Introduction
Colonic cancer has become the most common cancer, as
well as a general cause of human mortality from cancer. Mortality
rates from colonic cancer are predicted to increase substantially
in Costa Rica, Australia, the United States, Ireland and Canada,
with the mortality rate expected to rise by 60% until 2035
(1). It was also identified that
left and right-sided colon cancer differs significantly with
reference to histological, epidemiological and clinical parameters
(2,3). In addition, patients suffering from
right-sided colonic cancer usually have worse prognosis than those
with the other types of cancer (4). These variations may be due to genetic
differences that induce distinct carcinogenic and biological
behaviors. The impact of these findings on screening and treatment
remains to be elucidated (2).
Thus, it is crucial to further investigate the disparity between
two types of colon cancers, which may be beneficial in suppressing
the spread of primary cancer.
In order to further identify the variable genetic
expression of the two colon cancers, the GSE44076 (5), GSE31595 (6) and GSE26906 (7) datasets were downloaded from the
database of Gene Expression Omnibus (GEO). We then analyzed the
data to screen differential expression of genes (DEGs) and
construct a protein-protein interaction (PPI) network. Through
database analysis, we found several genes that were significantly
upregulated or downregulated, respectively, in left and right-sided
colonic cancer.
Several lines of evidence indicated that COL11A1 was
upregulated in various cancers, including colorectal (8–10),
pancreatic (11), non-small cell
lung (12), breast (13), ovarian (14), and head region and neck squamous
cell cancer (15), suggesting an
oncogenic role for COL11A1. Regarding the formation and progression
of tumors, COL11A1 was confirmed to facilitate the progression of
ovarian, and head and neck squamous cell carcinoma (14–16).
It was also reported as a possible biomarker of colon cancer
(12,17).
The expression of TWIST1, a key factor in the
conversion of epithelial-mesenchymal transition (EMT), is closely
associated with the high probability of tumorigenesis,
chemotherapeutic resistance, poor prognosis and metastasis
(18–20). Furthermore, TWIST1 was be found to
be overexpressed in various malignant tumors, such as cancer of the
thyroid (21), esophagus (22), breast (23), lung (24), stomach (25), pancreas (26), hepatocytes (27), colon (28) and cervix (29). Thus, the expression of TWIST1 is
often used as a potential target for cancer treatment (27,30–32).
Through database analysis, we determined COL11A1,
TWIST1, insulin-like 5 (INSL5) and chromogranin A (CHGA) to be hub
proteins for upregulation, and 3β-hydroxysteroid dehydrogenase
(HSD3B2) was a hub protein for downregulation. To further verify
the importance of these genes in left- and right-sided colon
cancer, we selected and determined the expression levels of COL11A1
and TWIST1 in various tissue samples obtained from our department.
Through the combination of database and experimental analyses, the
present study is the first to identify genetic dissimilarities
between these two colon cancer types to the best of our knowledge.
Our findings also revealed that the mRNA and protein expression
levels of COL11A1 and TWIST1 were upregulated in right-sided
colonic cancer than in left-sided tumor samples. This suggested
that COL11A1 and TWIST1 may be potential prognostic markers and/or
molecular targets for the treatment of right-sided colon
cancer.
Materials and methods
Ethics statement
The present study was approved by the Ethics
Committee of Xiamen University. In total, 17 left-sided colonic
cancer and 13 right-sided colonic cancer samples were collected
from the Zhongshan Hospital of Xiamen University between August
2014 and August 2015 (the patients and samples are characteristics
in Table I). This research was
conducted in accordance with the Declaration of Helsinki (33), the International Ethics Standards
for Human Biomedical Research (34) and the relevant provisions of the
National Natural Science Foundation of China, jointly supported by
the World Health Organization and the International Council of
Medical Science Organizations (35).
 | Table I.Clinicopathological characteristics
in colon cancer samples. |
Table I.
Clinicopathological characteristics
in colon cancer samples.
|
| Tumor location |
|
|---|
|
|
|
|
|---|
| Clinicopathological
parameters | Left | Right | P-value |
|---|
| Age, years |
|
| 0.269 |
|
≥50 | 10 | 5 |
|
|
<50 | 7 | 8 |
|
| Sex |
|
| 0.004 |
|
Male | 13 | 3 |
|
|
Female | 4 | 10 |
|
| TNM stage |
|
| 0.633 |
| Tis
+I | 2 | 0 |
|
| II | 10 | 8 |
|
|
III | 4 | 4 |
|
| IV | 1 | 1 |
|
| Histology
grade |
|
| 0.432 |
| Well +
moderate | 12 | 10 |
|
|
Poor | 3 | 3 |
|
| Mucinous
adenocarcinoma | 2 | 0 |
|
Gene expression profile data
We downloaded and used three datasets from the GEO
database and the data of patients enrolled in this study. The
inclusion criteria were set as follows: i) Patients with clinical
information, including the site of resection or biopsy; and ii)
patients with complete cancer data and normal data.
GSE44076 is a dataset containing 246 samples
provided by Solé et al (5),
utilizing Affymetrix Human Genome U219 Array platform. The dataset
comprised 66 colon resected left-sided colonic cancer samples and
38 right-sided samples from colonic carcinoma tissues, as well as
98 normal samples for colonic tissues.
The GSE31595 dataset constituted 37 samples provided
by Thorsteinsson et al (6),
utilizing the Affymetrix Human Genome U133 Plus 2.0 Array platform.
This dataset comprised 14 colon resected left-sided colonic cancer
samples and 23 right-sided samples from colonic carcinoma
tissues.
The GSE26906 dataset contained 90 samples provided
by Birnbaum et al (7),
utilizing Affymetrix Human Genome U219 Array platform. The dataset
comprised 65 colon resected left-sided colonic cancer samples and
25 right-sided samples from colonic carcinoma tissues.
Data pre-processing and DEG
analysis
We used the Affy package (36) to preprocess the three separate
datasets, correcting their background and transforming them from
probe level into gene symbol. Robust multiarray average was used to
normalize the values of probe level intensity and the signal
estimates of generated probe set. Subsequently, the DEGs between
left and right-sided colonic cancer of three separate datasets were
conducted by utilizing the limma package (37) in R. We calculated the fold-change
(FC) of relative genetic expression, and the threshold criteria of
DEG selection were P<0.05 and |log2FC|≥1. Subsequently, on the
basis of limma package in R, we selected DEGs between colon cancer
and normal samples with the dataset of GSE44076 (37). We also calculated the FC in
relative gene expression. This threshold criteria used for DEG
selection were P<0.05, |log2FC|≥2.
PPI network construction of the DEGs
between colonic cancers and normal tissue samples
The potential interactions among the DEGs between
colonic cancers and normal tissue samples in the GSE44076 dataset
was predicted by the Search Tool for the Retrieval of Interacting
Genes (STRING) database (version 10) (38,39).
The proteins from database were entered in STRING. With the
criterion of a combined score of >0.4, and as visualized with
Cytoscape (version 3.2.1), only interactions which contained at
least one DEG were selected to establish the PPI network (40).
Identification of the hub gene between
left and right-sided colonic cancer
We obtained the intersection of total DEGs of
left-sided compared with right-sided colonic cancer in three
separate datasets, and the DEGs of GSE44076 between colon cancers
and normal tissues with the criterion of a combined score of
>0.4. The DEGs contained in the intersection were identified as
the hub gene.
Prognoscan database analysis and
literature review
We utilized the PrognoScan database (http://www.abren.net/PrognoScan/) to analyze
potential associations between DEGs and survival in colon cancer
using Kaplan-Meier analysis (41).
Furthermore, the patients were compared on the basis of high and
low expression to analyze survival according to the PrognoScan
database. In order to select oncogenes for further analysis, a
literature search of PubMed was performed for each gene using the
term ‘GENE NAME]’, ‘cancer’.
Tumors
In the present study, mRNA Droplet Digital™-PCR was
run on the pane of 17 fresh-frozen left-sided colonic cancer
tissues and 13 right-sided samples collected at the Zhongshan
Hospital of Xiamen University between 2014 and 2015 (Table I). All samples were frozen at −80°C
immediately after collection by surgeons who directly took part in
the study and removed the surgical specimens. The mean age of the
17 patients was 64.82 years (ranging from 43–83 years-old) and the
maximum tumor diameter ranged from 2.7–12 cm (mean, 5.476 cm) in
left sided colon cancer. The mean age of the 13 patients was 61.15
years (ranging from 39–82 years-old) and the maximum tumor diameter
ranged from 2–10 cm (mean, 5.362 cm) in right sided colon cancer.
There were no significant differences between the two groups in age
(P=0.4281) and maximum tumor diameter (P=0.8824).
Cell culture and transfection
The colon cancer HCT116 cell line was purchased from
the Institute of Cell Biology and cultured in RPMI-1640 (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Excell
Bio, Shanghai, China), 100 U/ml penicillin and 100 µg/ml
streptomycin at 37°C in a humidified incubator containing 5%
CO2. In total, 5 µg Flag-TWIST1 or COL11A1 plasmids
(Public Protein/Plasmid Library) were transfected into HCT116
cells; after 36 h, the cells were collected. TWIST1 or COL11A1
expression was analyzed using a Flag-antibody. The expression
levels of E-cadherin and N-cadherin or inhibitor of NF-κB (IKKβ)
were determined using reverse transcription-quantitative (RT-q)
PCR, and the expression levels were normalized to GAPDH. The
results are presented as the mean ± SD of three independent
experiments. Plasmid DNA transfections were performed with
Turbofect reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's instructions.
