Introduction
The mammalian target of rapamycin (mTOR) plays a
central role in cell physiology and controls several cellular
functions, including proliferation, growth, survival, autophagy and
metabolism (1). mTOR has emerged
as a critical effector of cell-signalling pathways commonly
upregulated in several types of human cancer and is a major target
for cancer therapy (2).
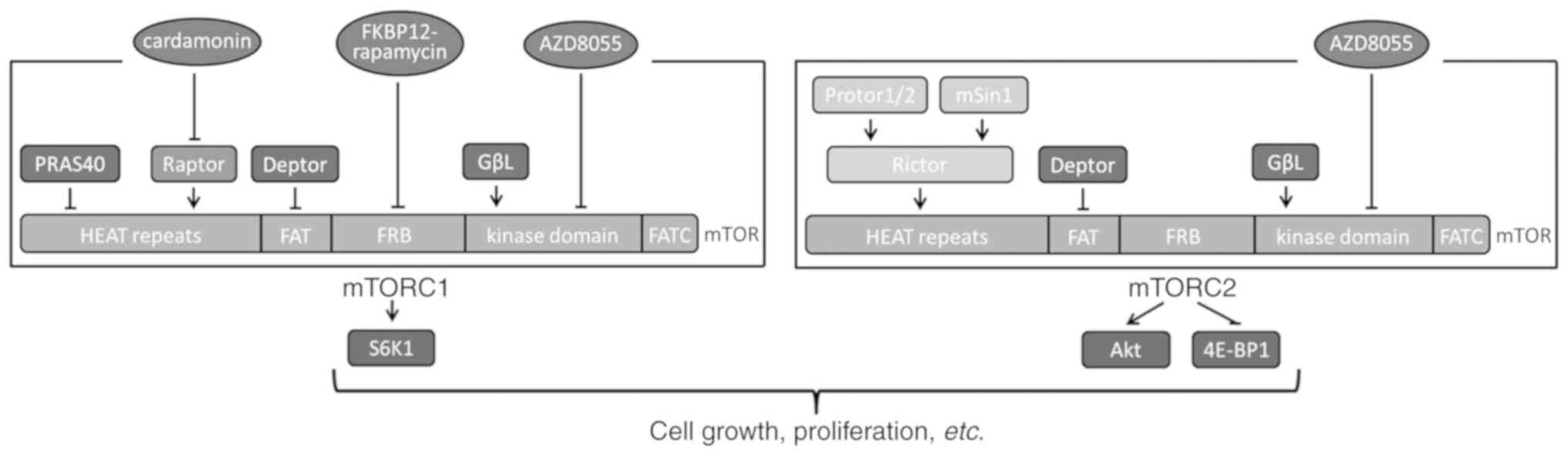
mTOR exists in two functionally and structurally
distinct multiprotein complexes termed mTOR complex (mTORC) 1 and
mTORC2 (Fig. 1). mTORC1 contains
the regulatory-associated protein of mTOR (Raptor), proline-rich
protein kinase B (Akt) substrate 40 kDa, G-protein β-subunit-like
protein/LST8 (GβL) and DEP domain containing mTOR-interacting
protein (DEPTOR). Raptor is an essential component of mTORC1 and
recruits ribosomal protein S6 kinase B1 (S6K1) to mTOR for
phosphorylation (3). mTORC2 is
mainly comprised of a rapamycin-insensitive companion of mTOR, GβL
and DEPTOR. mTORC2 phosphorylates Akt on Ser 473 and eukaryotic
translation initiation factor 4E binding protein 1 (EIF4EBP1) on
Thr 37/46 (4).
Two classes of mTOR inhibitors are currently in
clinical use or undergoing clinical trials for cancer treatment
(5). Rapamycin was the first mTOR
inhibitor to be identified. The rapalogs (rapamycin and its
analogues) form a gain-of-function complex with 12-kDa
FK506-binding protein (FKBP12), which binds to the
FKBP12/rapamycin-binding (FRB) domain of mTOR (6). The rapamycin/FKBP12 complex
allosterically inhibits kinase activity of mTOR and disrupts the
association of Raptor with mTORC1 (7). Additionally, it inhibits the
phosphorylation of S6K1, but has a lesser impact on the
phosphorylation of EIF4EBP1 (Thr 37/46) (8). Rapalogs may be used to treat a wide
range of malignancies and numerous clinical trials have been
performed in cancer patients (9,10).
However, the efficacy of rapalogs as monotherapy for patients with
breast cancer, kidney cancer and pancreatic neuroendocrine tumours
is not as promising as initially expected, as only a subset of
patients exhibit objective responses to rapalogs and the responses
are frequently short-lived (10).
Acquired resistance has emerged as a barrier to the antineoplastic
activity of this class of mTOR inhibitors (11,12).
The second generation of mTOR inhibitors includes mTORC1/mTORC2
dual inhibitors, such as AZD8055, torin 1 and PP242. These
inhibitors target the ATP-binding site of the mTOR kinase domain
and are collectively called ATP-competitive mTOR inhibitors
(13). AZD8055 exhibited more
powerful antiproliferative and proapoptotic effects, as well as
more complete inhibition of mTORC1, compared with rapalogs in
preclinical studies, which likely results from its additional
inhibitory effect on Akt and EIF4EBP1 phosphorylation (14). Several ATP-competitive mTOR
inhibitors have been or are currently being investigated in
clinical trials for a wide variety of malignancies (11,15).
Unfortunately, primary or acquired resistance has begun to emerge
(16). A previous study
investigating a xenografted model of human breast cancer revealed
that AZD8055 treatment completely inhibited tumour growth. However,
after 11 days of treatment, tumour regrowth was observed (17).
Targeted cancer therapy exerts selective pressure on
tumour cells, which leads to the preferential growth of resistant
subpopulations and necessitates the development of next generation
therapies to treat the resistant cancer. Cardamonin is as an mTOR
inhibitor that has been shown to decrease the proliferation of
various cancer cells (18–20). Recent studies have demonstrated
that cardamonin and its analogues decrease the proliferation of
non-small-cell lung cancer cells and prevent metastasis by
inhibiting the mTOR signalling pathway (21,22).
Furthermore, Jin et al (23) revealed that cardamonin modulates
cell metabolism by repressing the activities of mTOR and S6K1 in
breast cancer cells. Cardamonin, unlike rapamycin, inhibits mTOR
without the involvement of FKBP12 (20). Additionally, cardamonin has no
effect on the phosphorylation of Akt, which is decreased by AZD8055
(24). Cardamonin has previously
been shown to inhibit the mTORC1 signalling pathway by decreasing
the protein level of Raptor (24,25).
