Introduction
Hepatitis B virus (HBV) infection exhibits a wide
spectrum of clinical liver manifestations (1). Primary human hepatocytes (PHHs) are
currently considered to be the ‘gold standard’ cell model for
studying HBV infection (2).
However, poor availability, variation between batches and ethical
issues regarding PHHs limit their application. Primary hepatocytes
from non-human primates, such as chimpanzees and monkeys, are
expensive, difficult to obtain and also associated with ethical
concerns (3). Primary
tupaia hepatocytes (PTHs) from the treeshrew genus Tupaia
belangeri are less expensive, but also difficult to obtain, and
HBV infection of PTHs is different compared with that of PHHs, as
the addition of dimethyl sulfoxide (DMSO) plus polyethylene glycol
8000 (PEG8000) during infection does not significantly promote HBV
infectivity (3).
The discovery of the HBV functional receptor human
sodium taurocholate co-transporting polypeptide (hNTCP) (4) has enabled the development of primary
cell models and immunocompetent animal models based on easily
obtainable laboratory animals. However, complementation of mouse
and rat hepatocytes with hNTCP does not result in susceptibility to
HBV infection (4–6), which indicates that additional host
factors may be needed for the post-entry HBV lifecycle in mouse and
rat hepatocytes. Therefore, other easily obtained laboratory
animals that do not pose the same ethical concerns as primates may
be a promising alternative solution for primary cell and animal
models of HBV infection.
In the presence of hNTCP, hepatocytes from primates
are susceptible to HBV infection, whereas mouse and rat hepatocytes
resist HBV infection (4–6). In the present study, it was presumed
that animals with short evolutionary distance to primates may
facilitate the HBV post-entry lifecycle. The generation of an
easily obtained primary cell model for HBV infection based on other
laboratory animals may be valuable for fundamental research on HBV
infection and antiviral drugs. To achieve this goal, evolutionary
distance was assessed based on single-copy homologous genes of
seven species. The hepatocytes from candidate animals (pigs and
rabbits) were then isolated, cultured and infected with
hNTCP-recombinant lentivirus, followed by HBV infection. Anti-HBV
drugs were also screened in the hNTCP-complemented hepatocytes.
Materials and methods
Cell isolation and culture
Ethical approval for the study was granted by the
Institutional Bioethics Committee of Shenzhen Second People's
Hospital, First Affiliated Hospital of Shenzhen University
(Shenzhen, China). All animal experiments were performed in
accordance with the Ministry of Health Guidelines for the Care and
Use of Laboratory Animals (no. GB 14925-2001) and the ARRIVE
guidelines.
Primary pig hepatocytes (PPHs) were isolated from
the liver tissues of Wuzhishan minipigs (n=3; estimated age, 8
weeks) provided by the Beijing Genomics Institute, China. Primary
rabbit hepatocytes (PRHs) were isolated from the liver tissues of
New Zealand rabbits (n=4; estimated age, 4 weeks) provided by
Guangdong Medical Animal Center, China. Animals were euthanized by
cutting the inferior vena cava under anaesthesia by intraperitoneal
injection of 30 mg/kg pentobarbital sodium (7). Death was confirmed by respiratory
arrest, cardiac arrest and reflex deficiency. The liver was removed
as quickly as possible, perfused using a two-step collagenase
perfusion procedure and plated as previously described (8,9).
Briefly, cell viability was 78–93%, as determined by trypan blue
staining and counting. Primary hepatocytes were seeded at a density
of 2×105 viable cells/cm2 in 12-well tissue
culture plates pre-coated with collagen (Gibco; Thermo Fisher
Scientific, Inc.) in dimethyl sulfoxide (DMSO)-free primary human
hepatocyte maintenance medium (PMM) (4) containing 5% v/v fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc.) for 4–6 h. Afterwards,
the medium was replaced with fresh PMM. For the PRH culture, PMM
containing 10% v/v FBS was essential during plating and
maintenance. The cells were maintained at 37°C in a 5%
CO2 incubator, and the medium was replaced every 2
days.
Huh7D cells (10)
were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS, 100 U/ml
penicillin and 100 µg/ml streptomycin maintained at 37°C in a 5%
CO2 incubator. Huh7D cells were induced using inducing
medium (DMEM supplemented with 5% FBS, 100 U/ml penicillin, 100
µg/ml streptomycin and 2.5% DMSO) for 24–72 h prior to HBV
infection.
HBV and lentivirus production
HepAD38 cells were kindly provided by Professor Chen
Xulin (Wuhan Institute of Virology, CAS), with the permission of
Professor Robert W. King (Institute for Cancer Research, Fox Chase
Cancer Center, Philadelphia, PA, USA) (11). HepAD38 cells were maintained in
Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS, 100 U/ml penicillin
and 100 µg/ml streptomycin maintained at 37°C in a 5%
CO2 incubator. For the production of HBV particles,
HepAD38 cells were maintained in Williams E medium supplemented
with 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin, 5
µg/ml insulin, 18 ng/ml hydrocortisone and 2–2.5% DMSO. The culture
media was collected at an interval of 3–4 days. Concentrated HBV
infectious particles were repaired as previously reported (12). Briefly, the supernatant was
centrifuged to remove dead cells and debris at 1,000 × g for 5 min
at 4°C. The supernatant was then filtered through a 0.45-µm filter
to remove any further debris. Subsequently, the supernatant was
loaded into an ultrafiltration device with an intercepted molecular
weight of 100 kDa (EMD Millipore) and centrifuged at 1,000 × g for
30–60 min. The final volume for ultrafiltration was 250–500 µl,
with a starting volume of 15 ml. The concentrated supernatant was
collected and stored at −80°C. The hNTCP-recombinant lentivirus was
produced as previously described (10). The concentrated lentivirus was
stored at −80°C until use in the experiments.
The pooled patient serum used for this study was
reported elsewhere (8,9), of which personal permissions were
obtained from HBV-infected patients. HBV particles purified from
patient serum were described previously (12). Briefly, Nycodenz (Takeda
Pharmaceutical Company, Ltd.) was dissolved in Hepato-STIM
Hepatocyte Defined Medium (BD Biosciences). Serum from patients
with HBV (viral load >108 copies/ml) was loaded onto
the Nycodenz gradient (range, 8–50%) in 11×34 mm polycarbonate
centrifuge tubes (Beckman Coulter, Inc.). The samples were
centrifuged in a TLS55 swing-out rotor (Beckman Coulter, Inc.) for
45 min at 200,000 × g at 20°C. Subsequently, 155 µl aliquots were
consecutively removed (fractions 1 to 8). Fractions 6 and 7 were
pooled with 8.4×108 HBV copies/ml, as measured by
quantitative PCR (qPCR).