Western blotting
The colon cancer samples were lysed with cell lysis
buffer 20 mM Tris-HCl (pH 7.5), 20 mM β-glycerophosphate, 150 mM
NaCl, 1 mM sodium orthovanadate, 1 mM PMSF, 10 mM NaF, 10 µg/ml
leupeptin, 2 µg/ml aprotinin,1% Triton X-100 and 1 mM EDTA].
Protein concentration was determined using the bicinchoninic acid
method and 80 µg of total protein was used for western blotting.
Protein lysates were separated using 12% SDS-PAGE and transferred
to nitrocellulose membranes. The membranes were blocked for 1 h in
5% BSA and incubated with primary antibodies overnight at 4°C,
washed, and incubated with secondary antibodies (1:5,000) for 1 h.
The membranes were then washed and the protein bands were
visualized using the Immobilon Western Chemiluminescent HRP
Substrate Kit (EMD Millipore).
Western blotting was performed with the following
primary antibodies: actin monoclonal antibody (1:5,000; cat. no.
A5441; Sigma-Aldrich; Merck KGaA), Twist1 (1:1,000; cat. no. 46702;
Cell Signaling Technology, Inc.) and COL11A1 (1:1,000; cat. no.
42818; Cell Signaling Technology, Inc.). The horseradish
peroxidase-conjugated secondary goat anti-rabbit and goat
anti-mouse (1:5,000; cat. nos. 31466 and A16078; Thermo Fisher
Scientific, Inc.). densitometry was performed using Quantity One
software (Bio-Rad Laboratories, Inc.).
RNA isolation, reverse
transcription-quantitative PCR (RT-qPCR)
In the present study, total RNA was isolated using
the RNA Simple Total RNA Kit (Biotek, Inc.), according to the
manufacturer's instructions, and first-strand cDNA was obtained by
utilizing First-strand cDNA Synthesis Kit (Biotek, Inc.). The RT
reaction was performed at 42°C for 60 min and 85°C for 5 min, the
cDNA was stored at −20°C. We amplified cDNA samples via qPCR and
the SYBR Green Real-Time PCR Master Mix (Toyobo Life Science)
utilizing an ABI 7500 Real-Time PCR System (Applied Biosystems;
Thermo Fisher Scientific, Inc.) with the following primers: GAPDH
forward, 5′-TCTCCTCTGACTTCAACAGCGA-3′ and GAPDH reverse,
5′-GTCCACCACCCTGTTGCTGT-3′; TWIST1 forward,
5′-GGCCAGGTACATCGACTTCC-3′, TWIST1 reverse,
5′-TCCAGACCGAGAAGGCGTAG-3′; COL11A1 forward,
5′-CAACTCAGCCATCCTGACTT-3′, COL11A1 reverse,
5′-GTCTCACCACCAGATGTGAA-3′. The thermocycling conditions for the
qPCR were as follows: 15 min at 95°C, followed by 10 cycles of 30
sec at 94°C, 90 sec at 72°C; and 20 cycle of 30 sec at 94°C, 90 sec
at 58°C and 60 sec at 72°C, with a final incubation for 60 min at
72°C.
Statistical analysis
Data are presented the mean ± SD of at lowest three
individual experiments. All statistical tests were performed using
Graphpad Prism 6.0 (GraphPad Software, Inc.). One-way ANOVA
followed by a Bonferroni's post-test for manifold comparisons, and
pair-wise comparisons with Student's test (two-tailed) were
conducted. The correlation analyses were proceeded with Pearson's
test. P<0.05 was considered to indicate a statistically
significant difference.
Confirmed by continually changing the cut-off values
in the analysis of all our results, receiver operating curves (ROC)
curves were established by all potential specificity/sensitivity
pairs with an particular detection method (42). In our study, ROC curves were used
to identify the hub genes that would be highly predictive of the
tissue samples and could distinguish between left and right-sided
colon cancer tissues.
Functional enrichment analysis
To investigate the functions that may be changed by
the DEGs identified, functional Gene Ontology (GO) enrichment
analysis was performed using Database for Annotation,
Visualization, and Integrated Discovery (version 6.8; http://david.ncifcrf.gov/) (43) to determine the biological functions
