Introduction
Varicose veins (VVs) is one of the most common
diseases with chronic venous insufficiency that affects ~73%
females and 56% males worldwide (1). A number of factors are involved in
the occurrence of VVs, such as constrictive underclothes, standing
for long periods of time and suffering from diabetes mellitus
and/or hypertension, which can lead to blood accumulation in the
lower extremities of the body (2).
Recent advances have suggested that reflux induced by venous valve
dysfunction (3), intimal
hyperplasia, alterations in the extracellular matrix (ECM),
abnormal glucose metabolism, local inflammation, local hypoxia,
endothelial cell injury, smooth muscle cell proliferation and
apoptosis, and increased vein wall tension (4–6) may
be associated with the mechanistic changes resulting in the
development and progression of VVs. However, these advancements
account for a limited number of putative mechanisms and thus, it is
important to further investigate the molecular mechanisms involved
in the pathogenesis of VVs and improve the existing therapeutic
strategies.
tRNA-derived fragments (tRFs), which are 15–50
nucleotides long, are members of the small RNA family that are
derived as unique sequences from mature tRNAs or precursor tRNAs.
With the advances in next-generation sequencing and bioinformatics
tools, tRFs have been successfully detected in eukaryotic cells and
demonstrated to participate in various pathological processes
(7,8). tRFs serve key roles in physiological
processes, such as regulating mRNA stability (9,10),
inhibiting translation initiation and elongation (11,12),
regulating ribosome biogenesis (13), functioning as an epigenetic factor
(14), regulating RNA reverse
transcription as a guide RNA (15), preventing apoptosis by binding to
cytochrome c (16), immune
regulation (17) and degradation
of target RNAs, such as mRNAs and microRNAs (miRNAs) (18–20).
Several studies have reported that tRFs are involved in a number of
diseases, including cancer (21,22),
neurodegenerative diseases (12),
acquired metabolic diseases (23)
and infectious diseases (24).
Additionally, tRFs have been demonstrated to function as biomarkers
in the diagnosis of these diseases (25). However, the potential molecular
mechanism of tRFs in the occurrence and progression of VVs remains
poorly understood.
In the present study, the key tRFs and their
potential molecular mechanisms in VVs were investigated, the
differentially expressed tRFs (DETs) in VVs were identified using
small RNA sequencing (RNA-seq), and the potential functional roles
of these tRFs were further analyzed using bioinformatic tools.
Materials and methods
Patients and samples
This study was approved by the Ethics Committee of
Shanghai East Hospital (Shanghai, China) and all patients enrolled
in the present study provided written informed consent. All
experiments were performed following the Code of Ethics of the
World Medical Association. Primary VVs were obtained from 24
patients who underwent open surgery or endovenous laser treatment.
Phlebectomy was performed to obtain vascular tissue exhibiting
signs of VVs and matched adjacent normal vascular tissue (ANV) from
patients undergoing surgery at Shanghai East Hospital between July
2017 and January 2018. The clinical information of patients with
VVs is presented in Table I. The
vascular tissues of patients with VV and the matched ANVs were
stored at −80°C until experimental use.
 | Table I.Clinicopathological characteristics
of patients with varicose veins. |
Table I.
Clinicopathological characteristics
of patients with varicose veins.
| Characteristic | Patients
(n=24) |
|---|
| Age (years) | 64.2±9.9 |
| Female | 13 |
| Male | 11 |
| Hypertension | 13 |
| Diabetes
Mellitus | 1 |
| Left limb
operation | 13 |
| Right limb
operation | 11 |
| CEAP (grade) | 3.7±0.7 |
Library preparation and RNA-seq of
small RNA
Samples used for sequencing were selected randomly.
Total RNA from VVs and matched ANVs (N=4) was extracted from the
vascular tissue. Small RNAs (135–170 bp) were isolated using a
QIAquick gel extraction kit (Qiagen, Inc.). A miRNA library was
constructed by Ying Biotech from the isolated purified small RNA
using NEBNext® Multiplex Small RNA Library Prep kit
(cat. no. E7560S; New England Biolabs, Inc.), followed by analysis
of the small RNA-seq data. Briefly, miRNA was bound with 3′ and
5′-adapters, and cDNA constructs were created by RT-PCR. The miRNA
library was submitted for deep sequencing on Illumina HiSeq™ 2500
platform (Illumina Inc.).
Bioinformatics analysis
The raw sequencing data was used to filter low
quality and short reads (<15 nucleotides). To identify known
small RNAs, all small RNA clean reads were aligned to the miRBase
database (http://www.mirbase.org). Unmapped reads
with miRBase database were aligned to Genomic tRNA database
(http://gtrnadb.ucsc.edu) prior to alignment to
tRFdb (http://genome.bioch.virginia.edu/trfdb) and MintBase
(http://cm.jefferson.edu/MINTbase/).
Eventually, tRFs were obtained by mapping these databases. Unmapped
reads from the Genomic tRNA database were aligned to other
corresponding databases of small RNAs (piRNA, small nuclear RNA,
small nucleolar RNA and other small RNAs). The tRFs with
significantly different expression levels between tissues of
patients with VVs and matched ANVs were selected based on
statistical values of log2 fold change
(log2FC) >1 or <-1 and false discovery rate (FDR)
<0.05 using EBSeq (https://www.biostat.wisc.edu/~kendzior/EBSEQ/) based
on negative binomial distributions. mRNAs and target genes of the
differently expressed tRFs were predicted using miRanda (http://www.microrna.org/microrna/home.do) and
RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid).
The intersection of miRanda (score >150; energy <-20) and
RNAhybrid (energy <-25) was revealed to be significant. The
functions and pathways of the majority of the target genes were
classified using the Gene Ontology (GO; http://www.geneontology.org/), and Kyoto Encyclopedia
of Genes and Genomes (KEGG; http://www.genome.jp/kegg) databases. GO terms were
classified as biological processes (BP), molecular functions (MF)
and cell components (CC). In addition, a network between tRFs,
target genes and pathways was performed using cytoscape 3.6.1
(https://cytoscape.org) (26).
Log2(FC) was used to represent the signal
difference between the experimental group and the control group
calculated by the logarithm to base 2, and FDR was used to
determine the multiple hypothetical testing error misjudgment
rates. Lower FDR represented fewer errors.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from vascular tissues
exhibiting VVs and the matched ANVs of 20 patients using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. Quality and
concentration of RNA was assessed using NanoDrop ND1000
spectrophotometer (Thermo Fisher Scientific, Inc.). Isolated RNA
was reverse transcribed into cDNA using RevertAid™ Fist Strand cDNA
Synthesis kit (cat. no. K1622; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. qPCR was performed using
SYBR Green PCR kit (Qiagen, Inc.) and detected on an ABI Q6
detection system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The thermocycling conditions were as follow: 95°C for 10 min
followed by 45 cycles of 95°C for 15 sec, 60°C for 1 min; the
melting conditions were 95°C for 10 sec, 60°C for 1 min, 95°C for
15 sec for one cycle. U6 was used as the internal control, and
relative quantification and calculations were done using the
2−ΔΔCq method (27).
