Introduction
Prostate cancer is one of the most common human
malignancies and the second leading cause of cancer-associated
mortality in men in western countries (1,2).
According to an updated estimate, prostate cancer has the highest
incidence amongst male tumors in 2018, accounting for 19% of male
malignant tumors, while this disease has been reported as the
second leading cause of cancer-associated mortality (3). In recent years, the incidence of
prostate cancer has greatly increased in China (4,5).
Furthermore, since the average life span is increasing and due to
increased westernization of lifestyles, the incidence of prostate
cancer is also increasing annually in China; notably, prostate
cancer has been listed as one of the fastest growing tumors in
China in the 21st century (6).
Prostate cancer predominantly occurs in older men, and the median
age of patients is ~72 years old; in addition, prostate cancer is
the third most common major malignant tumor of the urogenital
system, following bladder and renal tumors worldwide (7). The morphological characteristics and
structure of prostate cancer are usually complex, and there is no
obvious clinical symptom in the early stages; therefore, it is very
difficult to make a clear diagnosis of prostate cancer (8). At present, the determination of
prostate-specific antigen (PSA) in clinical serum samples is the
most important diagnostic tool for prostate cancer. However, it has
previously been reported that PSA is not the gold standard for
diagnosis of prostate cancer. Although the sensitivity is high
(~78.7%), the specificity is only 59.2%, the false negative rate is
38–48% and the false positive rate is 25%; therefore, prostate
cancer can easily escape diagnosis or be misdiagnosed (9). Notably, rectal examination, imaging
examination and other examination methods are also used in the
clinical diagnosis of prostate cancer; however, these examinations
do not achieve the desired results for the diagnosis of early
prostate cancer, and may cause unnecessary injury and pain to
patients (10,11). At present, there are various
methods for the treatment of prostate cancer, including surgical
treatment, hormone therapy, radiotherapy, etc. Although these
methods are widely used in clinical treatment, there are several
limitations in the treatment and prognosis of prostate cancer
(12–14). Therefore, research has recently
paid more attention to the prevention and treatment of prostate
cancer. Exploration of the molecular mechanisms underlying the
development of prostate cancer, and identification of novel methods
for the treatment of prostate cancer, have recently been focuses of
research.
Numerous studies have reported that microRNAs
(miRNAs/miRs) serve an irreplaceable role in the progression and
development of prostate cancer, and have significant impacts on the
biological behaviors of prostate cancer, including proliferation,
differentiation, invasion and hormone dependence (15,16).
It has important application value and potential to provide
reliable molecular markers for the diagnosis and prognosis of
prostate cancer, which has unlimited potential as a target for
treatment. miRNAs, which contain 17–25 nucleotides, suppress gene
expression by binding to the 3′-untranslated regions (3′UTRs) of
target genes, and have roles in various biological processes,
including development, differentiation, cell proliferation and
apoptosis (17–19). Distinct miRNA expression profiles
have been identified in human prostate cancer tissues and cell
lines, and it has been reported that miRNAs may act as either tumor
suppressors or oncogenes in prostate cancer, depending on the
functions of their target genes (20,21).
Therefore, it is important to investigate the abnormal expression
of miRNAs and their functions in prostate cancer, in order to
provide effective and novel therapeutic targets for anti-prostate
cancer therapy.
miR-589-5p has previously been reported to be
frequently abnormally expressed in numerous types of human cancer
(22,23). However, to the best of our
knowledge, the expression and biological effects of miR-589-5p in
prostate cancer have yet to be elucidated. Therefore, the present
study aimed to investigate the expression levels of miR-589-5p in
prostate cancer tissues and cell lines. In addition, the functions
and underlying molecular mechanisms in prostate cancer were
explored.
Materials and methods
Prostate cancer tissues
The present study was approved by the Ethical
Committee of Nanjing Medical University (Huai'an, China). All
patients provided written informed consent. A total of 30 prostate
cancer tissues and corresponding adjacent normal tissues were
obtained from patients who underwent surgical resection between
June 2016 and April 2017 at the Affiliated Huai'an No. 1 People's
Hospital of Nanjing Medical University (Huai'an, China); their
average age was 45±7 years. old; none of the patients received
pre-operative chemotherapy or radiotherapy. All fresh tissues were
stored at −80°C for further study.
Cell culture and transfection
The DU-145 and PC3 human prostate cancer cell lines,
and RWPE-1 normal prostate epithelial cell line were purchased from
the Shanghai Institute of Biochemistry and Cell Biology of the
Chinese Academy of Sciences (Shanghai, China). All cells were
maintained in Dulbecco's modified Eagle's medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10%
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.),
100 U/ml penicillin, and 100 µg/ml streptomycin (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) at 37°C in an incubator containing
5% CO2.
DU-145 and PC3 cells were seeded into 6-well plates
at a density of 4×105 cells/well. Once cells reached
60–70% confluence, transfection with synthesized miR-589-5p mimics
negative control (NC), miR-589-5p mimics, miR-589-5p inhibitors NC,
miR-589-5p inhibitors, chemokine (C-C motif) ligand 5 (CCL-5) small
interfering RNAs (si-CCL-5) and negative control siRNA (siRNA NC)
were synthesized by Invitrogen; Thermo Fisher Scientific, Inc and
was performed with Lipofectamine® 2000 Reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) at a final
concentration of 50 nM for 24 h, according to the manufacturer's
protocols. The sequences were as follows: miR-589-5p mimics
forward, 5′-TTTTGGAGTTTTTTTTGGTTTTT-3′ and reverse,
5′-CAAAAACAAAACCAA-AATCA-3′; miR-589-5p inhibitors forward,
5′-TGTTGGAGAGCCAAGTGGTATTT-3′ and reverse,
5′-CCTGAACAGAACCGGACTCA-3′; miR-589-5p mimics NC or miR-589-5p
inhibitors NC forward, 5′-GGCTGCATTGGCTGGCGAAACCCGUC-3′ and
reverse, 5′-ATGCGUGCCCTGCTGTTGC-TCCATGTCG-3′; si-CCL5 forward
5′-GCGTCGAGTTTGTCACGAGA-3′, reverse 5′-TGACACTCCTTTACTGTGCT-3′.
Transfected cells were cultured for 24, 48 or 72 h under the same
conditions as aforementioned.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from prostate cancer tissues
and cell lines using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) at 37°C for 30 min, according to the
manufacturer's protocol. For detection of miR-589-5p expression,
TaqMan MicroRNA Reverse Transcription kit (Takara Biotechnology
Co., Ltd., Dalian, China) was used to synthesize cDNA according to
the manufacturer's protocol. The expression levels of miR-589-5p
were examined using TaqMan MicroRNA PCR kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. To quantify CCL5 mRNA expression, the PrimeScript RT
reagent kit (Applied Biosystems; Thermo Fisher Scientific, Inc.)
was used to synthesize cDNA according to the manufacturer's
protocol. Subsequently, CCL-5 mRNA was amplified using SYBR Premix
Ex Taq (Applied Biosystems; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. U6 and GAPDH were used as
internal controls for miR-589-5p and CCL-5, respectively. The PCR
primers used in the present study were as follows: miR-589-5p,
forward 5′-CGAGGTCAGCGTGATTTCATGG-3′, reverse
5′-TGTGTCCAAGTCCCAGCCAGAG-3′; U6, forward 5′-CTCGCTTCGGCAGCACA-3′,
reverse 5′-AACGCTTCACGAATTTGCGT-3′; CCL-5, forward
5′-CAGTCGTCTTTGTCACCCGA-3′, reverse 5′-TGTAACTGCTGCTGTGTGGT-3′; and
GAPDH, forward 5′-ACAACTTTGGTATCGTGGAAGG-3′ and reverse
5′-GCCATCACGCCACAGTTTC-3′. The thermocycling conditions comprised
one cycle at 95°C for 30 sec, followed by 42 cycles of
amplification (95°C for 3 sec and 60°C for 30 sec). Fold-changes in
miR-589-5p and CCL-5 mRNA expression were calculated using the
2−ΔΔCq method (24),
and RT-qPCR reactions were performed in triplicate.