You et al (26)
demonstrated that cardamonin exerts cardioprotective effects in
left ventricular remodelling by disrupting the mTOR-Raptor
association, suggesting that cardamonin is a specific mTORC1
inhibitor.
In the present study, resistant MCF-7 breast cancer
cells and HeLa cervical cancer cells were generated by exposing the
parental cells to gradually increasing concentrations of rapamycin
or AZD8055. The inhibitory effect of cardamonin on the
proliferation and the mTOR signaling pathway in the rapamycin- and
AZD8055-resistant cells was subsequently investigated.
Materials and methods
Chemical reagents
Cell culture supplies were purchased from Gibco;
Thermo Fisher Scientific, Inc. AZD8055 was obtained from
AstraZeneca Pharmaceuticals. Cardamonin, rapamycin and MTT were
purchased from Sigma-Aldrich; Merck KGaA. Antibodies against mTOR
(rabbit mAb; cat. no. 2972), phosphorylated (p)-mTOR (Ser 2448;
rabbit mAb; cat. no. 2971), S6K1 (rabbit mAb; cat. no. 9202),
p-S6K1 (Thr 389; rabbit mAb; cat. no. 9205), Akt (rabbit mAb; cat.
no. 9272), p-Akt (Ser 473; rabbit mAb; cat. no. 4060), EIF4EBP1
(rabbit mAb; cat. no. 9452), p-EIF4EBP1 (T37/46; rabbit mAb; cat.
no. 9459), Raptor (rabbit mAb; cat. no. 2280), β-actin (rabbit mAb;
cat. no. 4970) and the secondary antibody (anti-rabbit IgG, mouse
horseradish peroxidase-linked; cat. no. 7074) were purchased from
Cell Signalling Technologies, Inc.
Cell culture
MCF-7 and HeLa cells were obtained from The Cell
Bank of the Type Culture Collection of the Chinese Academy of
Sciences. MCF-7 cells were cultured in a 1:1 mixture of DMEM:F12
medium (HyClone; GE Healthcare Life Sciences) supplemented with 4
mM glutamine. HeLa cells were cultured in RPMI 1640 media (HyClone;
GE Healthcare Life Sciences). Both media were supplemented with 10%
FBS (HyClone; GE Healthcare Life Sciences), penicillin (100 U/ml)
and streptomycin (100 µg/ml). The cells were incubated at 37°C with
5% CO2.
Selection of drug resistant
clones
Resistant MCF-7 breast cancer and HeLa cervical
cancer cells were generated by exposing the parental cells to a
gradually increasing concentration (5–500 nM) of either rapamycin
or AZD8055 over 8 months. The media was replaced weekly. The
resistant cells were subsequently generated through a single cell
clone selection and tested for sensitivity to rapamycin or AZD8055.
The resistant cells were passaged in drug-free media over 12
months. The sensitivity of the two clones with the greatest
resistance (i.e., the highest IC50 to rapamycin and
AZD8055, respectively) to rapamycin or AZD8055 was assessed.
Cell viability assay
The MTT assay was used to analyse the effect of
cardamonin on cell viability as previously described (20). Parental MCF-7, HeLa and mTOR
inhibitor resistant MCF-7, HeLa cells (5×103 cells per
well) were seeded in 96-well plates and cultured overnight. A total
of 20 µl rapamycin or AZD8055 at the indicated concentrations (0.1,
0.3, 1, 3, 10, 30, 100, 300, 1,000, 3,000 and 10,000 nM) or
cardamonin at the indicated concentrations (1, 1.8, 3.2, 5.6, 10,
18, 32, 56, 100, 180 and 320 µM) was added to each well and the
cells were incubated for 48 h. A total of 10 µl MTT (5 mg/ml)
solution was added to each well and the cells were incubated for an
additional 4 h. The purple formazan crystals were dissolved using
DMSO and the number of surviving cells was assessed by determining
the optical density at a wavelength of 570 nm using a microplate
reader. The IC50 was determined by fitting to a standard
4-parameter logistic using GraphPad Prism software (version 5;
GraphPad Software, Inc.).
Clonogenic survival assay
The clonogenic survival assay was performed as
previously described (20).
Parental MCF-7, HeLa and mTOR inhibitor resistant MCF-7, HeLa cells
(1×103/well) were seeded in a 6-well plate and incubated
overnight. The cells were treated with rapamycin or AZD8055 for 48
h, followed by two washes with their respective media. The cells
were subsequently cultured for 7 days. Cells were fixed with
ethanol (75%) at room temperature for 15 min and stained with 1%
crystal violet at room temperature for 60 min. Colonies (>30
cells/colony) were counted using a Leica DMIL LED microscope
(magnification, ×200; Leica Microsystems GmbH) in triplicate wells.
Five independent experiments were performed.
Western blotting
The parental MCF-7, HeLa and mTOR inhibitor
resistant MCF-7, HeLa cells were treated with rapamycin or AZD8055
for 48 h. The cells were subsequently washed twice with ice-cold
PBS and lysed using radioimmunoprecipitation assay lysis buffer [20
mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM
EGTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium
pyrophosphate, 1 mM β-glycerophosphate, 1 mM
Na3VO4 and 1 µg/ml leupeptin] and 1 mM
phenylmethylsulfonyl fluoride for 20 min at 4°C. The lysates were
centrifuged at 15,000 × g at 4°C for 20 min. The total protein
concentration in the supernatant was determined using a
bicinchoninic acid assay and 40 µg protein/lane was separated via
6–12% SDS-PAGE. The separated proteins were subsequently
transferred onto a polyvinylidene difluoride membrane and blocked
for 1 h at room temperature with 5% bovine serum albumin (Cell
Signalling Technologies, Inc.) in 1X TBST (0.1% Tween 20). The
membrane was incubated overnight at 4°C with the primary antibodies
(all used at a 1:1,000). The membrane was subsequently incubated
the horseradish peroxidase-conjugated goat anti-rabbit
immunoglobulin (1:2,000) at room temperature for 1 h. Protein bands
were visualized using enhanced chemiluminescence (SignalFire ECL
reagent; cat. no. 6883; Cell Signalling Technologies, Inc) and an
X-ray film.
