Introduction
There has been a dramatic increase in the prevalence
of allergic diseases, which are common and frequently present in
clinical practice (1,2). Dermatophagoides pteronyssinus
and Dermatophagoides farinae are the major house dust mite
(HDM) species, and are the most significant sources of indoor
allergens for inhalation, causing allergic diseases, including
allergic asthma and allergic rhinitis atopic dermatitis (3,4).
Desensitization treatment with HDM extracts is currently one of the
most effective treatments against allergies to HDMs (5). However, it is difficult to ensure the
consistency of the natural extracts of HDMs because of their
complex components, including inflammatory molecules (ceramides,
kallikreins and endotoxin), that are usually responsible for side
effects and poor efficacy (6).
Therefore, recombinant allergens are given prior to their natural
counterparts in HDM-specific immunotherapy to improve the efficacy
and safety of presentation, diagnosis and clinical immunotherapy
(7).
Decades of research have revealed and characterized
>30 different allergens in D. farinae; group 1 and group
2 allergens are the most clinically relevant, as they possess
IgE-binding activity in >80% of patients with HDM allergies
(8–10). These allergens induce T helper 2
immune responses by encoding cysteine proteases and epididymal
proteins (9,11). Translation elongation factors
(TEFs) are expressed in Cladosporium, Rubber and HDM (12–14).
The allergenicity of the TEF 2 in D. farinae has not been
defined. Therefore, understanding the allergenicity of TEF 2 will
be important for the detection and treatment of allergic
responses.
Currently, with the development of bioinformatics,
the concept of precision medicine is emerging. Bioinformatics can
predict key target spots of a disease, which can provide critical
references for the drug and therapy development (15,16).
Previously, epitope peptide vaccines were considered to have
excellent potential application prospects, therefore, it is
critical to predict the epitope of novel allergens (17–19).
Novel vaccines, synthesized based on B-cells of the T-cell epitope,
can activate the produce specific antibodies from human B cells,
and can also eliminate infected cells by activating cytotoxic
lymphocytes (17). Therefore, the
present study cloned, expressed and purified TEF 2, and evaluated
its allergenicity. The properties of TEF 2 were predicted by using
bioinformatics tools, providing valuable information for further
vaccine development.
Materials and methods
Sera and skin prick test (SPT)
The written informed consent was obtained from each
participant for the use of peripheral blood samples and SPT. The
serum and SPT of 37 allergic patients (22 female, 15 male; 8–86
years) were from The First Affiliated Hospital of Guangzhou Medical
University. The sera of 3 healthy subjects (3 male, 8–15 years)
were recruited from Shenzhen Children's Hospital. The 11 children
among the subjects were approved by legal guardians. The samples
were collected between January 2014 and December 2015. the present
study was approved by the ethic Committee of the Institutional
Review Board of the School of Medicine, Shenzhen University.
Obtaining the gene encoding TEF 2
In a previous study, the draft genome of
Dermatophagoides farinae were assembled using
high-throughput sequencing (20).
The gene sequence of Der f TEF 2 was obtained by high-throughput
sequencing from Dermatophagoides farinae (Hong Kong
Bioinformatics Centre, Hong Kong) and then the gene of Der f TEF 2
was synthesizing by Sangon Biotech in Shanghai. The 18 µl cDNA
mixed with 2 µl (1/10 volume) of 10X DNA loading buffer (Beijing
Solarbio Science & Technology Co., Ltd.) were added to the
sample tank and the electrophoresis started. After the
electrophoresis, the gel was stained by ethidium bromide, and
images were captured under a UV lamp with a wavelength of 302
nm.
Expression and purification of
recombinant TEF 2
The cDNA of Der f TEF 2 was inserted into a pMD-18T
vector (Takara Bio, Inc.), cloned into the BamHI and
HindIII sites and the plasmids were heat-transformed into
E. coli Top10 (Invitrogen; Thermo Fisher Scientific, Inc.
The bacteria were incubated in LB medium (Beijing Solarbio Science
& Technology Co., Ltd.) with 100 mg/l ampicillin and cultured
overnight at 37°C. The recombinant colonies were transferred into
LB medium with ampicillin to expand the culture, followed by
plasmid extraction using plasmid DNA extraction kit (Omega Bio-Tek,
Inc). The recombinant plasmid was characterized using BamHI
and sequenced by BGI Group. After sequencing, the correct clone was
ligated into the pET-32a expression vector (Novagen; Merck KGaA) at
37°C for 4 h. Initial cloning was performed with the E. coli
strain Top10 cultured overnight at 37°C; a single colony was
picked, and the plasmid was extracted for identification using
BamHI and HindIII restriction enzymes. pET-32a-TEF 2
was transformed into E. coli BL21 (Invitrogen; Thermo Fisher
Scientific, Inc.) cells for expression. The cells were grown in LB
medium supplemented with 50 µg/ml ampicillin until the logarithmic
phase was reached (A600 nm =0.6–0.9). Expression of TEF 2 was
induced by adding 20 µl of isopropyl-D-thiogalactopyranoside (IPTG;
1 mol/l) to the LB medium. After 4 h of continued incubation at
37°C, the bacteria were harvested via centrifugation at 11,180 × g
at room temperature for 5 min and 1 ml of the bacterial culture
resuspended in 100 µl deionized water and mixed with 20–25 µl 10X
protein SDS-PAGE loading buffer (Beijing Solarbio Science &
Technology Co., Ltd.). The expression of the recombinant proteins
was analyzed using 12% SDS-PAGE electrophoresis.
Following the induced expression of the recombinant
protein, the samples were lysed using ultrasonic treatment at 4°C
and an amplitude of 38% for 5 min (1 sec pulse on and 0.1 sec pulse
off). The supernatant was purified using a balanced Ni-NTA column
(ShangHai, Sangon Biotech, cat. no. C600792, 5 ml, 90 µm) at a
speed of 2 ml/min and 4°C. After washing with a washing buffer (50
mM Tris, 40 mM imidazole and 0.5 M NaCl, pH 8), the protein was
eluted slowly using an elution buffer (50 mM Tris, 0.3 M imidazole
and 0.2 M NaCl, pH 8). The elution peak was collected by elution
buffer (50 mM Tris, 0.3 M imidazole and 0.2 M NaCl, pH 8).
ELISAs
The HDMs allergic sera IgE antibodies specific to
TEF 2 were measured indirectly using ELISAs. Briefly, 96-well
microtiter plates were coated with 100 ng/well of TEF 2 at a
concentration of 1 µg/ml in carbonate buffered solution (15 mM
Na2CO3 and 35 mM NaHCO3, pH 9.5)
at 4°C overnight and then blocked with 200 µl 3% BSA in PBS at 37°C
for 120 min. Each well was prepared with the serum (diluted 1:5;
100 µl/well) as the primary antibody or 3% BSA (as a negative
control, Beijing Solarbio Science & Technology Co., Ltd.) and
incubated at 37°C for 60 min. The plates were incubated with
peroxidase-conjugated goat anti-human IgE (1:2,000, A18793,
Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 60 min.
Each incubation step was followed by three washes with PBS-0.05%
Tween 20 (PBST). Color was developed by adding 100 µl/well TMB at
37°C for 10 mins and terminated by the addition of 2 M H2SO4 (50
µl/well). Absorbance was determined using a microplate reader at
450 nm.