Cell viability assay
Cell viability was evaluated using the Cell Counting
kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc.,
Rockville, MD, USA). Briefly, DU-145 and PC3 cells at a density of
3×103 cells/well were cultured in a 96-well plate and
transfected with miR-589-5p mimics NC, miR-589-5p mimics,
miR-589-5p inhibitors NC or miR-589-5p inhibitors. After 24, 48 or
72 h at 37°C and 5% CO2, 10 µl CCK-8 reagent was added
to each well and the cells were incubated for another 4 h. The
optical density was observed at a wavelength of 450 nm using a
microplate reader (LNB, Shanghai, China).
Cell apoptosis
DU-145 and PC3 cells at a density of
1×105 cells/well were cultured in a 6-well plate for 48
h post-transfection. Subsequently, the cells were detached, washed
with PBS and centrifuged at 12,000 × g for 5 min. The cells were
then incubated with 500 µl buffer, 5 µl fluorescein
isothiocyanate-Annexin V and 5 µl propidium iodide (PI) for 1 h at
37°C, according to the manufacturer's protocols (Shanghai Yeasen
Biotechnology, Co., Ltd., Shanghai, China). Cell apoptosis were
analyzed using a flow cytometer (FACSCalibur; BD Biosciences, San
Jose, CA, USA).
Wound-healing assay
For the wound-healing assay, DU-145 and PC3 cells
were seeded into a 6-well plate at a density of 1×105
cells/well post-transfection for 24 h. Once cell confluence reached
90%, the cell monolayers were wounded in the surface of the plates
using a 200 µl pipette tip and washed with serum-free medium to
remove floating cells. The cells were then cultured in a serum-free
medium for cell recovery. Images of the cells were captured at 0
and 48 h under an inverted microscope.
Transwell invasion assay
The Transwell invasion assay was performed to
evaluate the invasive capacity of prostate cancer cells. Following
transfection for 24 h, DU-145 and PC3 cells at a density of
5×104 cells/well in 100 µl serum-free medium were seeded
into the upper chambers of a Transwell system (8 µm; Corning
Corporation, Corning, NY, USA), which were coated with Matrigel (BD
Pharmingen; BD Biosciences). Culture medium containing 20% FBS was
added to the lower chambers. After 48 h at 37°C, the cells
remaining on the upper compartments were removed, whereas the
invaded cells were fixed with 4% paraformaldehyde at room
temperature for 30 min and stained with 10% crystal violet at room
temperature for 15 min. Cell numbers were obtained from five fields
per membrane under a light microscope (Olympus Corporation, Tokyo,
Japan). The following equations were used to analyze findings:
Invasion rate (%) = invasive cells/total cells.
Target prediction for miR-589-5p and
luciferase reporter assay
The candidate target genes of miR-589-5p were
analyzed using the miRNA target prediction programs PicTar
(pictar.mdc-berlin.de/) and TargetScan
(www.targetscan.org/). The 3′UTR
sequences of CCL-5 containing the putative binding site for
miR-589-5p were amplified from human genomic DNA, and cloned into
the pGL3 vector (Promega Corporation, Madison, WI, USA), in order
to obtain the recombinant vectors pGL3-CCL-5-wild-type (wt) and
pGL3-CCL-5-mutant (mut). Subsequently, DU-145 and PC3 cells
(1×105) were plated in 48-well plates for 24 h, and
psiCheck-2 with the wild-type (WT) or mutant (MUT) 3′-untranslated
region (UTR) of CCL-5 was cotransfected with miR-589-5p mimics or
miR-NC at a concentration of 50 nM using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After
transfection for 48 h, cells were collected and subjected to a
luciferase assay using the Dual-Luciferase Reporter Assay kit
according to the manufacturer's protocol (Promega Corporation).
Western blotting
Total protein was extracted from tissues and
transfected cells using radioimmunoprecipitation assay buffer
(Beijing Solarbio Science & Technology Co., Ltd., Beijing,
China). Subsequently, the supernatants were collected following
centrifugation at 12,000 × g for 20 min at 4°C, and the protein
concentrations were determined using a Bicinchoninic Acid Protein
Assay kit (Vazyme, Piscataway, NJ, USA). Loading buffer was added
to the supernatant of samples and the proteins were denatured at
100°C for 5 min. Equal amounts of protein (30 µg) were then
separated by 12% SDS-PAGE and transferred onto polyvinylidene
difluoride membranes. The membranes were blocked with 5% non-fat
milk for at room temperature for 1 h, washed four times with
Tris-buffered saline plus 0.1% Tween (TBST; 15 min/wash), and
incubated overnight at 4°C with primary antibodies. The following
primary antibodies were used: Anti-CCL-5 (cat. no. ab189841;
1:1,000), B-cell lymphoma 2 (Bcl-2)-associated X protein (Bax; cat.
no. ab32503; 1:1,000), Bcl-2 (cat. no. ab196495; 1:1,000),
caspase-3 (cat. no. ab90437; 1:1,000), caspase-9 (cat. no. ab25758;
1:1,000), matrix metalloproteinase (MMP)-2 (cat. no. ab37150;
1:1,000), MMP-9 (cat. no. ab73734; 1:1,000) and GAPDH (cat. no.
ab9485; 1:1,000) (all from Abcam, Cambridge, UK). Subsequently, the
membranes were washed with TBST three times (15 min/wash) and
incubated with horseradish peroxidase-conjugated secondary
antibodies (cat. no. ab6721; 1:2,000; Abcam) for 2 h at room
temperature. Protein bands were examined using the Bio-Rad Imaging
system (Hercules, CA, United States) with an enhanced
chemiluminescence western blotting substrate kit (ProteinTech
Group, Inc., Wuhan, China) and the results were measured using
ImageJ software 1.48 (National Institutes of Health, Bethesda, MD,
USA). GAPDH were used as a loading control.
Statistical analysis
All data are expressed as the means ± standard
deviation, and all experiments were repeated at least three times.
