4) have
made numerous attempts to establish conditional knockout mice but
failed to obtain Ppm1bflox/flox mice, which are required
if Ppm1b gene is to be deleted conditionally. Because a
haploinsufficiency of Ppm1b resulted in a decreased number of
ovulated oocytes by superovulation (Fig. 4), agents that activate Ppm1b may be
a beneficial agent for increasing the number of ovulated oocytes
and consequently litter sizes. Ppm1b reportedly dephosphorylates
Pax2, which encodes the protein required for the development of
several organs in embryos, and results in switching Pax2 from a
transcriptional activator to a repressor protein (41). The ablation of Ppm1b sustains the
phosphorylation of the tumor suppressor gene, the retinoblastoma
gene (RB1), and down regulates U2OS osteosarcoma cell growth
(42). Thus, Ppm1b appears to
support the proliferation of tumor cells. On the contrary, the
knockdown of Ppm1b activates Rho GTPase, which results in the
invasion and migration of breast cancer cells (43), suggesting that Ppm1b has a
suppressive function in these cells. Thus, the functional
consequence of Ppm1b largely depends on the types of cells, and a
complete understanding of its roles are awaited for the purpose of
improved female reproduction and cancer therapy.
In conclusion, we report herein that a heterozygous
deficiency of the Ppm1b gene in female ICR mice results in a
deteriorated response in hormone-stimulated ovulation. The
identification of the target proteins of Ppm1b should provide clues
to our understanding of the phosphorylation-dephosphorylation
reactions that regulate the ovulation process in the ovary in
response to hormonal stimuli.
Acknowledgements
The authors would like to thank Mr. Tsunekata Ito
(Animal Center, Institute for Promotion of Medical Science
Research, Faculty of Medicine, Yamagata University, Yamagata,
Japan) for providing advice on animal handling.
Funding
This work was partly supported by the Cooperative
Research Project Program of the Joint Usage/Research Center at the
Institute of Development, Aging and Cancer, Tohoku University
(grant no. 2012-4).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors contributions
NI performed the majority of the experiments. TH, RW
and NK assisted with some experiments. MO and TK established the
Ppm1b+/− mice and provided them for this study. JF
designed the study, interpreted data, drafted the manuscript and
revised it critically for important intellectual content. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study included animal experiments only. Animal
experiments were performed in accordance with the Declaration of
Helsinki under a protocol approved by the Animal Research Committee
of Yamagata University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Shi Y: Serine/threonine phosphatases:
Mechanism through Structure. Cell. 139:468–484. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lifschitz-Mercer B, Sheinin Y, Ben-Meir D,
Bramante- Schreiber L, Leider-Trejo L, Karby S, Smorodinsky NI and
Lavi S: Protein phosphatase 2Calpha expression in normal human
tissues: An immunohistochemical study. Histochem Cell Biol.
116:31–39. 2001.PubMed/NCBI
|
|
3
|
Lin X, Duan X, Liang YY, Su Y, Wrighton
KH, Long J, Hu M, Davis CM, Wang J, Brunicardi FC, et al: PPM1A
functions as a Smad phosphatase to terminate TGF-beta signaling.
Cell. 125:915–928. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sasaki M, Ohnishi M, Tashiro F, Niwa H,
Suzuki A, Miyazaki J, Kobayashi T and Tamura S: Disruption of the
mouse protein Ser/Thr phosphatase 2Cbeta gene leads to early
pre-implantation lethality. Mech Dev. 124:489–499. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Takekawa M, Maeda T and Saito H: Protein
phosphatase 2Calpha inhibits the human stress-responsive p38 and
JNK MAPK pathways. EMBO J. 17:4744–4752. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Duan X, Liang YY, Feng XH and Lin X:
Protein serine/threonine phosphatase PPM1A dephosphorylates Smad1
in the bone morphogenetic protein signaling pathway. J Biol Chem.
281:36526–36532. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dai F, Shen T, Li Z, Lin X and Feng XH:
PPM1A dephosphorylates RanBP3 to enable efficient nuclear export of
Smad2 and Smad3. EMBO Rep. 12:1175–1181. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hanada M, Kobayashi T, Ohnishi M, Ikeda S,
Wang H, Katsura K, Yanagawa Y, Hiraga A, Kanamaru R and Tamura S:
Selective suppression of stress-activated protein kinase pathway by
protein phosphatase 2C in mammalian cells. FEBS Lett. 437:172–176.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hanada M, Ninomiya-Tsuji J, Komaki K,
Ohnishi M, Katsura K, Kanamaru R, Matsumoto K and Tamura S:
Regulation of the TAK1 signaling pathway by protein phosphatase 2C.