Primers were designed by Ying Biotech. The primer sequences are
listed in Table II.
 | Table II.Primer sequences. |
Table II.
Primer sequences.
| Name | Primer
sequences |
|---|
| U6-F |
5′-CGATACAGAGAAGATTAGCATGGC-3′ |
| U6-R |
5′-AACGCTTCACGAATTTGCGT-3′ |
|
tRF-36-F900BY4D84KRIMEF |
5′-ACATGGTCTAGCGGTTAGGATTC-3′ |
|
tRF-36-F900BY4D84KRIMER |
5′-AGTGCGTGTCGTGGAGTCG-3′ |
|
tRF-23-87R8WP9IYF |
5′-CGCAGTCCCTGGTGGTCTAGT-3′ |
|
tRF-23-87R8WP9IYR |
5′-AGTGCGTGTCGTGGAGTCG-3′ |
|
tRF-40-86J8WPMN1E8Y7Z2RF |
5′-AGCTCACAAGAACTGCTAACTCATG-3′ |
|
tRF-40-86J8WPMN1E8Y7Z2RR |
5′-AGTGCGTGTCGTGGAGTCG-3′ |
Statistical analysis
Statistical analysis was performed using SPSS
version 12.0 statistical software (SPSS, Inc.) and the data were
visualized using GraphPad Prism 5.0 (GraphPad Software, Inc.). The
RT-qPCR data were presented as the mean ± standard deviation and
analyzed using paired Student's t-test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Conventional characteristics of
tRFs
To study the potential function of tRFs in VVs,
small RNA-seq was performed on the resected VV and matched ANV
tissues from patients with VVs (N=4). To further elucidate the
common characteristics of tRFs in human VVs, a detailed preliminary
analysis of the sequencing data was performed. The results
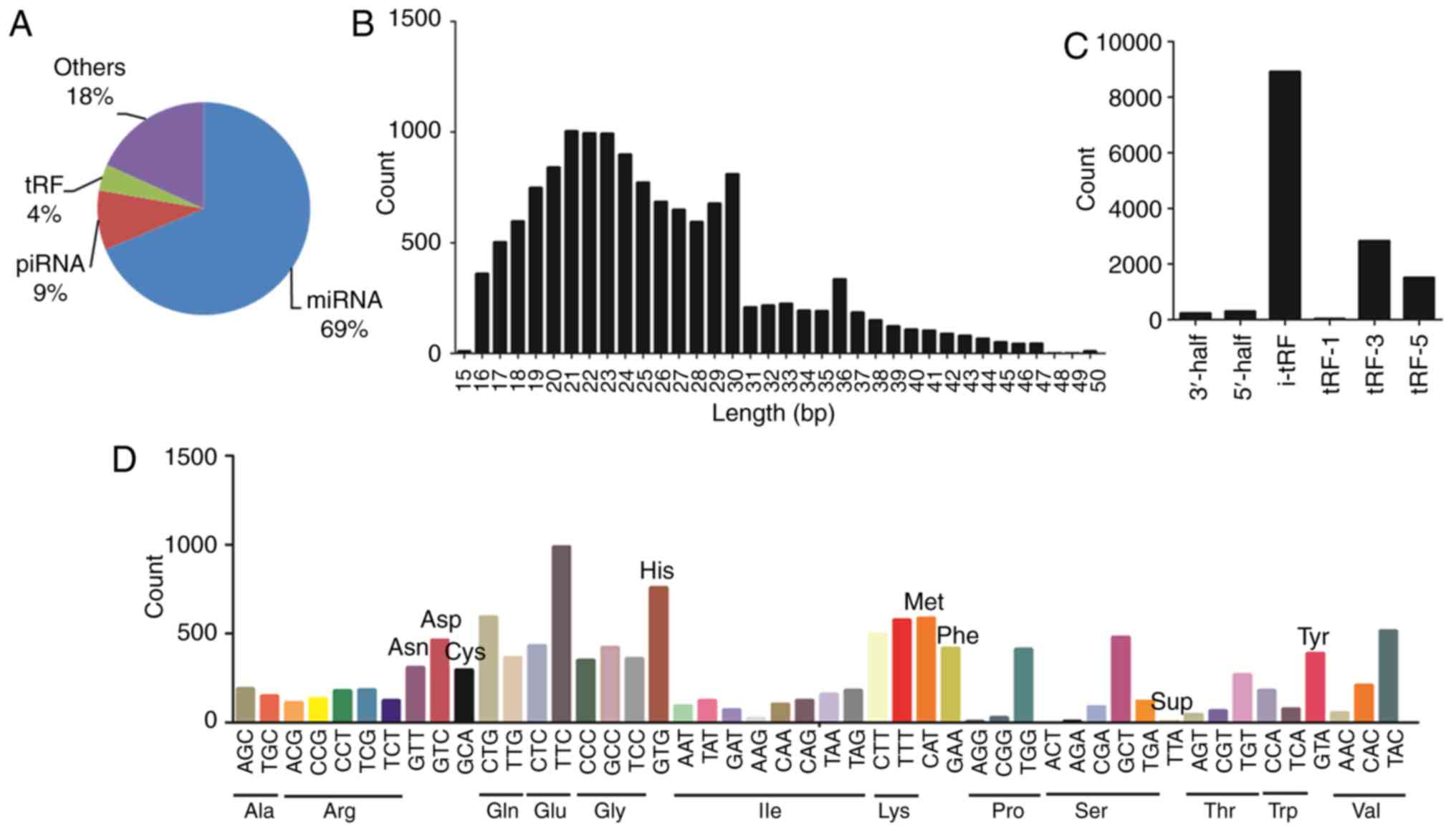
demonstrated that tRFs accounted for 4% of the total small RNAs
(Fig. 1A). The length of tRFs was
observed to be 15–50 nucleotides (Fig.
1B). A total of 13,789 tRFs were identified, which comprised
the following subtypes: 3′-half (N=226), 5′-half (N=293), internal
tRF (i-tRF; N=8,908), tRF-1 (tRFs derived from the 3′trailer
fragment of precursor tRNAs; N=29), tRF-3 (tRFs derived from the
extreme 3′ends of mature tRNAs; N=2,831) and tRF-5 (tRFs from the
extreme 5′ends of mature tRNAs; N=1,502), as presented in Fig. 1C. The analysis revealed that most
tRFs subtypes were transcribed from i-tRF. Fig. 1D shows the that the tRFs derived
from tRNA(Glu) were the most abundant, followed by that from
tRNA(Gly) and tRNA(Lys).
Identification of DETs in VVs
To increase the reliability of differential
expression detected from the expression profile between VV and
matched ANV tissues, tRFs with low signal intensities or those
which were not consistently expressed in all the tissues were
filtered out. From the remaining signals, tRFs that exhibited at
least a 2-fold expression change and P<0.05 were considered
differentially expressed and were selected for further analysis.
Overall, the small RNA-seq analysis revealed 45 DETs in VVs
compared with the matched ANV tissues (Fig. 2A). Of these, 14 tRFs were
identified to be significantly upregulated and 31 tRFs were
significantly downregulated (Fig.
