Introduction
Gastric cancer is one of the most common malignant
tumors in humans. Despite its morbidity and mortality exhibiting a
steady downward trend worldwide, gastric cancer remains a great
burden to human health as the third leading cause of
cancer-associated mortality (1,2). A
report revealed that the region with the highest incidence of
gastric cancer is East Asia, particularly China, with an annual
incidence of approximately 4–6 cases per 10,000 individuals
(3). In addition, the National
Cancer Institute data indicated that the incidence and diagnosis of
this disease is mainly between the ages of 60–70 years, and the
majority of patients have a poor prognosis due to diagnosis in the
late stages of the disease (4).
Approximately 90% of stomach tumors are adenocarcinomas, which are
classified into two main histologic types: High differentiated or
intestinal-type gastric cancer; and undifferentiated or
diffuse-type gastric cancer. Intestinal-type gastric cancer is
pertinent to atrophy of the gastric mucosa and intestinal
metaplasia, while diffuse-type tumors usually arise in tissue
without atrophic total gastritis (5).
MicroRNA (miR), as a type of non-encoding RNA with a
length of approximately 22 bp, regulates gene expression at the
post-transcriptional level. The known miRNAs regulate approximately
30% of the genes in the human genome, which participate in human
development, cell proliferation and differentiation, hematopoiesis
and cell apoptosis (6). Nearly 50%
of the miRNAs are positioned on the tumor-associated genomic
regions, including fragile sites, chromosomal amplification and
loss of heterozygosity chromosome region (7,8).
Thus, miRNAs can significantly affect the entire process from tumor
occurrence to tumor metastasis. Abnormal miRNA expression is also
associated with the occurrence and development of gastric cancer,
and can regulate the expression of multiple oncogenes or
anti-oncogene, thereby influencing tumor cell proliferation,
migration, invasion, apoptosis and other biological characteristics
(9).
Low expression of miR-539 has been reported in
osteosarcoma tissues, and this was negatively correlated with the
clinical stage, recurrence and metastasis of the disease; thus, low
expression of miR-539 may be an independent prognostic indicator of
overall survival in patients with osteosarcoma (10). In hepatocellular carcinoma (HCC),
miR-539 was reported to function as a tumor suppressor, and its
expression was downregulated in HCC tissues and cell lines
(11). However, forced expression
of miR-539 was able to induce the apoptosis of HepG2 cells and
enhance the sensitivity of HCC cells to arsenic trioxide (11). In nasopharyngeal carcinoma (NPC),
miR-539 targeted cyclin-dependent kinase 4 to induce cell cycle
arrest, resulting in the inhibition of NPC cell growth (12). In non-small cell lung cancer
(13) and thyroid cancer, miR-539
inhibited the proliferation, migration and invasion of cancer cells
by targeting CARMA1 (14).
Although an increasing number of studies have reported that the
role of miR-539 in cancer is mainly as a tumor suppressor, the
specific role of miR-539 in gastric cancer remains unknown.
Therefore, the present study focused on the role of
miR-539 in the process of gastric cancer. Initially, it was
demonstrated that miR-539 was poorly expressed in gastric cancer
tissues and cell lines. Subsequently, it was observed that miR-539
inhibited the proliferation and migration of gastric cancer cells,
and this effect may be achieved partly by targeting SRY-box 5
(SOX5). Overall, the study reveals that miR-539 serves an important
role in the process of gastric cancer, indicating its potential
application in the treatment of gastric cancer.
Materials and methods
Collection of clinical tissue
samples
Between August 2015 and July 2016, 30 gastric cancer
patients receiving surgical treatment were recruited into the
present study from The First People's Hospital of Changde City
(Changde, China). The inclusion criteria were as follows: Gastric
cancer was confirmed with pathologic diagnosis (15); and the patients had not undergone
any radiotherapy and chemotherapy prior to surgical treatment.
Tumor and adjacent tissue samples were collected from the patients
and stored at −80°C. The proportion of malignant cells in the
cancer tissue samples was >80%, while the adjacent tissue
samples included <10% of malignant cells. The study was approved
by the Ethics Committee of Changde Vocational Technical College
Research (Changde, China). Patients provided signed informed
consent prior to participation in the study.
Cell lines and cell culture
Gastric cancer cell lines SGC7901, MGC803 and
HGC-27, as well as the normal gastric mucosa cell line GES-1, were
purchased from American Type Culture Collection (Manassas, VA, USA)
and preserved until cell recovery. The cells were cultured in 5%
CO2 at 37°C with Dulbecco's modified Eagle's medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
containing 10% fetal bovine serum (FBS), penicillin (100 U/ml) and
streptomycin (100 µg/ml).
Cell transfection
At 1 day before cell transfection, cells in the
logarithmic growth phase were selected and seeded into a 96-well
plate (5×103). miR-539 mimic, miR-539 inhibitor and
scramble mimic negative control (miR-NC) were synthesized by
GenePharma Co., Ltd., (Shanghai, China). The cells were transfected
with 20 nmol/l miR-539 mimic or miR-NC, and the miR-539 inhibitor
(high concentration solution, 20 µm; diluted in ddH2O)
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. For the
rescue experiments, miR-539 mimics and SOX5 overexpression with
miR-539 mimics were transfected into SGC7901 and MGC803 cells.