HBV infection
PPHs and PRHs were seeded and maintained at 100%
confluence. Huh7D cells (10) were
seeded at 50% confluence and cultured to 80% confluence prior to
induction with 2.5% DMSO. Cells were infected with
hNTCP-recombinant lentivirus in the presence of 6 µg/ml polybrene
(Sigma-Aldrich; Merck KGaA). The medium was replaced at 12 h. To
ensure expression of hNTCP protein, cells were maintained for at
least 72 h prior to HBV infection. At 3 days post-lentivirus
infection, concentrated HBV stock solution diluted in PMM (10% v/v
FBS was added for PRHs) supplemented with 4% (w/v) PEG8000 was
added directly to the cells with a multiplicity of infection (MOI)
of 1,000. For HBV infection with purified patient serum, a MOI of
200 was used. Following incubation overnight, cells were washed
thoroughly three times with phosphate-buffered saline, and the
media were refreshed. Cells were maintained at 37°C in a 5%
CO2 incubator, and the medium was changed every 2
days.
For the competing infection assays, the cells were
pre-incubated with different concentrations of the HBV entry
inhibitor myr-preS12-47 at 37°C for 30 min in a 5%
CO2 incubator, and infectious medium containing
myr-preS12-47 was added. For the blocking infection
assays, infectious media were pre-incubated with different
concentrations of the HBV entry inhibitor 4B10 at 37°C for 30 min
in a 5% CO2 incubator, and the infectious medium
(containing 4B10) was added to the cells. For the inhibition of HBV
replication assays, different concentrations of Lamivudine were
incubated with cells post-HBV infection.
Cytotoxicity assay
For cytotoxicity of patients' pool serum, the pool
serum was incubated with hNTCP-expressing PPH or heat-inactivated
at 56°C for 30 min before incubation. Cell viabilities were
measured by Cell Counting Kit-8 from Yeasen Biotechnology
(Shanghai) Co., Ltd., according to the manufacturer's instructions.
For cytotoxicity of inhibitors, different concentrations of
inhibitors (myr-preS12-47, 4B10 and Lamivudine) were
incubated with hNTCP-expressing PPH for 48 h. Cell viabilities were
measured by Cell Counting Kit-8 according to the manufacturer's
instructions.
ELISA
Secreted hepatitis B surface antigen (HBsAg) and
hepatitis B e-antigen (HBeAg) were detected in medium collected
from HBV-infected cells using a HBsAg/HBeAg ELISA kit (Shanghai
Kehua Bio-engineering Co., Ltd), according to the manufacturer's
instructions. The results were assessed using a Multiskan MK3
microplate spectrophotometer (Thermo Fisher Scientific, Inc.). The
optical density (OD) values are presented as the mean ± standard
deviation of OD450-OD630. The cut-off value
was calculated according to the manufacturer's instructions.
PCR analysis
HBV covalently closed circular DNA (cccDNA) was
extracted as described by Köck et al (13). Briefly, ~2×106 cells in
a single well in a 6-well culture plate were lysed with 500 µl
NP-40 lysis buffer (50 mM Tris pH 8.0, 140 mM NaCl and 0.5% NP-40)
for 5–10 min until the cells detached from the well. Cells were
thoroughly rinsed several times with the NP-40 lysis buffer. The
lysate (nuclei and cell debris) was centrifuged for 5 min at 1,500
× g, and the supernatant was removed. A total of 500 µl NP-40 lysis
buffer, 5 µl ethylenediaminetetraacetic acid (EDTA; 0.5 M) and 5 µl
proteinase K (20 mg/ml; Tiangen Biotech Co., Ltd.) were added to
the nuclear pellet and incubated at 56°C for 2 h. The mixture was
loaded onto a QIAshredder column (Qiagen GmbH) and centrifuged at
room temperature for 1 min at 15,800 × g. The flow-through was
incubated at 56°C for 1 h, the DNA was purified by
phenol-chloroform extraction and ethanol precipitation, and the
pellet was solubilized in ddH2O. The genomic DNA, linear
viral DNA and relaxed circular DNA were digested for 8 h at 37°C by
plasmid-safe ATP-dependent deoxyribonuclease DNase (Epicentre;
Illumina, Inc.). Following inactivation at 70°C for 30 min, cccDNA
was detected with the primers ccc-1580 and ccc-2314 (Table I) (4,14). A
10-fold dilution series of plasmid pGEM-3Z-1.3×HBV (15) was used as the standard.
 | Table I.Primers used for PCR. |
Table I.
Primers used for PCR.
| Primer | Sequence
(5′-3′) | Thermocycling
conditions |
|---|
| ccc1580 | F:
TGCACTTCGCTTCACCT | 45 cycles: 95°C for
10 sec, 61°C for 20 sec, 72°C for 40 sec |
| ccc2314 | R:
AGGGGCATTTGGTGGTC |
|
| hNTCP | F:
CTCAAATCCAAACGGCCACAATAC | 40 cycles: 95°C for
30 sec, 60°C for 20 sec, 72°C for 30 sec |
|
| R:
CACACTGCACAAGAGAATGATGATC |
|
| HBVp | F:
ACCAATCGCCAGTCAGGAAG | 40 cycles: 95°C for
30 sec, 60°C for 30 sec, 72°C for 30 sec |
|
| R:
ACCAGCAGGGAAATACAGGC |
|
| β-actin | F:
ATCGTGCGTGACATTAAGGAG | 40 cycles: 95°C for
30 sec, 60°C for 20 sec, 72°C for 30 sec |
|
| R:
GGAAGGAAGGCTGGAAGAGT |
|
The hNTCP mRNA level was detected as described
previously (9). Briefly, total
intracellular RNA was extracted using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), and cDNA was
synthesized using the PrimeScript RT kit with gDNA Eraser (Takara
Biotechnology Co., Ltd.). The cDNA products were subjected to qPCR
with primers for hNTCP and β-actin (Table I). β-actin served as the internal
control for sample normalization. The relative hNTCP mRNA
expression levels were calculated using the 2−ΔΔCq
method (16) and the expression of
each group was calculated as relative to the levels of Huh7D cells.
All samples were measured in triplicate and all experiments were
repeated independently three times.
For quantification of HBV copies, HBV DNA was
extracted using QIAamp DNA Blood Mini kit (Qiagen GmbH), according
to the manufacturer's instructions. qPCR was performed using
FastStart Universal SYBR® Green Master Mix (Roche
Diagnostics). A pCH9-3091 (17)
plasmid (containing 1.05X HBV genomic DNA) was used as the
standard. The exact HBV copies were calculated with molecular
weight of plasmid, plasmid concentration and Avogadro constant. The
primer sequences and the thermocycling conditions for all qPCR
experiments are presented in Table
I.