Determination of inhibition using
ELISA
The serum of patients allergic to TEF 2 was used for
inhibition experiments to determine the cross reactivity among TEF
2 and HDMs. Patients' sera (supplied pre-diluted in 2% BSA in PBST)
were pre-incubated with purified Der f TEF 2 or dust mite extract
(DME) obtained from mites that were cryopreserved in liquid
nitrogen and then ground to a powder (final concentration: 0.00001,
0.0001, 0.001, 0.01 or 1 g/ml) at 4°C overnight. Pre-incubated Der
f TEF 2 or DME as the primary antibody were added to the microtiter
plates pre-coated with TEF 2 or DME (0.1 µg/well) and the ELISA was
performed as in the previous section. According to ELISA test
procedures, data from three experiments were collected. The
inhibition rates were calculated according to the following
formula: Inhibition
(%)=(OD0-ODinhibitor)/(OD0-ODBSA),
where: OD0=the optical density of antigen without any
inhibitor, ODinhibitor=the optical density after adding
an inhibitor (0.00001, 0.0001, 0.001, 0.01 or 1 g/ml TEF2 or DME),
and ODBSA=the optical density with only BSA in the
plate.
SPT of recombinant TEF 2
SPTs were performed with purified and
endotoxin-removed TEF 2 (0.01 mg/ml). dissolved in phosphate buffer
(pH 7.4, 50 mM phosphate buffer, 100 mM NaCl). Glycerin was added
to a final concentration of 50%. The sample contained histamine
phosphate (5 mg/ml) as the positive control or saline as a negative
control. The results were checked 20 min after the SPT. The results
were classified as follows: The prick spot became a wheal and fleck
surrounding the wheal, it was positive (+); 3+, the response was
the same as or stronger that of the histamine control; 2+, the
response was weaker than that of the histamine control but stronger
than that of the negative control; 1+, the response was
significantly weaker than that of the histamine control but
slightly stronger than that of the negative control; negative, no
response (21).
Analysis of immunogenicity by
immunoblotting
SPTs and sera from non-allergic children were used
to assess immunogenicity. TEF 2 was subjected to 12% SDS-PAGE and
transferred to a nitrocellulose membrane for immunoblot analysis.
The membrane was blocked with 3% BSA for 1 h at 37°C and incubated
with SPTs and sera from non-allergic children (diluted 1:5 with 5%
BSA-PBST, PBST: PBS containing 0.05% Tween 20; 100 µl/well) for 1 h
at room temperature. After washing five times with TBST, the
membrane was incubated with a secondary antibody
(peroxidase-conjugated goat anti-human IgE, 1:2,000, A18793,
Invitrogen; Thermo Fisher Scientific, Inc.) for 1 h at 37°C.
Finally, after washing three times with TBS-T, the membrane was
developed with a DAB kit (Invitrogen; Thermo Fisher Scientific,
Inc.).
Bioinformatics analysis of TEF 2
The open reading frame (ORF) of TEF 2 was analyzed