RNA extraction and quantitative
polymerase chain reaction (qPCR)
Total RNA was extracted from the tissues or cultured
cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and a miRNeasy Mini kit (Qiagen GmbH, Hilden, Germany)
according to the manufacturers' protocol. TaqMan MicroRNA Assay
(Invitrogen; Thermo Fisher Scientific, Inc.) was used to perform
qPCR in order to qualify miRNA expression. RNA was converted into
complementary DNA by reverse transcription using mRNA/miRNA reverse
transcription kits (for mRNA, RevertAid RT Reverse Transcription
kit; Fermentas; Thermo Fisher Scientific, Inc.; and for miRNA,
All-in-One™ qRT-PCR Detection kit; Fumeng, Guangzhou, China), with
the following temperature protocols: For mRNA, 42°C for 1 h and
70°C for 10 min; and for miRNA, 37°C for 1 h and 85°C for 5 min.
Then qPCR was subsequently performed using SYBR Premix DimerEraser
(Takara Biotechnology Co., Ltd., Dalian, China) on a 7900HT Fast
Real-Time PCR system (Thermo Fisher Scientific, Inc.). The qPCR
thermal cycling conditions were as follows: 95°C for 15 sec; 40
cycles at 95°C for 5 sec; 55°C for 34 sec; 72°C for 30 sec;
followed by melting curve analysis. The level of mature miR-539 was
normalized to that of the U6 endogenous control, while
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an
internal control for the detection of SOX mRNA expression. Fold
changes were calculated using the comparative threshold cycle value
(2−ΔΔCq) method (16).
The experiments were performed in triplicate, and the primers used
are listed in Table I.
 | Table I.Primers used in quantitative
polymerase chain reaction. |
Table I.
Primers used in quantitative
polymerase chain reaction.
| Gene | Primer sequences
(5′-3′) |
|---|
| miR-539 sense |
GGAGAAATTATCCTTGGTGTG |
| U6 sense |
ATTGGAACGATACAGAGAAGATT |
| SOX5 forward |
AGCATGCTTACTGACCCTGATTTA |
| SOX5 reverse |
GGGAGTCCTATGGCCACAAGTCT |
| GAPDH forward |
TCATGGGTGTGAACCATGAGAA |
| GAPDH reverse |
GGCATGGACTGTGGTCATGAG |
Cell proliferation test
Cells were seeded into 96-well plates at a density
of 2,000 cells per well and cultured for 1, 2, 3, 4 and 5 days
after transfection. In order to determine cell proliferation, Cell
Counting Kit-8 (CCK-8; Sangon Biotech Co., Ltd., Shanghai, China)
assay was performed. Cells were incubated with 10% CCK-8 reagent at
37°C until visual color conversion occurred. Subsequently, the
absorbance at 450 nm was measured using a microplate reader (Thermo
Fisher Scientific, Inc.). All experiments were conducted in
triplicate.
Transwell assay
Transwell assay was performed to detect cell
migration and invasion. A total of 1.0×105 cells were
suspended in serum-free medium and plated in the upper chambers of
transwell inserts in a 6-well plate (Corning Incorporated, Corning,
NY, USA). Medium containing 10% FBS was added to the lower chamber,
serving as a chemoattractant. Following incubation for 24 h at 37°C
in 5% CO2, cells in the lower chamber were fixed in
methanol for 15 min and stained with 0.05% crystal violet in PBS
for 15 min before counting under a microscope (Olympus Corporation,
Tokyo, Japan). Cells that had not migrated from the upper chamber
were removed by wiping the surface with a cotton swab, and invasive
cells were fixed with 4% formaldehyde in PBS and subsequently
stained with 1% crystal violet in 2% ethanol. Images of the cells
on the lower surface of the filter were obtained under a light
microscope (magnification, ×200).
Dual-luciferase reporter assay
TargetScan (www.targetscan.org/) and miRanda (www.microrna.org/microrna/home.do) were
initially used to predict the potential target gene of miR-539,
which was revealed to be SOX5; a luciferase assay was performed to
confirm this. A fragment of the human SOX5 3′-untranslated region
(3′UTR) containing the miR-539 binding site was amplified by PCR
and cloned downstream of the firefly luciferase gene in the
pMIR-REPORT vector (Ambion; Thermo Fisher Scientific, Inc.) to
obtain the wild-type (WT) SOX5 reporter vector. The mutation
fragment was cloned into the pMIR-REPORT vector to produce the
pMIR-SOX5-mut (Mut) 3′UTR vector. Cells (2×104) were
seeded into 24-well plates on the day before transfection, and then
cotransfected with the constructs and miR-539 mimic or miR-NC.
Luciferase activity was measured after 48 h using the
Dual-Luciferase Reporter Assay system (Promega Corporation,
Madison, WI, USA).
Statistical analysis
The quantitative data between groups were compared
and analyzed by Student's t-test (two tailed) or by one-way
analysis of variance followed by Tukey's test. Data are expressed
as the mean ± standard deviation. P<0.05 was considered to
indicate a statistically significant difference.