Statistical analysis
All data are presented as the mean ± standard
deviation. Statistical analyses were performed using SPSS software
(version 19; IMB Corp.). The one-way analysis of variance followed
by the Tukey-Kramer multiple comparison test was used to compare
the different groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
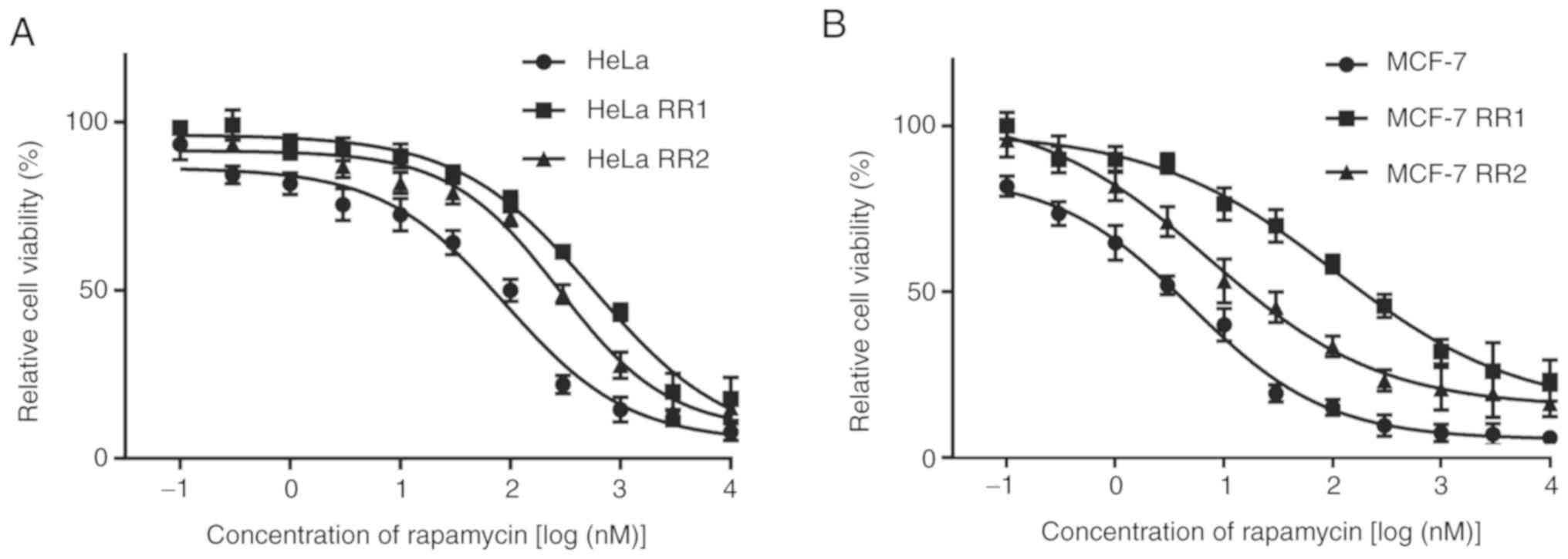
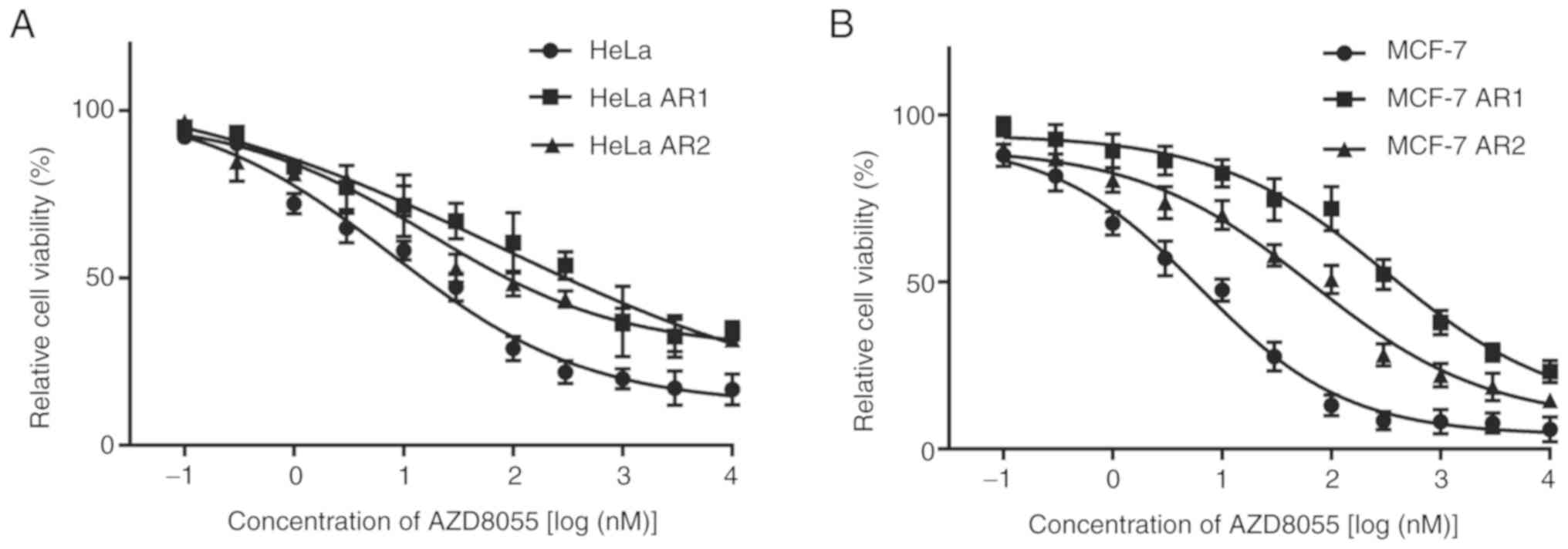
Generation of HeLa and MCF-7 clones
with acquired resistance to rapamycin or AZD8055
Rapamycin-resistant (RR) and AZD8055-resistant (AR)
HeLa and MCF-7 cells were generated by culturing parental HeLa and
MCF-7 cells with a gradually increasing concentration of rapamycin
or AZD8055 for 8 months. Parental HeLa and MCF-7 cells were
sensitive to rapamycin and AZD8055. The resistance of the selected
cell clones was determined using the MTT assay. The sensitivity of
the two clones with the greatest resistance (i.e., the highest
IC50 to rapamycin and AZD8055, respectively) to
rapamycin or AZD8055 was assessed. RR and AR clones were
significantly less sensitive to their respective drugs than the
parental cells at 48 h (Figs. 2
and 3; Tables I and II). The clone with the highest
resistance was selected for subsequent experiments. These clones
retained resistance to rapamycin or AZD8055 when passaged in
drug-free media over 12 months, demonstrating that the acquired
resistance was not transient.
 | Table I.IC50 of rapamycin in the
cells. |
Table I.