Western blotting
Western blotting was performed as previously
described (8). The primary
antibodies used were rabbit anti-human hNTCP polyclonal antibody
(cat. no. HPA042727; Sigma-Aldrich; Merck KGaA; dilution 1:1,000),
and mouse anti-human β-actin monoclonal antibody (cat. no.
60008-1-Ig Proteintech Group, Inc.; dilution 1:1,000). The
secondary antibodies used were HRP-conjugated Affinipure Goat
Anti-Rabbit IgG(H+L) (cat. no. SA00001-2; Proteintech Group, Inc.;
dilution 1:5,000), HRP-conjugated Affinipure Goat Anti-Mouse
IgG(H+L) (cat. no. SA00001-1; Proteintech Group, Inc.; dilution
1:5,000).
Immunofluorescence assay
Immunofluorescence assay was performed as described
previously (8). The primary
antibody used was anti-rhodopsin antibody (cat. no. ab5417; Abcam;
dilution 1:1,000) rabbit anti-human hNTCP polyclonal antibody (cat.
no. HPA042727; Sigma-Aldrich; Merck KGaA; dilution 1:750). The
secondary antibody used was Alexa Fluor Plus 488-conjugated Goat
anti-Rabbit IgG (H+L) (cat. no. A32731; Thermo Fisher Scientific,
Inc.; dilution 1:1,000). The nuclei were stained with
4′,6-diamidino-2-phenylindole (DAPI; Roche Diagnostics; dilution
1:2,000), and the cells were examined under a fluorescence
microscope (Leica Microsystems GmbH), magnification, ×200. The
images were processed using Adobe Photoshop CS5 (Adobe Systems,
Inc.).
Divergence tree
A divergence tree was created based on the
single-copy homologous genes of the following species: Homo
sapiens (human), Macaca mulatta (rhesus macaque), Sus
scrofa (domestic pig), Tadarida brasiliensis (bat),
Oryctolagus cuniculus (rabbit), Mus musculus (mouse)
and Rattus norvegicus (rat) using the OrthoMCL v2.0
(http://orthomcl.org/orthomcl/), Muscle
(https://www.ebi.ac.uk/Tools/msa/muscle/), PHYML v3.0
(http://www.atgc-montpellier.fr/phyml/), and MCMCTree
v4.4 (http://abacus.gene.ucl.ac.uk/software/paml.html)
software.
Statistical analysis
The results are presented as the mean ± SD.
Statistical analysis was performed on GraphPad Prism 6 (GraphPad
Software, Inc.) using Student's t-test or one-way ANOVA followed by
Least Significant Difference post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
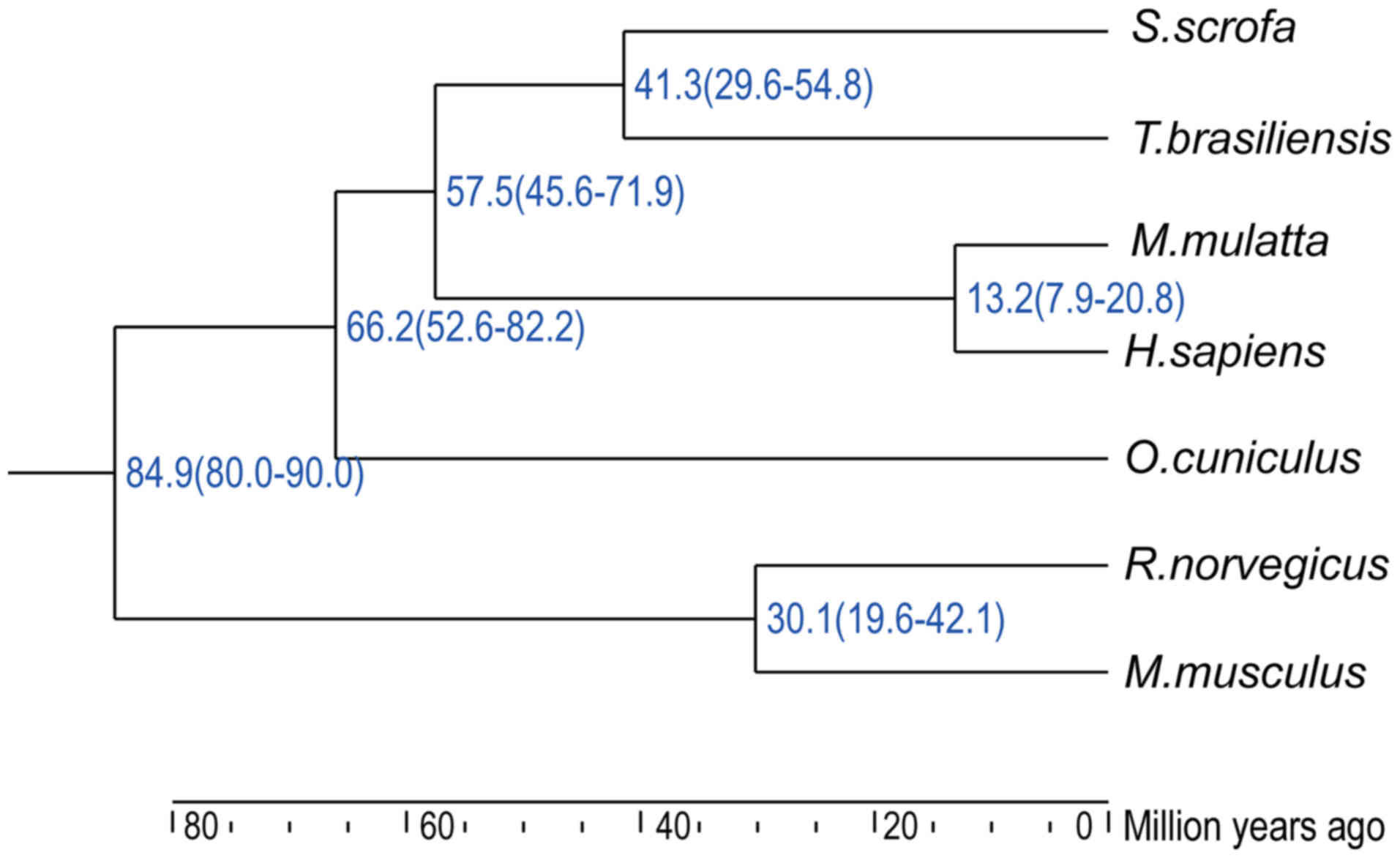
Evolutionary distance based on
single-copy homologous genes
The use of animals close to H. sapiens and
M. mulatta in the divergence tree for the isolation of
primary hepatocytes may facilitate the HBV post-entry lifecycle.