Results
Expression of miR-539 in gastric
cancer
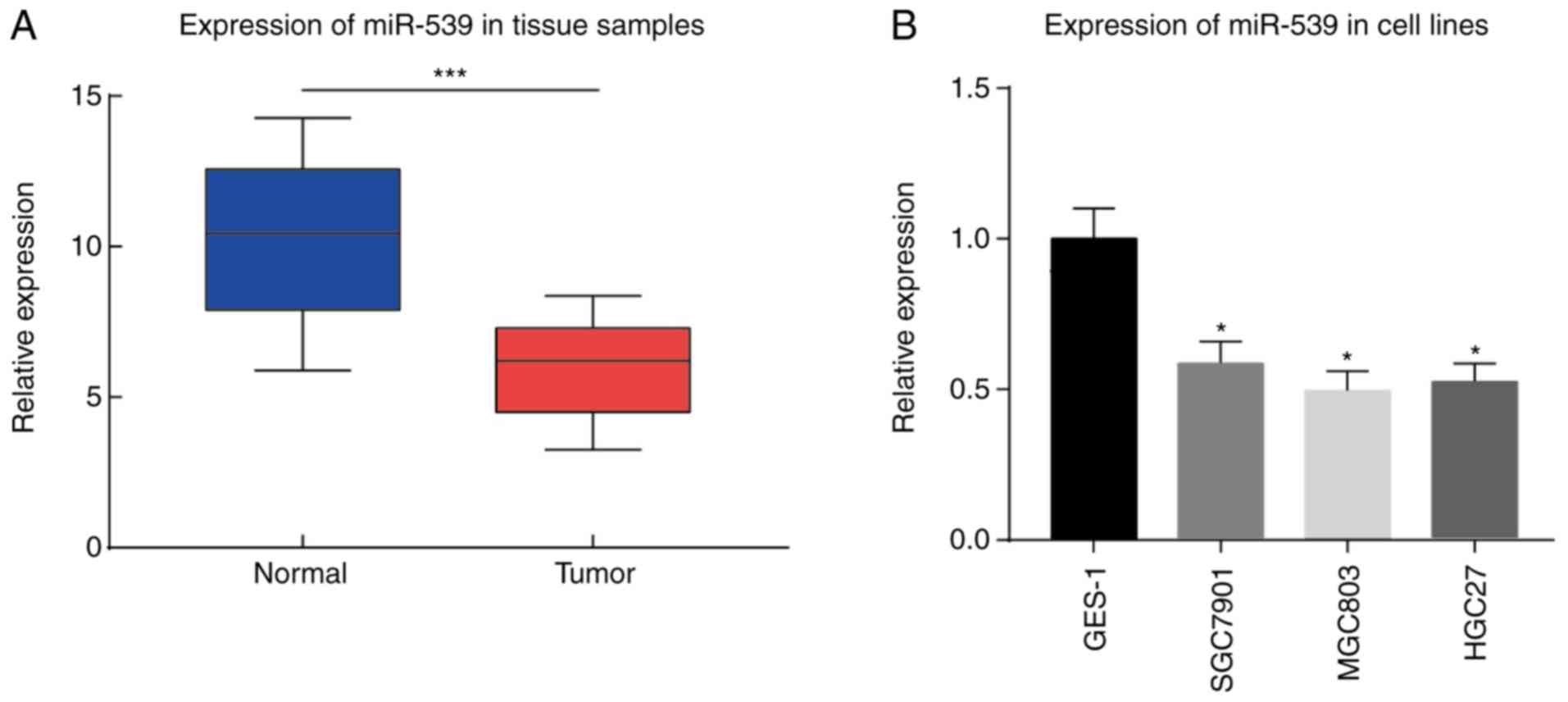
miR-539 expression in gastric cancer cell lines,
gastric mucosa cell line GES-1, and tumor and adjacent tissue
samples from 30 cases gastric cancer patients was determined by
qPCR. The results demonstrated that miR-539 exhibited a
significantly lower expression in the gastric tumor tissue samples
compared with that in the adjacent tissue samples (P<0.001;
Fig. 1A). Furthermore, in gastric
cancer cell lines SGC7901, MGC803 and HGC-27, the expression of
miR-539 was markedly reduced as compared with that in the gastric
mucosa cell line GES-1 (P<0.05; Fig. 1B).