using the NCBI database (http://www.ncbi.nlm.nih.gov/), its physicochemical
properties were predicted using the ProtParam Tool (http://web.expasy.org/protparam/), and the amino
acid sequence was predicted using the Translate Tool (http://web.expasy.org/translate/) (22,23).
NetPhos3.1 (http://www.cbs.dtu.dk/services/NetPhos/) was used to
predict phosphorylation sites, and Subcellular localization of
cathepsin D was predicted by CELLO 2.5 (24). ProtScale (http://web.expasy.org/protscale/) was used to assess
the hydrophilicity, average flexibility and the relative mutability
(25,26). Secondary structural elements were
obtained using DNAStar. InterPro5.0 (http://www.ebi.ac.uk/interpro/), ScanProsite
(http://prosite.expasy.org/scanprosite/) and MotifScan
(http://myhits.isb-sib.ch/cgi-bin/motif_scan) was used
to assess functional sites. The phylogenetic tree was constructed
using BLAST, MEGA 5.0 and ClustalX 2.1 software (27). The Bioinformatics Predicted
Antigenic Peptides (BPAP) system (http://imed.med.ucm.es/Tools/antigenic.pl), DNAStar
Protean system (DNAStar Inc, Madison, WI, USA) and the BepiPred 1.0
server (http://www.cbs.dtu.dk/services/BepiPred/) were used to
predict the B cell epitopes of TEF 2 (28–30).
Furthermore, the Immune Epitope Database, Propre, SYFPEITHI,
Net-MHCII 2.2 server and NetMHCIIpan-3.0 server were used to study
the T cell epitopes of Der f TEF 2 (28–30).
Statistical analysis
All data are presented as the mean ± SD and analyzed
SPSS 18.0 statistical software (SPSS, Inc.). Student's t-test was
used for the mean differences between two groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
Synthesizing and analysis of amino
acid sequence homology, alignment and molecular evolution
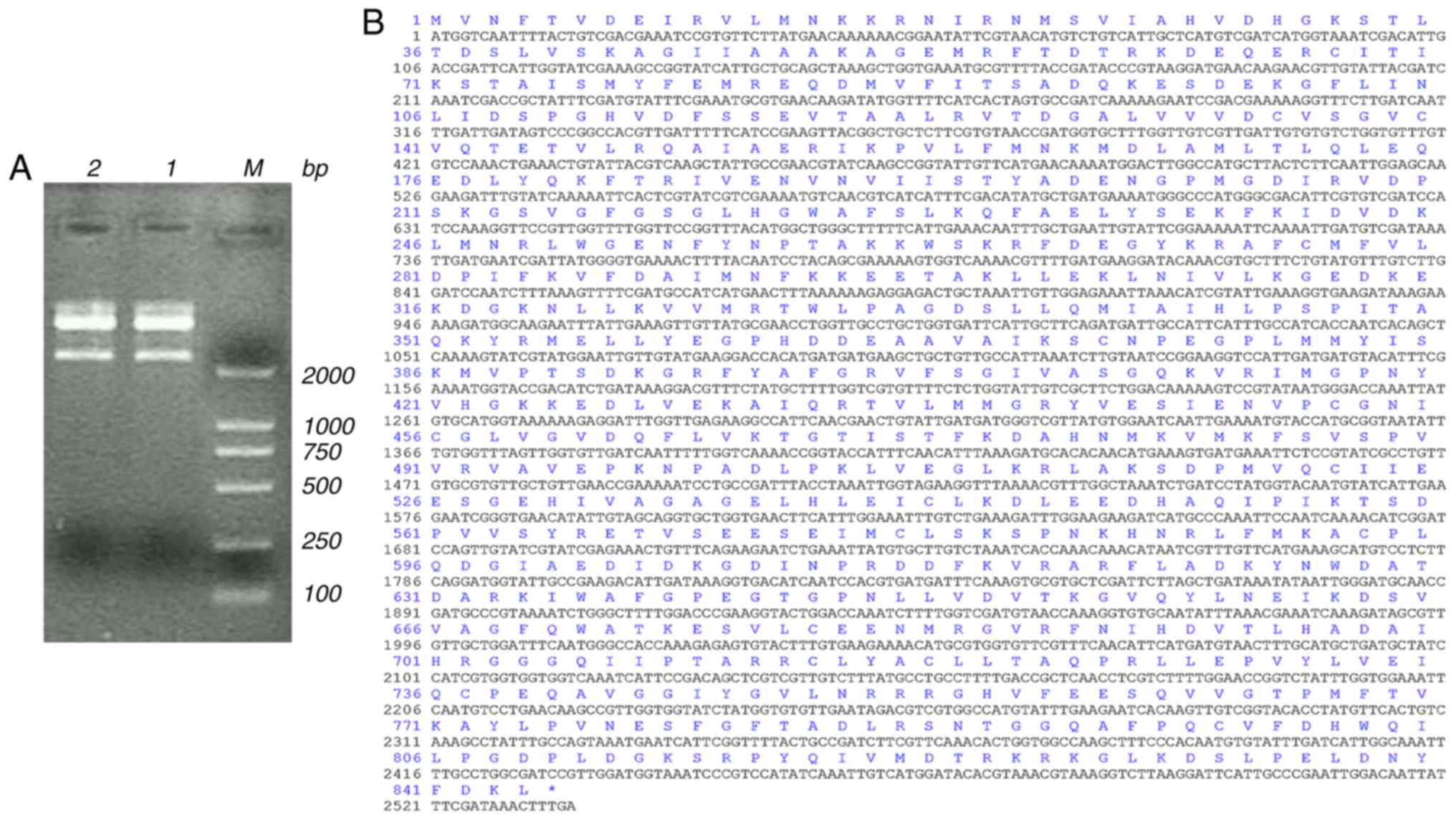
The cDNA of Der f TEF 2 showed bright bands at
~2,000 bp after double enzymatic digestion (Fig. 1A). The sequencing results showed
that one ORF of Der f TEF 2 is 2,535 bp long and encodes 844 amino
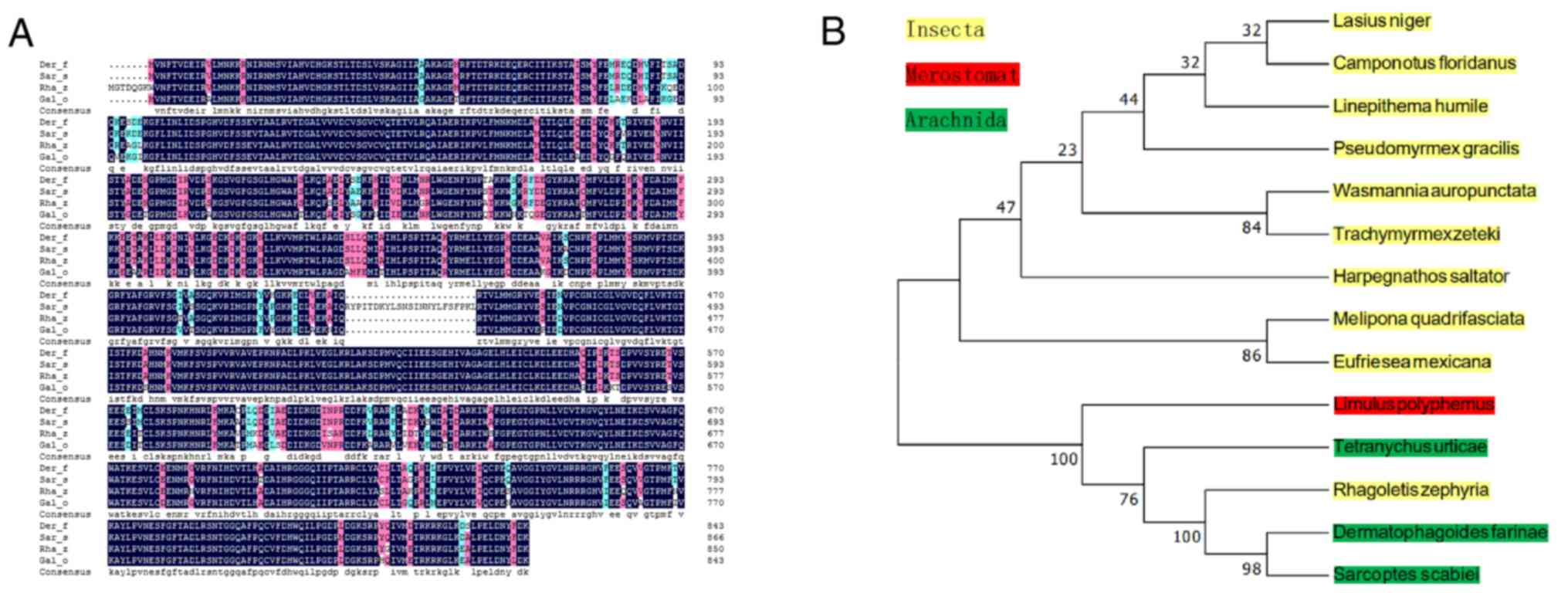
acids from the ATG start codon to the TAA stop codon (Fig. 1B). Alignment between Der f TEF 2
and the homologous amino acid sequences from different species of
Insecta, Merostomata and Arachnida was conducted using BLAST. Der f
TEF 2 is 94, 93 and 87% similar to Sarcoptes scabiei
(GenBank: KPM11996.1), Rhagoletis zephyria (GenBank:
XP_017490130.1) and Galendromus occidentalis (GenBank:
XP_003744110.1), respectively, indicating high homology (Fig. 2A). The homologous sequence was
output in FASTA format after using MEGA 5.0 and ClustalX 2.1
software to construct the molecular evolutionary tree. The results
showed that the TEF 2 gene from Der f has a relatively close
relationship with the TEF 2 gene from Lasius niger, D. farina,
Camponotus floridanus, Harpegnathos saltator, Melipona
quadrifasciata and Sarcoptes scabiei (Fig. 2B).