Based on the single-copy homologous genes of seven species
(18), a divergence tree was
generated (Fig. 1). The divergence
years were estimated to be in the following order: M.
musculus and R. norvegicus (84.9 million years) >
O. cuniculus (69.8 million years) > H. sapiens and
M. mulatta (59.9 million years) > S. scrofa and
T. brasiliensis. Primary hepatocytes isolated from H.
sapiens and M. mulatta fully support the HBV post-entry
lifecycle, whereas hepatocytes from M. musculus and R.
norvegicus are resistant (4–6). In
addition, bats, such as T. brasiliensis, carry pathogenic
hepadnaviruses that are antigenically related to HBV and are
capable of infecting human hepatocytes (19), indicating that T.
brasiliensis hepatocytes may also facilitate the HBV post-entry
lifecycle as hepadnaviruses are highly host selective. Notably,
hepadnavirus has been detected in S. scrofa (domestic pigs)
(20,21), and O. cuniculus (rabbits),
which may also be infected by an HBV-like virus (22). Therefore, easily obtained
laboratory animals, such as pigs and rabbits, may be suitable as
potential hosts for HBV infection after hNTCP complementation.
Culture and hNTCP complementation of
PPHs and PRHs
PPHs and PRHs were isolated by a two-step
collagenase digestion and plated on collagen-coated culture plates.
PPHs were maintained in PMM (4)
for >2 weeks and exhibited a typical highly differentiated
morphology, such as spherical bright nuclei and a polarized shape
(Fig. 2A). However, PRHs rapidly
deteriorated in PMM and died within 2 days (data not shown).
Following optimization, PRHs could be maintained in PMM
supplemented with 10% FBS for approximately 10–14 days and
exhibited a typical highly differentiated morphology (Fig. 2A). The difference in maintenance
medium between PPHs and PRHs indicated that PPHs may more closely
resemble PHHs compared with PRHs, which matched the evolutionary
distances identified in the divergence tree (Fig. 1).
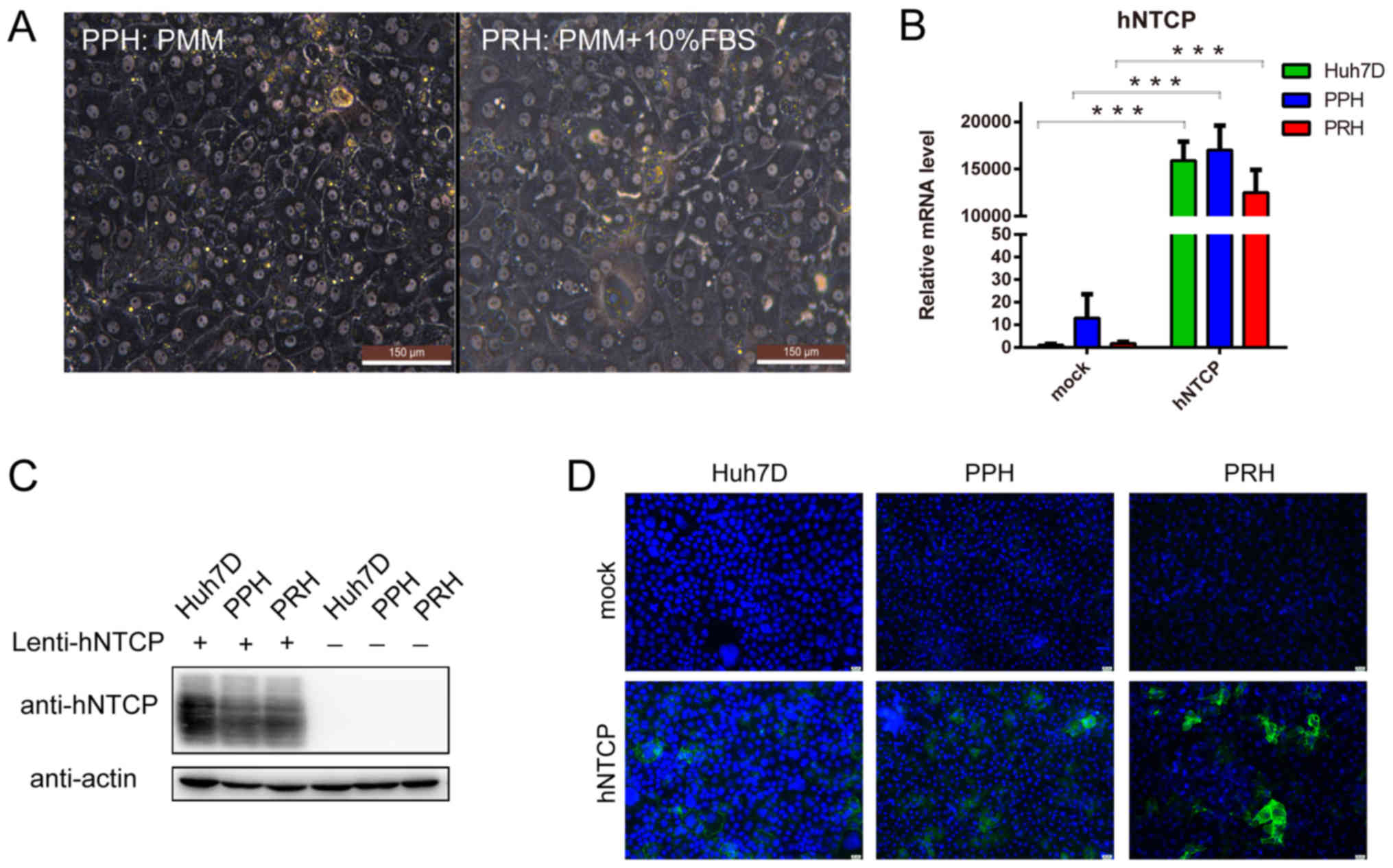 | Figure 2.Culture and hNTCP complementation of
PPHs and PRHs. (A) The morphology of PPHs and PRHs was assessed at
4 days post-seeding using a phase-contrast microscope. Scale bar,
150 µm. The maintenance medium is indicated. (B) The different cell
groups were subjected to reverse transcription-quantitative PCR to
determine the relative hNTCP mRNA expression levels. (C) Lysates of
the different cell groups were subjected to western blotting to
detect hNTCP protein expression levels. (D) At 4 days
post-lentivirus infection, the different cell types groups were
analyzed by immunofluorescence. Green, hNTCP staining; blue,
nucleus. Scale bar, 25 µm. ***P<0.001, with comparisons
indicated by lines. hNTCP, human sodium taurocholate
co-transporting polypeptide; PPH, primary pig hepatocyte; PRH,
primary rabbit hepatocyte; PMM, primary human hepatocyte
maintenance medium; FBS, fetal bovine serum. |
As a positive control for HBV infection, the human
cell line Huh7D exhibits increased susceptibility to HBV infection
in the presence of hNTCP compared with the HepG2 cell line
(10); therefore, the Huh7D cell
line was selected in the present study as a positive control for
subsequent experiments. PPHs and PRHs, as well as Huh7D cells, were
infected with the hNTCP-recombinant lentivirus. Cells were
harvested 4 days post-lentiviral infection to determine the hNTCP
mRNA expression levels by qPCR and protein expression levels by
western blotting, or were fixed to detect hNTCP expression and
location by immunofluorescence assay. hNTCP expression was only
observed in cells infected with hNTCP-recombinant lentivirus
(Fig. 2B and C). Although the same
MOI of the lentivirus (MOI=1) was used, the estimated percentage of
hNTCP-positive PPHs and Huh7D cells was 70–80%, whereas the
estimated percentage of hNTCP-positive PRHs was slightly lower at
50–60% (Fig. 2D). These
hNTCP-expressing cells were subsequently subjected to HBV
infection.