Effect of miR-539 on the proliferation
and migration of gastric cancer cells
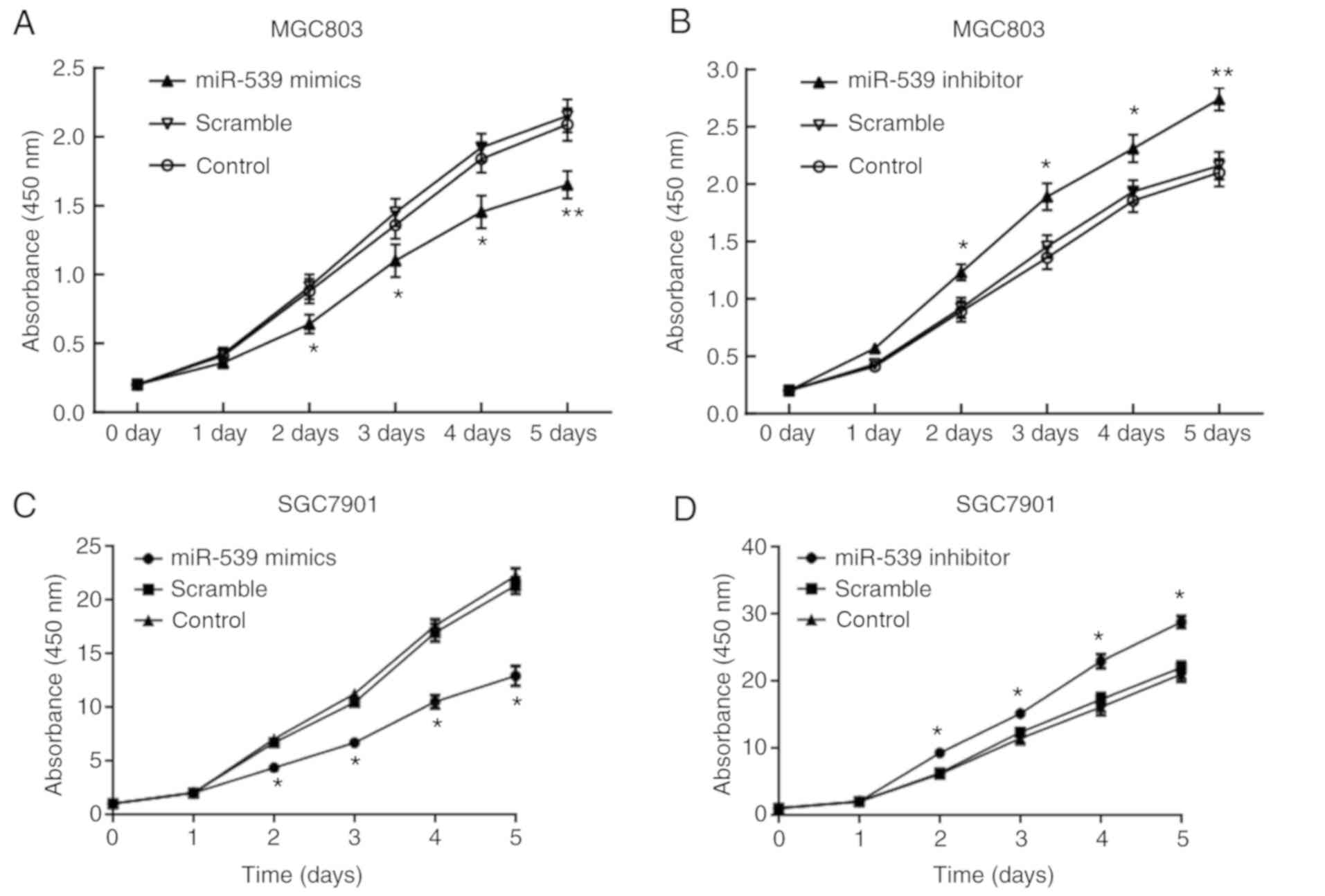
The results of CCK-8 assay demonstrated that,
compared with the cells in the scramble group, the cell
proliferation was significantly decreased from day 2 of
transfection with miR-539 mimic in the gastric cancer cell lines
MGC803 (Fig. 2A and B) and SGC7901
(Fig. 2C and D). By contrast, cell
proliferation was significantly promoted after transfection with
miR-539 inhibitor. In the transwell assay, gastric cancer cell
lines transfected with miR-539 mimic had significantly decreased
migration compared with the control group, which was contrary to
the enhanced migration observed in cells transfected with miR-539
inhibitor (P<0.05 and P<0.01; Fig. 2E and F).