Statistical analyses were performed using SPSS 20.0 (IBM Corp.,
Armonk, NY, USA). Multiple group comparisons were conducted using
one-way analysis of variance and Tukey's test. Student's t-test was
used to compare only two groups. The association between CCL-5 and
miR-589-5p expression in prostate cancer was evaluated using
Spearman's correlation coefficient. P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-589-5p is downregulated in
prostate cancer tissues and cell lines
In order to explore the role of miR-589-5p in
prostate cancer, RT-qPCR was performed to evaluate its expression
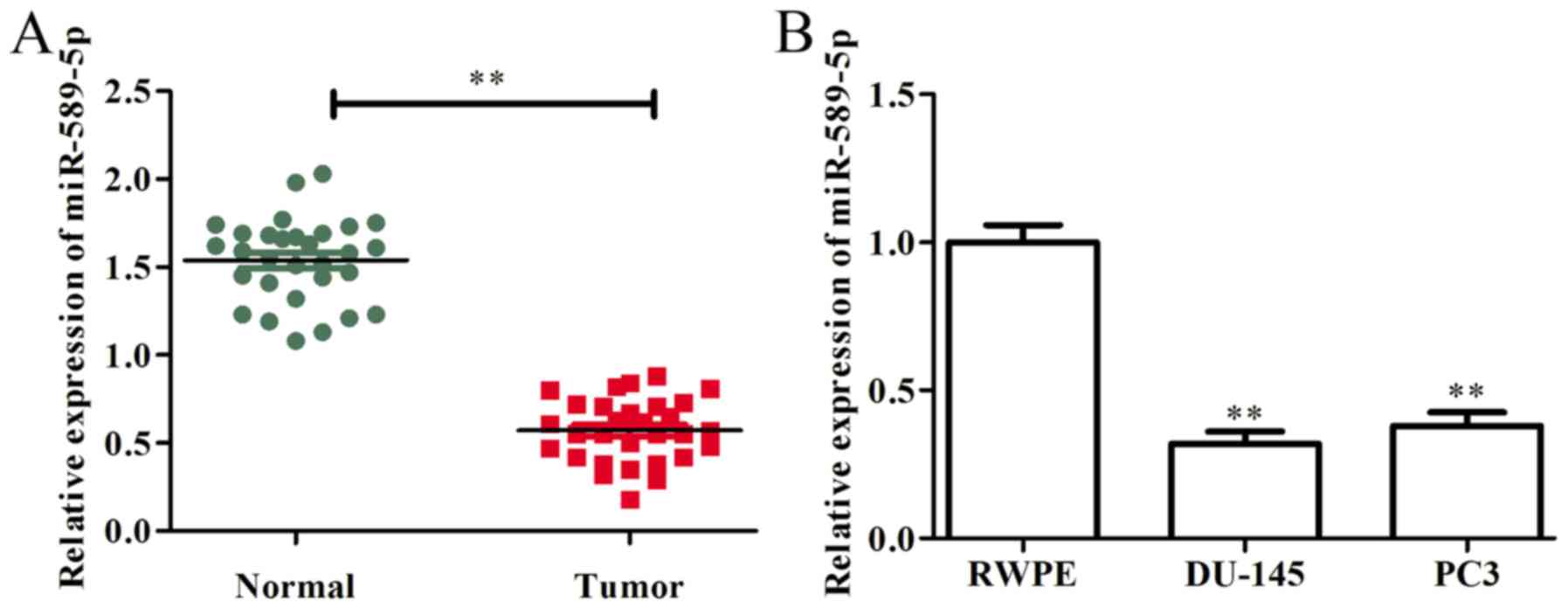
in prostate cancer tissues and cells. As presented in Fig. 1A, miR-589-5p was markedly
downregulated in prostate cancer tissues compared with in
corresponding adjacent normal tissues. In addition, the expression
levels of miR-589-5p were examined in prostate cancer cells (DU-145
and PC3) and in RWPE normal prostate epithelial cells. The results
demonstrated that, compared with in RWPE cells, the expression
levels of miR-589-5p were significantly decreased in DU-145 and PC3
cells (Fig. 1B). These findings
indicated that miR-589-5p may act as a tumor suppressor in prostate
cancer progression and development.
miR-589-5p inhibits viability and
induces apoptosis of prostate cancer cells
The present study demonstrated that miR-589-5p was
downregulated in prostate cancer cells. To further explore the role
of miR-589-5p in the progression and development of prostate
cancer, miR-589-5p mimics NC, miR-589-5p mimics, miR-589-5p
inhibitors NC and miR-589-5p inhibitors were transfected into
DU-145 and PC3 cells. After transfection for 48 h, transfection
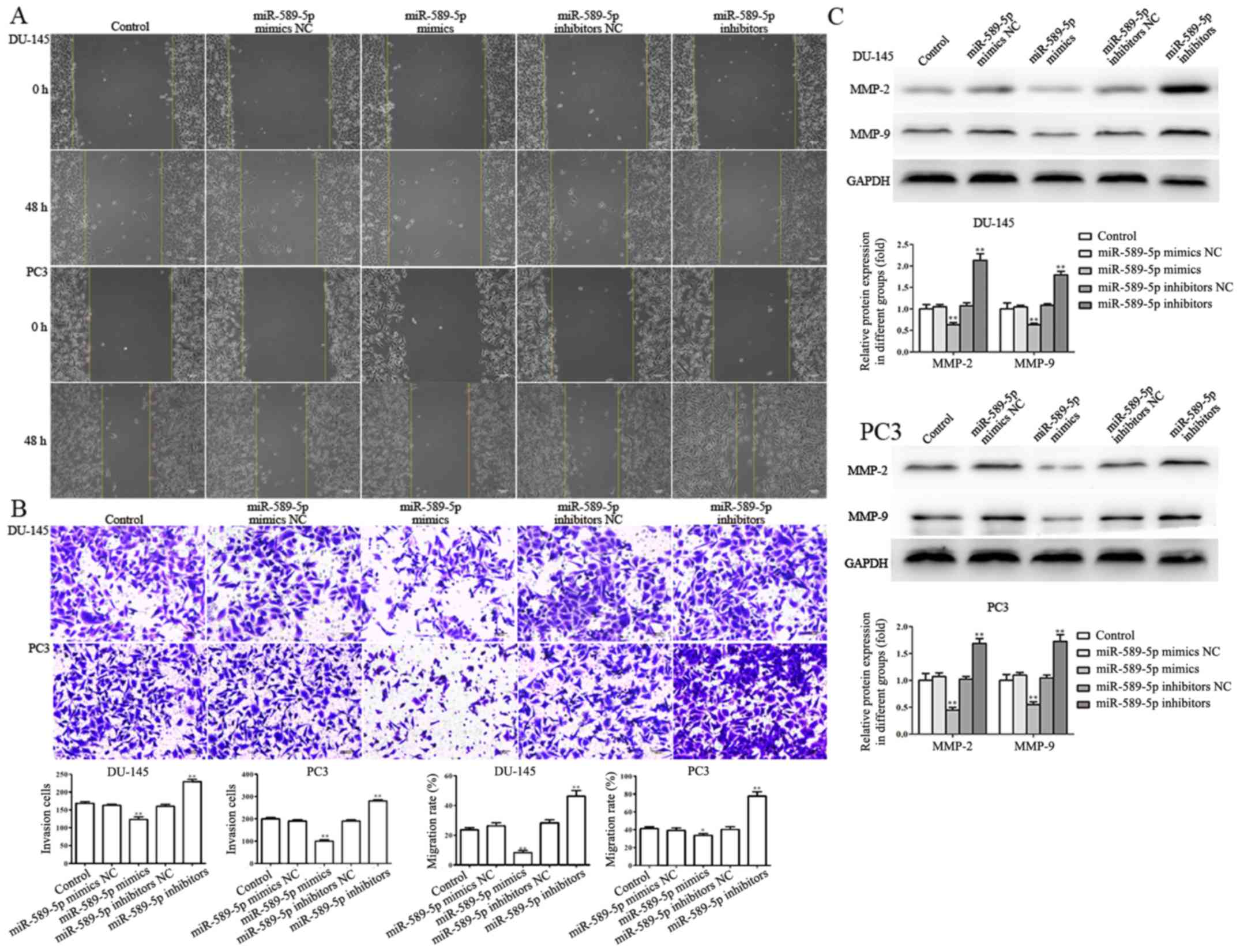
efficiency was determined by RT-qPCR. As shown in Fig. 2A, the expression levels of
miR-589-5p were significantly upregulated following transfection
with miR-589-5p mimics, whereas they were downregulated
post-transfection with miR-589-5p inhibitors, as compared with in
the NC-transfected DU-145 and PC3 cells.