HBV infection of primary
hepatocytes
HBV infection was performed using concentrated viral
stocks prepared from HepAD38 cells (20). According to our preliminary data,
the hNTCP-complemented PPHs could be maintained for >2 weeks
post-HBV infection, whereas hNTCP-complemented PRHs could be
maintained for 1 week (data not shown). Therefore, the experimental
period was limited to 7 days post-infection (dpi). Following
infection with HBV (MOI=1,000), all cell types without hNTCP
exhibited either negative or reduced secretion of HBsAg/HBeAg,
indicating that these cells were resistant to HBV infection.
Following hNTCP complementation, Huh7D and PPHs, but not PRHs, were
positive for HBsAg/HBeAg secretion, with continuously increased
secretion of HBsAg and HBeAg until the end of culture (Fig. 3A and B). Of note, HBsAg and HBeAg
secretion in hNTCP-complemented PPHs was significantly higher
compared with the levels in hNTCP-complemented Huh7D cells,
although hNTCP mRNA and protein levels were similar in these cells
(Fig. 2B and C).
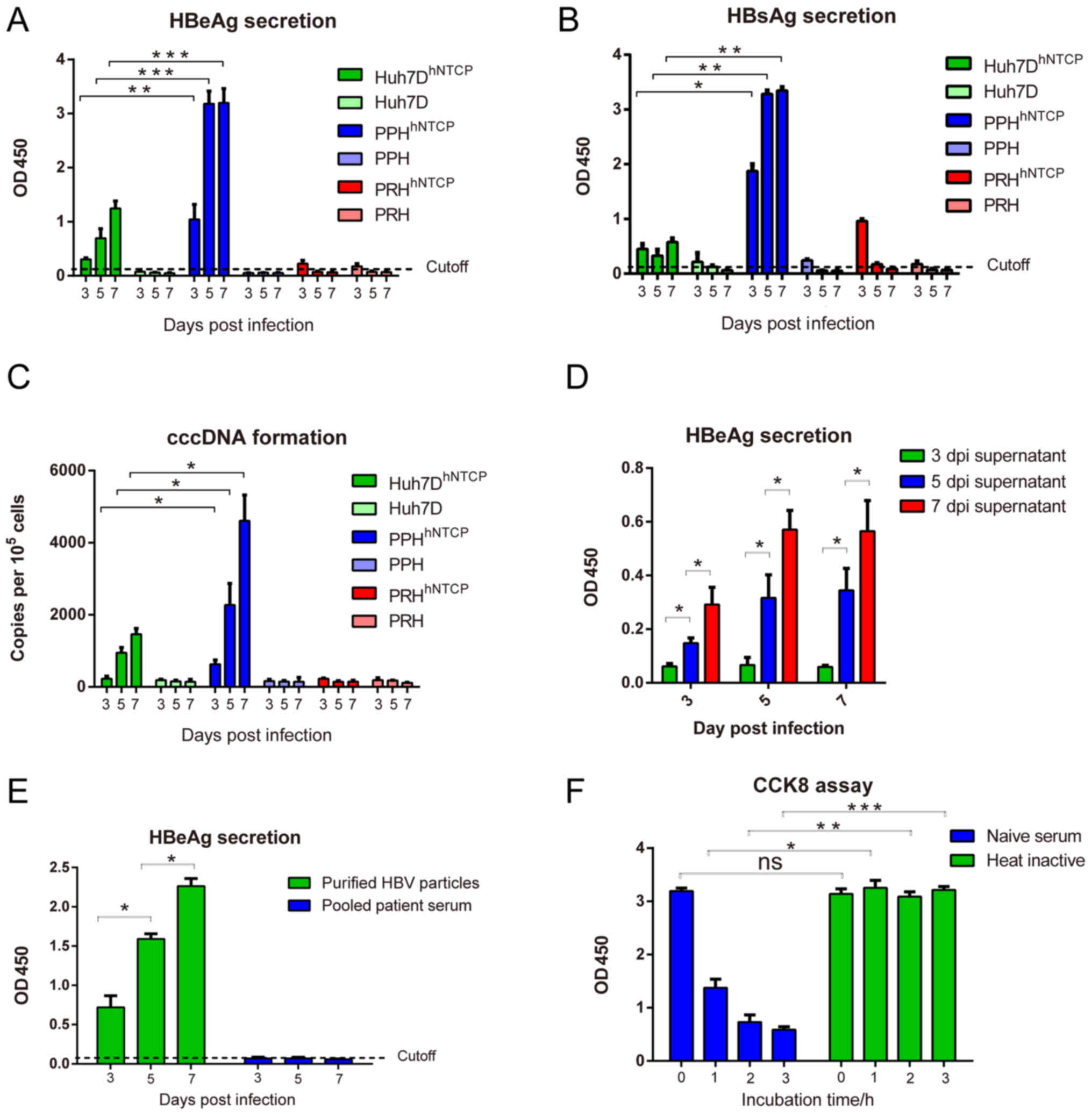 | Figure 3.HBV infection of hNTCP-complemented
hepatocytes. (A-C) Cells were infected with HBV stocks for 16–24 h,
and media were collected at 3, 5 and 7 dpi for detection of (A)
HBeAg and (B) HBsAg by ELISA, and (C) cccDNA by quantitative PCR.