Targeting association between miR-539
and SOX5 gene
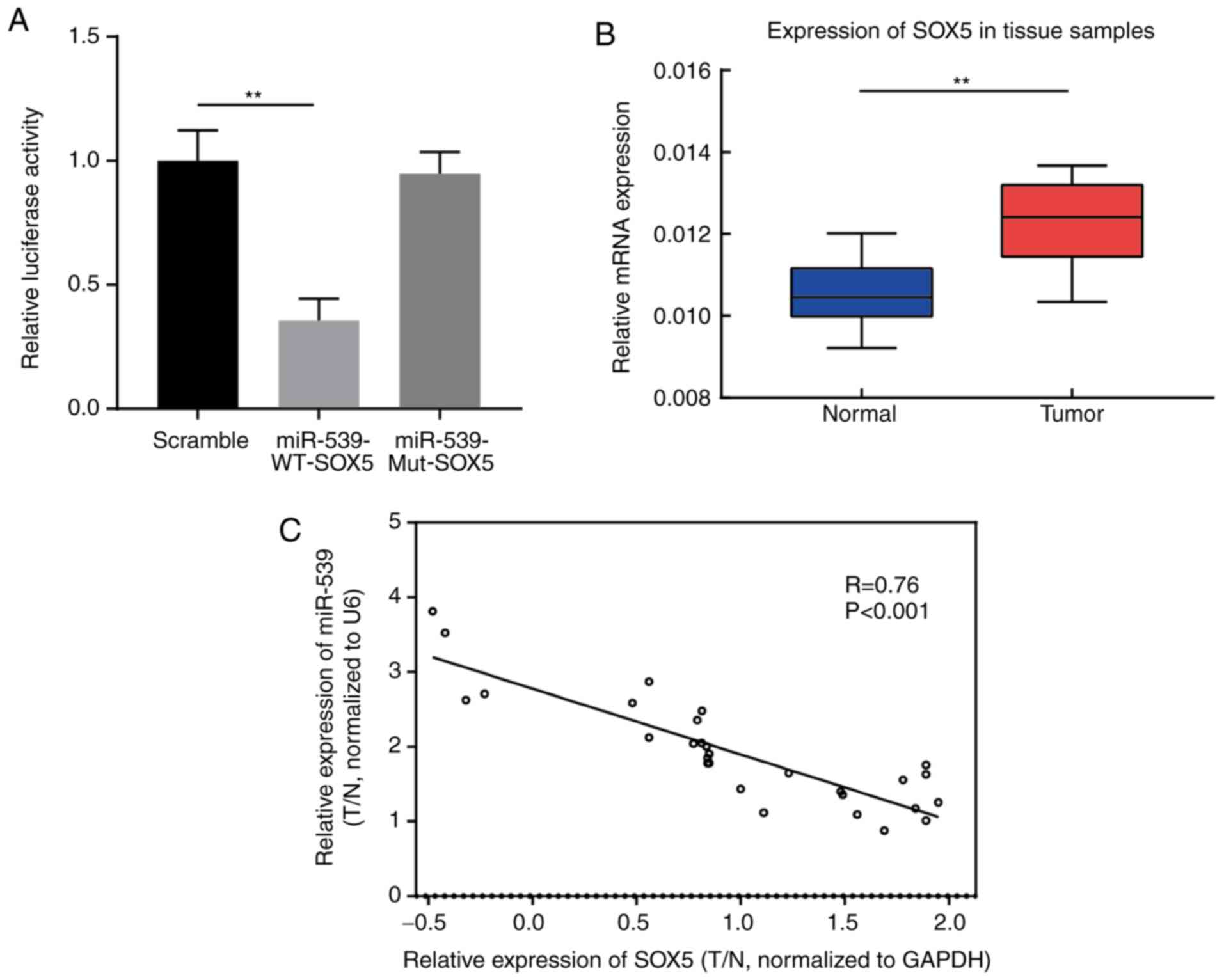
The association between miR-539 and SOX5 gene was
verified in gastric cancer cells using a luciferase assay. The
results suggested that, compared with the SOX5 Mut group and the
control group, the luciferase activity of SOX5 WT reporter vector
was significantly reduced in MGC803 cells transfected with miR-539
mimic, indicating that miR-539 directly targets SOX5 gene (Fig. 3A). In addition, the results of qPCR
revealed that SOX5 expression in gastric tumor tissues was markedly
higher in comparison with that in adjacent tissues (Fig. 3B). Spearman's correlation analysis
further demonstrated that there was a negative correlation between
miR-539 and SOX5 gene expression levels in clinical tissue samples
of gastric cancer (R=−0.76; P<0.001; Fig. 3C).
SOX5 regulates the proliferation and
metastasis of gastric cancer cells
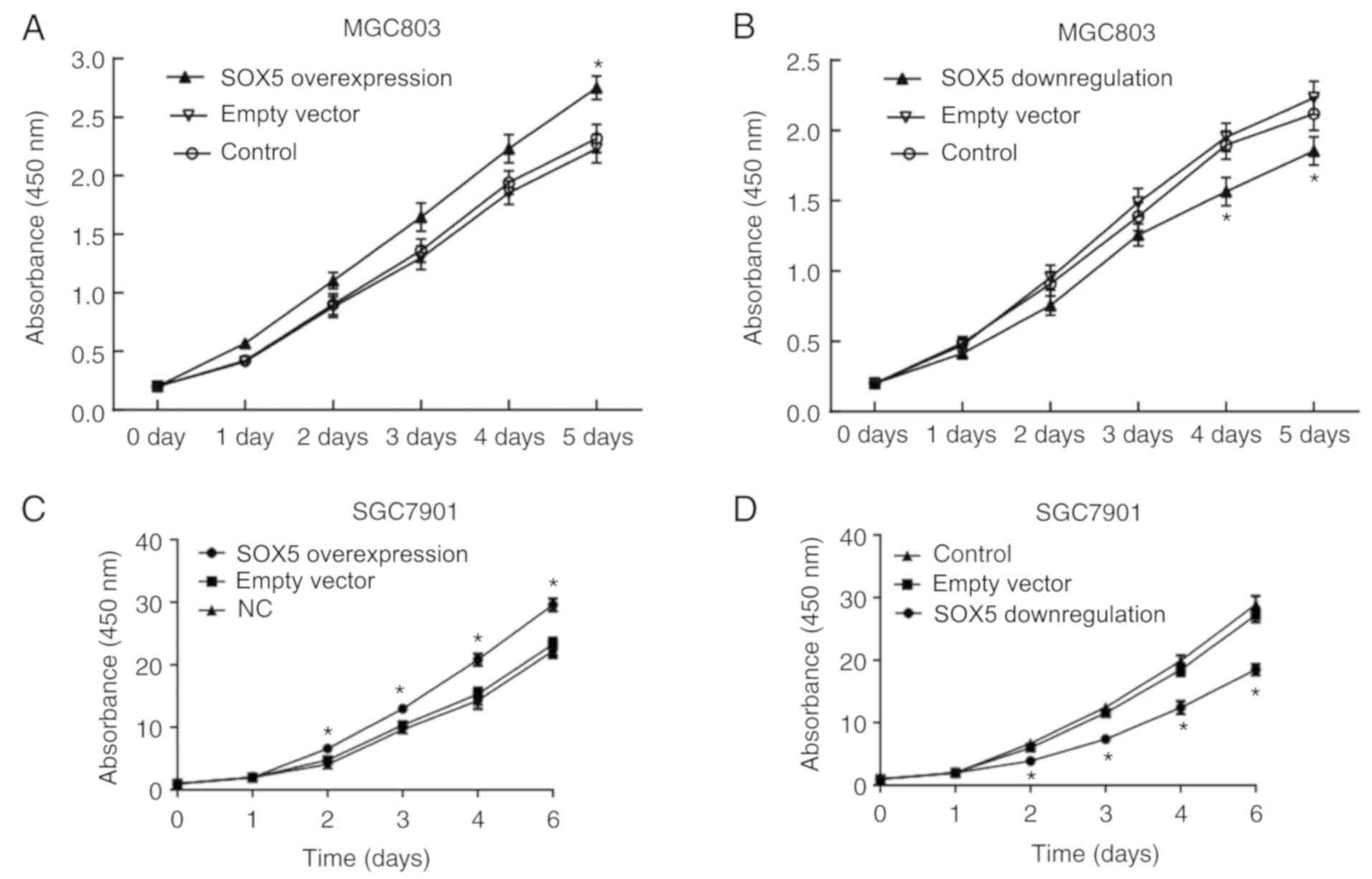
CCK-8 assay demonstrated that, compared with the
cells transfected with empty vector, MGC803 cells with SOX5
overexpression exhibited significantly increased proliferation
(P<0.05); while MGC803 cells with SOX5 inhibition had notably
decreased proliferation (P<0.05; Fig. 4A and B). The same results were
confirmed in SGC7901 cells (Fig. 4C
and D). In the transwell experiment, compared with the control
group, the migration of MGC803 cells was significantly higher
following SOX5 overexpression, while it was significant lower
subsequent to SOX5 inhibition (Fig.
4E-H). The same results were confirmed in SGC7901 cells
(Fig. 4I and J). These results
suggest that SOX5 is also involved in cell proliferation and
migration in gastric cancer.
SOX5 restores the effect of miR-539 on
the proliferation and migration of gastric cancer cells
The aforementioned results suggested that the
proliferation and migration of gastric cancer cells were inhibited