IC50 of rapamycin in the
cells.
| Cells | IC50
(µM) |
|---|
| HeLa | 91.22±13.21 |
| HeLa RR1 |
569.8±32.25a |
| HeLa RR2 |
279.5±20.54a |
| MCF-7 | 5.01±0.61 |
| MCF-7 RR1 |
204.4±28.43b |
| MCF-7 RR2 |
27.32±5.13b |
 | Table II.IC50 of AZD8055 in the
cells. |
Table II.
IC50 of AZD8055 in the
cells.
| Cells | IC50
(µM) |
|---|
| HeLa | 9.75±1.29 |
| HeLa AR1 |
247.4±21.25a |
| HeLa AR2 |
76.8±9.05a |
| MCF-7 | 6.28±1.64 |
| MCF-7 AR1 |
325.6±22.65b |
| MCF-7 AR2 |
66.78±7.43b |
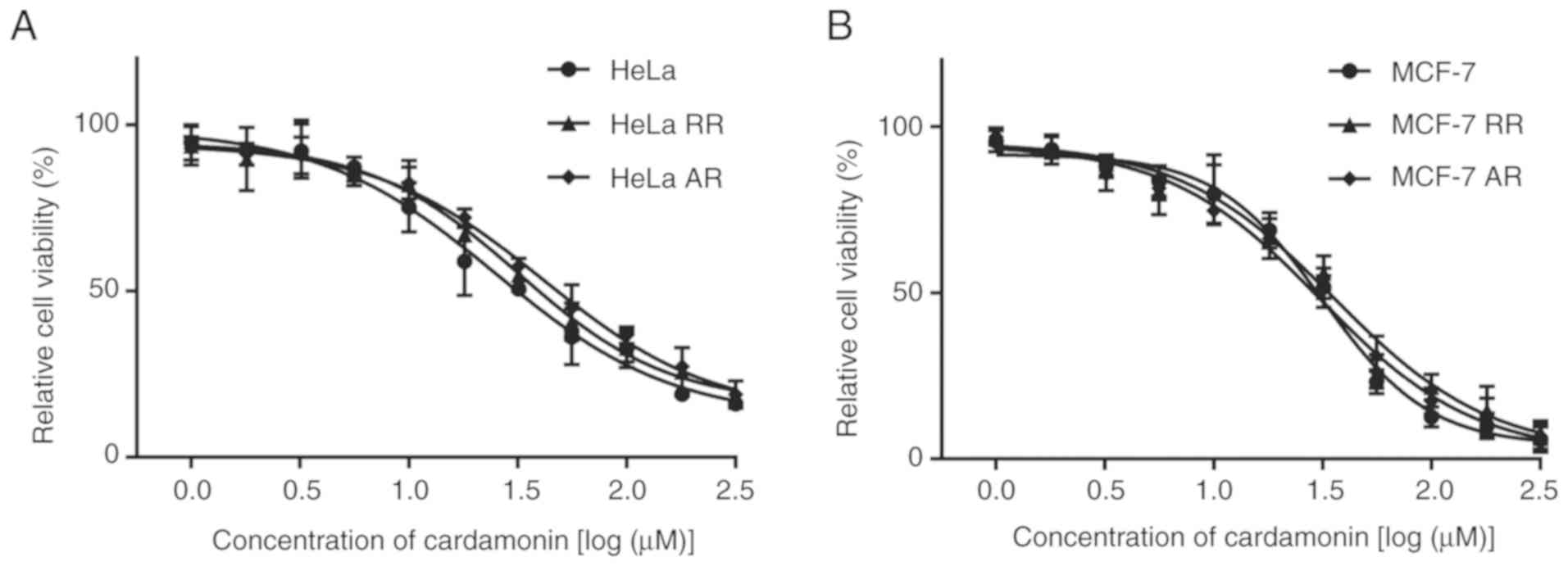
Cardamonin inhibits proliferation of
the mTOR inhibitor-resistant cells
The inhibitory activity of cardamonin in the
parental and resistant cells was investigated. Cardamonin inhibited
the proliferation of HeLa and MCF-7 cells. However, unlike
rapamycin and AZD8055, cardamonin also had an inhibitory effect on
the growth of RR and AR HeLa and MCF-7 cells (Fig. 4). The inhibitory efficacy of
cardamonin on the resistant cells was similar to the parental cells
(Table III).
 | Table III.IC50 of cardamonin in the
cells. |
Table III.
IC50 of cardamonin in the
cells.