Values over the cut-off line were considered positive. (D) HBV
infections were performed using PEG8000-concentrated supernatants
at 3, 5 and 7 dpi, and HBeAg secretion was measured by ELISA at the
indicated time points. (E) Raw HBV-positive serum and purified HBV
particles served as viral sources to infect hNTCP-complemented
PPHs, followed by HBeAg detection. (F) hNTCP-complemented PPHs were
incubated for the indicated time points with raw patient serum
(naïve serum) and heated patient serum, and cell viabilities were
detected by Cell Counting Kit-8. *P<0.05, **P<0.01 and
***P<0.001, with comparisons indicated by lines. hNTCP, human
sodium taurocholate co-transporting polypeptide; PPH, primary pig
hepatocyte; PRH, primary rabbit hepatocyte; HBV, hepatitis B virus;
dpi, days post-infection; HBsAg, hepatitis B surface antigen;
HBeAG, hepatitis B e-antigen; ns, not significant. |
To confirm the formation of cccDNA, the infected
cells were lysed, and nuclear DNA was extracted for qPCR. As
presented in Fig. 3C,
hNTCP-complemented PPHs produced the highest cccDNA levels among
the three types of cells, followed by hNTCP-complemented Huh7D
cells, whereas the cccDNA levels in hNTCP-complemented PRHs were
comparable to the controls. These results were similar with the
results of HBeAg and HBsAg secretion. To further confirm the
secretion of infectious HBV particles, the new virions in the
culture medium at 3, 5, and 7 dpi were concentrated by PEG8000 and
added to a new batch of hNTCP-complemented PPHs (Fig. 3D). HBeAg secretion in infected
hNTCP-complemented PPHs gradually increased in a time-dependent
manner, indicating that new virions were infectious and could be
passaged (Fig. 3D).
Serum from HBV-positive patients is frequently used
as a viral source for infection, as it is related to clinical viral
strains. hNTCP-complemented PPHs were infected at MOI=200 with raw
pooled patient serum and purified HBV particles. As presented in
Fig. 3E, HBeAg secretion was
positive and continuously increased during HBV infection with
purified HBV particles, indicating that hNTCP-complemented PPHs
could be infected by clinical viral strains. Raw pooled patient
serum was cytotoxic to PPHs, as the cells died in the presence of
this serum (data not shown). This cytotoxicity was significantly
reduced by heating the raw pooled patient serum at 56°C for 30 min
(Fig. 3F), indicating that natural
human anti-pig antibodies (such as anti-Gal or anti-Neu5Gc
antibodies) and the complement system were the major cause of
cytotoxicity. Therefore, purification is essential for the
establishment of HBV infection using human serum as the viral
source.
Anti-HBV drugs may be screened in the
hNTCP-complemented PPHs
Lamivudine, Myr-preS12-47 and 4B10 are
well-characterized inhibitors of HBV replication and/or HBV entry
(23–25) and were tested in HBV-infected
hNTCP-complemented PPHs in the present study. The three inhibitors
were non-cytotoxic to hNTCP-complemented PPHs (Fig. 4D). Myr-preS12-47 and
4B10, the peptide HBV entry inhibitors, exhibited
concentration-dependent inhibition of HBsAg and HBeAg secretion and
HBV DNA in the supernatants as higher concentration of inhibitors
significantly reduced antigens/virion secretion compared with the
lower concentration although there is a plateau, with a half
maximal inhibitory concentration (IC50) of 0.5–1 nM
(Fig. 4A-C), which matches the
reported data regarding HBV infection in the PHH model (23). Lamivudine, an inhibitor of HBV
replication, also exhibited concentration-dependent inhibition of
HBsAg and HBeAg secretion and HBV DNA in the supernatants, with an
IC50 of ~50 nM. The less effective inhibition of HBV
infection by lamivudine possibly occurred due to the failure of
lamivudine to block HBV entry. These results indicated that
HBV-infected hNTCP-complemented PPHs may be applicable for anti-HBV
drug screening.
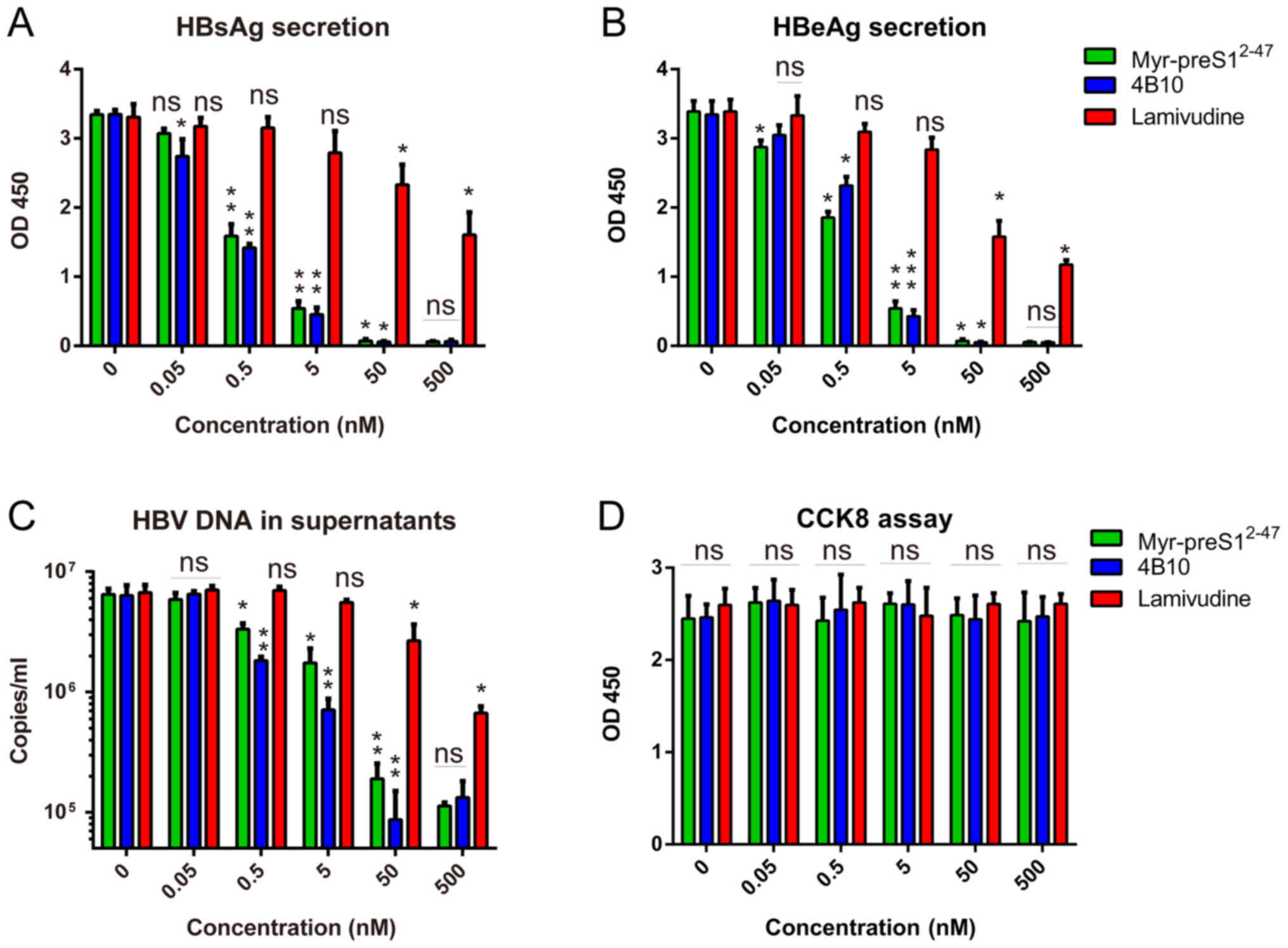 | Figure 4.Anti-HBV drugs tested in HBV-infected
hNTCP-complemented primary PPHs. HBV infections of HepAD38 cells
were performed in the presence of different doses of Lamivudine,
Myr-preS12-47 and 4B10. (A) Relative HBsAg and (B) HBeAg
secretion in the culture supernatant 7 days post-infection was
measured by ELISA. (C) HBV copies in the supernatant were measured
by quantitative PCR. (D) Lamivudine, Myr-preS12-47 and
4B10 were not cytotoxic to hNTCP-complemented PPHs, as evidenced by
Cell Counting Kit-8 assay. *P<0.05, **P<0.01 and
***P<0.001, with comparisons indicated by lines. hNTCP, human
sodium taurocholate co-transporting polypeptide; PPH, primary pig
hepatocyte; HBV, hepatitis B virus; HBsAg, hepatitis B surface
antigen; HBeAg, hepatitis B e-antigen; ns, not significant. |
Discussion
The results of the present study indicated species
tropism regarding the HBV post-entry lifecycle. S. scrofa
was hypothesized to facilitate the HBV post-entry lifecycle, as it
is close to T. brasiliensis in the divergence tree, both of
which are close to H. sapiens and M. mulatta. S.