by miR-539 and promoted by SOX5 gene overexpression. Therefore, it
was hypothesized that the regulation of miR-539 on gastric cancer
cells may be achieved by SOX5. Therefore, rescue experiments were
conducted to further identify the underlying mechanism. MGC803 and
SGC7901 cells were transfected with miR-NC, miR-539 mimic, or
co-transfected with miR-539 and SOX5 overexpression vectors. The
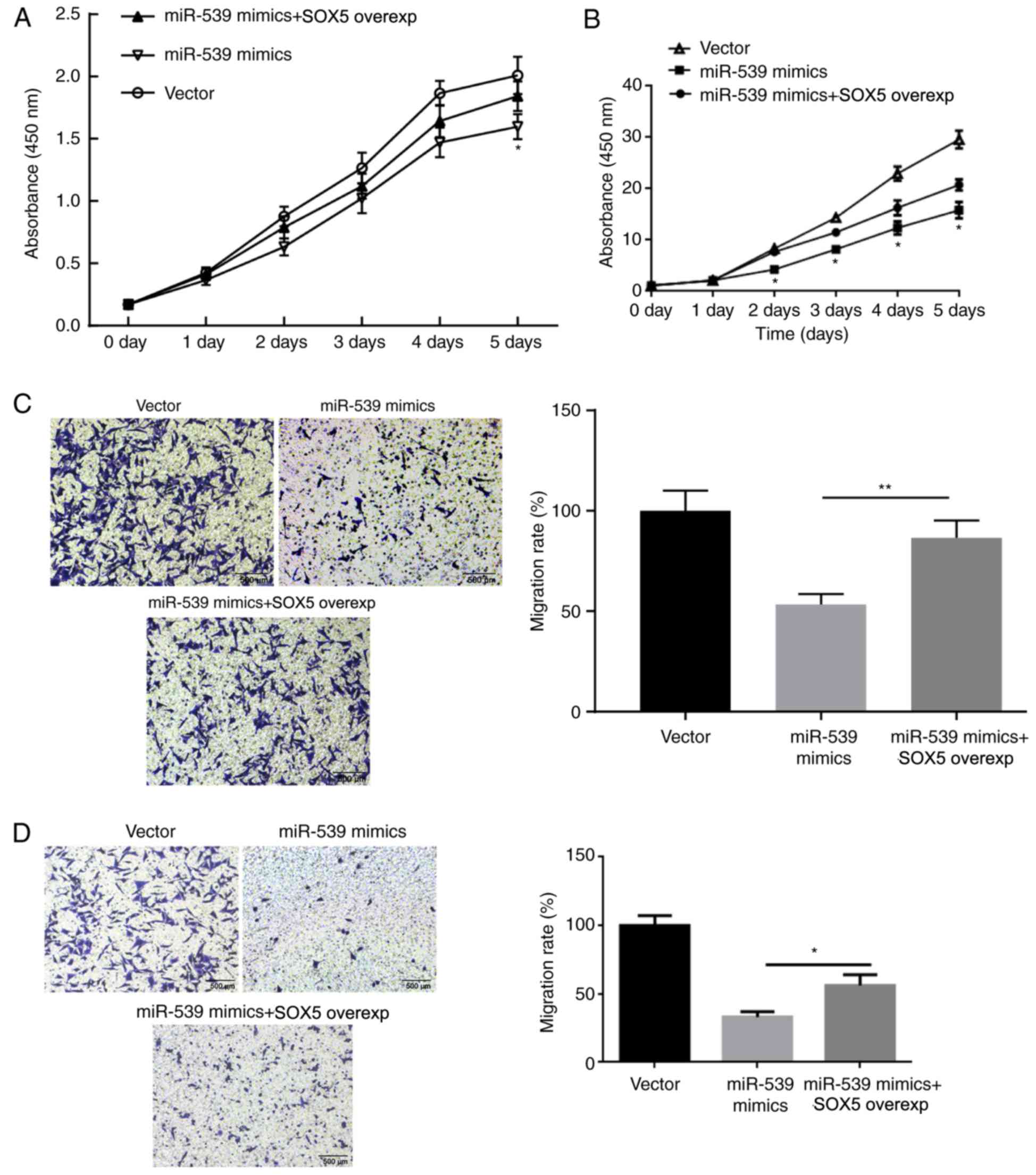
results of CCK-8 assay revealed that, compared with the group
transfected with miR-539 mimic alone, MGC803 (Fig. 5A) and SGC7901 (Fig. 5B) cells transfected with both
miR-539 and SOX5 overexpression vectors exhibited significantly
increased proliferation. Furthermore, transwell assay results
demonstrated that the migration of MGC803 (Fig. 5C) and SGC7901 (Fig. 5D) cells in miR539 + SOX5 group was
also significantly improved. In combination with the previous
experimental results, it is confirmed that the regulation of
miR-539 on gastric cancer cells was achieved by SOX5.
Discussion
Gastric cancer is a heterogeneous disease, whose
occurrence and development are associated with multiple
environmental factors and cancer pathways. The external factors
include Helicobacter pylori and Epstein-Barr virus
infection, alcohol abuse, body mass index and physical activity
(17), while the internal factors
are manifested in genetic inheritance (18). In the treatment of early gastric
cancer, surgical resection remains the main therapeutic strategy.
During the postoperative period, other comprehensive treatment
measures, such as adjuvant chemotherapy, can be applied. However,
the overall outcome of gastric cancer remains unfavorable. In
recent years, molecular targeted therapy has gradually a research
focus, and increasing molecular target drugs have been applied in
clinical trials. Thus, exploration of new molecular markers or