Immunogenicity of recombinant TEF
2
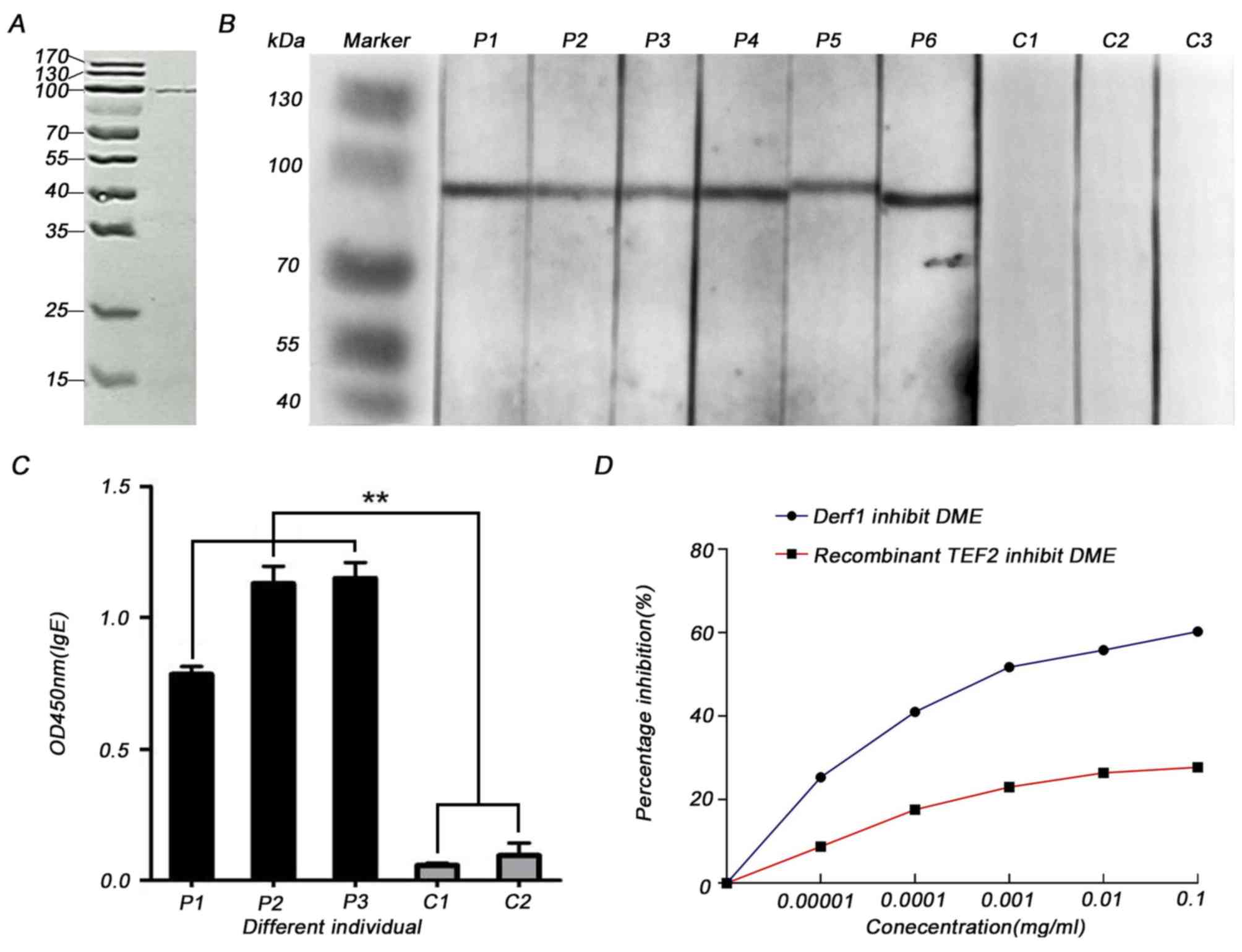
The SDS-PAGE analysis showed that TEF 2 was
successfully expressed had a molecular weight of ~100 kDa as
predicted (Fig. 3A). To determine
the allergenicity of recombinant TEF 2, immunoblot analysis was
used to determine if the recombinant protein could react with serum
from patients that are allergic to HDMs; the IgE-binding band had a
molecular weight of ~100 kDa as predicted (Fig. 3B). The serum IgE from patients
allergic to HDMs that was bound by recombinant TEF 2 was >6-fold
higher compared with the serum IgE from healthy controls when
quantified via ELISA (Fig. 3C).
For ELISA inhibition assays, dilutions of recombinant TEF 2 or DME
were incubated with the serum. The incubation inhibited the IgE
binding of the serum obtained from patients to the coated DME or
recombinant TEF 2 in a dose-dependent manner. The inhibition rate
of recombinant TEF 2 against DME was ~60% at 0.1 mg/ml (Fig. 3D). These results suggested that TEF
2 may be a novel allergen from the HDM allergen family that leads
to type I hypersensitivity.
Clinical SPTs
SPTs were performed with Der f TEF2 in 37 patients
sensitized to HDMs; 6 patients (16.2%) showed a positive reaction
(Table I).
 | Table I.Results of skin prick tests. |
Table I.
Results of skin prick tests.
|
|
|
| Net wheal size
(mm), level |
|---|
|
|
|
|
|
|---|
| Subject | Sex/age | Diagnosis | DME | Histamine | PS | r-Der f TEF 2 |
|---|
| 1 | Female/47 | BA, AR, FA | 4.5, ++ | 8 | 0 | 0 |
| 2 | Female/37 | AR | 2.5, ++ | 5 | 0 | 0 |
| 3 | Female/33 |
| 1.5, + | 5.75 | 0 | 0 |
| 4 | Male/24 | BA, AR | 2, + | 5 | 0 | 1.5, + |
| 5 | Male/33 | BA | 10, +++ | 6.5 | 0 | 0 |
| 6 | Male/36 | AR | 7.5, +++ | 5 | 0 | 0 |
| 7 | Female/43 | BA | 3.5, ++ | 5 | 0 | 0 |
| 8 | Female/51 | BA, AR | 2.5, ++ | 4.5 | 0 | 0 |
| 9 | Female/18 | BA, AR, DA | 3.5, +++ | 2 | 0 | 0 |
| 10 | Female/42 | BA, AR, FA | 3.5, ++ | 5 | 0 | 2, + |
| 11 | Female/47 | BA, AR, FA | 3, ++ | 6 | 0 | 0 |
| 12 | Male/48 | BA, AR | 5.5, +++ | 4.5 | 0 | 0 |
| 13 | Male/15 | BA, AR | 6, ++ | 7.75 | 0 | 0 |
| 14 | Female/39 | AR | 12.5, +++ | 8.5 | 0 | 0 |
| 15 | Male/63 | BA, DA | 8.5, +++ | 8 | 0 | 0 |
| 16 | Male/15 | BA, AR, FA | 6.5, +++ | 6 | 0 | 0 |
| 17 | Male/12 | BA, AR | 11, +++ | 6 | 0 | 0 |
| 18 | Female/18 | BA, AR | 5.5, +++ | 4.5 | 0 | 0 |
| 19 | Female/25 | AR | 6.5, +++ | 6.5 | 0 | 0 |
| 20 | Female/40 | BA, AR | 6.5, +++ | 5 | 0 | 0 |
| 21 | Male/75 | BA, AR, FA | 2.5, + | 6.5 | 0 | 0 |
| 22 | Male/11 | BA, AR, DA | 6.5, +++ | 5.5 | 0 | 0 |
| 23 | Female/26 | BA | 1, + | 5.5 | 0 | 0 |
| 24 | Male/15 | BA, AR, FA | 7, ++ | 8 | 0 | 2, + |
| 25 | Female/14 | BA, AR | 6.5, +++ | 6.5 | 0 | 0 |
| 26 | Male/8 | BA, AR, FA | 1, + | 4.5 | 0 | 0 |
| 27 | Female/43 | BA | 2, + | 4.5 | 0 | 0 |
| 28 | Male/47 | BA, AR | 3.5, ++ | 5.5 | 0 | 0 |
| 29 | Male/45 | AR | 2, + | 7.5 | 0 | 0 |
| 30 | Female/52 | AR, BA | 1.5, + | 5.5 | 0 | 0 |
| 31 | Male/20 | BA | 1.5, + | 5 | 0 | 0 |
| 32 | Female/20 | BA | 2, + | 4 | 0 | 0 |
| 33 | Female/13 | AR | 1.25, +++ | 6 | 0 | 2, + |
| 34 | Female/86 | BA, AR | 9, +++ | 5.5 | 0 | 2.75, ++ |
| 35 | Female/64 | BA | 2.25, + | 6 | 0 | 0 |
| 36 | Female/41 | BA, AR | 3.5, ++ | 5.5 | 0 | 0 |
| 37 | Female/44 | BA, AR | 4.5, +++ | 4.5 | 0 | 2, + |
Structural and functional
prediction