scrofa was experimentally demonstrated to facilitate the HBV
post-entry lifecycle in the present study. Although O.
cuniculus is closer to H. sapiens and M. mulatta
compared with M. musculus and R. norvegicus, PRHs
were unable to facilitate the HBV post-entry lifecycle in the
present study. According to the present results, R.
norvegicus and O. cuniculus may lack one or more
limiting factors for the HBV post-entry lifecycle. Identification
of these limiting factors may help in the development of
immunocompetent small animal models and identification of novel
antiviral targets.
As demonstrated in the present study, the
differentiation status is an important limiting factor in
productive HBV infection and replication. According to viral
antigen secretion and cccDNA formation, hNTCP-complemented PPHs may
be a superior cell model for HBV infection compared with the
dedifferentiated human hepatoma cell line Huh7D (10). The Huh7 cell line can be induced by
DMSO to a higher differentiation state compared with untreated
cells. Our previous study has also demonstrated this phenomenon
(10). However, the
differentiation state of well-differentiated Huh7 is still lower
compared with that of primary human hepatocytes. For example,
well-differentiated Huh7 is resistant to HBV infection. According
to our previous unpublished data, the expression levels of
differentiation-related genes are lower in well-differentiated Huh7
cells compared with primary human hepatocyte. As pig is close to
human in the divergence tree, PPHs complemented with hNTCP may be a
superior model for HBV infection compared with the dedifferentiated
human hepatoma cell line.
hNTCP expression in PPHs is controlled by a
relatively stable promoter (human elongation factor-1α); this is
different from the endogenous promoter that induces hNTCP
expression in PHHs, which leads to a rapid decrease in hNTCP
expression, thus causing PHHs to be resistant to HBV infection
(9). Therefore, hNTCP-complemented
PPHs may be more susceptible to HBV infection compared with PHHs.
However, hNTCP-complemented PPHs have certain limitations, such as
genomic differences compared with PHHs and limited genomic
annotation. One of these differences is the xenoantigens expressed
on pig cells (e.g., Gal and Neu5Gc), which can be recognized by
natural anti-pig antibodies in human blood and may lead to
complement-dependent cytotoxicity (26). This may explain why
hNTCP-complemented PPHs were fragile when raw patient serum was
used as the viral source in the present study. However,
hNTCP-complemented PPHs may partially replace the roles of PHHs in
HBV-related fundamental research and drug screening.
The MOI for HBV infection of PPHs using HepAD38
cells as viral source was different compared with that using
patient serum as viral source. The majority of viral particles from
HepaAD38 are naked, whereas the majority of viral particles in the
patient serum are intact. Therefore, different MOI was used in HBV
infection with different viral sources, which has been suggested
for successful HBV infection in our previous publications (8–10).
Lempp et al (27) have reported that hNTCP is the
limiting factor in HBV infection in pig hepatocytes, which was also
demonstrated in the present study. In addition, the possibility of
O. cuniculus facilitating the HBV post-entry lifecycle was
assessed and subsequently excluded from the present study, which
supplemented the literature on potential animal hosts for HBV
infection.
In conclusion, hNTCP-complemented PPHs may be a
valuable tool for investigating HBV infection and antiviral drugs.
The results also indicated that the pig is a promising candidate in
the development of an immunocompetent animal model.
Acknowledgements
The authors would like to thank Professor Chen Xulin
from Wuhan Institute of Virology, CAS (Wuhan) for kindly providing
HepAD38. They are also grateful to Professor M Nassal (University
of Freiburg) for kindly providing the protocol of cccDNA
extraction.
Funding
This work was supported by the National Natural
Science Foundation of China (grant. no. 81601760), the General
Financial Grant from the China Postdoctoral Science Foundation
(grant. no. 2016M602587), the Shenzhen Foundation of Science and
Technology (grant. no. JCYJ20160425104534335 to MZ), the Health and
Family Planning Commission of Shaoxing, Zhejiang Province, China
(grant. no. 2017QN001), the Science Technology Bureau of Shaoxing,
Zhejiang Province, China (grant. no. 2017B70022 to BQ), the Sanming
Project of Medicine in Shenzhen (grant. no. SZSM201412020 to LSM)
and Special Funds for the Construction of High Level Hospitals in
Guangdong Province (2019).
Availability of data and materials
The datasets generated and/or analysed during the
current study are available from the corresponding authors on
reasonable request.