2B). A detailed list of the 45 DETs identified in the present
study is presented in Table III.
The results suggested that the expression pattern of tRFs in VVs
was distinctly different compared with that in ANVs.
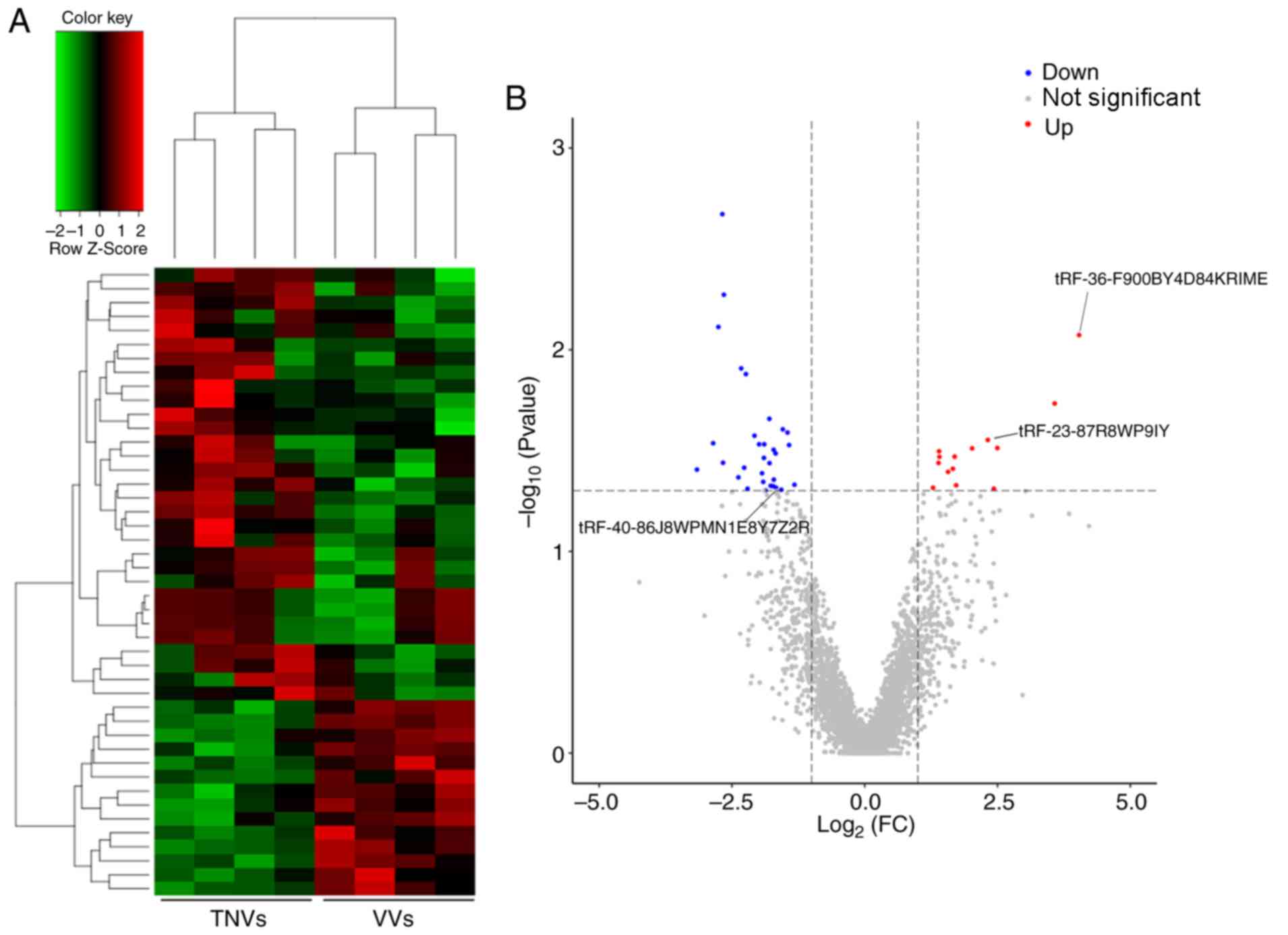 | Figure 2.Comparison of DETs between healthy
control and VV tissues. (A) Hierarchical cluster analysis of the 45
identified DETs. Red, high relative expression; black, medium
relative expression; green, low relative expression. (B) Volcano
plot for the tRFs from the RNA-seq analysis. Red, significantly
upregulated tRFs; blue, significantly downregulated tRFs; gray, no
significant change. DETs, differentially expressed tRFs; VV,
varicose vein; ANV, adjacent normal vein; tRFs, tRNA-derived
fragments. |
 | Table III.Top 10 downregulated and upregulated
tRNA fragments. |
Table III.
Top 10 downregulated and upregulated
tRNA fragments.
| A, Top 10
downregulated tRFs |
|---|
|
|---|
| Acc ID | Subtype | Anticodons | Length (bp) | FC | P-value |
|---|
|
tRF-37-86J8WPMN1E8Y7Z2 | tRF-5 | GluTTC | 37 | −1.723 | 0.048 |
|
tRF-36-MIF91SS2P4FINOE | i-tRF | LysCTT | 36 | −1.715 | 0.031 |
|
tRF-35-86J8WPMN1E8Y7Z | 5′-half | GluTTC | 35 | −1.712 | 0.044 |
|
tRF-37-MIF91SS2P4FINO5 | i-tRF | LysCTT | 37 | −1.678 | 0.033 |
|
tRF-40-86J8WPMN1E8Y7Z2R | tRF-5 | GluTTC | 40 | −1.669 | 0.048 |
|
tRF-34-PS5P4PW3FJHPEQ | 5′-half | LysTTT | 34 | −1.571 | 0.049 |
|
tRF-38-HPDE8P7SER9I69DV | tRF-3 | ArgACG | 38 | −1.542 | 0.025 |
|
tRF-18-RPM83004 | tRF-5 | LeuTAG, LeuAAG | 18 | −1.454 | 0.026 |
| tRF-17-RPM830P | tRF-5 | LeuTAG, LeuAAG | 17 | −1.424 | 0.030 |
| tRF-17-RPM830K | tRF-5 | LeuTAG, LeuAAG | 17 | −1.323 | 0.047 |
|
| B, Top 10
upregulated tRFs |
|
| Acc ID | Subtype |
Anticodons | Length
(bp) | FC | P-value |
|
|
tRF-21-2MK8J7K1B | tRF-3 | AlaTGC, AlaCGC | 21 | 1.407 | 0.040 |
|
tRF-31-ROD8N0X0JYOYE | 5′-half | TyrGTA | 31 | 1.570 | 0.039 |
|
tRF-18-PNR8YP04 | tRF-5 | GlyCCC, GlyGCC | 18 | 1.659 | 0.034 |
|
tRF-23-87R8WP9IY | tRF-5 | GluTTC | 23 | 1.693 | 0.047 |
|
tRF-33-LBINRXURYRSIV | tRF-3 | MetCAT | 33 | 1.722 | 0.031 |
|
tRF-41-Z4D7OOJ0QXL13XZ5D | i-tRF | HisGTG | 41 | 2.021 | 0.028 |
|
tRF-25-690M0RHB26 | tRF-3 | PheGAA | 25 | 2.317 | 0.049 |
|
tRF-30-DKNPNOYB7JXN | 5′-half | AlaTGC | 30 | 2.431 | 0.031 |
|
tRF-30-PW5SVP9N15WV | tRF-5 | HisGTG | 30 | 2.497 | 0.018 |
|
tRF-36-F900BY4D84KRIME | i-tRF | SerGCT | 36 | 3.575 | 0.008 |
GO and KEGG pathway enrichment
analysis of target genes of DETs
tRFs regulate gene expression and degradation of
target RNAs, such as mRNA and miRNA (18–20).