of these DEGs between tumor and normal tissues. An enriched gene
count >2 and cut-off criteria were selected based on P<0.05
in our study.
Results
Screening of DEGs
To further identify the differential gene expression
between these two types colonic cancers, we selected and downloaded
the GSE44076, GSE31595 and GSE26906 gene expression datasets from
the GEO database for DEG screening (Fig. 1). The GSE44076 dataset, amounting
to 48 DEGs, revealed 20 upregulated and 28 downregulated DEGs. The
GSE31595 dataset, amounting to 46 DEGs, contained 21 upregulated
and 25 downregulated DEGs. The GSE26906 dataset, amounting to 34
DEGs, revealed 23 upregulated and 11 downregulated DEGs.
Additionally, these DEGs of tumor and normal tissues were employed
for further analysis using the GSE44076 dataset (Table II).
 | Table II.DEGs were identified from three
profile datasets, including up-regulated genes and down-regulated
genes in which the colon cancer tissues compared with normal colon
tissues and right-sided colon cancer tissues compared to left-sided
colon cancer. |
Table II.
DEGs were identified from three
profile datasets, including up-regulated genes and down-regulated
genes in which the colon cancer tissues compared with normal colon
tissues and right-sided colon cancer tissues compared to left-sided
colon cancer.
| DEGs | Upregulated
genes | Downregulated
genes |
|---|
| GSE44076 tumor and
normal | IL8, MYC, MMP9,
CCNB1, AGT, BUB1, CENPA, CXCL1, TPX2, CEP55, UBE2C, NUF2, CXCL11,
CXCL5, GNG4, TIMP1, SPP1, MMP3, MND1, CXCL2, TRIP13, MMP7, ATAD2,
MMP1, CXCL3, COL1A1, ANLN, ERCC6L, CENPH, SKA3, DSCC1, SPC25, SOX9,
LGR5, CDCA7, CKAP2, BMP7, RNASEH2A, FOXA2, FAM54A, FCGR3A, MMP10,
DIAPH3, COL11A1, TWIST1, ETV4, LCN2, MYBL2, SULT2B1, COMP, COL10A1,
FABP6, MTHFD1L, ORC6L, MSX2, FAP | CXCL12, SST, NPY,
PYY, CCL19, GCG, ADRA2A, INSL5, CD36, CA2, CHGA, FABP4, CCL13,
HSD3B2, VIP, KLF4, CCL23, PCSK2, PDE6A, FABP1, MUC2, PDE9A, HPGD,
OASL, CTSG, RERGL, SGK1, AKR1C1, CFD, SLC4A4, DES, MMP28, HSD17B2,
DARC, PCK1, UGT2B15, UGT2B17, ADH1B, MYH11, PTGDR |
| GSE44076 right and
left | TTPA, TRIM54,
PNLIPRP2, ELAVL2, G0S2, RBP2, MSLN, EREG, ZNF43, TNNC1, PRAC, ODAM,
QPRT, PCP4, COL9A3, LY6G6D, BEX2, SLC26A3, GAL, CKMT2 | MB, HOXC6, HOXB6,
ARX, AIM2, SLC28A3, B3GNT6, L1TD1, DMRTA2, GABRP, GBA3, ART3,
CLDN2, FOXA1, CYP4X1, KRT6B, CA9, HEPACAM2, FOXD1, HOXB8, MS4A8B,
C4BPA, DMBT1, MT1H, MUC1, IDO1, KLK1, PIGR |
| GSE31595 right and
left | FGD1, PRAC1,
C11orf70, HS3ST3A1, PKIA, PI3, INSL5, HOXB13, PHACTR3, ASB9, SYNE4,
RLN2, FLJ41455, TWIST1, CKMT2, SLC35D3, CCDC113, COL11A1, PPBP,
EPYC, CPE | IL10, USP30-AS1,
SLC51A, HAR1A, CRTAM, NKG7, NPY6R, NR1H4, TREH, BLNK, MEP1B, GDPD3,
PITX2, CYP2C18, AIM2, PP7080, CD69, GBA3, GRAMD1C, CCL8, HSD3B2,
CCL5, HMGCS2, RARRES3, SLC20A1 |
| GSE26906 right and
left | PRAC1, CLDN5,
SERPINE2, PCDH19, CHGA, PRELP, SERPINA6, PLA1A, SCNN1B, STMN2,
MAP7D2, CHRDL1, MUC12, PCP4, HSD17B6, MN1, ARMCX1, CHRDL2, CLDN10,
CCL11, AOC3, CLC, BEX2 | PLA2G3, RIBC2,
CAPN6, PIWIL1, SLC28A3, GMPR, TMEM171, REG3A, L1TD1, FOXD1,
CLDN2 |
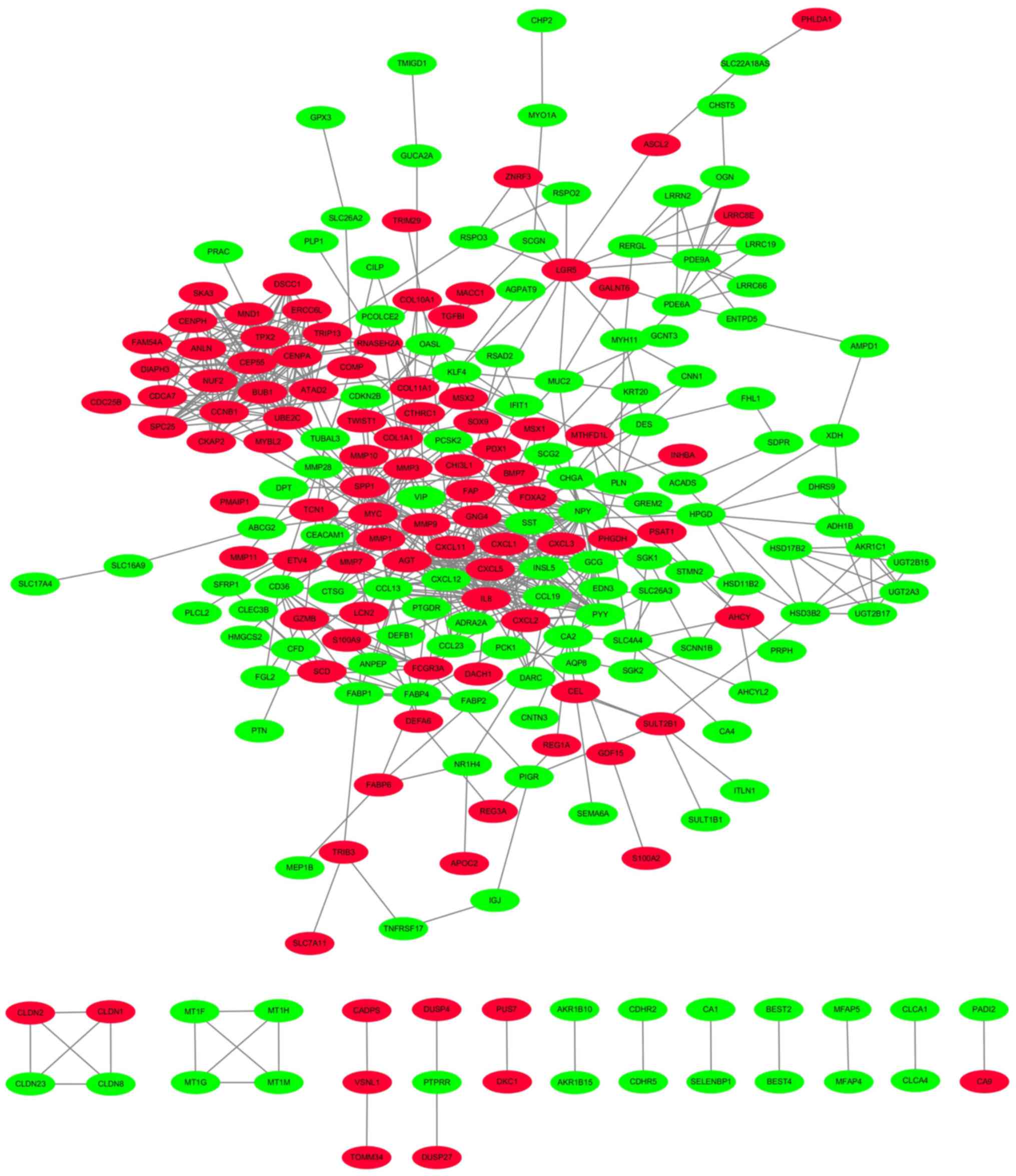
PPI network
A PPI network with these DEGs between tumor and
normal tissues was constructed from pre-processing of the GSE44076
dataset, via analysis with STRING and Cytoscape software. Proteins
and related interactions were symbolized by nodes and lines
respectively in the PPI network, which consisted of GSE44076,
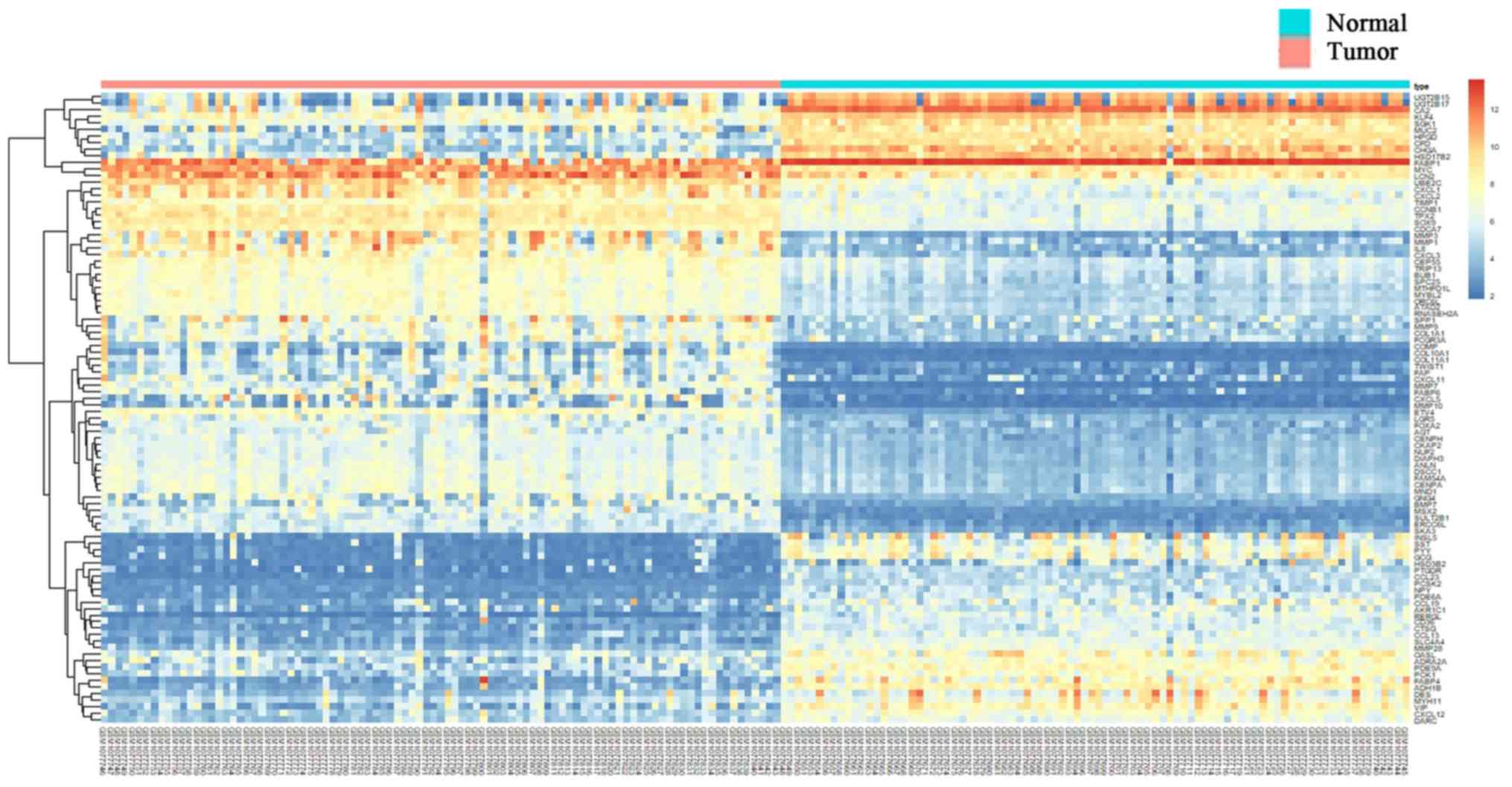
including 228 proteins and 683 interactions (Fig. 2). In the PPI network, the hub
proteins should be the nodes with an average connective degree ≥5.
From PPI network analysis, we could identify network of various
proteins in colon cancer.