| Cells | IC50
(µM) |
|---|
| HeLa | 23.71±3.96 |
| HeLa RR | 28.56±2.43 |
| HeLa AR | 30.40±3.08 |
| MCF-7 | 35.52±3.87 |
| MCF-7 RR | 37.35±3.92 |
| MCF-7 AR | 38.14±2.04 |
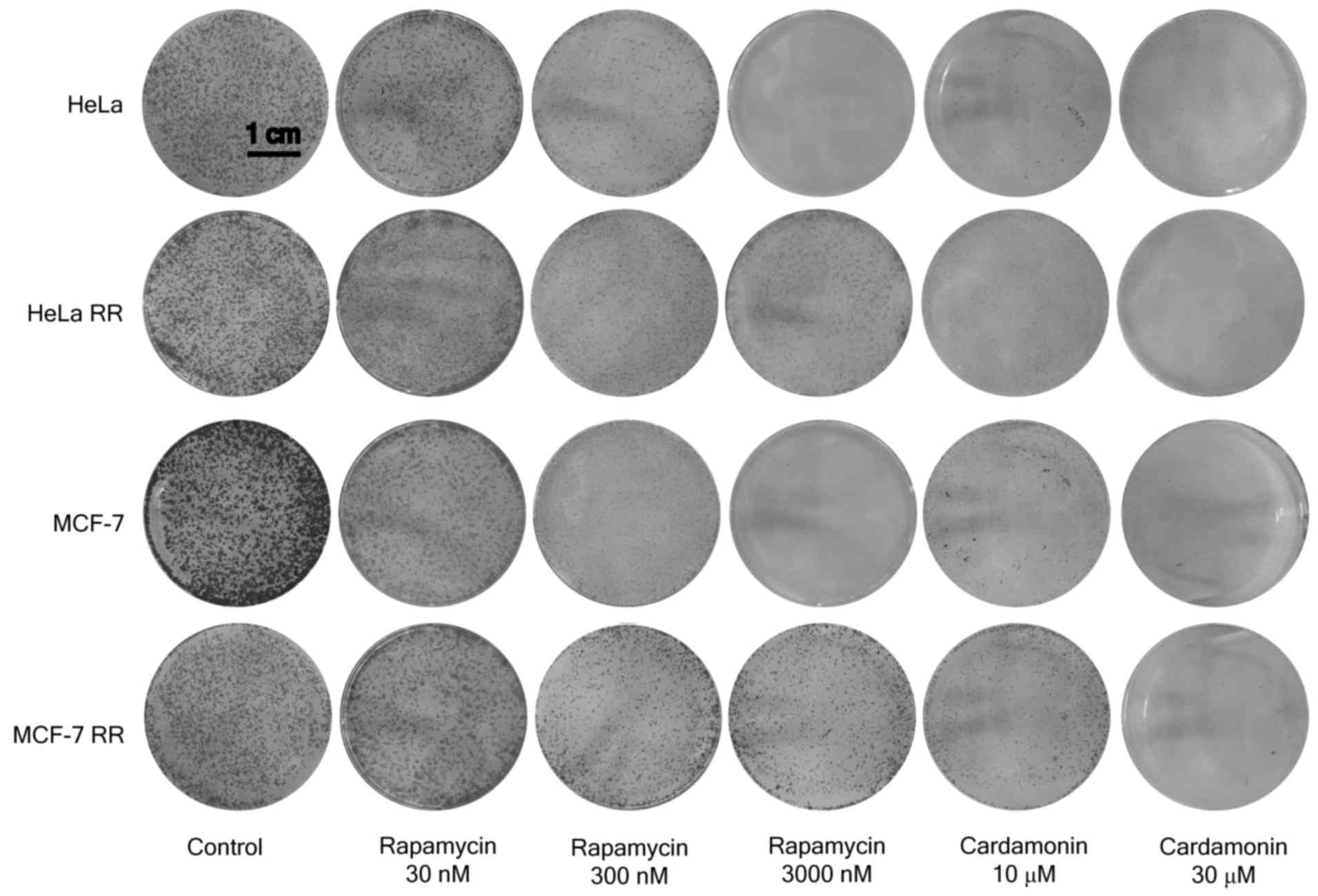
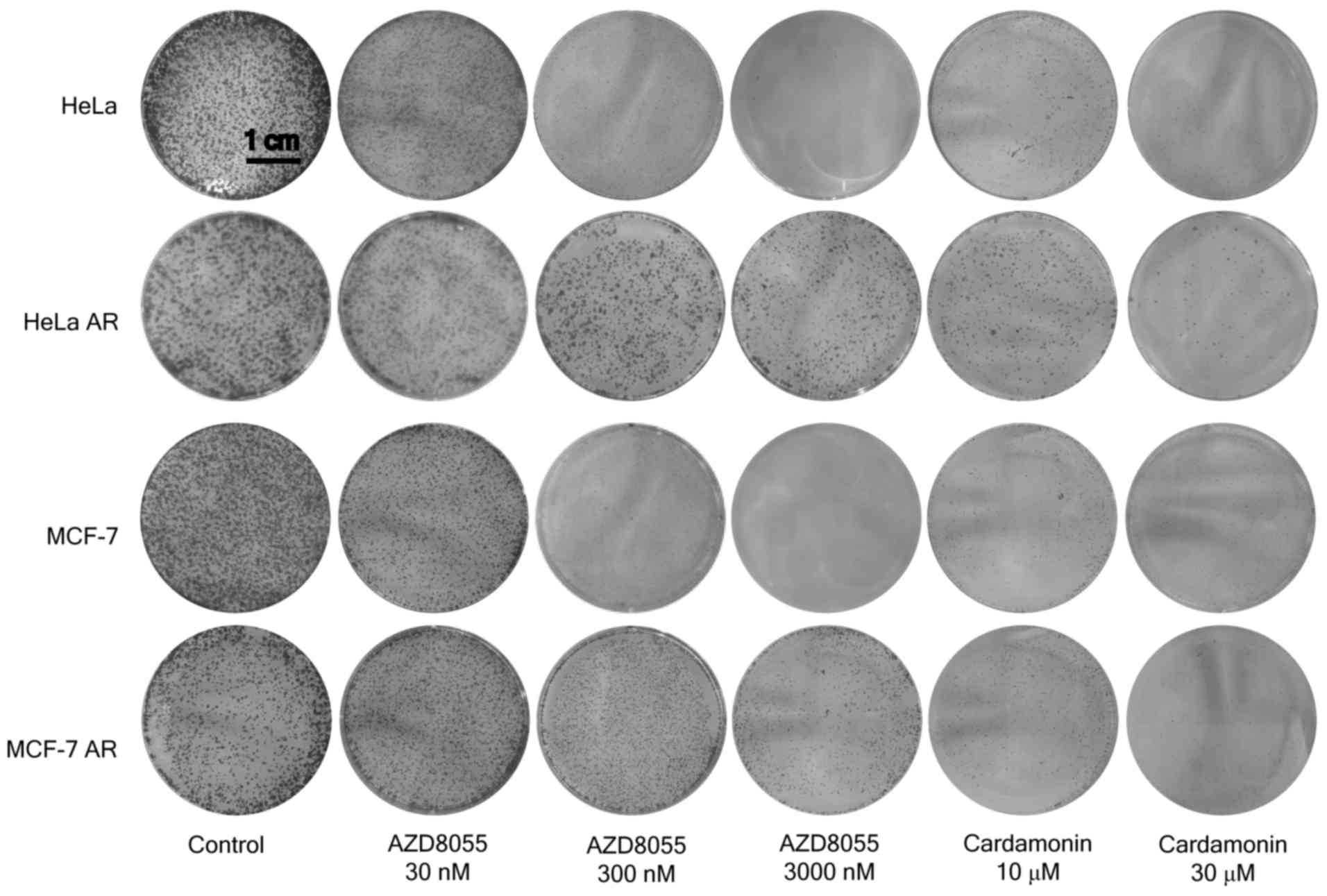
Cardamonin inhibits clone formation of
the mTOR inhibitor-resistant cells
Consistent with the results of the proliferation
assay, the RR and AR MCF-7 and HeLa cells were less sensitive to
rapamycin and AZD8055 in the clone formation assay when compared
with the parental cells. The resistant cells maintained full
sensitivity to cardamonin (Figs. 5
and 6).