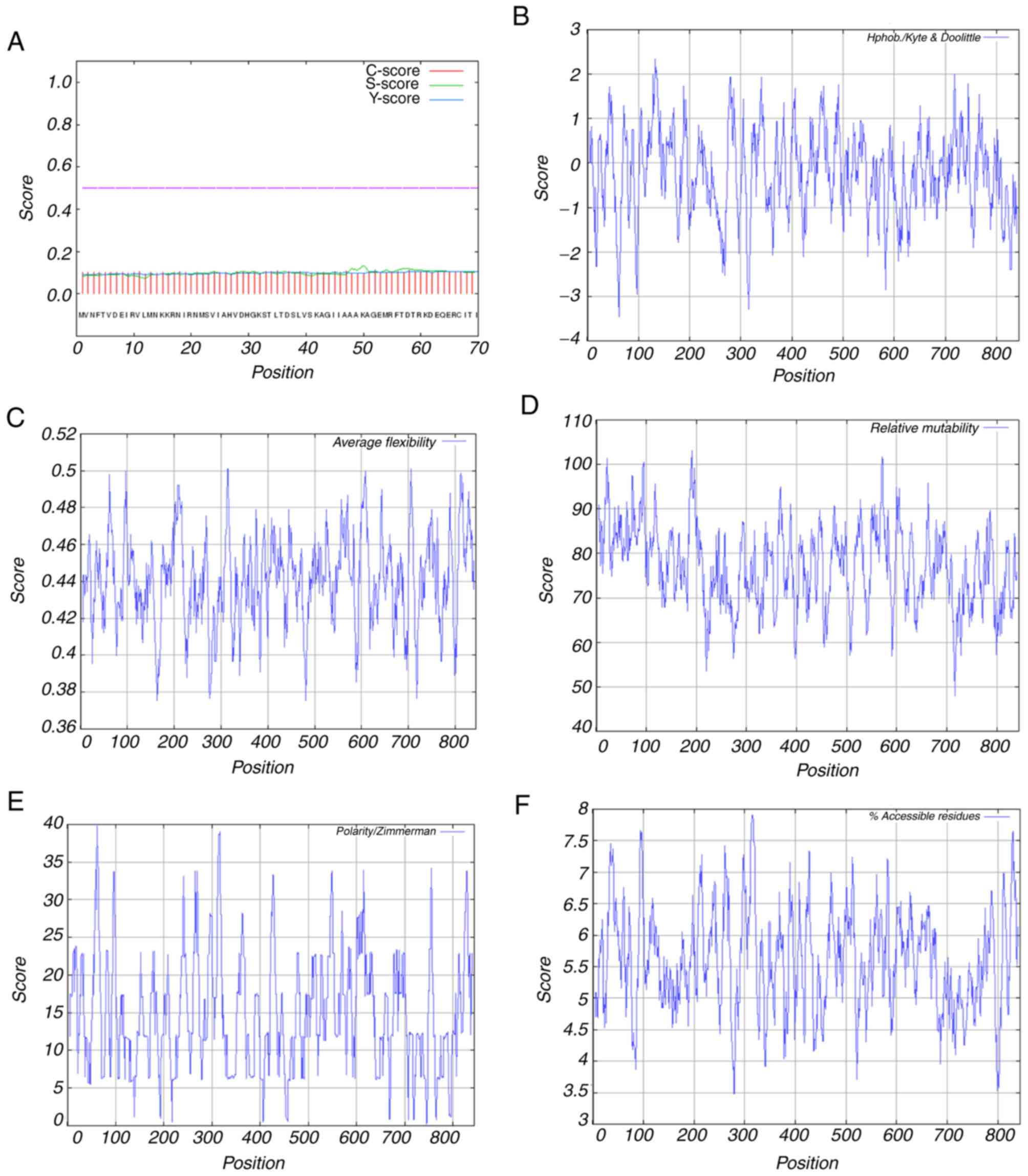
As shown, there was no cleavage site in the amino
acid sequence as predicted using SignalP 4.1 (Fig. 4A). Prediction of the
physicochemical properties of Der f TEF 2 using the ProtParam Tool
estimated that the molecular formula was
C4,216H6,703N1,141O1,240S47,
with a molecular weight of 94,722.32 Da and a theoretical
isoelectric point of 6.25. The grand average of hydrophobicity
(GRAVY) was predicted to be −0.222, the aliphatic index was 87.86
and the instability index was predicted to be 34.13. According to
the definition provided by ProtParam, when the instability
coefficient is <40, the protein is predicted to be stable;
therefore, the TEF 2 protein was classified as stable. Combined
with the analysis from the ProtScale software scale ‘Hphob./Kyte
& Doolittle’, these results indicated the hydrophilicity of the
protein. It showed that the most hydrophilic site of Der f TEF 2
appeared at amino acid position 62 with a score of −3.3456, and the
most hydrophobic site appeared at amino acid position 132 with a
score of 2.333 (Fig. 4B). The
maximum score for the average flexibility was calculated to be
0.501 at amino acid position 706; the minimum score at amino acid
position 481 was 0.376 (Fig. 4C).
The relative mutability was calculated to have a maximum score of
103.000 at amino acid position 191; the minimum score, at amino
acid position 716, was 47.889 (Fig.
4D). Using the Polarity/Zimmerman scale, the individual values
were scored at a maximum of 39.766 at residue 62 and at a minimum
of 0.237 at residue 745 (Fig. 4E).
The accessible residues (%) were calculated as a minimum score of
3.478 at residues 279 and 280, and a maximum score of 7.900 at
residues 314–316 (Fig. 4F).
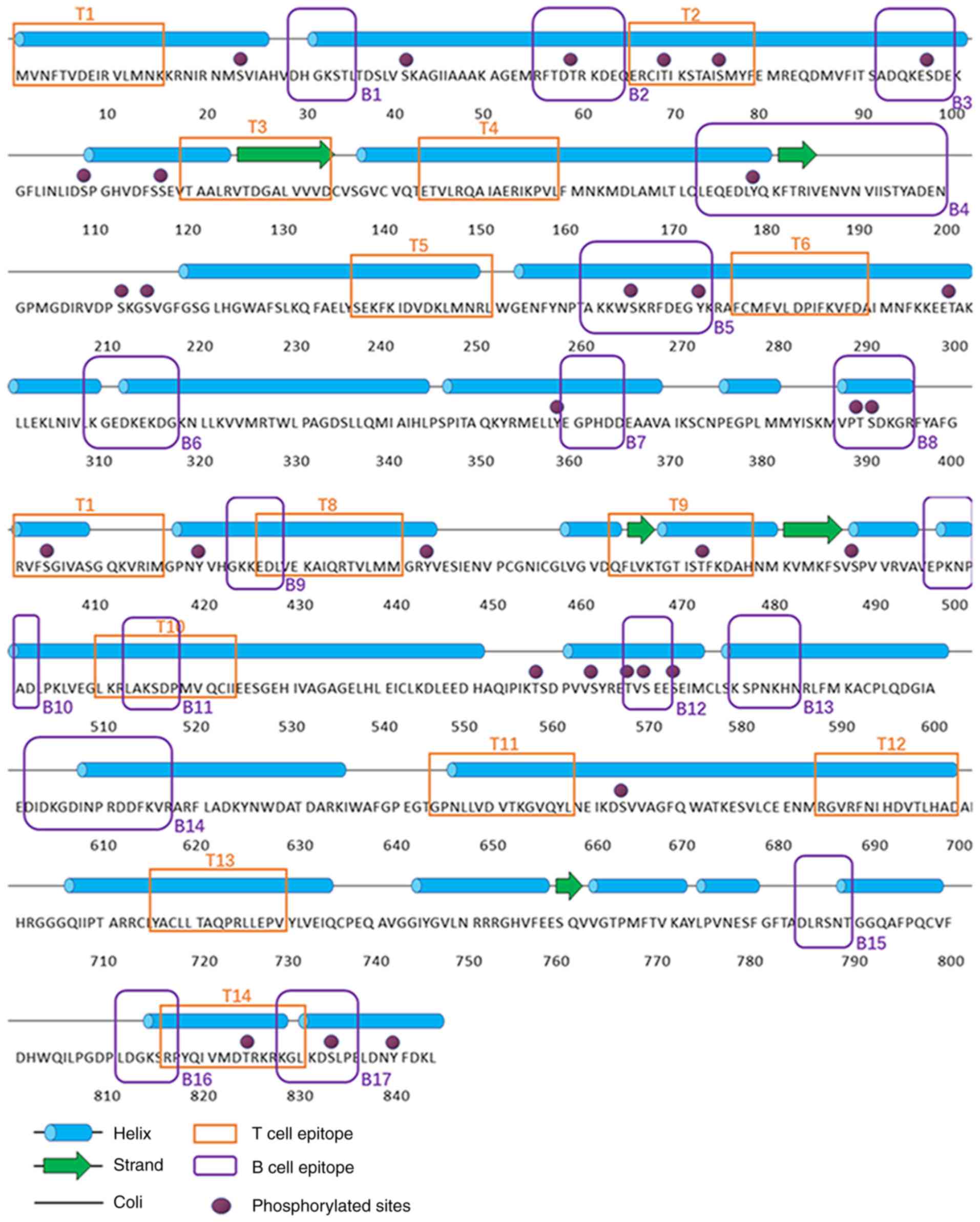
NetPhos3.1 identified 31 phosphorylation sites. In
total, 17 Ser residues (23, 41, 76, 97, 109, 116, 211, 214, 264,
391, 404, 488, 564, 570, 573, 664 and 833), 8 Thr residues (59, 69,
298, 390, 473, 558, 568, and 824) and 6 Tyr residues (179, 271,
359, 420, 443 and 840) were identified as potential phosphorylation
sites (Fig. 5). The secondary
structure prediction of Der f TEF 2 with DNAStar predicted 27
α-helices and 6 β-sheets in the protein (Fig. 5). The subcellular localization of
Der f TEF 2 was predicted to be in the plasma membrane and
extracellular space (Table II).
Moreover, Der f TEF 2 was predicted to be a P-loop-containing
nucleoside triphosphate hydrolase, Translation protein family
member with G_TR_2 and G_TR_1 peptidase domains (Table III).
 | Table II.Protein subcellular localization. |
Table II.
Protein subcellular localization.
| Support vector
machine | Localization | Reliability |
|---|
| Amino Acid
compartment | Cytoplasmic | 0.973 |
| N-peptide
compartment | Cytoplasmic | 0.704 |
| Partitioned seq.
compartment | Cytoplasmic | 0.854 |
| Physico-chemical
compartment | Cytoplasmic | 0.373 |
| Neighboring seq.
compartment CELLO Prediction: | Cytoplasmic | 0.942 |
|
| Cytoplasmic | 3.847 |
|
| Inner membrane | 0.580 |
|
| Periplasmic | 0.442 |
|
| Outer membrane | 0.088 |
|
| Extracellular | 0.044 |
 | Table III.Functional sites or motifs in the
allergen. |
Table III.
Functional sites or motifs in the
allergen.
| Prediction
tool | Functional sites or
motifs | Amino acid
position |
|---|
| InterPro 5.0 | P-loop containing
nucleoside triphosphate hydrolase | 4–345 |