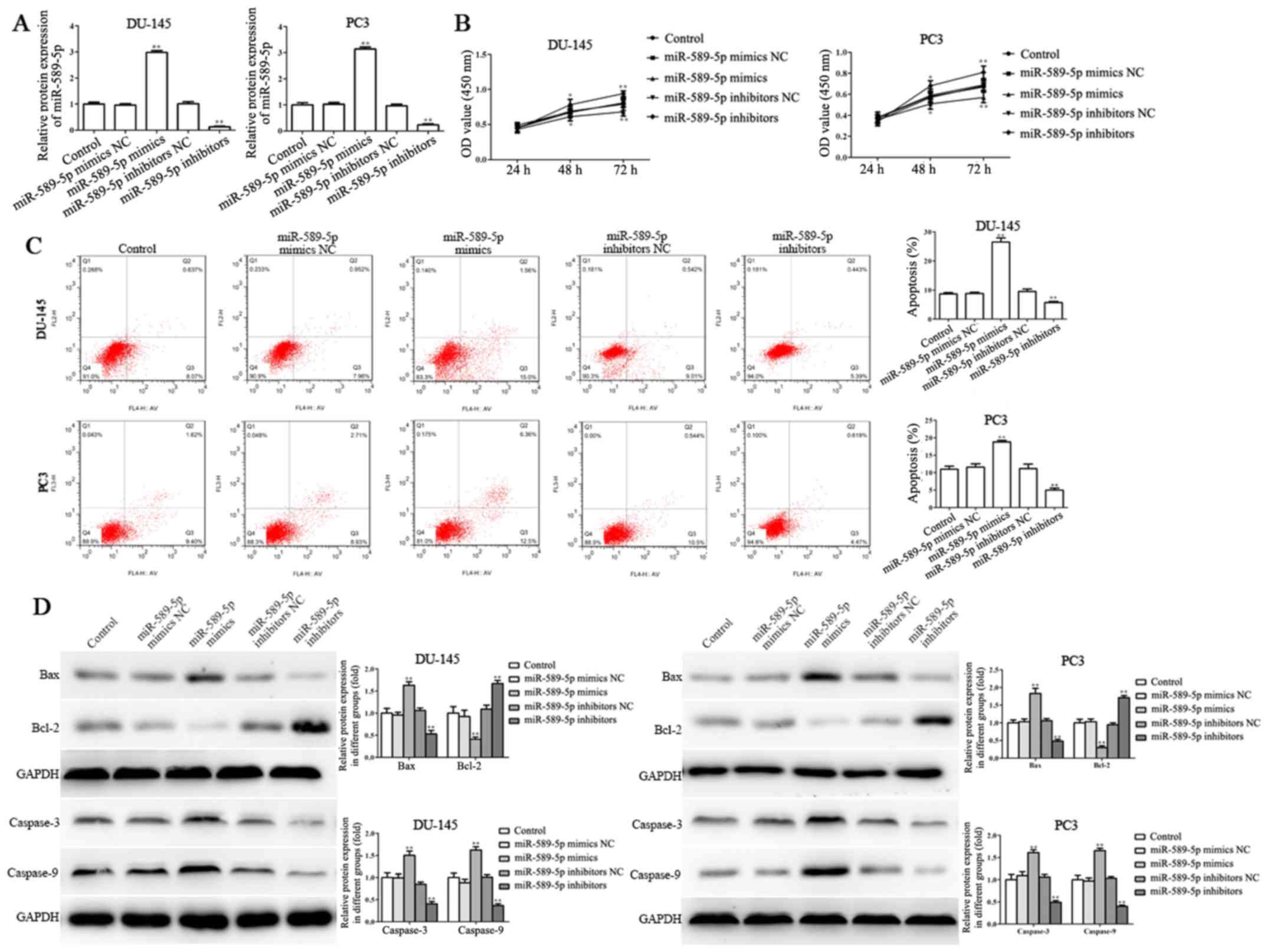 | Figure 2.miR-589-5p inhibits viability and
induces apoptosis of prostate cancer cells. (A) Transfection
efficiency was assessed by reverse transcription-quantitative
polymerase chain reaction post-transfection with miR-589-5p mimics
or miR-589-5p inhibitors in DU-145 and PC3 cells. (B) Viability of
DU-145 and PC3 cells was determined post-transfection with
miR-589-5p mimics or miR-589-5p inhibitors for 24, 48 and 72 h
using the Cell Counting kit-8 assay. (C) Apoptosis assays were
performed by flow cytometry post-transfection with miR-589-5p
mimics or miR-589-5p inhibitors for 48 h in DU-145 and PC3 cells.
(D) Protein expression levels of Bax, Bcl-2, caspase-3 and
caspase-9 were examined in DU-145 and PC3 cells transfected with
miR-589-5p mimics or miR-589-5p inhibitors for 48 h; band intensity
was semi-quantified using ImageJ software. Data are presented as
the means ± standard deviation of three independent experiments,
each experiment was performed in triplicate. *P<0.05,
**P<0.01 compared with the control group. Bax, Bcl-2-associated
X protein; Bcl-2, B-cell lymphoma 2; miR-589-5p, microRNA-589-5p;
NC, negative control; OD, optical density. |
CCK-8 assay was performed to evaluate the effects of
miR-589-5p on the viability of DU-145 and PC3 cells; the results
revealed that the viability of DU-145 and PC3 cells transfected
with miR-589-5p mimics was markedly suppressed compared with in
cells transfected with the NC, whereas miR-589-5p inhibitors
significantly promoted the viability of DU-145 and PC3 cells
compared with the NC group (Fig.
2B).
The induction of apoptosis is a critical process
through which miRNAs exert anticancer effects (25). Therefore, the association between
miR-589-5p and DU-145 and PC3 cell apoptosis was revealed by flow
cytometry. Post-transfection with miR-589-5p mimics NC, miR-589-5p
mimics, miR-589-5p inhibitors NC and miR-589-5p inhibitors, PI and
Annexin V double staining was conducted to evaluate apoptosis. As
shown in Fig. 2C, miR-589-5p
mimics promoted apoptosis, whereas miR-589-5p inhibitors inhibited
apoptosis of DU-145 and PC3 cells compared with in the NC
groups.
Tumor cell apoptosis is mediated by various
molecules that inhibit (e.g. Bcl-2) or induce (e.g. Bax and
caspases) cell death (26).
Therefore, the present study detected the protein expression levels
of Bcl-2, Bax, caspase-3 and caspase-9. The results demonstrated
that pro-apoptotic proteins, including Bax, caspase-3 and
caspase-9, were increased, whereas the expression levels of the
anti-apoptotic protein Bcl-2 were significantly decreased in DU-145
and PC3 cells transfected with miR-589-5p mimics (Fig. 2D). Conversely, transfection with
miR-589-5p inhibitors exerted the opposite effects on DU-145 and
PC3 cells.
miR-589-5p suppresses the migration
and invasion of prostate cancer cells
Wound-healing and Transwell invasion assays were
performed to investigate the effects of miR-589-5p on migration and
invasion of DU-145 and PC3 cells transfected with miR-589-5p mimics
NC, miR-589-5p mimics, miR-589-5p inhibitors NC and miR-589-5p
inhibitors. The results indicated that the migratory capacity of
DU-145 and PC3 cells transfected with miR-589-5p mimics was
markedly suppressed compared with cells transfected with miR-589-5p
mimics NC, whereas transfection of DU-145 and PC3 cells with
miR-589-5p inhibitors notably increased cell migration into the
wound (Fig. 3A). Furthermore, a
Transwell invasion assay revealed that miR-589-5p mimics markedly
suppressed DU-145 and PC3 cell invasion. However, when DU-145 and
PC3 cells were transfected with miR-589-5p inhibitors, invasion was
significantly increased compared with in cells in the miR-589-5p
inhibitors NC group (Fig. 3B).
During the progression and development of metastasis
in prostate cancer, MMPs serve a significant role (27). In the present study, the protein
expression levels of MMP-2 and MMP-9 were detected in DU-145 and
PC3 cells transfected with miR-589-5p mimics NC, miR-589-5p mimics,
miR-589-5p inhibitors NC and miR-589-5p inhibitors. The results
indicated that miR-589-5p mimics markedly suppressed the protein
expression levels of MMP-2 and MMP-9, whereas the expression levels
of MMP-2 and MMP-9 were significantly enhanced following
transfection with miR-589-5p inhibitors (Fig. 3C).