Analysis the association between DEGs
and survival
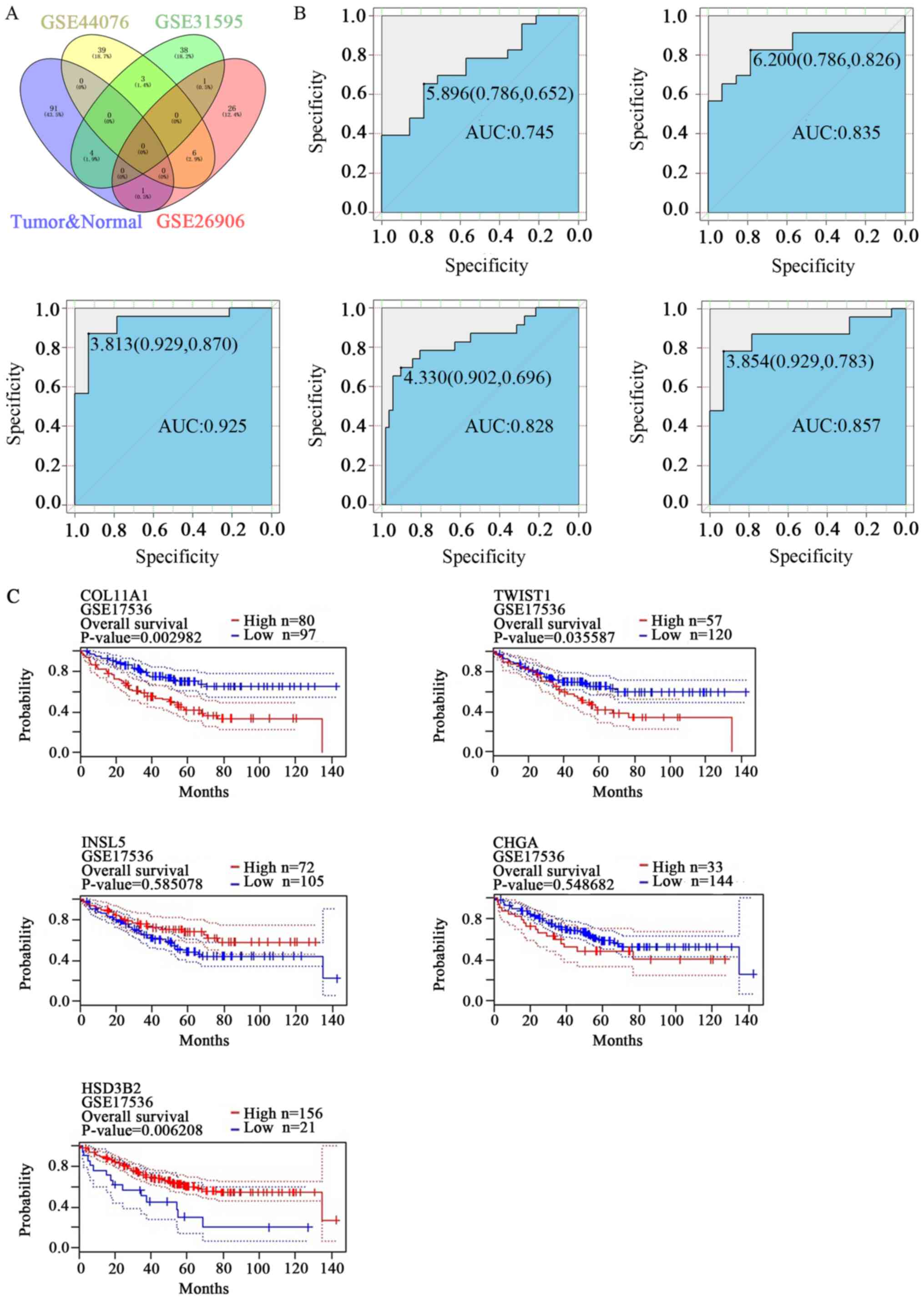
Subsequently, we obtained the intersection of DEGs
between left and right-sided colonic cancers obtained from the
three aforementioned databases and DEGs of tumor and normal samples
(Fig. 3A). COL11A1, TWIST1, INSL5
and CHGA were determined to be upregulated proteins, while HSD3B2
was downregulated protein in right-sided colonic cancer samples
than left-sided tissues. Among them, COL11A1, TWIST1 and HSD3B2
were in accordance with the DEG profiles of tumor and normal
tissue. We plotted ROC curves for these proteins. The closer to
upper-left corner the ROC curves are, the more accurate the test
is. The genes we screened exhibited a typical trend (Fig. 3B). In addition, through
Kaplan-Meier analysis, upregulated COL11A1 and TWIST1 was
associated with poor survival rates in colon cancer. By contrast,
the downregulation of HSD3B2 exhibited generally poor survival
rates in colon cancer. INSL5 and CHGA were not associated with a
statistically significant difference in survival rates (Fig. 3C). We performed a literature search
of PubMed for each gene using the terms ‘GENE NAME]’, ‘cancer’. Of
the five genes, according to a previous study COL11A1, and TWIST1
had been described as being candidate oncogene (44); therefore, in subsequent analyses,
we selected two of them (COL11A1 and TWIST1) for the further
verification.
Verification of the DEGs (COL11A1 and
TWIST1) between left and right-sided colonic cancer with western
blotting analysis and RT-qPCR
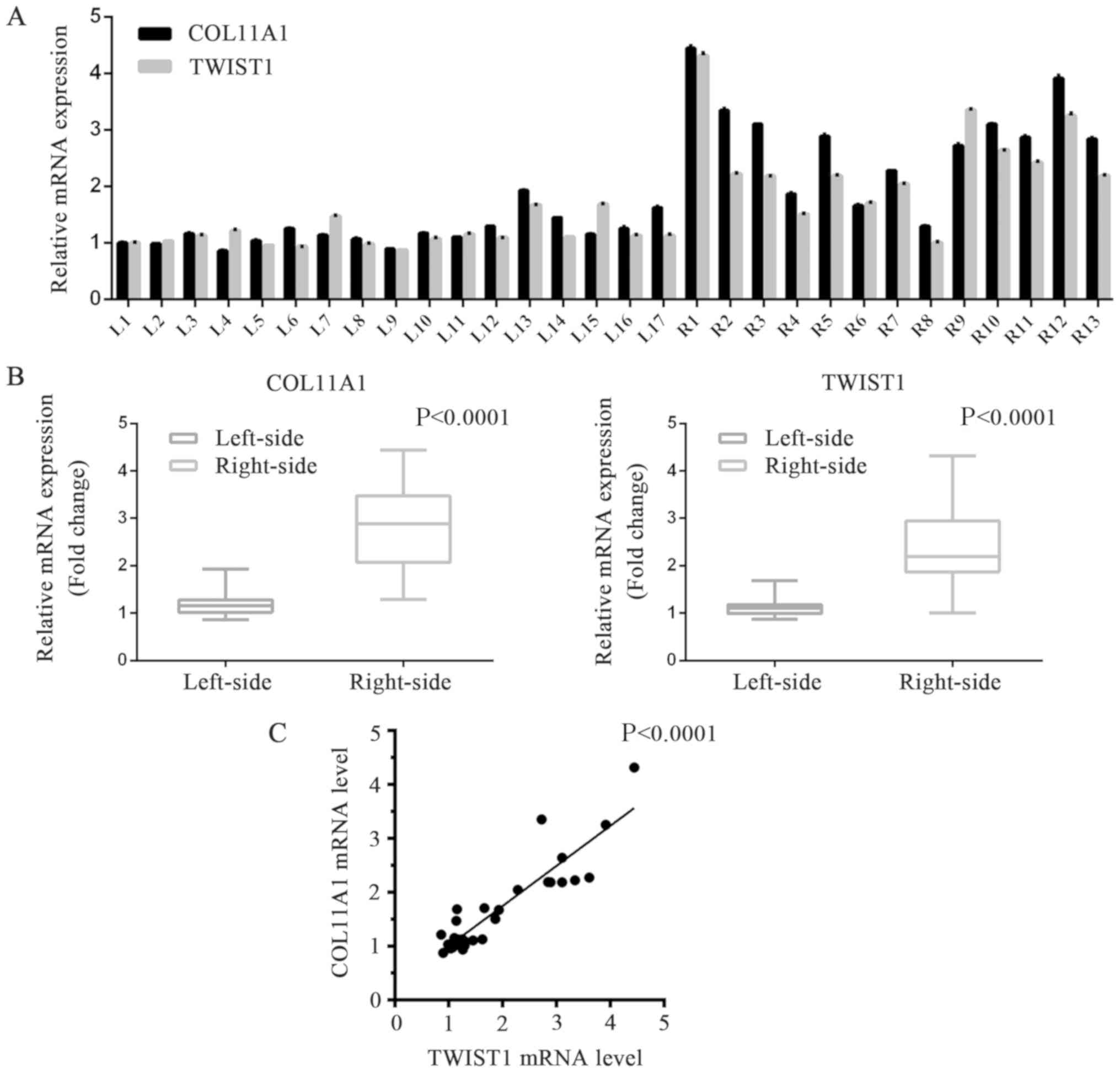
In order to estimate the levels of COL11A1 and
TWIST1 expression that are implicated in these two sorts of colon
cancer, we performed RT-qPCR analysis of 17 fresh frozen left-sided
colonic cancer samples and 13 right-sided ones obtained from our
department (Table III). It was
shown that the levels of COL11A1 and TWIST1 expression in cell
samples of right-sided colonic cancer samples were significantly
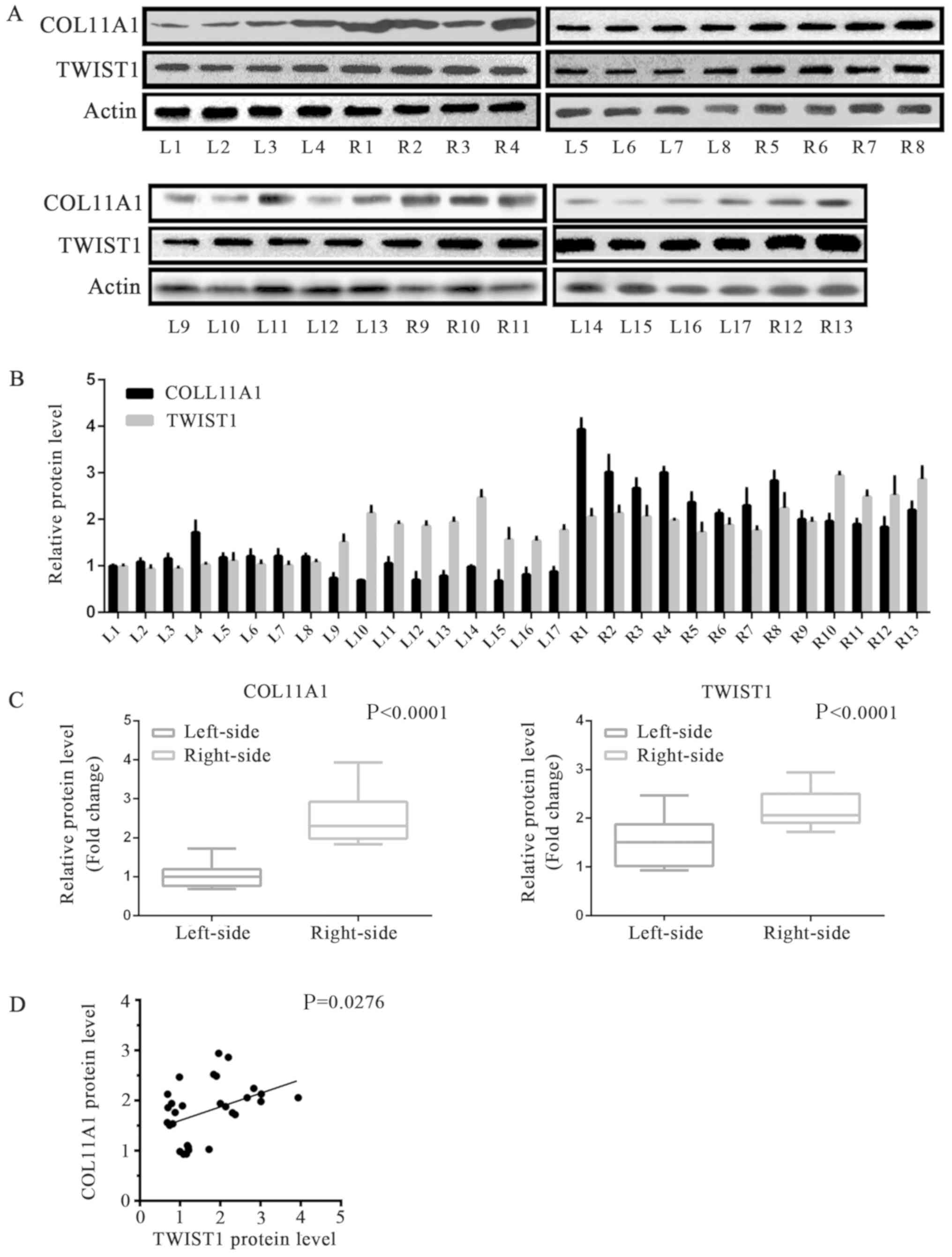
higher than that in the left side samples (Tables III and IV, Fig. 4A
and B). The protein expression levels of COL11A1 and TWIST1 in
the two types of cancer tissues were detected via western blotting
analysis. In accordance with the RT-qPCR results, significantly
increased levels of COL11A1 and TWIST1 expression were observed in
right-sided colon cancer tissues than in left-sided tumor samples
(Tables III and IV, Fig.
5A-C). We also analyzed the correlation between the expression
of COL11A1 and TWIST1 proteins in colonic cancer samples. The
expression of COL11A1 and TWIST1 proteins exhibited a positive
correlation (Figs. 4C and 5D), indicating that the two proteins
could serve key roles during the formation and progression of these
two types colonic cancers.
 | Table III.Clinicopathological characteristics
and COL11A1 mRNA expression in colon cancer samples. |
Table III.
Clinicopathological characteristics
and COL11A1 mRNA expression in colon cancer samples.
| Clinicopathological
parameters | No. of cases | COL11A1 expression
(2−ΔCq, mean) | P-value |
|---|
| Tumor location |
|
|
|
|
Left | 17 | 1.1993 | <0.0001 |
|
Right | 13 | 2.7947 |
|
| TNM stage |
|
|
|
| Tis +
I | 2 | 1.1530 | 0.7026 |
| II | 18 | 1.9950 |
|
|
III | 8 | 1.9223 |
|
| IV | 2 | 1.5617 |
|
| Histology
grade |
|
|
|
| Well +
moderate | 22 | 1.8974 | 0.4455 |
|
Poor | 6 | 2.1369 |
|
| Mucinous
adenocarcinoma | 2 | 1.0769 |
|
 | Table IV.Clinicopathological characteristics
and TWIST1 mRNA expression in colon cancer samples. |
Table IV.
Clinicopathological characteristics