|
| Small GTP-binding
protein domain | 20–174 |
|
| Translation
protein, β-barrel domain | 346–483 |
|
| Translation
elongation factor EFTu-like, domain 2 | 396–471 |
|
| EFG domain
III/V-like | 486–561 |
|
| Ribosomal protein
S5 domain 2-type fold | 563–727 |
|
| Ribosomal protein
S5 domain 2-type fold, subgroup | 580–720 |
|
| Translation
elongation factor EFG/EF2, domain IV | 608–723 |
|
| Elongation factor
EFG, domain V-like | 725–812 |
|
| Tr-type G domain,
conserved site | 58–73 |
| ScanProsite | Tr-type G domain
profile(G_TR_2) | 17–348 |
|
| Tr-type G domain
signature(G_TR_1) | 58–73 |
BPAP, DNAStar Protean and BepiPred 1.0 were used to
predict the B cell epitopes of Der f TEF 2. The hydrophilicity,
surface accessibility and flexibility of proteins play an important
role in the formation of antigens. When the hydrophilic score is
>0, the antigen index is >0 and surface accessibility is
>l, an epitope is likely to be formed. Consolidating the
predictions of these three tools, 17 potential B cell epitopes were
identified on Der f TEF 2 (amino acids 29–35, 55–64, 92–99,
173–200, 259–272, 311–318, 360–365, 388–395, 422–428, 496–502,
512–518, 567–572, 580–586, 602–617, 785–790, 811–817 and 827–836;
Fig. 5). The SYFPEITHI, NetMHCII
2.2 server, NetMHCIIpan-3.0 server, Immune Epitope Database and
Propred were applied to predict the T cell epitopes of Der f TEF 2.
Based on the results of these five immunoinformatics tools, the
potential epitopes of Der f TEF 2 were predicted to comprise 14
peptide sequences (amino acids 1–15, 65–79, 120–134, 144–159,
236–250, 275–289, 404–418, 426–440, 463–477, 510–524, 644–658,
684–698, 716–730 and 816–830; Table
IV and Fig. 5).
 | Table IV.Epitope prediction. |
Table IV.
Epitope prediction.
| Epitope, amino
acid | Sequence | SYFPEITHI | NetMHCII | NetMHCIIpan | IEDB | Preprod |
|---|
| 1–15 |
MVNFTVDEIRVLMNK | + + | + + | + + | + + | + + |
| 65–79 |
ERCITIKSTAISMYF | + + | + + | + + | + + | + + |
| 120–134 |
TAALRVTDGALVVVD | + + | + + | + + | + + | + + |
| 144–159 |
ETVLRQAIAERIKPVL | + + | + + | + + | + + | + + |
| 236–250 | EKFKIDVDKLMNRL | + + | + + | + + | + + | + + |
| 275–289 |
FCMFVLDPIFKVFDA | + + | + + | + + | + + | + + |
| 404–418 |
SGIVASGQKVRIMGP | + + | + + | + + | + + | + + |
| 426–440 |
EDLVEKAIQRTVLMM | + + | + + | + + | + + | + + |
| 463–477 |
QFLVKTGTISTFKDA | + + | + + | + + | + + | + + |
| 510–524 |
LKRLAKSDPMVQCII | + + | + + | + + | + + | + + |
| 644–658 |
GPNLLVDVTKGVQYL | + + | + + | + + | + + | + + |
| 684–698 |
RGVRFNIHDVTLHAD | + + | + + | + + | + + | + + |
| 716–730 | ACLLTAQPRLLEPV | + + | + + | + + | + + | + + |
| 816–830 |
RPYQIVMDTRKRKGL | + + | + + | + + | + + | + + |
Discussion
The present study used high-throughput sequencing
analysis of the HDM genome and transcriptome to study an HDM gene
that may be involved in the allergic response (20). Previous studies have shown that HDM
allergens are diverse, and numerous mite allergens have not been
completely identified or characterized (31). The identification and
characterization of novel HDM allergens may promote the development
of improved diagnostic tools and immunotherapeutic vaccines. TEF 2
is one of the most abundant proteins in eukaryotes and plays an
important role in various cellular processes, such as the
regulation of cellular energy metabolism and apoptosis (32,33).