To further explore the potential functions of tRFs in VVs, the
target genes of DETs were theoretically predicted using miRanda and
RNAhybrid. The results demonstrated that 8,114 genes were
associated with the 45 DETs in VVs, which involve 22,405 binding
sites (Fig. 3A). To further
understand the biological functions of these target genes, GO and
KEGG pathway enrichment analysis was performed. In the GO analysis,
the enrichment score was calculated as -log10 (P-value)
(P<0.05). A total of 656 GO terms were enriched in GO-BP. The
dataset included BPs such as ‘epidermal growth factor receptor
signaling pathway’ (GO: 0007173; P=2.975×10−9) and
‘vascular endothelial growth factor (VEGF) receptor signaling
pathway’ (GO: 0048010; P=5.593×10−9), which are
associated with vascular disease (Fig.
3B) (28,29). In GO-MF, 208 terms were
significantly enriched, which included ‘protein binding’ (GO:
0005515; P=1.848×10−21) and ‘transcription factor
binding’ (GO: 0008134; P=2.454×10−6; Fig. 3C); in GO-CC, 131 terms were
significantly enriched, including ‘cytoplasm’ (GO: 0005737;
P=1.61×10−16) and ‘nucleus’ (GO: 0005634;
P=1.083×10−10; Fig.
3D). KEGG pathway enrichment analysis revealed that these
target genes were significantly enriched in 6,401 signaling
pathways, such as ‘Wnt signaling pathway’ (PATH: 04310;
P=4.31×10−10), ‘calcium signaling pathway’ (PATH: 04020;
P=5.36×10−10) and ‘MAPK signaling pathway’ (PATH: 04010;
P=8.395×10−8; Fig. 3E).
These pathways were also associated with vascular disease. Thus,
the results suggested that tRFs may serve a role in VVs by
regulating Wnt, calcium and MAPK signaling pathways.
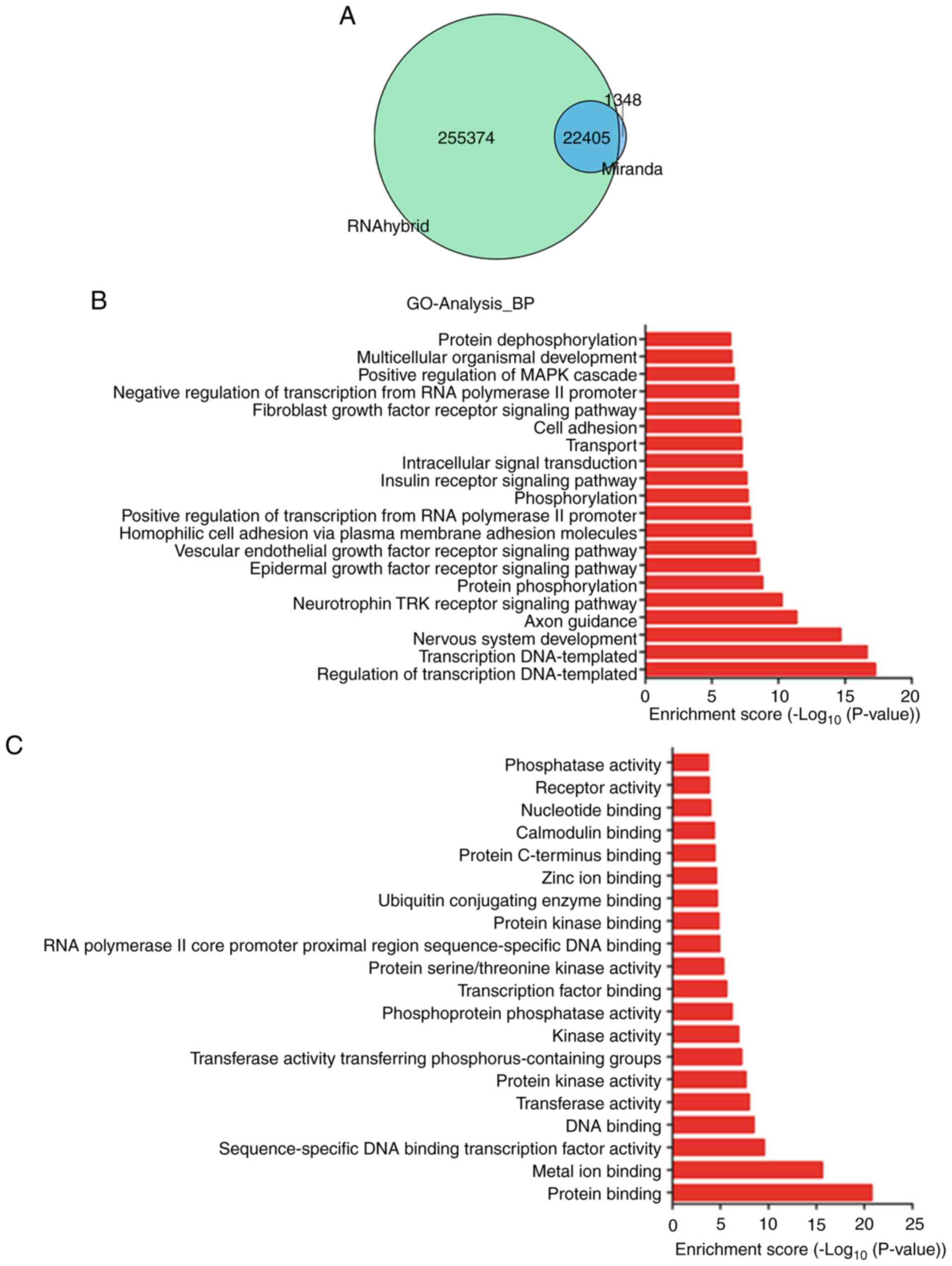 | Figure 3.GO term and KEGG pathway analysis of
target mRNAs. (A) The number of the predicted mRNA-target genes of
the differentially expressed tRFs. Blue, the number of target mRNAs
in the miRanda database; Green, the number of target mRNAs in the
RNAhybrid database. There were 22,405 shared target mRNAs in
miRanda and RNAhybrid. (B) Top 20 GO terms from the target gene
enrichment analysis for BP. (C) Top 20 GO terms from the target
gene enrichment analysis for MF. The circle size represents the
number of genes involved in the pathway. Green, low relative
expression; red, high relative expression. GO, Gene ontology; KEGG,
Kyoto Encyclopedia of Genes and Genomes; tRFs, tRNA-derived
fragments; BP, biological process; MF, molecular function; CC,
cellular component. (D) Top 20 GO terms from the target gene
enrichment analysis for CC. The circle size represents the number
of genes involved in the pathway. Green, low relative expression;
red, high relative expression. GO, Gene ontology; KEGG, Kyoto
Encyclopedia of Genes and Genomes; tRFs, tRNA-derived fragments;
BP, biological process; MF, molecular function; CC, cellular
component. (E) Top 20 terms from KEGG pathway enrichment analysis
of the target genes of the differentially expressed tRFs. The
circle size represents the number of genes involved in the pathway.