J Biol Chem. 276:5753–5759. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Prajapati S, Verma U, Yamamoto Y, Kwak YT
and Gaynor RB: Protein phosphatase 2Cbeta association with the
IkappaB kinase complex is involved in regulating NF-kappaB
activity. J Biol Chem. 279:1739–1746. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun W, Yu Y, Dotti G, Shen T, Tan X,
Savoldo B, Pass AK, Chu M, Zhang D, Lu X, et al: PPM1A and PPM1B
act as IKKbeta phosphatases to terminate TNFalpha-induced
IKKbeta-NF-kappaB activation. Cell Signal. 21:95–102. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zuo S, Xue Y, Tang S, Yao J, Du R, Yang P
and Chen X: 14-3-3 epsilon dynamically interacts with key
components of mitogen-activated protein kinase signal module for
selective modulation of the TNF-alpha-induced time course-dependent
NF-kappaB activity. J Proteome Res. 9:3465–3478. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hardie DG: AMP-activated protein kinase:
An energy sensor that regulates all aspects of cell function. Genes
Dev. 25:1895–1908. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Carling D, Mayer FV, Sanders MJ and
Gamblin SJ: AMP-activated protein kinase: Nature's energy sensor.
Nat Chem Biol. 7:512–518. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chida T, Ando M, Matsuki T, Masu Y,
Nagaura Y, Takano- Yamamoto T, Tamura S and Kobayashi T:
N-Myristoylation is essential for protein phosphatases PPM1A and
PPM1B to dephosphorylate their physiological substrates in cells.
Biochem J. 449:741–749. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tasdelen I, van Beekum O, Gorbenko O,
Fleskens V, van den Broek NJF, Koppen A, Hamers N, Berger R, Coffer
PJ, Brenkman AB and Kalkhoven E: The serine/threonine phosphatase
PPM1B (PP2Cβ) selectively modulates PPARγ activity. Biochem J.
451:45–53. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tontonoz P and Spiegelman BM: Fat and
beyond: The diverse biology of PPARgamma. Annu Rev Biochem.
77:289–312. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Park JH, Smith RJ, Shieh SY and Roeder RG:
The GAS41-PP2Cbeta complex dephosphorylates p53 at serine 366 and
regulates its stability. J Biol Chem. 286:10911–10917. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Park JH, Hale TK, Smith RJ and Yang T:
PPM1B depletion induces premature senescence in human IMR-90
fibroblasts. Mech Ageing Dev. 138:45–52. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wenk J, Trompeter HI, Pettrich KG, Cohen
PT, Campbell DG and Mieskes G: Molecular cloning and primary
structure of a protein phosphatase 2C isoform. FEBS Lett.
297:135–138. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Terasawa T, Kobayashi T, Murakami T,
Ohnishi M, Kato S, Tanaka O, Kondo H, Yamamoto H, Takeuchi T and
Tamura S: Molecular cloning of a novel isotype of Mg(2+)-dependent
protein phosphatase beta (type 2C beta) enriched in brain and
heart. Arch Biochem Biophys. 307:342–349. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hou EW, Kawai Y, Miyasaka H and Li SS:
Molecular cloning and expression of cDNAs encoding two isoforms of
protein phosphatase 2C beta from mouse testis. Biochem Mol Biol
Int. 32:773–780. 1994.PubMed/NCBI
|
|
23
|
Kato S, Terasawa T, Kobayashi T, Ohnishi
M, Sasahara Y, Kusuda K, Yanagawa Y, Hiraga A, Matsui Y and Tamura
S: Molecular cloning and expression of mouse mg(2+)-dependent
protein phosphatase beta-4 (type 2C beta-4). Arch Biochem Biophys.
318:387–393. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kato S, Kobayashi T, Kusuda K, Nishina Y,
Nishimune Y, Yomogida K, Yamamoto M, Sakagami H, Kondo H, Ohnishi
M, et al: Differentiation-dependent enhanced expression of protein
phosphatase 2Cbeta in germ cells of mouse seminiferous tubules.