Cardamonin inhibits the activity of
mTORC1 in the mTOR inhibitor-resistant cells
In the rapamycin-resistant cells, phosphorylation of
the rapamycin-sensitive site on S6K1 (T389) was unaffected even at
higher concentrations of rapamycin (300 nM). Phosphorylation of the
key mTORC2 effectors, Akt and EIF4EBP1, was unaffected by rapamycin
but strongly reduced by AZD8055. In the AR cells, phosphorylation
of Akt and EIF4EBP1 was less sensitive to AZD8055. As expected, in
the parental and resistant cells, the phosphorylation of mTOR and
its substrate S6K1 was inhibited by cardamonin while that of Akt
and EIF4EBP1 was not affected. Interestingly, compared with AZD8055
and rapamycin, cardamonin decreased the protein level of Raptor in
RR and AR cells (Figs. 7 and
8).
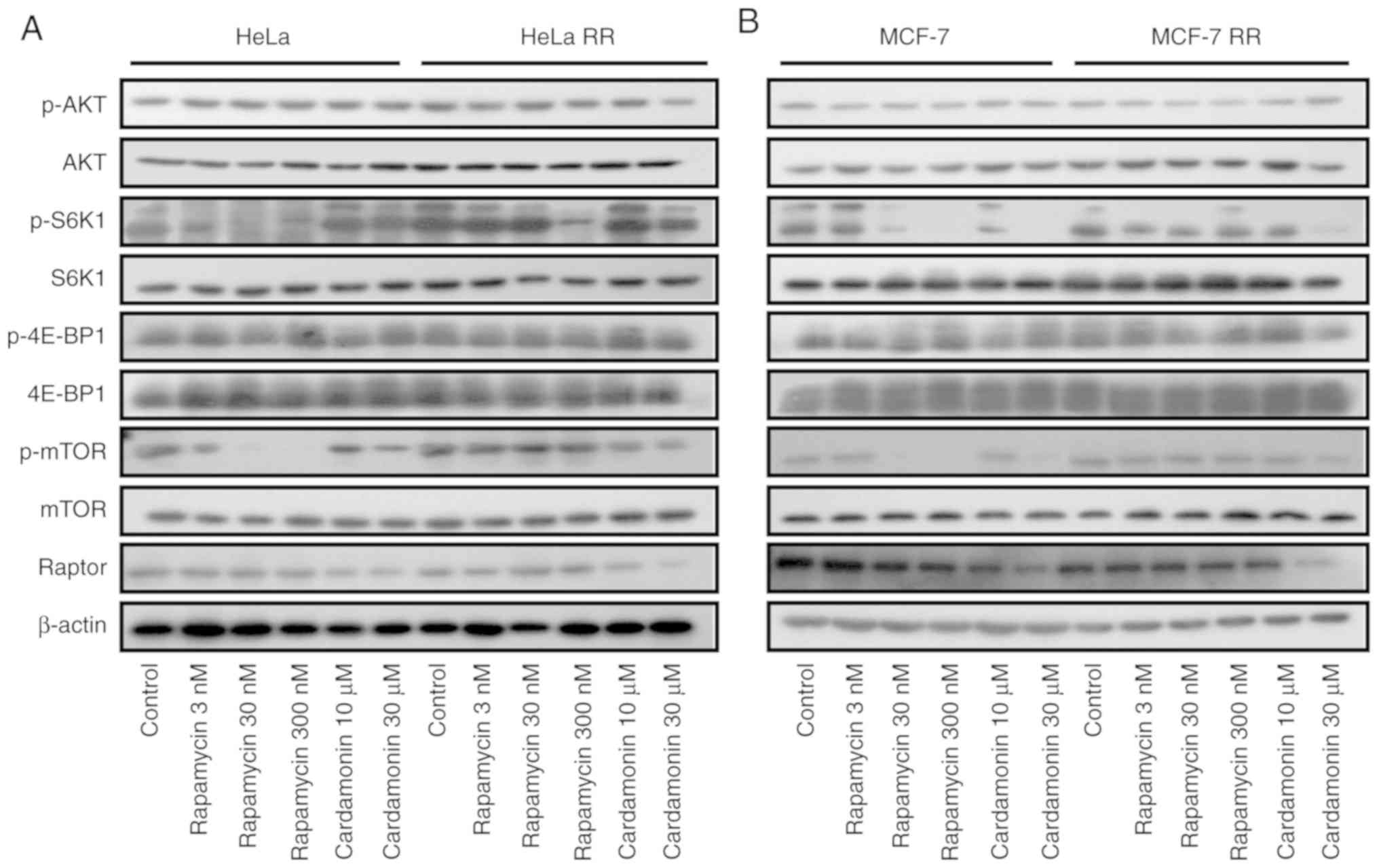 | Figure 7.Cardamonin inhibits the mTORC1
signalling pathway in rapamycin-resistant cells. (A) HeLa, HeLa RR
and (B) MCF-7, MCF-7 RR cells were treated with rapamycin for 48 h.
The protein levels of Akt, p-Akt, S6K1, p-S6K1, 4E-BP1, p-4E-BP1,
mTOR, p-mTOR and Raptor were determined by western blotting.