Green, low relative expression; red, high relative expression. GO,
Gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; tRFs,
tRNA-derived fragments; BP, biological process; MF, molecular
function; CC, cellular component. |
Experimental validation of tRF
expression using RT-qPCR
A precise list of the DETs identified in the present
study based on their comprehensive raw signal intensities, and the
final FC >2 and P<0.05 criteria, is provided in Table III. Based on their high abundance
and fold changes, three tRFs were shortlisted, which comprised two
upregulated tRFs (tRF-36-F900BY4D84KRIME and tRF-23-87R8WP9IY) and
one downregulated tRF (tRF-40-86J8WPMN1E8Y7Z2R). To independently
confirm the differential expression of these tRFs between VV and
matched ANV samples, RT-qPCR was performed to determine their
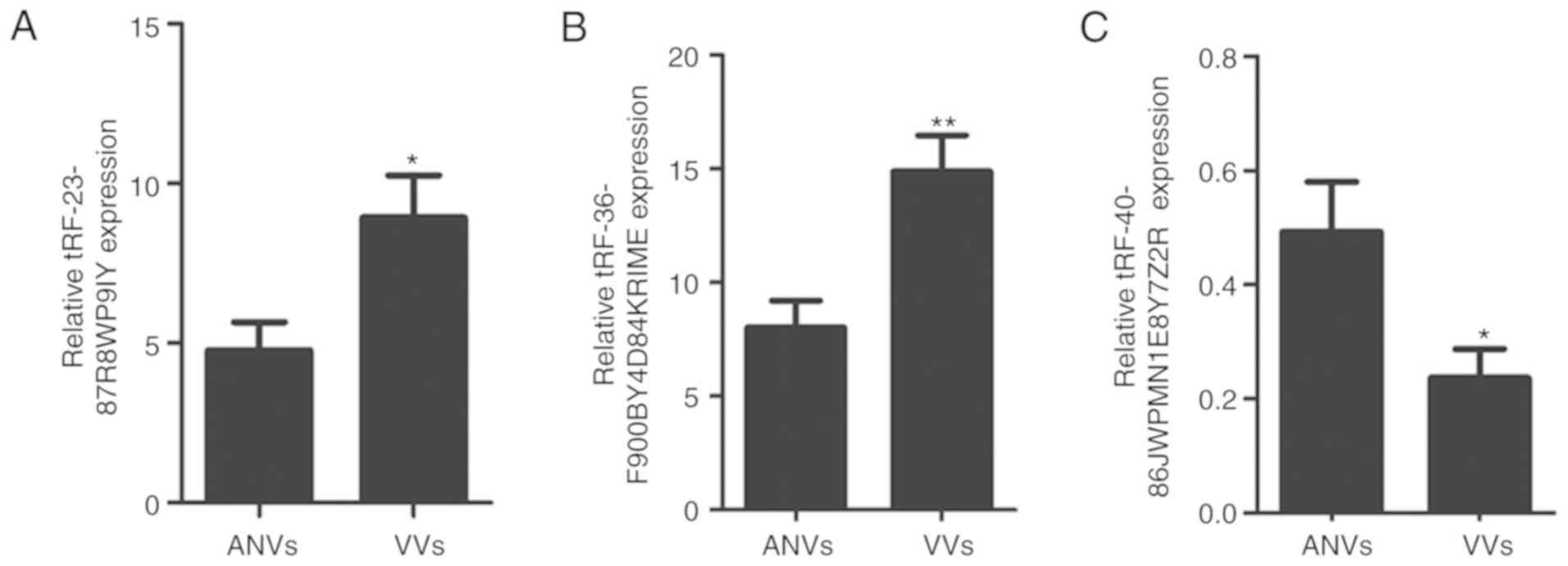
expression levels (N=20). The results revealed that in VV tissues,
tRF-23-87R8WP9IY and tRF-36-F900BY4D84KRIME were significantly
upregulated (P=0.001 and P=0.013, respectively), while
tRF-40-86J8WPMN1E8Y7Z2R was significantly downregulated (P=0.016)
compared with matched ANV tissues (Fig. 4). The results were consistent with
those of the small RNA-seq analysis, confirming the overall
validity of the RNA-seq data, and highlighting the potential gene
regulatory roles of the three DETs in VVs.