and TWIST1 mRNA expression in colon cancer samples.
| Clinicopathological
parameters | No. of cases | TWIST1 expression
(2−ΔCq, mean) | P-value |
|---|
| Tumor location |
|
| <0.0001 |
|
Left | 17 | 1.1546 |
|
|
Right | 13 | 2.3867 |
|
| TNM stage |
|
| 0.7767 |
| Tis +
I | 2 | 1.2984 |
|
| II | 18 | 1.7548 |
|
|
III | 8 | 1.7536 |
|
| IV | 2 | 1.2214 |
|
| Histology
grade |
|
| 0.3601 |
| Well +
moderate | 22 | 1.6988 |
|
|
Poor | 6 | 1.9118 |
|
| Mucinous
adenocarcinoma | 2 | 0.9055 |
|
Functional enrichment analysis
The GO analysis suggested that COL11A1 was involved
in ‘proteinaceous extracellular matrix’, ‘collagen trimer’ and
‘chondrocyte development’, and TWIST1 was involved in ‘osteoblast
differentiation’, transcription factor activity of RNA polymerase
II, ‘positive regulation of transcription from RNA polymerase II
promoter’, ‘sequence-specific DNA binding’ and ‘nucleus’ (Table V).
 | Table V.Functional enrichment analysis for
differentially-expressed genes between tumor and normal contained
TWIST1 or COL11A1 based on GO. |
Table V.
Functional enrichment analysis for
differentially-expressed genes between tumor and normal contained
TWIST1 or COL11A1 based on GO.
| Category | Term | Description | P-value | Genes |
|---|
|
GOTERM_CC_DIRECT | GO:0005578 | Proteinaceous
extracellular matrix | 0.0022724 | MMP9, COMP,
COL11A1, MMP1, COL10A1 |
|
GOTERM_BP_DIRECT | GO:0001649 | Osteoblast
differentiation | 0.0028232 | MSX2, COL1A1,
TWIST1, SPP1 |
|
GOTERM_BP_DIRECT | GO:0045944 | Positive regulation
of transcription from RNA polymerase II promoter | 0.0096635 | CKAP2, FOXA2,
ATAD2, BMP7, MYC, ETV4, TWIST1 |
|
GOTERM_MF_DIRECT | GO:0000981 | RNA polymerase II
transcription factor activity, sequence-specific DNA binding | 0.0108839 | FOXA2, MYBL2, ETV4,
TWIST1 |
|
GOTERM_CC_DIRECT | GO:0005581 | Collagen
trimer | 0.0135131 | COL1A1, COL11A1,
COL10A1 |
|
GOTERM_BP_DIRECT | GO:0002063 | Chondrocyte
development | 0.04367 | MSX2, COL11A1 |
|
GOTERM_CC_DIRECT | GO:0005634 | Nucleus | 0.0498886 | FOXA2, NUF2, TPX2,
MND1, ATAD2, UBE2C, MYBL2, CENPH, MSX2, SPC25, CDCA7, CENPA, MYC,
ETV4, TWIST1 |
Discussion
In 1990, Bufill was the first to report that
distinct biological pathways may be associated with left and right
colonic carcinoma (45). Since
then, many studies have revealed significant differences in
epidemiological, clinical and histological parameters between the
two regions of colonic cancers (2–4,46).
Irrespective of age and gender, synchronous/metachronous, the level
of BRAF, microsatellite instability (MSI) and consensus molecular
subtype (CMS), the location of a primary tumor is considered as an
important individual prognostic factor for colonic cancer (47). Patients with left-sided colonic
cancer usually have better prognoses than those with right-sided
colonic cancer (4). Furthermore,
there are varying responses in palliative chemotherapy, as well as
cetuximab and bevacizumab owing to tumor location (47–50).
Primary left-sided colon cancer patients may benefit from the
chemotherapy with additional cetuximab in both first and
second-line treatments for metastatic colorectal cancer (49). It has been reported by Venook et
al (47) that in treating
patients with right-side colonic cancer, bevacizumab was more
effective than cetuximab. Increasing evidence manifested that
molecular profiles of cancers may also differ across these sites
(51–54). The incidence rate of CpG island
methylator phenotype-high, BRAF mutation, and MSI-high in
colorectal cancer increased gradually along the bowel from the
rectum to the ascending colon (54,55).