targets will be helpful for diagnosis in gastric cancer.
Accumulating studies have demonstrated that
microRNAs can function as oncogenes or tumor suppressor genes by
differential expression in a variety of tumor cells. Some microRNAs
can promote the proliferation, invasion or metastasis of tumor
cells in malignant tumors through multiple pathways. For instance,
miR-539 is able to inhibit the invasion and migration of osteogenic
sarcoma cells by targeting matrix metalloproteinase-8 (18). In HCC, low expression of miR-539
was reported in cancer tissues and cells, and this miRNA was able
to induce HCC cell apoptosis by mediating signal transducer and
activator of transcription 3 pathway (11).
In the present study, miR-539 was observed to be
downregulated in gastric cancer tissues and cell lines. In
combination with the findings of a previous study (13), it is hypothesized that miR-539
functions as a tumor suppressor in gastric cancer. In the
subsequent experiments, the proliferation of MGC803 and SGC7901
cells transfected with miR-539 mimic was found to be significantly
decreased. The results of transwell assay indicated that miR-539
promoted the migration of gastric cancer cells. Next, the potential
target gene of miR-539 was predicted by TargetScan and miRanda, and
it was observed that SOX5 was a potential target of miR-539, which
was then verified by luciferase assay. Compared with Mut SOX5
reporter vector, miR-539 mimic significantly reduced the luciferase
activity of WT SOX5 reporter vector.
Previous studies have demonstrated that SOX5
participates in the process of tumor epithelial-mesenchymal
transition (EMT). EMT is accompanied by marked changes in cell
morphology and behavior, such as in cell proliferation,
differentiation, adhesion and migration. In addition, EMT can
enhance the migration and invasion properties of cancer cells, and
induce stem cell properties in tumor cells (19,20).