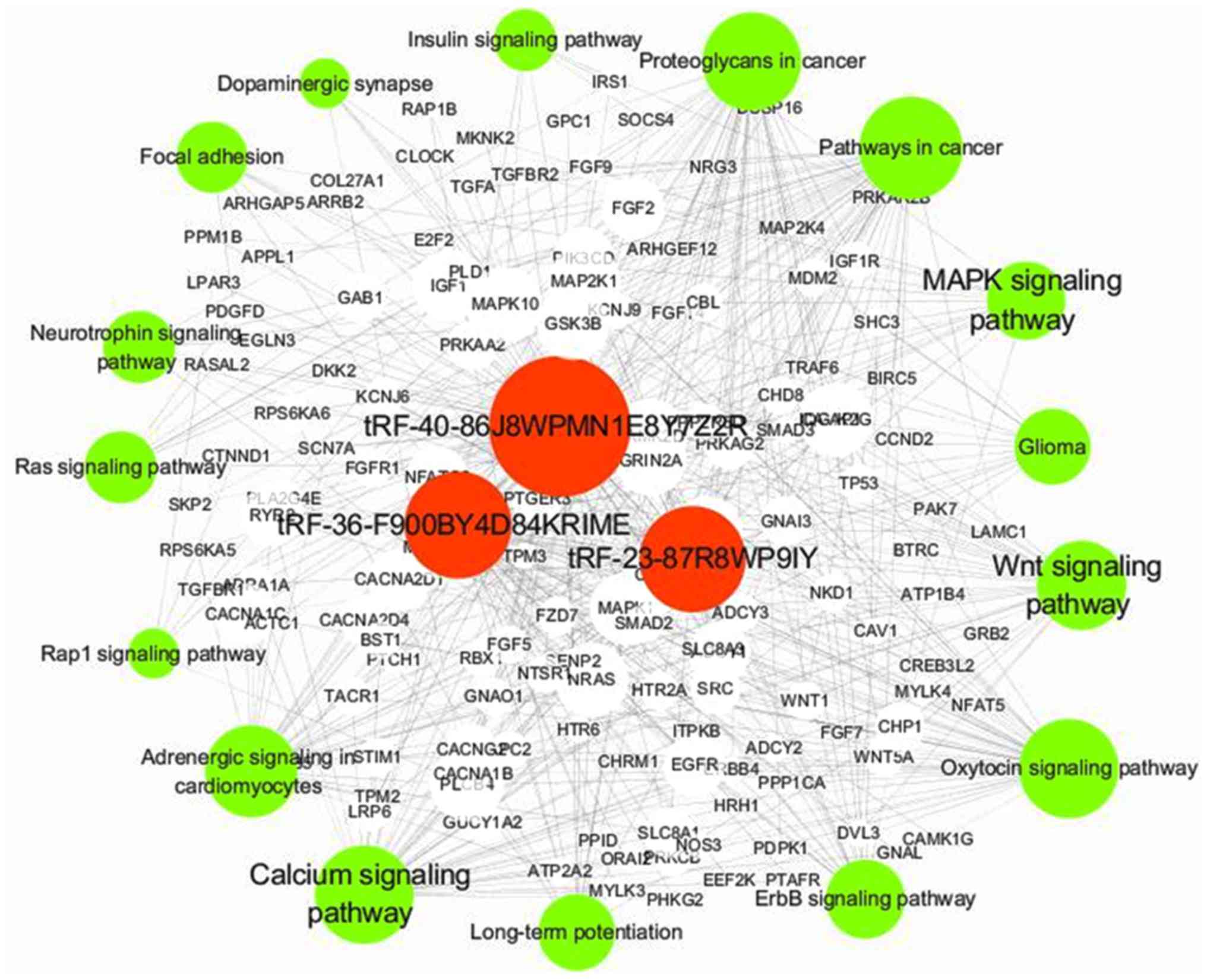
Signaling pathway regulation network
of DETs
To further understand the regulatory mechanism of
the three DETs in VVs, a regulation network of the target genes of
the shortlisted tRFs was constructed (Fig. 5). The network was integrated and
analyzed using Cytoscape. The network demonstrated that the target
genes of the three tRFs may be involved in VVs by regulating Wnt,
calcium and MAPK signaling pathways, which are associated with
vascular diseases (30–32). Moreover, stearoyl-CoA desaturase
(SCD), FK506 binding protein 5 (FKBP5) and toll-like receptor 6,
which have been shown to be involved in inflammation (33–36),
were regulated by the differentially expressed tRFs
(tRF-36-F900BY4D84KRIME, tRF-23-87R8WP9IY and
tRF-40-86J8WPMN1E8Y7Z2R; Table
SI).
Thus, tRF-36-F900BY4D84KRIME, tRF-23-87R8WP9IY and
tRF-40-86J8WPMN1E8Y7Z2R may serve important roles in the molecular
mechanism of VVs. These findings may help elucidate the potential
function of tRFs in the occurrence and progression of VVs.
Discussion
In the present study, 45 DETs, which included 14
upregulated and 31 downregulated tRFs, were identified to be
significantly altered in VVs compared with in ANVs. The target
genes of the DETs were significantly enriched in Wnt, calcium and
MAPK signaling pathways. In addition, two upregulated tRFs and one
downregulated tRF were shortlisted, which may regulate Wnt, calcium
and MAPK signaling pathways. The results of the present study
suggest that tRFs may serve significant roles in the occurrence and
progression of VVs through the regulation of Wnt, calcium and MAPK
signaling pathways.
Several studies have reported that Wnt inhibitory
factor 1 can inhibit angiogenesis of human umbilical vein
endothelial cells (HUVECs) under hypoxic conditions (37), and activation of the Wnt/β-catenin
pathway can enhance VEGF-dependent angiogenesis, thus promoting the
stability of VEGF-induced new blood vessels in vivo
(30,38). In addition, Krüppel-like factor
2/Wnt family member 9b (Klf2/Wnt9b) signaling protects endothelial
cells that are sheared by fluid forces and thereby acts directly on
the complex cellular processes of heart valve development (39). Previous studies have reported that
calcium-independent signaling is responsible for the
endothelium-dependent vasodilating activity in HUVECs (40). Tumor-associated calcium signal
transducer 2 regulates neovascularization of non-small-cell lung
cancer by activating the ERK1/2 signaling pathway (31). In addition, novel calcitonin
gene-related peptide/chitosan-strontium-calcium phosphate cement
enhances the proliferation of HUVECs (41). Furthermore, increasing
extracellular Ca2+ significantly triggers TNF-α-induced
vascular cell adhesion molecule 1 activation and monocyte adhesion
in HUVECs (42). Several reports
have revealed that the p38 MAPK inhibitor (CBS3830) restricts
vascular re-modeling in arterialized vein grafts (35). Suppressing the MAPK/NF-κB signaling
pathway protects HUVECs from lipopolysaccharide-induced oxidative
stress and inflammation (32).
Advanced glycation end products (AGE) impair the functions of
saphenous vein through the AGE receptor/MAPK signaling pathway in
diabetes (43). Studies have also
demonstrated that the activation of the MAPK/ERK pathway can
enhance the proliferation and migration of HUVECs (44), whereas disrupting MAPK signaling
pathway inhibits angiogenesis in HUVECs (45). In addition, CBS3830 restricts
vascular remodeling in arterialized vein grafts (35). These reports suggest that Wnt,
calcium and MAPK signaling pathways are associated with vascular
disease. In the present study, the signaling pathway regulation
network revealed that the target genes of the three identified tRFs
(tRF-36-F900BY4D84KRIME, tRF-23-87R8WP9IY and
tRF-40-86J8WPMN1E8Y7Z2R) were associated with the three signaling
pathways. This suggests that alteration in the expression of the
three shortlisted tRFs may facilitate the development of novel
strategies to manipulate the occurrence and progression of VVs.
Inflammation has been shown to play an important
role in VVs. For instance, polymorphisms of genes associated with
inflammation can influence the risk of VVs (46). Lattimer et al (47) found that the concentrations of
IL-6, IL-8 and MCP-1 in blood from the site of VVs was increased,
which indicated that there may be inflammation in the tissues
drained via the VVs. In the present study, several inflammatory
factors, including SCD, FKBP5 and ILR6 were found to be regulated
by tRF-36-F900BY4D84KRIME, tRF-23-87R8WP9IY and
tRF-40-86J8WPMN1E8Y7Z2R, respectively. The high expression of SCD
is associated with metabolic diseases, including obesity and
insulin resistance, however, SCD is also involved in regulating
inflammation and stress in different cell types, such as β-cells,
fat cells, macrophages, endothelial cells and myocytes (33,34).
Laminar shear stress plays an important role in vascular
homeostasis by increasing the expression of SCD1 in vascular
endothelial cells through peroxisome proliferator-activated
receptor-γ (48). FKBP5 is an
FK506 binding protein; FK506 binding proteins are part of the
immunophilins family that can bind to immunosuppressive drugs,
including rapamycin and FK506 (49). ILR6 is the receptor for IL-6, which
is involved in inflammatory reactions (50). The ectopic upregulation of IL6R has
been shown to trigger vascular remodeling by regulating
inflammatory cell infiltration (51). Therefore, tRFs may serve a
regulatory role in VVs by mediating genes involved in
inflammation.