Authors' contributions
LSM and MZ conceived and designed the study. MZ and
BQ completed data acquisition, analysis and interpretation in this
study. MZ wrote the manuscript. XSD, XLZ, BQ, YL, ZGH and CCW
performed the experiments. CCW reviewed and edited the manuscript.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Institutional
Bioethics Committee of Shenzhen Second People's Hospital, First
Affiliated Hospital of Shenzhen University, Shenzhen, China. All
animal experiments were performed in accordance with the Ministry
of Health Guidelines for the Care and Use of Laboratory Animals
(no. GB 14925-2001), and all procedures were approved by the
Laboratory Animal Ethics Committee of the First Affiliated Hospital
of Shenzhen University. All experiments were performed in
accordance with the relevant guidelines and regulations. The
manuscript was prepared and revised according to the ARRIVE
guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Beasley RP: Rocks along the road to the
control of HBV and HCC. Ann Epidemiol. 19:231–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sheahan T, Jones CT and Ploss A: Advances
and challenges in studying hepatitis C virus in its native
environment. Expert Rev Gastroenterol Hepatol. 4:541–550. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Weizsäcker FV and Roggendorf M: Models of
viral hepatitisBasel; New York: Karger; 2005
|
|
4
|
Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z,
Huang Y, Qi Y, Peng B, Wang H, et al: Sodium taurocholate
cotransporting polypeptide is a functional receptor for human
hepatitis B and D virus. Elife. 1:e000492012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He W, Ren B, Mao F, Jing Z, Li Y, Liu Y,
Peng B, Yan H, Qi Y, Sun Y, et al: Hepatitis D virus infection of
mice expressing human sodium taurocholate co-transporting
polypeptide. PLoS Pathog. 11:e10048402015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ni Y, Lempp FA, Mehrle S, Nkongolo S,
Kaufman C, Fälth M, Stindt J, Königer C, Nassal M, Kubitz R, et al:
Hepatitis B and D viruses exploit sodium taurocholate
co-transporting polypeptide for species-specific entry into
hepatocytes. Gastroenterology. 146:1070–1083. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yuan W, Wu JY, Zhao YZ, Li J, Li JB, Li ZH
and Li CS: Comparison of early sequential hypothermia and delayed
hypothermia on neurological function after resuscitation in a swine
model. Am J Emerg Med. 35:1645–1652. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou M, Zhao F, Li J, Cheng Z, Tian X, Zhi
X, Huang Y and Hu K: Long-term maintenance of human fetal
hepatocytes and prolonged susceptibility to HBV infection by
co-culture with non-parenchymal cells. J Virol Methods.
195:185–193. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou M, Huang Y, Cheng Z, Zhao F, Li J,
Zhi X, Tian X, Sun W and Hu K: Revival, characterization, and
hepatitis B virus infection of cryopreserved human fetal
hepatocytes. J Virol Methods. 207:29–37. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou M, Zhao K, Yao Y, Yuan Y, Pei R, Wang
Y, Chen J, Hu X, Zhou Y, Chen X and Wu C: Productive HBV infection
of well-differentiated, hNTCP-expressing human hepatoma-derived
(Huh7) cells. Virol Sin. 32:465–475. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ladner SK, Otto MJ, Barker CS, Zaifert K,
Wang GH, Guo JT, Seeger C and King RW: Inducible expression of
human hepatitis B virus (HBV) in stably transfected hepatoblastoma
cells: A novel system for screening potential inhibitors of HBV
replication. Antimicrob Agents Chemother. 41:1715–1720. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Köck J, Nassal M, MacNelly S, Baumert TF,
Blum HE and von Weizsäcker F: Efficient infection of primary tupaia
hepatocytes with purified human and woolly monkey hepatitis B
virus. J Virol. 75:5084–5089. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Köck J, Rösler C, Zhang JJ, Blum HE,
Nassal M and Thoma C: Generation of covalently closed circular DNA
of hepatitis B viruses via intracellular recycling is regulated in
a virus specific manner. PLoS Pathog. 6:e10010822010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Glebe D, Aliakbari M, Krass P, Knoop EV,
Valerius KP and Gerlich WH: Pre-s1 antigen-dependent infection of
Tupaia hepatocyte cultures with human hepatitis B virus. J Virol.
77:9511–9521. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Doitsh G and Shaul Y: A long HBV
transcript encoding pX is inefficiently exported from the nucleus.
Virology. 309:339–349. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nassal M: The arginine-rich domain of the
hepatitis B virus core protein is required for pregenome
encapsidation and productive viral positive-strand DNA synthesis
but not for virus assembly. J Virol. 66:4107–4116. 1992.PubMed/NCBI
|
|
18
|
Fushan AA, Turanov AA, Lee SG, Kim EB,
Lobanov AV, Yim SH, Buffenstein R, Lee SR, Chang KT, Rhee H, et al:
Gene expression defines natural changes in mammalian lifespan.
Aging Cell. 14:352–365. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Drexler JF, Geipel A, König A, Corman VM,
van Riel D, Leijten LM, Bremer CM, Rasche A, Cottontail VM, Maganga
GD, et al: Bats carry pathogenic hepadnaviruses antigenically
related to hepatitis B virus and capable of infecting human
hepatocytes. Proc Natl Acad Sci USA. 110:16151–16156. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vieira YR, dos Santos DR, Portilho MM,
Velloso CE, Arissawa M, Villar LM, Pinto MA and de Paula VS:
Hepadnavirus detected in bile and liver samples from domestic pigs
of commercial abattoirs. BMC Microbiol. 14:3152014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li W, She R, Liu L, You H and Yin J:
Prevalence of a virus similar to human hepatitis B virus in swine.
Virol J. 7:602010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tang H, Zhou S, Zhao L, Wang J, Lei B and
Cao Z: Primary serologic investigation of HBV-like virus existence
in domestic animals and fowls. Hua Xi Yi Ke Da Xue Xue Bao.
21:181–184. 1990.(In Chinese). PubMed/NCBI
|
|
23
|
Ye X, Zhou M, He Y, Wan Y, Bai W, Tao S,
Ren Y, Zhang X, Xu J, Liu J, et al: Efficient inhibition of
hepatitis B virus infection by a preS1-binding peptide. Sci Rep.
6:293912016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Glebe D, Urban S, Knoop EV, Cag N, Krass
P, Grün S, Bulavaite A, Sasnauskas K and Gerlich WH: Mapping of the
hepatitis B virus attachment site by use of infection-inhibiting
preS1 lipopeptides and tupaia hepatocytes. Gastroenterology.
129:234–245. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Doong SL, Tsai CH, Schinazi RF, Liotta DC
and Cheng YC: Inhibition of the replication of hepatitis B virus in
vitro by 2′,3′-dideoxy-3′-thiacytidine and related analogues. Proc
Natl Acad Sci USA. 88:8495–8499. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Joziasse DH and Oriol R:
Xenotransplantation: The importance of the Galalpha1,3Gal epitope
in hyperacute vascular rejection. Biochim Biophys Acta.
1455:403–418. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lempp FA, Wiedtke E, Qu B, Roques P,
Chemin I, Vondran FWR, Le Grand R, Grimm D and Urban S: Sodium
taurocholate cotransporting polypeptide is the limiting host factor
of Hepatitis B Virus infection in macaque and pig hepatocytes.
Hepatology. 66:703–716. 2017. View Article : Google Scholar : PubMed/NCBI
|