It has been reported that Rubber Elongation Factor is the major
allergen in latex gloves, and triggers occupational latex allergy
in an increasing number of healthcare workers and patients
(34). The genes of Der f TEF 2
are highly conserved among different species, indicating that there
is cross-reactivity for this protein. Thus, it was important to
investigate the allergenicity of TEF 2 in D. farinae.
In the present study, Der f TEF 2 was identified and
characterized as a novel allergen from D. farinae via
cloning and expression of the recombinant protein. The recombinant
Der f TEF 2 is encoded by a 2,535-bp ORF that produces a predicted
sequence of 844 amino acids. Using a SPT, Der f TEF 2 was shown to
be an important allergen and was found to react with the sera from
16.2% of patients sensitized to HDMs. Furthermore, it was found
that the serum IgE from patients allergic to HDMs bound to the
recombinant TEF 2 protein and that the 6 patients with positive SPT
results showed high levels of recombinant TEF2-specific IgE in the
sera, as determined by ELISA and immunoblotting. Immune inhibition
assays showed IgE cross-reactivity between recombinant Der f TEF 2
and DME. These results suggested that Der f TEF 2 is a novel type
of allergen from HDMs.
Bioinformatics is an important technique for
predicting the corresponding sequences, structures, functional
properties and allergenicity of proteins (35). In the present study, the amino acid
physicochemical parameters, homology, phosphorylation sites,
secondary structure, and the T cell and B cell epitopes of Der f
TEF 2 were analyzed via bioinformatics tools. The predicted results
showed that Der f TEF 2 is highly homologous to various insect
species, contains 17 serine, 8 threonine and 6 tyrosine residues
that are potential phosphorylation sites, and is a relatively
stable protein. Using the software DNAStar to predict the secondary
structure of TEF 2, it was found that there are 27 α-helices and 5
β-sheets folds within the protein. In addition to these indices,
the average flexibility, relative mutability, polarity and the
accessible residues were determined using ProtScale tools. These
bioinformatic characterizations will facilitate the understanding
of the relationship between the structure and function of HDM
allergens. ProtParam predicted that the GRAVY score of Der f TEF 2
was −0.222 and the ProtScale prediction showed that the hydrophilic
peptide chains were distributed throughout the amino acid sequence,
and that there were significantly more hydrophilic than hydrophobic
peptide chains. These tools showed that Der f TEF 2 is hydrophilic
and soluble. The relative mutability indicates the probability that
an amino acid is replaced by another amino acid. The relative
mutability of Der f TEF 2 was found to have the highest score at
residue 716. The accessible residues are the sum of the
accessibility of the atoms, which, for Der f TEF 2, were predicted
with maximum scores at residues 314, 315 and 316; thus, the active
site is located on the surface of the protein.
Antigenic epitopes, also known as antigenic
determinants, are specific antigenic sites that can be recognized
by the corresponding antibody or antigen receptor and are the main
chemical substances of immune system recognition (36). According to the reverse vaccine
method, the use of bioinformatics technology for T cell epitope
prediction, and then effective immunological experiments to verify
the bioinformatics results, not only ensures the accuracy of the
results, but also can save the time and expense of generating
synthetic peptides (37).
To improve the accuracy of the predicted results,
three different tools were used (BPAP, DNAStar Protean and BepiPred
1.0) to synthesize the data and identify the B cell epitopes of Der
f TEF 2 (amino acids 29–35, 55–64, 92–99, 173–200, 259–272,
311–318, 360–365, 388–395, 422–428, 496–502, 512–518, 567–572,
580–586, 602–617, 785–790, 811–817 and 827–836). According to a
previous study, asthma is a disease associated with genetic
inheritance and the environment and HDM-induced asthma was
associated with the HLA-DRB1*0301 and HLA-DRB1*0401 alleles
(38). Therefore, the alleles
HLA-DRB1*0301 and HLA-DRB1*0401 were used to predict the T cell
epitopes of the D. farinae allergen TEF 2. Based on the
results of five algorithms (SYFPEITHI, NetMHCII 2.2 server,
NetMHCIIpan-3.0, IEDB and Propred), 14 HLA-DRB1*0301- and
HLA-DRB1*0401-restricted candidate T cell epitopes of Der f TEF 2
antigens were obtained (amino acid 1–15, 65–79, 120–134, 144–159,
236–250, 275–289, 404–418, 426–440, 463–477, 510–524, 644–658,
684–698, 716–730 and 816–830). The use of biotechnology to obtain
recombinant allergens can provide novel insight and strategies for
the development of specific immunotherapies and the treatment of
allergic diseases.
In summary, the present study showed that TEF 2
exhibits high immunogenicity. The preset study also analyzed the
structure and function of TEF 2 using bioinformatic technology,
which may provide safe and sensitive reagents for the diagnosis and
treatment of allergic diseases.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
Natural Science Foundation of China (grant nos. 91442118 and
91542104), the Science and Technology Project of Guangdong Province
(grant nos. 2013B031800023, 2014B090901041 and 2016A020216029), the
Shenzhen Scientific Technology Basic Research Projects (grant nos.
JCYJ20150525092941036 and JCYJ20160328144536436) and the PhD
Start-up Fund of the Natural Science Foundation of Guangdong
Province, China (grant no. 2014A030310192).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DC designed and performed experiments, analyzed
data, interpreted the results and was a major contributor in
writing the manuscript. QF and JL designed and performed
experiments, analyzed data, interpreted the results and wrote the
manuscript. CH, NH, KXC and BS provided key material and
interpreted the results. ZL interpreted the results, supervised the
study and edited the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Approval to conduct the present studies was obtained
from the Ethic Committee of the Institutional Review Board of the
School of Medicine, Shenzhen University. All participants,
including children approved by legal guardians, provided written
informed consent to participate in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Arlian LG: House-dust-mite allergens: A
review. Exp Appl Acarol. 10:167–186. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pawankar R, Canonica G and Holgate S:
World allergy organization (WAO) white book on allergy. Update.
2013.PubMed/NCBI
|
|
3
|
Valenta R, Linhart B, Swoboda I and
Niederberger V: Recombinant allergens for allergen-specific
immunotherapy: 10 years anniversary of immunotherapy with
recombinant allergens. Allergy. 66:775–783. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chapman MD, Smith AM, Vailes LD and Pomés
A: Recombinant allergens for immunotherapy. Allergy Asthma Proc.
23:5–8. 2002.PubMed/NCBI
|
|
5
|
Bousquet J and Michel FB: Specific
immunotherapy in asthma: Is it effective? J Allergy Clin Immunol.
94:1–11. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Thomas WR, Smith WA and Hales BJ: The
allergenic specificities of the house dust mite. Chang Gung Med J.
27:563–569. 2004.PubMed/NCBI
|
|
7
|
Valenta R and Niederberger V: Recombinant
allergens for immunotherapy. J Allergy Clin Immunol. 119:826–830.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Van Der Veen MJ, Jansen HM, Aalberse RC
and van der Zee JS: Der p 1 and Der p 2 induce less severe late
asthmatic responses than native Dermatophagoides pteronyssinus
extract after a similar early asthmatic response. Clin Exp Allergy.