Research has also revealed that SOX5 in breast cancer can directly
bind to the Twist1 promoter to activate the gene expression, while
it also controls the ETM process, suggesting that SOX5 participates
in the progression of breast cancer by activating the Twist1/ETM
axis (21). However, in osteogenic
sarcoma, SOX5 promotes cell migration and invasion, which may be
achieved by regulating the expression of Snail, so as to promote
EMT (22). Consistent with the
observations of previous studies investigating SOX5 in other
tumors, the present study suggested that SOX5 was highly expressed
in clinical cases of gastric cancer, and that SOX5 overexpression
repressed cell proliferation and migration, which was in contrast
to the effect of SOX5 silencing. The results of rescue experiments
demonstrated that cell proliferation and migration was
significantly increased following transfection with miR-539 and
SOX5 overexpression vector, implying that miR-539 inhibited the
proliferation and migration of gastric cancer cells by targeting
SOX5. Based on the research results, it is speculated that the
inhibitory effect of miR-539 on the proliferation and migration of
gastric cancer cells may be achieved by indirectly regulating EMT
through mediating SOX5 expression.
In conclusion, miR-539 was observed to inhibit the
proliferation and migration of gastric cancer cells by targeting
SOX5. miR-539 may function as a tumor suppressor and provide a
potential target for therapeutic strategies of gastric
carcinoma.
Acknowledgements
Not applicable.
Funding
The present study was supported by Scientific
Research Project of Education Department of Hunan Province (grant
no. 18c1220).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
YZ conceived and designed the study, and wrote the
manuscript. SD performed the expriments and analyzed the data. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Changde Vocational Technical College Research
(Changde, China). Patients provided signed informed consent prior
to participation in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Parkin DM: The global health burden of
infection-associated cancers in the year 2002. Int J Cancer.
118:3030–3044. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Morgan KA: Current controversies in cancer
care for the surgeon. Springer; 2016, View Article : Google Scholar
|
|
4
|
Soares FA, Coimbra FJ, Pelosof AG, Freitas
HC, Begnami MD, Costa Jr WL, Fannelli MF, Lopes de Mello CA, morim
M, Pizzi MP, et al: Genomics and epidemiology for gastric
adenocarcinomas. Appl Cancer Res. 37:72017. View Article : Google Scholar
|
|
5
|
Rugge M, Fassan M and Graham DY:
Epidemiology of gastric cancer. Gastric Cancer. Springer; pp.
23–34. 2015, View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bushati N and Cohen SM: microRNA
functions. Ann Rev Cell Dev Bio. 23:175–205. 2007. View Article : Google Scholar
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li Q, Li J, Dai W, Li YX and Li YY:
Differential regulation analysis reveals dysfunctional regulatory
mechanism involving transcription factors and microRNAs in gastric
carcinogenesis. Artif Intell Med. 77:12–22. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mirghasemi A, Taheriazam A, Karbasy SH,
Torkaman A, Shakeri M, Yahaghi E and Mokarizadeh A: Retraction
Note: Down-regulation of miR-133a and miR-539 are associated with
unfavorable prognosis in patients suffering from osteosarcoma.
Cancer Cell Int. 16:842016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu C, Zhou R, Zhou Q, Chang Y and Jiang
M: microRNA-539 suppresses tumor growth and tumorigenesis and
overcomes arsenic trioxide resistance in hepatocellular carcinoma.
Life Sci. 166:34–40. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lv LY, Wang YZ, Zhang Q, Zang HR and Wang
XJ: miR-539 induces cell cycle arrest in nasopharyngeal carcinoma
by targeting cyclin-dependent kinase 4. Cell Biochem Funct.
33:534–540. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Retraction, ; Xianzheng Gao, Shenglei Li,
Wencai Li, Guannan Wang, Wugan Zhao, Jing Han, Changying Diao,
Xiaohui Wang and Mingzhi Zhang: MicroRNA-539 suppresses tumor cell
growth by targeting the WNT8B gene in non-small cell lung cancer.
J. Cell. Biochem. Accepted Article doi.org/10.1002/jcb.26634. J
Cell Biochem. Dec 21–2017.(Epub ahead of print).
|
|
14
|
Gu L and Sun W: MiR-539 inhibits thyroid
cancer cell migration and invasion by directly targeting CARMA1.
Biochem Biophys Res Commun. 464:1128–1133. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen GD and Liu YL: Clinical, endoscopic
and pathologic analysis of 36 cases of early gastric cancer. China
J Endosc. 2012:(In Chinese).
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Singh S and Jha HC: Status of epstein-barr
virus coinfection with Helicobacter pylori in gastric
cancer. J Oncol. 2017:34562642017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jin H and Wang W: MicroRNA-539 suppresses
osteosarcoma cell invasion and migration in vitro and targeting
Matrix metallopeptidase-8. Int J Clin Exp Pathol. 8:8075–8082.
2015.PubMed/NCBI
|
|
19
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pei XH, Lv XQ and Li HX: Sox5 induces
epithelial to mesenchymal transition by transactivation of Twist1.
Biochem Biophys Res Commun. 446:322–327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang D and Liu S: SOX5 promotes
epithelial-mesenchymal transition in osteosarcoma via regulation of
Snail. J BUON. 22:258–264. 2017.PubMed/NCBI
|