In summary, 45 DETs were identified in VVs, which
included 14 upregulated and 31 downregulated tRFs compared with
their expression levels in ANVs. Alterations in the expression of
the three shortlisted tRFs, tRF-36-F900BY4D84KRIME,
tRF-23-87R8WP9IY and tRF-40-86J8WPMN1E8Y7Z2R, may facilitate in the
development of novel strategies for VV treatment. Further studies
will be required to confirm whether tRFs participate in the
occurrence and development of VVs through the Wnt, calcium and MAPK
signaling pathways. The present study provides a new evidence basis
for further investigation of the functional roles of tRFs in
VVs.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the Pudong New Area
Health and Family Planning Commission Technology Plan (grant no.
2017B-7 to BZ), the Fundamental Research Funds for the Central
Universities (grant no. 1507219069 to BZ) and Tongji University
Outstanding HR Plan (grant no. 22120180028 to CY). The funding
bodies had no role in study design, data collection and analysis,
decision to publish, or preparation of the manuscript.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
CY, XW, YH, GC, JG and BZ conceived and designed the
study. CY and XW performed the experiments. YH, GC, HC and JG
analyzed and interpreted the data. CY drafted the article. GC, JG
and BZ critically revised the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Shanghai East Hospital (Shanghai, China) and informed consent was
obtained from all patients enrolled. All procedures performed in
this study involving human participants were in accordance with the
ethical standards of the institutional research committee and with
the 1964 Helsinki Declaration and its later amendments or
comparable ethical standards.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
VVs
|
varicose veins
|
|
tRFs
|
tRNA-derived fragments
|
|
ANVs
|
matched adjacent normal vein
tissues
|
|
DETs
|
differentially expressed tRFs
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
References
|
1
|
Yetkin E, Ileri M and Waltenberger J:
Ecchymosis: A novel sign in patients with varicose veins. Clin
Hemorheol Microcirc. 68:413–419. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li X, Jiang XY, Ge J, Wang J, Chen GJ, Xu
L, Xie DY, Yuan TY, Zhang DS, Zhang H, et al: Aberrantly expressed
lncRNAs in primary varicose great saphenous veins. PLoS One.
9:e861562014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Labropoulos N, Tiongson J, Pryor L,
Tassiopoulos AK, Kang SS, Ashraf Mansour M and Baker WH: Definition
of venous reflux in lower-extremity veins. J Vasc Surg. 38:793–798.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lim CS, Kiriakidis S, Paleolog EM and
Davies AH: Increased activation of the hypoxia-inducible factor
pathway in varicose veins. J Vasc Surg. 55:1427–1439. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jacobs BN, Andraska EA, Obi AT and
Wakefield TW: Pathophysiology of varicose veins. J Vasc Surg Venous
Lymphat Disord. 5:460–467. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Raffetto JD and Khalil RA: Mechanisms of
varicose vein formation: Valve dysfunction and wall dilation.
Phlebology. 23:85–98. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li S, Xu Z and Sheng J: tRNA-Derived small
RNA: A novel regulatory small non-coding RNA. Genes (Basel).
9:E2462018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Anderson P and Ivanov P: tRNA fragments in
human health and disease. FEBS Lett. 588:4297–4304. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Luo S, He F, Luo J, Dou S, Wang Y, Guo A
and Lu J: Drosophila tsRNAs preferentially suppress general
translation machinery via antisense pairing and participate in
cellular starvation response. Nucleic Acids Res. 46:5250–5268.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Goodarzi H, Liu X, Nguyen H C, Zhang S,
Fish L and Tavazoie S F: Endogenous tRNA-derived fragments suppress
breast cancer progression via YBX1 displacement. Cell. 161:790–802.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sobala A and Hutvagner G: Small RNAs
derived from the 5′end of tRNA can inhibit protein translation in
human cells. RNA Biol. 10:553–563. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ivanov P, Emara MM, Villen J, Gygi SP and
Anderson P: Angiogenin-induced tRNA fragments inhibit translation
initiation. Mol Cell. 43:613–623. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Couvillion MT, Bounova G, Purdom E, Speed
TP and Collins K: A Tetrahymena Piwi bound to mature tRNA
3′fragments activates the exonuclease Xrn2 for RNA processing in
the nucleus. Mol Cell. 48:509–520. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Martinez G, Choudury SG and Slotkin RK:
tRNA-derived small RNAs target transposable element transcripts.
Nucleic Acids Res. 45:5142–5152. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ruggero K, Guffanti A, Corradin A, Sharma
VK, De Bellis G, Corti G, Grassi A, Zanovello P, Bronte V, Ciminale
V and D'Agostino DM: Small noncoding RNAs in cells transformed by
human T-cell leukemia virus type 1: A role for a tRNA fragment as a
primer for reverse transcriptase. J Virol. 88:3612–3622. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Saikia M, Jobava R, Parisien M, Putnam A,
Krokowski D, Gao XH, Guan BJ, Yuan Y, Jankowsky E, Feng Z, et al:
Angiogenin-cleaved tRNA halves interact with cytochrome c,
protecting cells from apoptosis during osmotic stress. Mol Cell
Biol. 34:2450–2463. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Z, Xiang L, Shao J and Yuan Z: The
3′CCACCA sequence of tRNAAla(UGC) is the motif that is important in
inducing Th1-like immune response, and this motif can be recognized
by Toll-like receptor 3. Clin Vaccine Immunol. 13:733–739. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hafner M, Landthaler M, Burger L, Khorshid
M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC,
Munschauer M, et al: Transcriptome-wide identification of
RNA-binding protein and microRNA target sites by PAR-CLIP. Cell.
141:129–141. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Q, Li T, Xu K, Zhang W, Wang X, Quan
J, Jin W, Zhang M, Fan G, Wang MB and Shan W: The tRNA-Derived
small RNAs regulate gene expression through triggering
sequence-specific degradation of target transcripts in the oomycete
pathogen phytophthora sojae. Front Plant Sci. 7:19382016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Haussecker D, Huang Y, Lau A, Parameswaran
P, Fire AZ and Kay MA: Human tRNA-derived small RNAs in the global
regulation of RNA silencing. RNA. 16:673–695. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pekarsky Y, Balatti V, Palamarchuk A,
Rizzotto L, Veneziano D, Nigita G, Rassenti LZ, Pass HI, Kipps TJ,
Liu CG and Croce CM: Dysregulation of a family of short noncoding
RNAs, tsRNAs, in human cancer. Proc Natl Acad Sci USA.
113:5071–5076. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Balatti V, Pekarsky Y and Croce CM: Role
of the tRNA-derived small RNAs in Cancer: New potential biomarkers
and target for therapy. Adv Cancer Res. 135:173–187. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi
J, Feng GH, Peng H, Zhang X, Zhang Y, et al: Sperm tsRNAs
contribute to intergenerational inheritance of an acquired
metabolic disorder. Science. 351:397–400. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Garcia-Silva MR, Cabrera-Cabrera F, Güida
MC and Cayota A: Novel aspects of tRNA-derived small RNAs with
potential impact in infectious diseases. Adv Bioscience Biotechnol.