31:705–714. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Trombone AP, Tobias KR, Ferriani VP,
Schuurman J, Aalberse RC, Smith AM, Chapman MD and Arruda LK: Use
of a chimeric ELISA to investigate immunoglobulin E antibody
responses to Der p 1 and Der p 2 in mite-allergic patients with
asthma, wheezing and/or rhinitis. Clin Exp Allergy. 32:1323–1328.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Meyer CH, Bond JF, Chen MS and Kasaian MT:
Comparison of the levels of the major allergens Der p I and Der p
II in standardized extracts of the house dust mite,
Dermatophagoides pteronyssinus. Clin Exp Allergy. 24:1041–1048.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shakib F, Ghaemmaghami AM and Sewell HF:
The molecular basis of allergenicity. Trends Immunol. 29:633–642.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen DX, Zhou PK and Ran PX: Screening and
identification of dominant allergens of Dermatophagoides
pteronyssinus and Dermatophagoides farina. China Trop Med.
39:431–436. 2009.
|
|
13
|
Walker C, Muniz MF, Rolim JM, Martins RR,
Rosenthal VC, Maciel CG, Mezzomo R and Reiniger LR: Morphological
and molecular characterization of Cladosporium cladosporioides
species complex causing pecan tree leaf spot. Genet Mol Res.
15:152016. View Article : Google Scholar
|
|
14
|
Berthelot K, Lecomte S, Estevez Y,
Coulary-Salin B, Bentaleb A, Cullin C, Deffieux A and Peruch F:
Rubber elongation factor (REF), a major allergen component in Hevea
brasiliensis latex has amyloid properties. PLoS One. 7:e480652012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen R and Snyder M: Promise of
personalized omics to precision medicine. Wiley Interdiscip Rev
Syst Biol Med. 5:73–82. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bahcall O: Precision medicine. Nature.
526:3352015. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ghaffari-Nazari H, Tavakkol-Afshari J,
Jaafari MR, Tahaghoghi-Hajghorbani S, Masoumi E and Jalali SA:
Improving Multi-epitope long peptide vaccine potency by using a
strategy that enhances CD4+ T help in BALB/c mice. PLoS
One. 10:e01425632015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nezafat N, Ghasemi Y, Javadi G, Khoshnoud
MJ and Omidinia E: A novel multi-epitope peptide vaccine against
cancer: An in silico approach. J Theor Biol. 349:121–134. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shahsavandi S, Ebrahimi MM, Sadeghi K and
Mahravani H: Design of a heterosubtypic epitope-based peptide
vaccine fused with hemokinin-1 against influenza viruses. Virol
Sin. 30:200–207. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chan TF, Ji KM, Yim AK, Liu XY, Zhou JW,
Li RQ, Yang KY, Li J, Li M, Law PT, et al: The draft genome,
transcriptome, and microbiome of Dermatophagoides farinae reveal a
broad spectrum of dust mite allergens. J Allergy Clin Immunol.
135:539–548. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lin JL, Wang YY, Xiao XJ, Wu YL, Sun BQ,
Gao AJ, Liu ZG, Li J, Yang PC and Liu XY: Characterization of a new
subtype of allergen in dermatophagoides farinae-Der f 28. J Thorac
Dis. 7:1842–1849. 2015.PubMed/NCBI
|
|
22
|
Lafarga T, O'Connor P and Hayes M:
Identification of novel dipeptidyl peptidase-IV and
angiotensin-I-converting enzyme inhibitory peptides from meat
proteins using in silico analysis. Peptides. 59:53–62. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Thirugnanasambantham K, Muralidaran S and
Mandal AK: Molecular cloning, computational and expression analysis
of anthocyanidin reductase in tea (Camellia sinensis). Appl Biochem
Biotechnol. 174:130–145. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu CS, Lin CJ and Hwang JK: Predicting
subcellular localization of proteins for Gram-negative bacteria by
support vector machines based on n-peptide compositions. Protein
Sci. 13:1402–1406. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lu YJ, Lu G, Shi DZ, Li LH and Zhong SF:
Bioinformatic analysis for structure and function of TCTP from
Spirometra mansoni. Asian Pac J Trop Med. 6:709–712. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Seddigh S: Comprehensive comparison of two
protein family of P-ATPases (13A1 and 13A3) in insects. Comput Biol
Chem. 68:266–281. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lin J, Wang H, Li M, Liang Z, Jiang C, Wu
Y, Liu Z, Yang P and Liu X: Characterization and analysis of a cDNA
coding for the group 29b (Der f 29b) allergen of Dermatophagoides
farinae. Am J Transl Res. 8:568–577. 2016.PubMed/NCBI
|
|
28
|
Chen H, Yang HW, Wei JF and Tao AL: In
silico prediction of the T-cell and IgE-binding epitopes of Per a 6
and Bla g 6 allergens in cockroaches. Mol Med Rep. 10:2130–2136.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li X, Yang HW, Chen H, Wu J, Liu Y and Wei
JF: In silico prediction of T and B cell epitopes of Der f 25 in
Dermatophagoides farinae. Int J Genomics. 2014:4839052014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang H, Chen H, Jin M, Xie H, He S and Wei
JF: Molecular cloning, expression, IgE binding activities and in
silico epitope prediction of Per a 9 allergens of the American
cockroach. Int J Mol Med. 38:1795–1805. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
An S, Chen L, Long C, Liu X, Xu X, Lu X,
Rong M, Liu Z and Lai R: Dermatophagoides farinae allergens
diversity identification by proteomics. Mol Cell Proteomics.
12:1818–1828. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Faller WJ, Jackson TJ, Knight JR, Ridgway
RA, Jamieson T, Karim SA, Jones C, Radulescu S, Huels DJ, Myant KB,
et al: mTORC1-mediated translational elongation limits intestinal
tumour initiation and growth. Nature. 517:497–500. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Leprivier G, Remke M, Rotblat B, Dubuc A,
Mateo AR, Kool M, Agnihotri S, El-Naggar A, Yu B, Somasekharan SP,
et al: The eEF2 kinase confers resistance to nutrient deprivation
by blocking translation elongation. Cell. 153:1064–1079. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Marchetti-Deschmann M and Allmaier G:
Allergenic compounds on the inner and outer surfaces of natural
latex gloves: MALDI mass spectrometry and imaging of proteinous
allergens. J Mass Spectrom. 44:61–70. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cui Y, Teng F, Yu L, Zhou Y, Zhang C and
Yang L: Dermatophagoides farinae allergen Der f 9: Cloning,
expression, purification, characterization and IgE-binding in
children with atopic asthma. Pediatr Pulmonol. 52:282–292. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yu S, Zhang H, Yao D, Liu W, Wang X, Chen
X, Wei Y, Zhang Z, Wang J, Yu L, et al: Identification of
CD4+ T-cell epitopes on iron-regulated surface
determinant B of Staphylococcus aureus. Microb Pathog. 89:108–113.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Masignani V, Rappuoli R and Pizza M:
Reverse vaccinology: A genome-based approach for vaccine
development. Expert Opin Biol Ther. 2:895–905. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Movahedi M, Moin M, Gharagozlou M,
Aghamohammadi A, Dianat S, Moradi B, Nicknam MH, Nikbin B and
Amirzargar A: Association of HLA class II alleles with childhood
asthma and Total IgE levels. Iran J Allergy Asthma Immunol.
7:215–220. 2008.PubMed/NCBI
|