β-actin was used as the loading control. n=3. HeLa RR,
rapamycin-resistant HeLa cells; MCF-7 RR, rapamycin-resistant
MCF-7cells. mTORC1, mammalian target of rapamycin complex 1; p,
phosphorylated; S6K1, ribosomal protein S6 kinase B1; 4EBP1,
eukaryotic translation initiation factor 4E binding protein 1; Akt,
protein kinase B. |
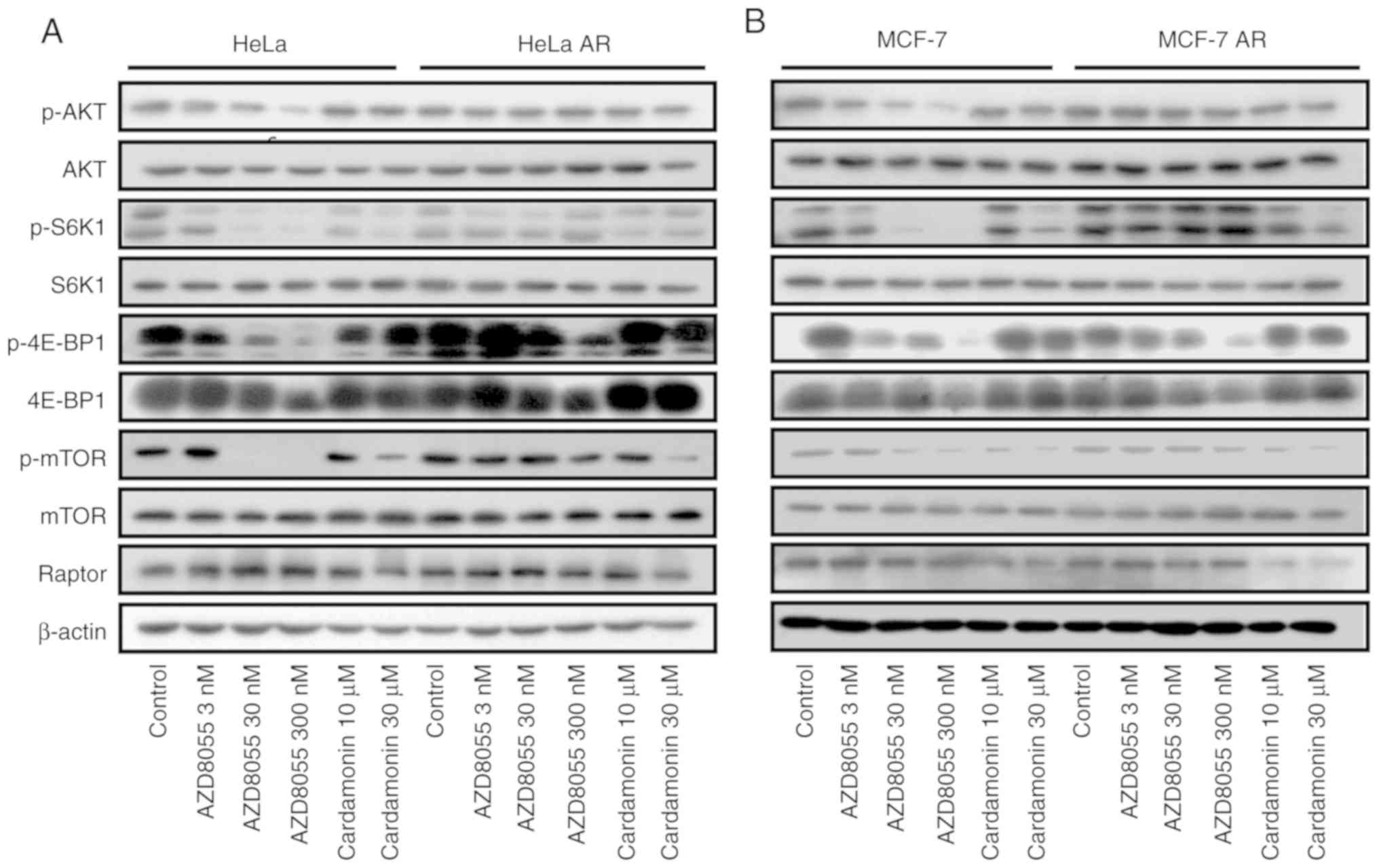 | Figure 8.Cardamonin inhibits the mTORC1
signalling pathway in AR cells. (A) HeLa, HeLa AR and (B) MCF-7,
MCF-7 AR cells were treated with AZD8055 for 48 h. The protein
levels of Akt, p-Akt, S6K1, p-S6K1, 4E-BP1, p-4E-BP1, mTOR, p-mTOR
and Raptor were determined by western blotting. β-actin was used as
the loading control. n=3. HeLa AR, AZD8055-resistant HeLa cells;
MCF-7 AR, AZD8055-resistant MCF-7 cells; mTORC1, mammalian target
of rapamycin complex 1; p, phosphorylated; S6K1, ribosomal protein
S6 kinase B1; 4EBP1, eukaryotic translation initiation factor 4E
binding protein 1; Akt, protein kinase B. |
Discussion
The PI3K/Akt/mTOR signalling pathway is commonly
upregulated in cancer and increasing evidence has demonstrated that
mTOR is a key signalling transduction node in this pathway.
Increased mTOR activity is a prominent feature of cancer cells
(5,27). Therefore, mTOR has emerged as an
important molecular target for the treatment of cancer (28). Several mTOR inhibitors have already
undergone clinical trials for various types of cancer; however, the
results of these trials are not satisfactory (29,30).
Rapalogs, including temsirolimus and everolimus,
have been approved by the FDA for the treatment of certain
advanced-stage tumours (31,32).
However, the efficacy of these agents is limited by the emergence
of resistance (11). Exposure of
MCF-7 breast cancer cells or HeLa cervical cancer cells to high
doses of either rapamycin or AZD8055 for 8 months results in the
emergence of resistant cells (33,34).
A previous study reported that the mTOR sequence of
AZD8055-resistant cells harboured mutations in the mTOR kinase
domain while that of rapamycin-resistant cells contained mutations
in the FRB domain (35). The
present study did not perform mTOR sequencing, however, the
resistant MCF-7 and HeLa cells generated were shown to be
insensitive to rapamycin or AZD8055. Furthermore, the proliferation
and clone formation of MCF-7 and HeLa cells were sensitive to
rapamycin and AZD8055 at clinically relevant levels, while the RR
and AR clones were significantly less so. In addition, in the RR
cells, phosphorylation of the rapamycin-sensitive sites on S6K1
(T389) was unaffected even at high concentrations of rapamycin. In
the AR cells, phosphorylation of Akt and EIF4EBP1 was less
sensitive to AZD8055. The present study revealed that cardamonin
significantly inhibited the viability and clone formation of
parental and resistant cells. The emergence of acquired resistance
is the main reason for the lack of efficacy of mTOR inhibitors in
clinical practice (35). However,
the role of mTOR inhibitors in cancer treatment continues to evolve
as novel compounds are continuously being developed.