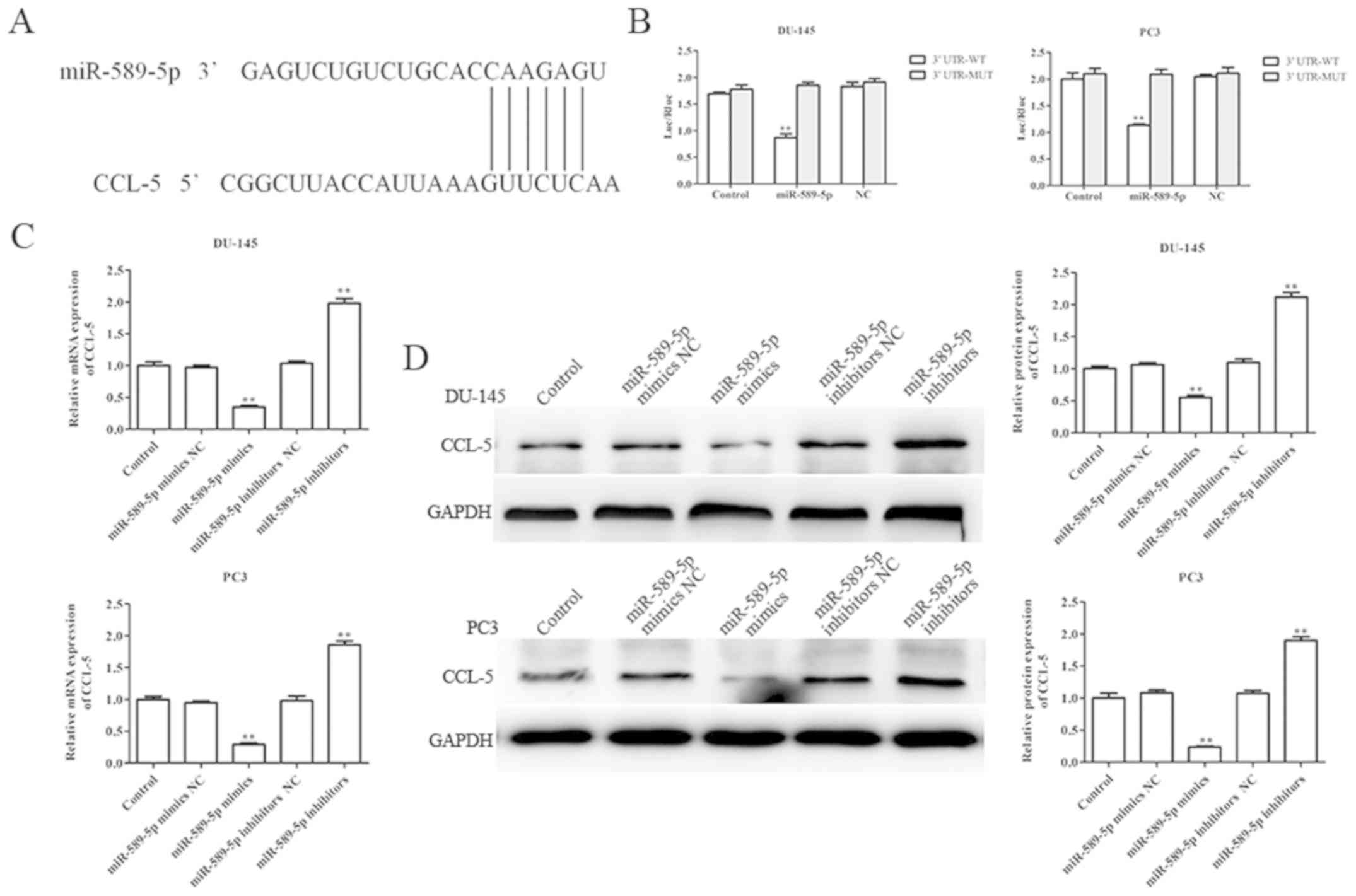
CCL-5 is a direct target of miR-589-5p
in prostate cancer cells
This study aimed to explore the molecular mechanisms
underlying the effects of miR-589-5p on prostate cancer cell
viability, apoptosis, migration and invasion. Firstly, PicTar and
TargetScan were used to predict the possible target gene of
miR-589-5p; the results demonstrated that CCL-5 may be a potential
target gene of miR-589-5p (Fig.
4A). Furthermore, a luciferase reporter assay was performed to
confirm whether miR-589-5p binds to the 3′UTR of CCL-5; the results
demonstrated that miR-589-5p directly binds to the 3′UTR of CCL-5
following co-transfection with miR-589-5p mimics and luciferase
reporter vector; ectopic expression of miR-589-5p decreased the
luciferase activity of CCL-5 3′UTR-wt, whereas it did not affect
that of CCL-5 3′UTR-mut (Fig.
4B).
To further confirm whether CCL-5 was a direct target
gene of miR-589-5p, RT-qPCR and western blotting were performed to
evaluate the mRNA and protein expression levels of CCL-5 in DU-145
and PC3 cells post-transfection with miR-589-5p mimics NC,
miR-589-5p mimics, miR-589-5p inhibitors NC and miR-589-5p
inhibitors. The results revealed that overexpression of miR-589-5p
significantly decreased the mRNA and protein expression levels of
CCL-5, whereas knockdown of miR-589-5p increased the mRNA and
protein expression levels of CCL-5 in DU-145 and PC3 cells
(Fig. 4C and D). These findings
strongly indicated that CCL-5 was the direct target gene of
miR-589-5p.
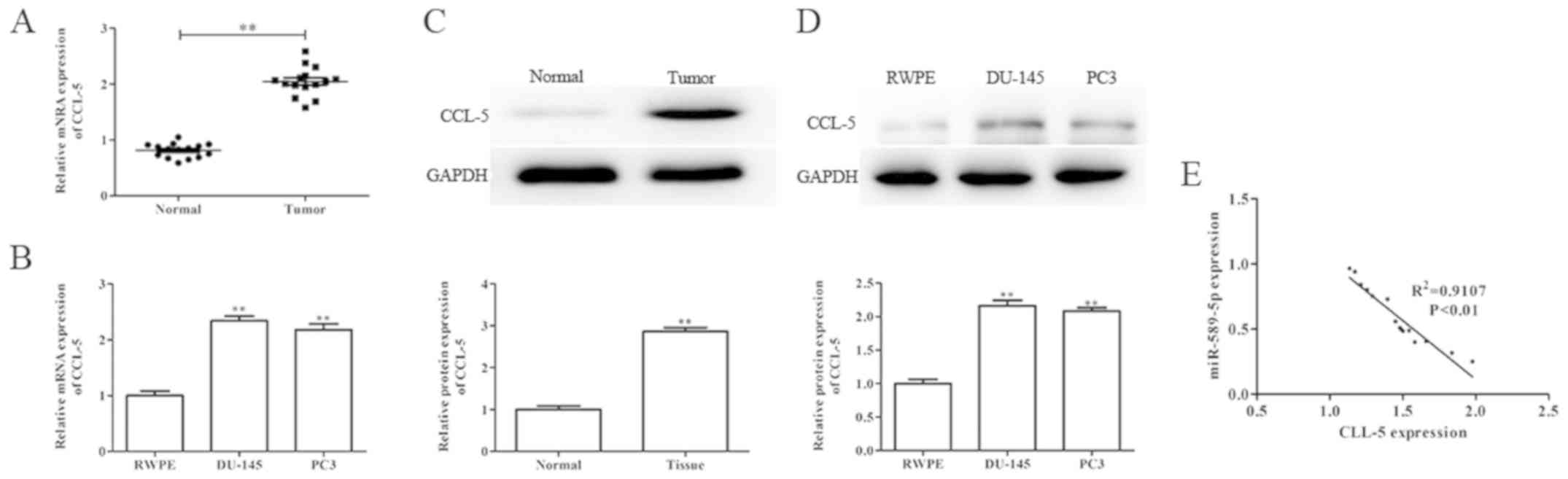
CCL-5 is upregulated in prostate
cancer tissues and cells
To further evaluate the role of CCL-5 in prostate
cancer, the mRNA and protein expression levels of CCL-5 were
determined in prostate cancer tissues and cells by RT-qPCR and
western blotting. As shown in Fig.