4:17–25. 2013. View Article : Google Scholar
|
|
25
|
Zheng LL, Xu WL, Liu S, Sun WJ, Li JH, Wu
J, Yang JH and Qu LH: tRF2Cancer: A web server to detect
tRNA-derived small RNA fragments (tRFs) and their expression in
multiple cancers. Nucleic Acids Res. 44:W185–W193. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Makki N, Thiel KW and Miller FJ Jr: The
epidermal growth factor receptor and its ligands in cardiovascular
disease. Int J Mol Sci. 14:20597–20613. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Olsson AK, Dimberg A, Kreuger J and
Claesson-Welsh L: VEGF receptor signalling-in control of vascular
function. Nat Rev Mol Cell Biol. 7:359–371. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen Y, Zhang Y, Deng Q, Shan N, Peng W,
Luo X, Zhang H, Baker PN, Tong C and Qi H: Inhibition of wnt
inhibitory factor 1 under hypoxic condition in human umbilical vein
endothelial cells promoted angiogenesis in vitro. Reprod Sci.
23:1348–1358. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gosgnach W, Boixel C, Névo N, Poiraud T
and Michel JB: Nebivolol induces calcium-independent signaling in
endothelial cells by a possible beta-adrenergic pathway. J
Cardiovasc Pharmacol. 38:191–199. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ge JJ, Zhao ZW, Zhou ZC, Wu S, Zhang R,
Pan FM and Abendroth DK: p38 MAPK inhibitor, CBS3830 limits
vascular remodelling in arterialised vein grafts. Heart Lung Circ.
22:751–758. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sampath H and Ntambi JM: The role of
stearoyl-CoA desaturase in obesity, insulin resistance, and
inflammation. Ann N Y Acad Sci. 1243:47–53. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu X, Strable MS and Ntambi JM: Stearoyl
CoA desaturase 1: Role in cellular inflammation and stress. Adv
Nutr. 2:15–22. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kim DW, Hwang HS, Kim DS, Sheen SH, Heo
DH, Hwang G, Kang SH, Kweon H, Jo YY, Kang SW, et al: Enhancement
of anti-inflammatory activity of PEP-1-FK506 binding protein by
silk fibroin peptide. J Microbiol Biotechnol. 22:494–500. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schöniger S, Böttcher D, Theuss T and
Schoon HA: Expression of Toll-like receptors 2, 4 and 6 in equine
endometrial epithelial cells: A comparative in situ and in vitro
study. Res Vet Sci. 112:34–41. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Staniszewska A, Onida S and Davies AH:
Compression therapy for uncomplicated varicose veins-Too little for
too much? Phlebology. 34:148–150. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Song ZY, Wang F, Cui SX and Qu XJ:
Knockdown of CXCR4 Inhibits CXCL-induced angiogenesis in HUVECs
through downregulation of the MAPK/ERK and PI3K/AKT and the
Wnt/β-catenin pathways. Cancerinvestigation. 36:10–18. 2018.
|
|
39
|
Birdsey GM, Shah AV, Dufton N, Reynolds
LE, Osuna Almagro L, Yang Y, Aspalter IM, Khan ST, Mason JC, Dejana
E, et al: The endothelial transcription factor ERG promotes
vascular stability and growth through Wnt/β-catenin signaling. Dev
Cell. 32:82–96. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Goddard LM, Duchemin AL, Ramalingan H, Wu
B, Chen M, Bamezai S, Yang J, Li L, Morley MP, Wang T, et al:
Hemodynamic forces sculpt developing heart valves through a
KLF2-WNT9B paracrine signaling axis. Dev Cell. 43:274–289.e5. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Guo X, Zhu X, Zhao L, Li X, Cheng D and
Feng K: Tumor-associated calcium signal transducer 2 regulates
neovascularization of non-small-cell lung cancer via activating
ERK1/2 signaling pathway. Tumour Biol. 39:10104283176943242017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lv T, Liang W, Li L, Cui X, Wei X, Pan H
and Li B: Novel calcitonin gene-related
peptide/chitosan-strontium-calcium phosphate cement: Enhanced
proliferation of human umbilical vein endothelial cells in vitro. J
Biomed Mater Res B Appl Biomater. 107:19–28. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li S, Ning H, Ye Y, Wei W, Guo R, Song Q,
Liu L, Liu Y, Na L, Niu Y, et al: Increasing extracellular
Ca2+ sensitizes TNF-alpha-induced vascular cell adhesion
molecule-1 (VCAM-1) via a TRPC1/ERK1/2/NFκB-dependent pathway in
human vascular endothelial cells. Biochim Biophys Acta Mol Cell
Res. 1864:1566–1577. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xuan H, Yuan W, Chang H, Liu M and Hu F:
Anti-inflammatory effects of Chinese propolis in
lipopolysaccharide-stimulated human umbilical vein endothelial
cells by suppressing autophagy and MAPK/NF-κB signaling pathway.
Inflammopharmacology. 27:561–571. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sun Y, Kang L, Li J, Liu H, Wang Y, Wang C
and Zou Y: Advanced glycation end products impair the functions of
saphenous vein but not thoracic artery smooth muscle cells through
RAGE/MAPK signalling pathway in diabetes. J Cell Mol Med.
20:1945–1955. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu YW, Zuo PY, Zha XN, Chen XL, Zhang R,
He XX and Liu CY: Octacosanol enhances the proliferation and
migration of human umbilical vein endothelial cells via activation
of the PI3K/Akt and MAPK/Erk pathways. Lipids. 50:241–251. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lattimer CR, Kalodiki E, Geroulakos G,
Hoppensteadt D and Fareed J: Are inflammatory biomarkers increased
in varicose vein blood? Clin Appl Thromb Hemost. 22:656–664. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Shadrina A, Tsepilov Y, Smetanina M,
Voronina E, Seliverstov E, Ilyukhin E, Kirienko A, Zolotukhin I and
Filipenko M: Polymorphisms of genes involved in inflammation and
blood vessel development influence the risk of varicose veins. Clin
Genet. 94:191–199. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Qin X, Tian J, Zhang P, Fan Y, Chen L,
Guan Y, Fu Y, Zhu Y, Chien S and Wang N: Laminar shear stress
up-regulates the expression of stearoyl-CoA desaturase-1 in
vascular endothelial cells. Cardiovasc Res. 74:506–514. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Van Snick J: Interleukin-6: An overview.
Annu Rev Immunol. 8:253–278. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tamura Y, Phan C, Tu L, Le Hiress M,
Thuillet R, Jutant EM, Fadel E, Savale L, Huertas A, Humbert M and
Guignabert C: Ectopic upregulation of membrane-bound IL6R drives
vascular remodeling in pulmonary arterial hypertension. J Clin
Invest. 128:1956–1970. 2018. View Article : Google Scholar : PubMed/NCBI
|