Loree et al (56) found
that, based on comparing the CMS level of an isolated cohort of
>600 patients, the sigmoid-rectal region appeared to differ from
the other sites, while the transverse colon differs from other
right-sided locations. Several studies revealed that distal colon
cancer has no significant differences compared with rectal cancer
with reference to disease-free survival, somatic alterations and
overall survival (57,58). Thus, it is crucial to investigate
the molecular variances between two types colonic cancers.
We first reported the differential expression of
COL11A1 and TWIST1 between left and right-sided colonic cancers. We
observed that COL11A1 and TWIST1 mRNA were upregulated in
right-sided colon cancer tissues than in left-sided tissues based
on RT-qPCR analysis, which was in accordance with our results of
bioinformatical analyses.
COL11A1 and TWIST1 mRNA expression was positively
related to tumor location. The results support the notion that
upregulated COL11A1 and TWIST1 in right-sided colonic cancer may
have notable impact on the invasiveness and worse prognosis of the
right-sided colonic cancer.
Furthermore, it has been known that cancer cells
have interactions with their surrounding stroma, which was
associated with the mechanism of tumor cell invasion (59). There are two chains of type XI
collagen involved and one of them is a smaller fibrous collagen and
encoded exactly by COL11A1 gene (60). Additionally, the extracellular
matrix (ECM) has been considered to be important in tumor behavior,
and forms the interstitial matrix conducive to regulating and
integrating cell behavior (61–63).
Thus, collagens, as major components of ECM, are related to
regulation of several important biological processes, including
cell proliferation, differentiation and migration (64–66).
In addition, the development of the COL11A1 mutations was linked
with Marshall syndrome and type II Stickler syndrome (67,68).
Based on our results, COL11A1 is mainly enriched in proteinaceous
extracellular matrix, collagen trimer and chondrocyte development;
COL11A1 was also the predominant node in the PPI network in our
study. Proteinaceous extracellular matrix-related genes that could
increase the aggression and change the metastatic properties of
cells in vivo colon cancer contribute to poor prognosis in
cancer patients (69). The strong
overexpression of COL11A1 has been discovered in various studies
for examining diversities between tumor and normal tissues, and
linked to the metastasis of tumor and poor prognosis (14). A previous study reported that the
expression of COL11A1 of colon tumors is related to
anaphase-promoting complex/β-catenin path of colon cancer
metastasis (11). We will continue
to investigate the mechanism of COL11A1 in the two sides of colon
cancer in future work.
According to the results of the GO and PPI analyses,
TWIST1 which was enriched in ‘osteoblast differentiation’,
transcription factor activity of RNA polymerase II, ‘positive
regulation of transcription from RNA polymerase II promoter’,
‘sequence-specific DNA binding’ and ‘nucleus’, was also the
predominant node in the PPI network. Increasing evidence has
indicated that cancer cells characterized as cancer stem cells
(CSC) or EMT have a strong potential to promote the progression,
aggression, metastasis and chemoresistance of cancer (70–73).
It has been shown that the EMT program is associated with the
self-renewal ability of cells and may efficiently induce tumors
(74,75). It is the essential helix-loop-helix
transcription factors (such as TWIST1) that crucial for EMT and
CSC; one of these factors (71)
suppresses the expression of E-cadherin and promotes EMT during the
progression of tumors (76).
Specifically, in the promoter of E-cadherin, TWIST1 combines with
E-box elements, suppresses the transcription of the cell-cell
adhesion molecule expression and promotes EMT, contributing to
metastasis during the progression of tumors (77). It has been known that TWIST1 has
wide expression in many human cancers, including colorectal cancer
(31), and its strong expression
has been linked to poor prognosis and chemotherapeutic resistance
(27,30–32,78).
In colorectal cancer patients, TWIST1 expression has been
associated with chemosensitivity to oxaliplatin and 5-fluorouracil
(78). Downregulation of TWIST1
expression induced apoptosis and enhanced the sensitivity of
chemotherapy (78). Generally,
TWIST1 can promote the chemotherapeutic resistance of cancer cells
(23), increase the number of CSCs
(74), as well as enhance
aggression and metastasis of cancer cells (77,79–85)
by suppressing their senescence and apoptosis induced by oncogenes.
A study suggested that TWIST1 may be used as a vital clinical
biomarker for antiangiogenic therapy as its polymorphisms were
related to the survival of patients with metastatic colorectal
cancer who were treated with first-line bevacizumab-based
chemotherapy (28). The present
study proposed the importance of TWIST1 upregulation in right-sided
colonic cancer tissues. Patients with right-sided colonic cancer
exhibited better outcomes of bevacizumab treatment than those with
left-sided colon cancer (86).
Through our results, we also speculated that TWIST1
may associated with COL11A1 to enhance the development of colon
cancer. A study showed that the level of TWIST1 mRNA had a positive
correlation with that of COL11A1 messenger RNA in ovarian cancers.
In addition, COL11A1 could promote the activation of nuclear
factor-κB by activating the transcription of IKKβ, and promote the
expression of TWIST1, thereby regulating chemoresistance and
apoptosis (44). In our
experiments, we also demonstrated that TWIST1 could promote the
progression of EMT, with the downregulation of E-Cadherin and the
upregulation of N-Cadherin, while COL11A may promote the
transcription of IKKβ in colon cancer cell (Fig. S1). This suggested the complexity
of the regulatory mechanism mediated by these two proteins in left
and right-sided colonic cancer. the limitations of our study
include the small cohort of patients enrolled for analysis.
In brief, our work suggested that the expression
levels of COL11A1 and TWIST1 differ between these two types colonic
cancers and may be promising therapeutic targets for treating
right-sided colonic cancer. However, further study is also
necessary to illustrate the potential molecular mechanisms, and
functions of COL11A1 and TWIST1 between these two types of colonic
cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
Foundation of Fujian Provincial Department of Science and
Technology (grant nos. 2019J01556, 2018-ZQN-87 and 2018-2-66) and
the Foundation of Xiamen Municipal Bureau of Science and Technology
(grant nos. 3502Z20174080 and 3502Z20184038).
Availability of data and materials
The data of four original microarray datasets
GSE44076, GSE31596 and GSE26906 were downloaded from NCBI-Gene
Expression Omnibus database (Available online: http://www.ncbi.nlm.nih.gov/geo).
Authors' contributions
JH conceived and designed the experiments. JZ, XH,
SY and YJ performed the experiments. CS analyzed the data. CS and
JH wrote the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Xiamen University. All samples were collected and
gathered at the Zhongshan Hospital of Xiamen University between
2014 and 2015. The research content of this subject strictly
follows the Helsinki Declaration, the International Ethics
Standards for Human Biomedical Research, and relevant provisions of
the National Natural Science Foundation of China, and the World
Health Organization and the International Council of Medical
Science Organizations.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Araghi M, Soerjomataram I, Jenkins M,
Brierley J, Morris E, Bray F and Arnold M: Global trends in
colorectal cancer mortality: Projections to the year 2035. Int J
Cancer. 144:2992–3000. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Benedix F, Kube R, Meyer F, Schmidt U,
Gastinger I and Lippert H; Colon/Rectum Carcinomas (Primary Tumor)
Study Group, : Comparison of 17,641 patients with right- and
left-sided colon cancer: Differences in epidemiology perioperative
course, histology, and survival. Dis Colon Rectum. 53:57–64. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Alexiusdottir KK, Möller PH, Snaebjornsson
P, Jonasson L, Olafsdottir EJ, Björnsson ES, Tryggvadottir L and
Jonasson JG: Association of symptoms of colon cancer patients with
tumor location and TNM tumor stage. Scand J Gastroenterol.
47:795–801. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hansen IO and Jess P: Possible better
long-term survival in left versus right-sided colon cancer-a
systematic review. Dan Med J. 59:A44442012.PubMed/NCBI
|
|
5
|
Solé X, Crous-Bou M, Cordero D, Olivares
D, Guinó E, Sanz-Pamplona R, Rodriguez-Moranta F, Sanjuan X, de Oca
J, Salazar R and Moreno V: Discovery and validation of new
potential biomarkers for early detection of colon cancer. PLoS One.
9:e1067482014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Thorsteinsson M, Kirkeby LT, Hansen R,
Lund LR, Sørensen LT, Gerds TA, Jess P and Olsen J: Gene expression
profiles in stages II and III colon cancers: Application of a
128-gene signature. Int J Colorectal Dis. 27:1579–1586. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Birnbaum DJ, Laibe S, Ferrari A, Lagarde
A, Fabre AJ, Monges G, Birnbaum D and Olschwang S; COL2 Project, :
Expression profiles in stage II colon cancer according to APC gene
status 1 2. Transl Oncol. 5:72–76. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fischer H, Stenling R, Rubio C and
Lindblom A: Colorectal carcinogenesis is associated with stromal
expression of COL11A1 and COL5A2. Carcinogenesis. 22:875–878. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fischer H, Salahshor S, Stenling R, Björk
J, Lindmark G, Iselius L, Rubio C and Lindblom A: COL11A1 in FAP
polyps and in sporadic colorectal tumors. Bmc Cancer. 1:172001.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bowen KB, Reimers AP, Luman S, Kronz JD,
Fyffe WE and Oxford JT: Immunohistochemical localization of
collagen type XI alpha1 and alpha2 chains in human colon tissue. J
Histochem Cytochem. 56:275–283. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Badea L, Herlea V, Dima SO, Dumitrascu T
and Popescu I: Combined gene expression analysis of whole-tissue
and microdissected pancreatic ductal adenocarcinoma identifies
genes specifically overexpressed in tumor epithelia.