5A-D, the mRNA and protein expression levels of CCL-5 were
significantly upregulated in prostate cancer tissues and cells
compared with in the corresponding adjacent normal tissues and
normal prostate epithelial cells, respectively. In addition, the
association between CCL-5 and miR-589-5p expression in prostate
cancer was evaluated using Spearman's correlation analysis; the
results demonstrated that CCL-5 mRNA expression was negatively
correlated with miR-589-5p expression in prostate cancer tissues
(Fig. 5E).
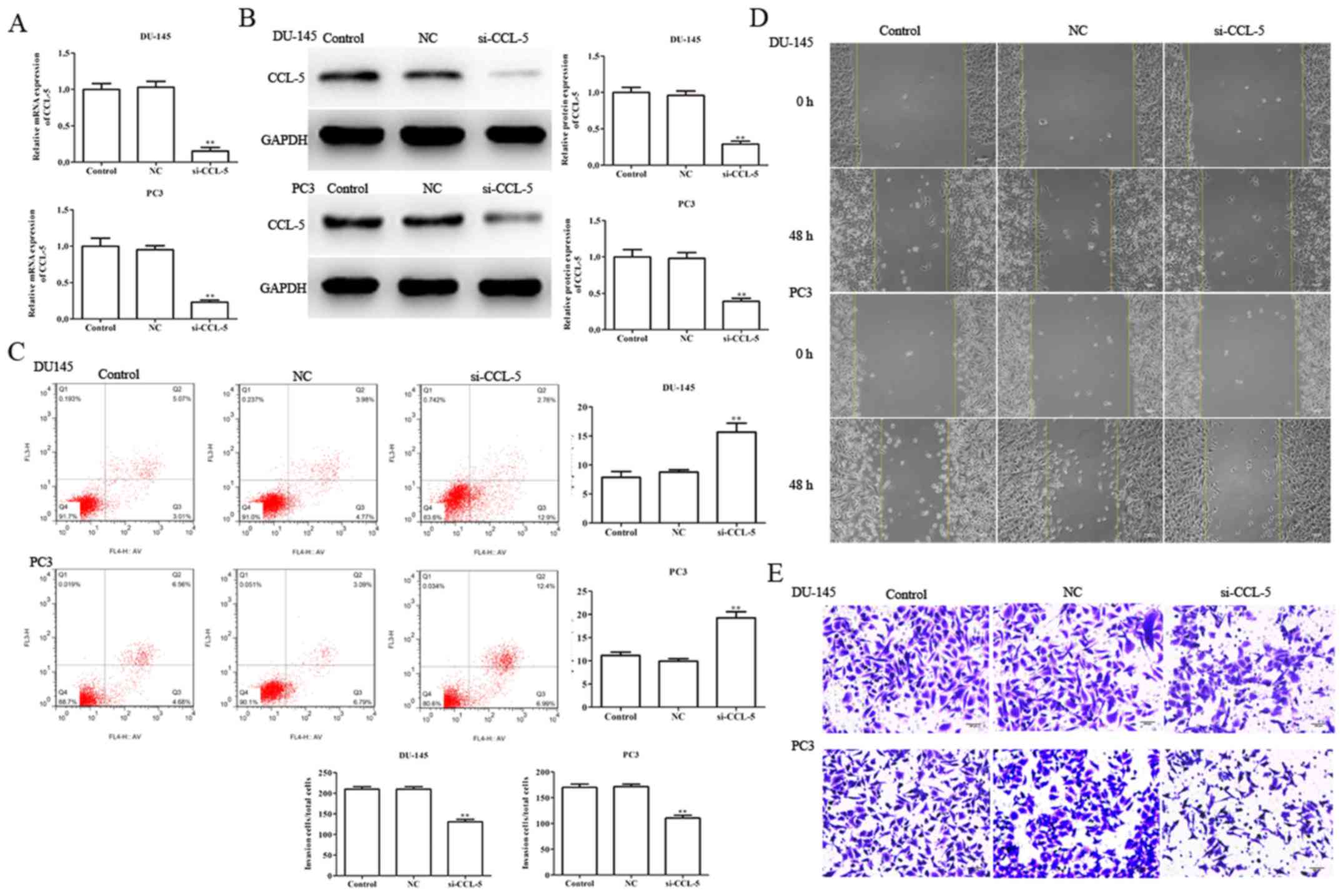
Knockdown of CCL-5 suppresses
viability, migration and invasion, and induces apoptosis of
prostate cancer cells
In order to investigate the role of CCL-5 in the
regulation of viability, apoptosis, migration and invasion in
DU-145 and PC3 cells, si-CCL-5 was transfected into DU-145 and PC3
cells using Lipofectamine® 2000 Reagent. Subsequently,
RT-qPCR was performed to determine transfection efficiency; the
results demonstrated that si-CCL-5 markedly inhibited the mRNA and
protein expression levels of CCL-5 in DU-145 and PC3 cells
(Fig. 6A and B). In addition, flow
cytometry was performed to assess the effects of CCL-5 on the
apoptosis of DU-145 and PC3 cells. The results verified that
si-CCL-5 markedly promoted the apoptosis of DU-145 and PC3 cells;
the proportions of apoptotic cells were 15.67±3.87 and 19.27±4.29
compared with in the control group (7.86±4.13 and 11.17±5.31),
respectively (Fig. 6C).
Furthermore, wound-healing, and Transwell invasion assays were
performed to examine the effects of CCL-5 on the migration and
invasion of DU-145 and PC3 cells. As shown in Fig. 6D and E, it was revealed that CCL-5,
as a target gene for miR-589-5p, may serve an essential role in the
progression and development of prostate cancer.
Discussion
Prostate cancer is the most common malignancy in
men, and is the second leading cause of cancer-associated mortality
in men. In addition, the incidence of prostate cancer increases
with age (28). There are no
symptoms during the early stage of prostate cancer, and symptoms
occur only when the tumor obstructs the urethra or bladder neck
(29). Therefore, early diagnosis
and treatment are important factors in determining the cure rates
and prognosis of tumor patients, and searching for reliable and
effective molecular markers to guide the diagnosis and prognosis of
prostate cancer is a popular topic in clinical research. miRNAs are
non-coding short-chain RNAs, which have significant roles as
oncogenes and tumor suppressor genes in the development of various
types of cancer (30,31). It has previously been reported that
miR-589-5p acts as a tumor suppressor in non-small cell lung cancer
(22). In addition, the long
non-coding RNA (lncRNA) putative fatty acid-binding protein 5-like
protein 3 (FABP5P3) can bind to miR-589-5p, which serves a
tumor-enhancing role in hepatocellular carcinoma; the lncRNA
FABP5P3/miR-589-5p/zinc finger MYND-type containing 19 axis
contributes to hepatocellular carcinoma cell proliferation,
migration and invasion (23). In
addition, miR-589-5p can regulate mitogen-activated protein kinase
kinase kinase 8 expression in hepatocellular carcinoma (32). However, the role and mechanism of
miR-589-5p in prostate cancer remain elusive. The present study
confirmed that the expression levels of miR-589-5p in prostate
cancer tissues and cell lines were abnormally downregulated, thus
suggesting that miR-589-5p may act as a tumor suppressor involved
in the progression and development of prostate cancer.