FEBS Lett. 396:293–297. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen W, Wu J, Li L, Zhang Z, Ren J, Liang
Y, Chen F, Yang C, Zhou Z, Su SS, et al: Ppm1b negatively regulates
necroptosis through dephosphorylating Rip3. Nat Cell Biol.
17:434–444. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Iuchi Y, Okada F, Tsunoda S, Kibe N,
Shirasawa N, Ikawa M, Okabe M, Ikeda Y and Fujii J: Peroxiredoxin 4
knockout results in elevated spermatogenic cell death via oxidative
stress. Biochem J. 419:149–158. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Myers M, Britt KL, Wreford NGM, Ebling FJP
and Kerr JB: Methods for quantifying follicular numbers within the
mouse ovary. Reproduction. 127:569–580. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kimura N, Tsunoda S, Iuchi Y, Abe H,
Totsukawa K and Fujii J: Intrinsic oxidative stress causes either
2-cell arrest or cell death depending on developmental stage of the
embryos from SOD1-deficient mice. Mol Hum Reprod. 16:441–451. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jabbour HN, Sales KJ, Catalano RD and
Norman JE: Inflammatory pathways in female reproductive health and
disease. Reproduction. 138:903–919. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Adhikari D and Liu K: Molecular mechanisms
underlying the activation of mammalian primordial follicles. Endocr
Rev. 30:438–464. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Adhikari D, Flohr G, Gorre N, Shen Y, Yang
H, Lundin E, Lan Z, Gambello MJ and Liu K: Disruption of Tsc2 in
oocytes leads to overactivation of the entire pool of primordial
follicles. Mol Hum Reprod. 15:765–770. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Adhikari D, Zheng W, Shen Y, Gorre N,
Hämäläinen T, Cooney AJ, Huhtaniemi I, Lan ZJ and Liu K: Tsc/mTORC1
signaling in oocytes governs the quiescence and activation of
primordial follicles. Hum Mol Genet. 19:397–410. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tong Y, Li F, Lu Y, Cao Y, Gao J and Liu
J: Rapamycin-sensitive mTORC1 signaling is involved in
physiological primordial follicle activation in mouse ovary. Mol
Reprod Dev. 80:1018–1034. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Light A and Hammes SR: Membrane receptor
cross talk in steroidogenesis: Recent insights and clinical
implications. Steroids. 78:633–638. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mizutani T, Ishikane S, Kawabe S, Umezawa
A and Miyamoto K: Transcriptional regulation of genes related to
progesterone production. Endocr J. 62:757–763. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yasui H, Hayashi S and Sakurai H: Possible
involvement of singlet oxygen species as multiple oxidants in p450
catalytic reactions. Drug Metab Pharmacokinet. 20:1–13. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rhee SG, Woo HA, Kil IS and Bae SH:
Peroxiredoxin functions as a peroxidase and a regulator and sensor
of local peroxides. J Biol Chem. 287:4403–4410. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Finkel T: Signal transduction by reactive
oxygen species. J Cell Biol. 194:7–15. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bae YS, Oh H, Rhee SG and Yoo YD:
Regulation of reactive oxygen species generation in cell signaling.
Mol Cells. 32:491–509. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Siddappa D, Kalaiselvanraja A, Bordignon
V, Dupuis L, Gasperin BG, Roux PP and Duggavathi R: Mechanistic
target of rapamycin (MTOR) signaling during ovulation in mice. Mol
Reprod Dev. 81:655–665. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Abraham S, Paknikar R, Bhumbra S, Luan D,
Garg R, Dressler GR and Patel SR: The Groucho-associated
phosphatase PPM1B displaces Pax transactivation domain interacting
protein (PTIP) to switch the transcription factor Pax2 from a
transcriptional activator to a repressor. J Biol Chem.
290:7185–7194. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Miller RE, Uwamahoro N and Park JH: PPM1B
depletion in U2OS cells supresses cell growth through RB1-E2F1
pathway and stimulates bleomycin-induced cell death. Biochem
Biophys Res Commun. 500:391–397. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cho HJ, Kim JT, Lee SJ, Hwang YS, Park SY,
Kim BY, Yoo J, Hong KS, Min JK, Lee CH, et al: Protein phosphatase
1B dephosphorylates Rho guanine nucleotide dissociation inhibitor 1
and suppresses cancer cell migration and invasion. Cancer Lett.
417:141–151. 2018. View Article : Google Scholar : PubMed/NCBI
|