Hepatogastroenterology. 55:2016–2027. 2008.PubMed/NCBI
|
|
12
|
Chong IW, Chang MY, Chang HC, Yu YP, Sheu
CC, Tsai JR, Hung JY, Chou SH, Tsai MS, Hwang JJ and Lin SR: Great
potential of a panel of multiple hMTH1, SPD, ITGA11 and COL11A1
markers for diagnosis of patients with non-small cell lung cancer.
Oncol Rep. 16:981–988. 2006.PubMed/NCBI
|
|
13
|
Feng Y, Sun B, Li X, Zhang L, Niu Y, Xiao
C, Ning L, Fang Z, Wang Y, Zhang L, et al: Differentially expressed
genes between primary cancer and paired lymph node metastases
predict clinical outcome of node-positive breast cancer patients.
Breast Cancer Res Treat. 103:319–329. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu YH, Chang TH, Huang YF, Huang HD and
Chou CY: COL11A1 promotes tumor progression and predicts poor
clinical outcome in ovarian cancer. Oncogene. 33:3432–3440. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sok JC, Lee JA, Dasari S, Joyce S,
Contrucci SC, Egloff AM, Trevelline BK, Joshi R, Kumari N, Grandis
JR and Thomas SM: Collagen type XI α1 facilitates head and neck
squamous cell cancer growth and invasion. Br J Cancer.
109:3049–3056. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu YH, Chang TH, Huang YF, Chen CC and
Chou CY: COL11A1 confers chemoresistance on ovarian cancer cells
through the activation of Akt/c/EBPβ pathway and PDK1
stabilization. Oncotarget. 6:23748–23763. 2015.PubMed/NCBI
|
|
17
|
Galván JA, García-Martínez J,
Vázquez-Villa F, García-Ocaña M, García-Pravia C,
Menéndez-Rodríguez P, González-del Rey C, Barneo-Serra L and de los
Toyos JR: Validation of COL11A1/procollagen 11A1 expression in
TGF-β1-activated immortalised human mesenchymal cells and in
stromal cells of human colon adenocarcinoma. BMC Cancer.
14:8672014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Brabletz T: EMT and MET in metastasis:
Where are the cancer stem cells? Cancer Cell. 22:699–701. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Eide T, Ramberg H, Glackin C, Tindall D
and Taskén KA: TWIST1, A novel androgen-regulated gene, is a target
for NKX3-1 in prostate cancer cells. Cancer Cell Int. 13:42013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Qin Q, Xu Y, He T, Qin C and Xu J: Normal
and disease-related biological functions of Twist1 and underlying
molecular mechanisms. Cell Res. 22:90–106. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Salerno P, Garcia-Rostan G, Piccinin S,
Bencivenga TC, Di Maro G, Doglioni C, Basolo F, Maestro R, Fusco A,
Santoro M and Salvatore G: TWIST1 plays a pleiotropic role in
determining the anaplastic thyroid cancer phenotype. J Clin
Endocrinol Metab. 96:E772–E781. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee KW, Kim JH, Han S, Sung CO, Do IG, Ko
YH, Um SH and Kim SH: Twist1 Is an independent prognostic factor of
esophageal squamous cell carcinoma and associated with its
epithelial-mesenchymal transition. Ann Surg Oncol. 19:326–335.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cheng GZ, Chan J, Wang Q, Zhang W, Sun CD
and Wang LH: Twist transcriptionally up-regulates AKT2 in breast
cancer cells leading to increased migration, invasion, and
resistance to paclitaxel. Cancer Res. 67:1979–1987. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hung JJ, Yang MH, Hsu HS, Hsu WH, Liu JS
and Wu KJ: Prognostic significance of hypoxia-inducible
factor-1alpha, TWIST1 and Snail expression in resectable non-small
cell lung cancer. Thorax. 64:1082–1089. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yan-Qi Z, Xue-Yan G, Shuang H, Yu C,
Fu-Lin G, Fei-Hu B, Shi-Ren S, Xu Feng W, Jie D and Dai-Ming F:
Expression and significance of TWIST basic helix-loop-helix protein
over-expression in gastric cancer. Pathology. 39:470–475. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Satoh K, Hamada S, Kimura K, Kanno A,
Hirota M, Umino J, Fujibuchi W, Masamune A, Tanaka N, Miura K, et
al: Up-regulation of MSX2 enhances the malignant phenotype and is
associated with twist 1 expression in human pancreatic cancer
cells. Am J Pathol. 172:926–939. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Matsuo N, Shiraha H, Fujikawa T, Takaoka
N, Ueda N, Tanaka S, Nishina S, Nakanishi Y, Uemura M, Takaki A, et
al: Twist expression promotes migration and invasion in
hepatocellular carcinoma. Bmc Cancer. 9:2402009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Matsusaka S, Zhang W, Cao S, Hanna DL,
Sunakawa Y, Sebio A, Ueno M, Yang D, Ning Y, Parekh A, et al:
TWIST1 polymorphisms predict survival in patients with metastatic
colorectal cancer receiving first-line bevacizumab plus
oxaliplatin-based chemotherapy. Mol Cancer Ther. 15:1405–1411.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Missaoui N, Hmissa S, Trabelsi A, Traoré
C, Mokni M, Dante R and Frappart L: Promoter hypermethylation of
CDH13, DAPK1 and TWIST1 genes in precancerous and cancerous lesions
of the uterine cervix. Pathol Res Pract. 207:37–42. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tran PT, Shroff EH, Burns TF, Thiyagarajan
S, Das ST, Zabuawala T, Chen J, Cho YJ, Luong R, Tamayo P, et al:
Twist1 suppresses senescence programs and thereby accelerates and
maintains mutant kras-induced lung tumorigenesis. PLoS Genet.
8:e10026502012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ruppenthal RD, Nicolini C, Filho AF,
Meurer R, Damin AP, Rohe A, Alexandre CO and Damin DC: TWIST1
promoter methylation in primary colorectal carcinoma. Pathol Oncol
Res. 17:867–872. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhao N, Sun BC, Zhao XL, Liu ZY, Sun T,
Qiu ZQ, Gu Q, Che N and Dong XY: Coexpression of Bcl-2 with
epithelial-mesenchymal transition regulators is a prognostic
indicator in hepatocellular carcinoma. Med Oncol. 29:2780–2792.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
World Medical Association Inc, .
Declaration of Helsinki. Ethical principles for medical research
involving human subjects. J Indian Med Assoc. 107:403–405.
2009.PubMed/NCBI
|
|
34
|
Council for International Organizations of
Medical Sciences, . International ethical guidelines for biomedical
research involving human subjects. Bull Med Ethics. 10:17–23.
2002.
|
|
35
|
Bankowski Z: Council for International
Organizations of Medical Sciences. J Med Imag Radiation Oncol.
15:83–87. 1971.
|
|
36
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Smyth GK: LIMMA: Linear models for
microarray dataBioinformatics and Computational Biology Solutions
Using R and Bioconductor. Gentleman R, Carey V, Dudoit S, Irizarry
R and Huber W: Statistics for Biology and Health. Springer; New
York, NY: pp. 397–420. 2005, View Article : Google Scholar
|
|
38
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9. 1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mizuno H, Kitada K, Nakai K and Sarai A:
PrognoScan: A new database for meta-analysis of the prognostic
value of genes. BMC Med Genomics. 2:182009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Weiss HL, Niwas S, Grizzle WE and
Piyathilake C: Receiver operating characteristic (ROC) to determine
cut-off points of biomarkers in lung cancer patients. Dis Markers.
19:273–278. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wu YH, Huang YF, Chang TH and Chou CY:
Activation of TWIST1 by COL11A1 promotes chemoresistance and
inhibits apoptosis in ovarian cancer cells by modulating
NF-κB-mediated IKKβ expression. Int J Cancer. 141:2305–2317. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bufill JA: Colorectal cancer: Evidence for
distinct genetic categories based on proximal or distal tumor
location. Ann Intern Med. 113:779–788. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Nitsche U, Stögbauer F, Späth C, Haller B,
Wilhelm D, Friess H and Bader FG: Right sided colon cancer as a
distinct histopathological subtype with reduced prognosis. Dig
Surg. 33:157–163. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Venook AP, Ou FS, Lenz HJ, Kabbarah O, QU
X and Niedzwiecki D: Primary (1°) tumor location as an independent
prognostic marker from molecular features for overall survival (OS)
in patients (pts) with metastatic colorectal cancer (mCRC):
Analysis of CALGB/SWOG 80405 (Alliance). J Clin Oncol. 35:3503.