Apoptosis is a programmed cell death controlled by
intracellular signals (33). There
are two classic apoptotic pathways: The death receptor-mediated
extrinsic pathway and the mitochondria-mediated intrinsic pathway.
Alterations in mitochondrial outer membrane permeabilization can
result in the release of various pro-apoptotic factors from the
intermembrane space to the cytoplasm, which further activates
caspases and induces cell apoptosis (34). Furthermore, upregulation of the
Bax:Bcl-2 ratio results in the release of cytochrome c from
the mitochondria and activates mitochondria-dependent caspases to
induce apoptotic cell death (35).
In the present study, overexpression of miR-589-5p markedly
inhibited the viability of DU-145 and PC3 prostate cancer cells,
whereas knockdown of miR-589-5p promoted DU-145 and PC3 cell
viability. In addition, miR-589-5p mimics increased the protein
expression levels of Bax, caspase-3 and caspase-9, and decreased
the protein expression levels of Bcl-2 in DU-145 and PC3 cells,
whereas miR-589-5p inhibitors exhibited the opposite effects.
Tumor invasion and metastasis are biological
characteristics of malignant tumors, and are the main causes of
mortality in patients with cancer (36). The present results confirmed the
inhibitory role of miR-589-5p in the invasion and metastasis of
DU145 and PC3 cells by wound-healing and Transwell invasion assays.
It has been reported that tumor invasion and metastasis are a
series of complex, multi-step and multi-factor dynamic process
between tumor cells and host cells or the extracellular matrix
(ECM) (37,38). A series of tissue barriers are
encountered in the process of tumor invasion and metastasis; these
barriers are made up of basement membrane, mesenchyme and matrix in
the ECM. Among the enzymes involved in the destruction of the ECM,
MMPs are the most closely associated with tumor invasion and
metastasis (39–41). The present study revealed that
miR-589-5p mimics significantly suppressed the expression levels of
MMP-2 and MMP-9, whereas the expression levels of MMP-2 and MMP-9
were promoted following transfection with miR-589-5p
inhibitors.
According to the strict complementary nature of base
pairs, miRNAs can target and bind mRNAs, and can inhibit the
expression of target genes through degrading target mRNAs or
inhibiting the translation progress. These functions of miRNAs
participate in the progression and development of cancer (42). CCL-5 is a chemokine that belongs to
the chemokine C-C family, which is mainly expressed in T
lymphocytes, macrophages, platelets, synovial fibroblasts, renal
tubular epithelial cells and certain types of tumor cells (43). Previous studies have detected CCL-5
expression in tissues and serum samples from patients with
pancreatic cancer, lung cancer, melanoma, etc., and it is
correlated with disease progression (44,45).
In addition, CCL-5, as a target gene, takes part in the development
of numerous types of tumor (46,47).
In this study, TargetScan and PicTar were used to analyze the
target genes of miR-589-5p, and a luciferase reporter assay
confirmed that CCL-5 may be a potential candidate of miR-589-5p.
Furthermore, the role of CCL-5 in the progression and development
of prostate cancer was evaluated. Upregulation of CCL-5 was
observed in prostate cancer tissues and cell lines, and si-CCL-5
had inhibitory effects on viability, apoptosis, migration and
invasion in DU145 and PC3 cells. In addition, the mRNA expression
levels of CCL-5 were negatively correlated with miR-589-5p
expression in prostate cancer tissues. These results confirmed that
CCL-5 acts as a target gene of miR-589-5p and may serve an
important role in the progression and development of prostate
cancer.
In conclusion, the present study confirmed that
miR-589-5p was downregulated in prostate cancer tissues and cell
lines. In addition, it was suggested that miR-589-5p may function
as a potential tumor suppressor in prostate cancer to inhibit cell
growth and metastasis by targeting CCL-5. Therefore, miR-589-5p may
be considered a reliable molecular marker for the diagnosis and
prognosis of prostate cancer. This study presented preliminary
research on the antitumor effects of miR-589-5p in prostate cancer.
At present, it is unclear whether miR-589-5p has anti-tumor effects
in other types of cancer and the underlying mechanism requires
further investigation; therefore, we aim to further explore the
signaling pathways that are involved in CCL-5-mediated regulation
of cell viability and metastasis in future studies.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BZ conceived and designed the present study. XJ and
FM performed the experiments. LJ and ZT acquired and analyzed the
data. All authors have read and approved the manuscript and agree
to be accountable for all aspects of the research in ensuring that
the accuracy and integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
This study was approved by the Ethical Committee of
Nanjing Medical University. All patients provided written informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kojima K, Fujita Y, Nozawa Y, Deguchi T
and Ito M: miR-34a attenuates paclitaxel-resistance of
hormone-refractory prostate cancer PC3 cells through direct and
indirect mechanisms. Prostate. 70:1501–1512. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Viticchiè G, Lena AM, Latina A, Formosa A,
Gregersen LH, Lund AH, Bernardini S, Mauriello A, Miano R, Spagnoli
LG, et al: miR-203 controls proliferation, migration and invasive
potential of prostate cancer cell lines. Cell Cycle. 10:1121–1131.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guan H, You Z, Wang C, Fang F, Peng R, Mao
L, Xu B and Chen M: MicroRNA-200a suppresses prostate cancer
progression through BRD4/AR signaling pathway. Cancer Med.
8:1474–1485. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fan H and Zhang YS: miR-490-3p modulates
the progression of prostate cancer through regulating histone
deacetylase 2. Eur Rev Med Pharmacol Sci. 23:539–546.
2019.PubMed/NCBI
|
|
5
|
Zhang FB, Du Y, Tian Y, Ji ZG and Yang PQ:
miR-1299 functions as a tumor suppressor to inhibit the
proliferation and metastasis of prostate cancer by targeting NEK2.
Eur Rev Med Pharmacol Sci. 23:530–538. 2019.PubMed/NCBI
|
|
6
|
Ota A, Tagawa H, Karnan S, Tsuzuki S,
Karpas A, Kira S, Yoshida Y and Seto M: Identification and
characterization of a novel gene, C13orf25, as a target for
13q31-q32 amplification in malignant lymphoma. Cancer Res.
64:3087–3095. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Doehn C, Böhmer T, Kausch I, Sommerauer M
and Jocham D: Prostate cancer vaccines: Current status and future
potential. BioDrugs. 22:71–84. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Oesterling JE: Prostate specific antigen:
A critical assessment of the most useful tumor marker for
adenocarcinoma of the prostate. J Urol. 145:907–923. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Moyer VA and U.S. Preventive Services Task
Force: Screening for prostate cancer: U.S. Preventive Services Task
Force recommendation statement. Ann Intern Med. 157:120–134. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tadayon F, Arezegar HR, Khorrami MH,
Hashemi Juzdani R, Shahdoost AA and Mellat M: Evaluation of
prostatic cancer prevalence in patients with prostatic-specific
antigen between 4 and 10 and normal digital rectal examination. Adv
Biomed Res. 5:1122016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zugor V, Kühn R, Engelhard K, Poth S,
Bernat MM, Porres D and Labanaris AP: The value of endorectal
magnetic resonance imaging of the prostate in improving the
detection of anterior prostate cancer. Anticancer Res.
36:4279–4283. 2016.PubMed/NCBI
|
|
12
|
Choi SK, Shim M, Kim M, Park M, Lee S,
Song C, Lee HL and Ahn H: Heterogeneous oncologic outcomes
according to surgical pathology in high-risk prostate cancer:
Implications for better risk stratification and preoperative
prediction of oncologic outcomes. J Cancer Res Clin Oncol.
143:1871–1878. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Murphy DG and Zargar H: Words of Wisdom.