2017. View Article : Google Scholar
|
|
48
|
von Einem JC, Heinemann V, von Weikersthal
LF, Vehling-Kaiser U, Stauch M, Hass HG, Decker T, Klein S, Held S,
Jung A, et al: Left-sided primary tumors are associated with
favorable prognosis in patients with KRAS codon 12/13 wild-type
metastatic colorectal cancer treated with cetuximab plus
chemotherapy: An analysis of the AIO KRK-0104 trial. J Cancer Res
Clin Oncol. 140:1607–1614. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Brulé SY, Jonker DJ, Karapetis CS,
O'Callaghan CJ, Moore MJ, Wong R, Tebbutt NC, Underhill C, Yip D,
Zalcberg JR, et al: Location of colon cancer (right-sided versus
left-sided) as a prognostic factor and a predictor of benefit from
cetuximab in NCIC CO.17. Eur J Cancer. 51:1405–1414. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shen H, Yang J, Huang Q, Jiang MJ, Tan YN,
Fu JF, Zhu LZ, Fang XF and Yuan Y: Different treatment strategies
and molecular features between right-sided and left-sided colon
cancers. World J Gastroenterol. 21:6470–6478. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kim GP, Colangelo LH, Wieand HS, Paik S,
Kirsch IR, Wolmark N and Allegra CJ; National Cancer Institute, :
Prognostic and predictive roles of high-degree microsatellite
instability in colon cancer: A national cancer institute-national
surgical adjuvant breast and bowel project collaborative study. J
Clin Oncol. 25:767–772. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Minoo P, Zlobec I, Peterson M, Terracciano
L and Lugli A: Characterization of rectal, proximal and distal
colon cancers based on clinicopathological, molecular and protein
profiles. Int J Oncol. 37:707–718. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Deng G, Kakar S, Tanaka H, Matsuzaki K,
Miura S, Sleisenger MH and Kim YS: Proximal and distal colorectal
cancers show distinct gene-specific methylation profiles and
clinical and molecular characteristics. Eur J Cancer. 44:1290–1301.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yamauchi M, Morikawa T, Kuchiba A, Imamura
Y, Qian ZR, Nishihara R, Liao X, Waldron L, Hoshida Y, Huttenhower
C, et al: Assessment of colorectal cancer molecular features along
bowel subsites challenges the conception of distinct dichotomy of
proximal versus distal colorectum. Gut. 61:847–854. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Phipps AI, Lindor NM, Jenkins MA, Baron
JA, Win AK, Gallinger S, Gryfe R and Newcomb PA: Colon and rectal
cancer survival by tumor location and microsatellite instability:
The Colon Cancer Family Registry. DisColon Rectum. 56:937–944.
2013. View Article : Google Scholar
|
|
56
|
Loree JM, Pereira AAL, Lam M, Willauer AN,
Raghav K, Dasari A, Morris VK, Advani S, Menter DG, Eng C, et al:
Classifying colorectal cancer by tumor location rather than
sidedness highlights a continuum in mutation profiles and consensus
molecular subtypes. Clin Cancer Res. 24:1062–1072. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Liu F, Li C, Jia H, Yang L, Wu Y, Zhao J,
Cai S, Zhu J and Xu Y: Is there a prognostic value of tumor
location among Chinese patients with colorectal cancer? Oncotarget.
8:38682–38692. 2017.PubMed/NCBI
|
|
58
|
Slattery ML, Curtin K, Wolff RK, Boucher
KM, Sweeney C, Edwards S, Caan BJ and Samowitz W: A comparison of
colon and rectal somatic DNA alterations. DisColon Rectum.
52:1304–1311. 2009. View Article : Google Scholar
|
|
59
|
Liotta LA, Rao CN and Barsky SH: Tumor
invasion and the extracellular matrix. Lab Invest. 49:636–649.
1983.PubMed/NCBI
|
|
60
|
Mio F, Chiba K, Hirose Y, Kawaguchi Y,
Mikami Y, Oya T, Mori M, Kamata M, Matsumoto M, Ozaki K, et al: A
Functional Polymorphism in COL11A1, which encodes the alpha 1 chain
of type XI collagen, is Associated with Susceptibility to Lumbar
Disc Herniation. Am J Hum Genet. 81:1271–1277. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Barkan D, Green JE and Chambers AF:
Extracellular matrix: A gatekeeper in the transition from dormancy
to metastatic growth. Eur J Cancer. 46:1181–1188. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hynes RO: The Extracellular Matrix: Not
Just Pretty Fibrils. Science. 326:1216–1219. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Raglow Z and Thomas SM: Tumor matrix
protein collagen XIα1 in cancer. Cancer Lett. 357:448–453. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Egeblad M, Rasch MG and Weaver VM: Dynamic
interplay between the collagen scaffold and tumor evolution. Curr
Opin Cell Biol. 22:697–706. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Prockop DJ, Kivirikko KI, Tuderman L and
Guzman NA: The biosynthesis of collagen and its disorders (second
of two parts). N Engl J Med. 301:77–85. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Li Y, Lacerda DA, Warman ML, Beier DR,
Yoshioka H, Ninomiya Y, Oxford JT, Morris NP, Andrikopoulos K,
Ramirez F, et al: A fibrillar collagen gene, Col11a1, is essential
for skeletal morphogenesis. Cell. 80:423–430. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Donoso LA, Edwards AO, Frost AT, Ritter R
III, Ahmad N, Vrabec T, Rogers J, Meyer D and Parma S: Clinical
variability of Stickler syndrome: role of exon 2 of the collagen
COL2A1 gene. Surv Ophthalmol. 48:191–203. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Annunen S, Körkkö J, Czarny M, Warman ML,
Brunner HG, Kääriäinen H, Mulliken JB, Tranebjaerg L, Brooks DG,
Cox GF, et al: Splicing mutations of 54-bp exons in the COL11A1
gene cause marshall syndrome, but other mutations cause overlapping
marshall/stickler phenotypes. Am J Hum Genet. 65:974–983. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ma C, Rong Y, Radiloff DR, Datto MB,
Centeno B, Bao S, Cheng AW, Lin F, Jiang S, Yeatman TJ and Wang XF:
Extracellular matrix protein betaig-h3/TGFBI promotes metastasis of
colon cancer by enhancing cell extravasation. Genes Dev.
22:308–321. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Prieto-García E, Díaz-García CV,
García-Ruiz I and Agulló-Ortuño MT: Epithelial-to-mesenchymal
transition in tumor progression. Med Oncol. 34:1222017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Kim A, Bae YK, Gu MJ, Kim JY, Jang KY, Bae
HI, Lee HJ and Hong SM: Epithelial-mesenchymal transition phenotype
is associated with patient survival in small intestinal
adenocarcinoma. Pathology. 45:567–573. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Kudo-Saito C, Shirako H, Takeuchi T and
Kawakami Y: Cancer metastasis is accelerated through
immunosuppression during Snail-induced EMT of cancer cells. Cancer
Cell. 15:195–206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Burk U, Schubert J, Wellner U, Schmalhofer
O, Vincan E, Spaderna S and Brabletz T: A reciprocal repression
between ZEB1 and members of the miR-200 family promotes EMT and
invasion in cancer cells. Embo Rep. 9:582–589. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wahl GM: BS1-1: Stem cells, cancer, and
cancer stem cells. Cancer Res. 71:BS12011.
|
|
76
|
Peinado H, Olmeda D and Cano A: Snail, Zeb
and bHLH factors in tumour progression: An alliance against the
epithelial phenotype? Nat Rev Cancer. 7:415–428. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Yang J, Mani SA, Donaher JL, Ramaswamy S,
Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A and
Weinberg RA: Twist, a master regulator of morphogenesis, plays an
essential role in tumor metastasis. Cell. 117:927–939. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zhu DJ, Chen XW, Zhang WJ, Wang JZ, Ouyang
MZ, Zhong Q and Liu CC: Twist1 is a potential prognostic marker for
colorectal cancer and associated with chemoresistance. Am J Cancer
Res. 5:2000–2011. 2015.PubMed/NCBI
|
|
79
|
Puisieux A, Valsesia-Wittmann S and
Ansieau S: A twist for survival and cancer progression. Br J
Cancer. 94:13–17. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Firulli AB and Conway SJ:
Phosphoregulation of Twist1 provides a mechanism of cell fate
control. Curr Med Chem. 15:2641–2647. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Fu J, Qin L, He T, Qin J, Hong J, Wong J,
Liao L and Xu J: The TWIST/Mi2/NuRD protein complex and its
essential role in cancer metastasis. Cell Res. 21:275–289. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Qin L, Liu Z, Chen H and Xu J: The steroid
receptor coactivator-1 (SRC-1) regulates twist expression and
promotes breast cancer metastasis. Cancer Res. 69:3819–3827. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Vernon AE and LaBonne C: Tumor metastasis:
A new twist on epithelial-mesenchymal transitions. Curr Biol.
14:R719–R721. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Karreth F and Tuveson DA: Twist induces an
epithelial-mesenchymal transition to facilitate tumor metastasis.
Cancer Biol Ther. 3:1058–1059. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Yang J, Mani SA and Weinberg RA: Exploring
a new twist on tumor metastasis. Cancer Res. 66:4549–4552. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Venook A, Niedzwiecki D, Lenz HJ, et al:
CALGB/SWOG 80405: Phase III trial of irinotecan/5-FU/leucovorin
(FOLFIRI) or oxaliplatin/5-FU/leucovorin (mFOLFOX6) with
bevacizumab (BV) or cetuximab (CET) for patients (pts) with KRAS
wild-type (wt) untreated metastatic adenocarcinoma of the colon or
rectum (MCRC). J Clin Oncol. 32:S182014. View Article : Google Scholar
|