Re: Chemohormonal therapy in metastatic hormone-sensitive prostate
cancer. Eur Urol. 69:5402016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Langius-Eklöf A, Christiansen M, Lindström
V, Blomberg K, Hälleberg Nyman M, Wengström Y and Sundberg K:
Adherence to report and patient perception of an interactive app
for managing symptoms during radiotherapy for prostate cancer:
Descriptive study of logged and interview data. JMIR Cancer.
3:e182017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cheng AM, Byrom MW, Shelton J and Ford LP:
Antisense inhibition of human miRNA and indications for an
involvement of miRNA in cell growth and apoptosis. Nucleic Acids
Res. 33:1290–1297. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu W, Sun M, Zou GM and Chen J: MicroRNA
and cancer: Current status and prospective. Int J Cancer.
120:953–960. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Eulalio A, Huntzinger E and Izaurralde E:
Getting to the root of miRNA-mediated gene silencing. Cell.
132:9–14. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bhayani MK, Calin GA and Lai SY:
Functional relevance of miRNA sequences in human disease. Mutat
Res. 731:14–19. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guan B, Wu K, Zeng J, Xu S, Mu L, Gao Y,
Wang K, Ma Z, Tian J, Shi Q, et al: Tumor-suppressive microRNA-218
inhibits tumor angiogenesis via targeting the mTOR component RICTOR
in prostate cancer. Oncotarget. 8:8162–8172. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mao A, Liu Y, Wang Y, Zhao Q, Zhou X, Sun
C, Di C, Si J, Gan L and Zhang H: miR-449a enhances
radiosensitivity through modulating pRb/E2F1 in prostate cancer
cells. Tumour Biol. 37:4831–4840. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu C, Lv D, Li M, Zhang X, Sun G, Bai Y
and Chang D: Hypermethylation of miRNA-589 promoter leads to
upregulation of HDAC5 which promotes malignancy in non-small cell
lung cancer. Int J Oncol. 50:2079–2090. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu Q, Luo Z, Lu G, Gui F, Wu J, Li F and
Ni Y: LncRNA FABP5P3/miR-589-5p/ZMYND19 axis contributes to
hepatocellular carcinoma cell proliferation, migration and
invasion. Biochem Biophys Res Commun. 498:551–558. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lin YC, Lin JF, Tsai TF, Chou KY, Chen HE
and Hwang TI: Tumor suppressor miRNA-204-5p promotes apoptosis by
targeting BCL2 in prostate cancer cells. Asian J Surg. 40:396–406.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang X, Yi Y, Lv Q, Zhang J, Wu K, Wu W
and Zhang W: Novel 1,3,5-triazine derivatives exert potent
anti-cervical cancer effects by modulating Bax, Bcl2 and caspases
expression. Chem Biol Drug Des. 91:728–734. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Powell WC, Knox JD, Navre M, Grogan TM,
Kittelson J, Nagle RB and Bowden GT: Expression of the
metalloproteinase matrilysin in DU-145 cells increases their
invasive potential in severe combined immunodeficient mice. Cancer
Res. 53:417–422. 1993.PubMed/NCBI
|
|
28
|
Heidenreich A, Bastian PJ, Bellmunt J,
Bolla M, Joniau S, van der Kwast T, Mason M, Matveev V, Wiegel T,
Zattoni F, et al: EAU guidelines on prostate cancer. Part 1:
Screening, diagnosis, and local treatment with curative
intent-update 2013. Eur Urol. 65:124–137. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Veltri RW and Christudass CS: Nuclear
morphometry, epigenetic changes, and clinical relevance in prostate
cancer. Adv Exp Med Biol. 773:77–99. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
He H, Jazdzewski K, Li W, Liyanarachchi S,
Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, et al:
The role of microRNA genes in papillary thyoid carcinoma. Proc Natl
Acad Sci USA. 102:19075–19080. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu M, Jolicoeur N, Li Z, Zhang L, Fortin
Y, L'Abbe D, Yu Z and Shen SH: Genetic variations of microRNAs in
human cancer and their effects on the expression of miRNAs.
Carcinogenesis. 29:1710–1716. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang X, Jiang P, Shuai L, Chen K, Li Z,
Zhang Y, Jiang Y and Li X: miR-589-5p inhibits MAP3K8 and
suppresses CD90+ cancer stem cells in hepatocellular
carcinoma. J Exp Clin Cancer Res. 35:1762016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Monteiro HP, Silva EF and Stern A: Nitric
oxide: A potential inducer of adhesion-related apoptosis-anoikis.
Nitric Oxide. 10:1–10. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Green DR and Reed JC: Mitochondria and
apoptosis. Science. 281:1309–1312. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Deveraux QL, Schendel SL and Reed JC:
Antiapoptotic proteins. The bcl-2 and inhibitor of apoptosis
protein families. Cardiol Clin. 19:57–74. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang H, Liu G, Shen D, Ye H, Huang J, Jiao
L and Sun Y: HOXA1 enhances the cell proliferation, invasion and
metastasis of prostate cancer cells. Oncol Rep. 34:1203–1210. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang WJ, Yang W, Ouyang ZH, Xue JB, Li XL,
Zhang J, He WS, Chen WK, Yan YG and Wang C: miR-21 promotes ECM
degradation through inhibiting autophagy via the PTEN/akt/mTOR
signaling pathway in human degenerated NP cells. Biomed
Pharmacother. 99:725–734. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cui Z, Tang J, Chen J and Wang Z:
Hsa-miR-574-5p negatively regulates MACC-1 expression to suppress
colorectal cancer liver metastasis. Cancer Cell Int. 14:472014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bar-Or A, Nuttall RK, Duddy M, Alter A,
Kim HJ, Ifergan I, Pennington CJ, Bourgoin P, Edwards DR and Yong
VW: Analyses of all matrix metalloproteinase members in leukocytes
emphasize monocytes as major inflammatory mediators in multiple
sclerosis. Brain. 126:2738–2749. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Westermarck J and Kähäri VM: Regulation of
matrix metalloproteinase expression in tumor invasion. FASEB J.
13:781–792. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kraiem Z and Korem S: Matrix
metalloproteinases and the thyroid. Thyroid. 10:1061–1069. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ventura A and Jacks T: MicroRNAs and
cancer: Short RNAs go a long way. Cell. 136:586–591. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Azenshtein E, Luboshits G, Shina S,
Neumark E, Shahbazian D, Weil M, Wigler N, Keydar I and Ben-Baruch
A: The CC chemokine RANTES in breast carcinoma progression:
Regulation of expression and potential mechanisms of promalignant
activity. Cancer Res. 62:1093–1102. 2002.PubMed/NCBI
|
|
44
|
Yaal-Hahoshen N, Shina S, Leider-Trejo L,
Barnea I, Shabtai EL, Azenshtein E, Greenberg I, Keydar I and
Ben-Baruch A: The chemokine CCL5 as a potential prognostic factor
predicting disease progression in stage II breast cancer patients.
Clin Cancer Res. 12:4474–4478. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kwon KH, Lee YC, Chung JH and Eun YG:
Association study of chemokine (C-C motif) ligand 5 gene
polymorphism and papillary thyroid cancer. J Invest Surg.
26:319–324. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kuo CH, Liu CJ, Lu CY, Hu HM, Kuo FC, Liou
YS, Yang YC, Hsieh MC, Lee OK, Wu DC, et al: 17β-estradiol inhibits
mesenchymal stem cells-induced human AGS gastric cancer cell
mobility via suppression of CCL5-Src/Cas/Paxillin signaling
pathway. Int J Med Sci. 11:7–16. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Swamydas M, Ricci K, Rego SL and Dréau D:
Mesenchymal stem cell-derived CCL-9 and CCL-5 promote mammary tumor
cell invasion and the activation of matrix metalloproteinases. Cell
Adh Migr. 7:315–324. 2013. View Article : Google Scholar : PubMed/NCBI
|