The mechanisms of acquired resistance to mTOR
inhibitors have not been fully elucidated but may include metabolic
alterations, S6K1-dependent feedback reactivation of the PI3K/Akt
signalling pathway and mTOR mutations (36). mTOR mutations have attracted
special interest and random mutagenesis screens in yeast
demonstrated that single amino acid changes in the FRB domain
conferred rapamycin resistance (37). The mutation in the FRB domain
disrupted the interaction of mTOR with the FKBP12-rapalogs complex
(35,38,39).
As mutations occur in the FRB domain rather than the kinase domain,
the mutant protein remains sensitive to inhibition by direct
ATP-competitive mTOR kinase inhibitors (35). mTOR mutations in the kinase domain
of AR clones may increase the understanding of the structure of the
mTOR kinase domain-kinase inhibitor complex. AZD8055 binds to mTOR
with similar affinities in both parental and mutant cancer cells;
however, mutations in the kinase domain increase the activity of
mTOR and cells with these mutations are still sensitive to rapalogs
(35). An increased understanding
of the mechanisms of acquired mTOR inhibitor resistance may lead to
the development of novel therapeutic strategies.
Cardamonin is a specific mTORC1 inhibitor, which
decreases the protein level of Raptor, disrupts the interaction of
mTOR and Raptor and interrupts the mTORC1 signalling cascade
(24–26). Therefore, cardamonin inhibits mTOR
independent of the FRB domain and the kinase domain. Recent studies
suggested that Raptor should be included in the pharmacodynamic
evaluation of mTOR inhibitor trials (40,41).
Everolimus-resistant breast cancer cells exhibited recovery of
mTORC1 signalling and Raptor upregulation (41). Earwaker et al (40) revealed that Raptor upregulation was
implicated in resistance to mTOR kinase inhibitors in renal cancer
cells. Therefore, the present study investigated whether cardamonin
could overcome mTOR resistance by decreasing Raptor in breast and
cervical cancer cells. As expected, the results demonstrated that
the phosphorylation of mTORC1 and S6K1, as well as the protein
level of Raptor, in RR and AR cells were decreased following
treatment with cardamonin. The potential application of cardamonin
in other types of cancer requires further investigation.
In conclusion, the results obtained in the present
study suggested that cardamonin may serve as a novel therapeutic
agent for the treatment of patients with cervical and breast cancer
that have acquired resistance to either rapalogs or ATP-competitive
inhibitors.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Foundation of Fujian Province (grant. no. 2019J01515), the
Innovative Medical Foundation of Fujian Provincial Health and
Family Planning Commission (grant. no. 2016-CX-13) and Foundations
of Fujian Provincial Maternity and Children's Hospital (grant. nos.
YCXM 18-05 and YCXQ 18-18).
Availability of data and materials
All data generated or analysed during this study are
included in this published article.
Authors' contributions
PN and DS conceived and designed the experiments. JL
performed the experiments. HC and JZ analysed the data. YZ prepared
the material and performed the experiments. PN and DS wrote the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Saxton RA and Sabatini DM: mTOR signaling
in growth, metabolism, and disease. Cell. 169:361–371. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mossmann D, Park S and Hall MN: mTOR
signalling and cellular metabolism are mutual determinants in
cancer. Nat Rev Cancer. 18:744–757. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sabatini DM: Twenty-five years of mTOR:
Uncovering the link from nutrients to growth. Proc Natl Acad Sci
USA. 114:11818–11825. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jordan NJ, Dutkowski CM, Barrow D, Mottram
HJ, Hutcheson IR, Nicholson RI, Guichard SM and Gee JM: Impact of
dual mTORC1/2 mTOR kinase inhibitor AZD8055 on acquired endocrine
resistance in breast cancer in vitro. Breast Cancer Res.
16:R122014. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Barra F, Evangelisti G, Ferro Desideri L,
Di Domenico S, Ferraioli D, Vellone VG, De Cian F and Ferrero S:
Investigational PI3K/AKT/mTOR inhibitors in development for
endometrial cancer. Expert Opin Investig Drugs. 28:131–142. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li J, Kim SG and Blenis J: Rapamycin: One
drug, many effects. Cell Metab. 19:373–379. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Oshiro N, Yoshino K, Hidayat S, Tokunaga
C, Hara K, Eguchi S, Avruch J and Yonezawa K: Dissociation of
raptor from mTOR is a mechanism of rapamycin-induced inhibition of
mTOR function. Genes Cells. 9:359–366. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yip CK, Murata K, Walz T, Sabatini DM and
Kang SA: Structure of the human mTOR complex I and its implications
for rapamycin inhibition. Mol Cell. 38:768–774. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Meng LH and Zheng XF: Toward rapamycin
analog (rapalog)-based precision cancer therapy. Acta Pharmacol
Sin. 36:1163–1169. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Owonikoko TK: Inhibitors of mTOR pathway
for cancer therapy, moving on from rapalogs to TORKinibs. Cancer.
121:3390–3392. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chiarini F, Evangelisti C, McCubrey JA and
Martelli AM: Current treatment strategies for inhibiting mTOR in
cancer. Trends Pharmacol Sci. 36:124–135. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wagle N, Grabiner BC, Van Allen EM, Hodis
E, Jacobus S, Supko JG, Stewart M, Choueiri TK, Gandhi L, Cleary
JM, et al: Activating mTOR mutations in a patient with an
extraordinary response on a phase I trial of everolimus and
pazopanib. Cancer Discov. 4:546–553. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schenone S, Brullo C, Musumeci F, Radi M
and Botta M: ATP-competitive inhibitors of mTOR: An update. Curr
Med Chem. 18:2995–3014. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
English DP, Roque DM, Carrara L, Lopez S,
Bellone S, Cocco E, Bortolomai I, Schwartz PE, Rutherford T and
Santin AD: HER2/neu gene amplification determines the sensitivity
of uterine serous carcinoma cell lines to AZD8055, a novel dual
mTORC1/2 inhibitor. Gynecol Oncol. 131:753–758. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Jia QA, Kadel D, Zhang XF and
Zhang QB: Targeting mTORC1/2 complexes inhibit tumorigenesis and
enhance sensitivity to 5-flourouracil (5-FU) in hepatocellular
carcinoma: A preclinical study of mTORC1/2-targeted therapy in
hepatocellular carcinoma (HCC). Med Sci Monit. 24:2735–2743. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu DQ, Toyoda H, Qi L, Morimoto M, Hanaki
R, Iwamoto S, Komada Y and Hirayama M: Induction of MEK/ERK
activity by AZD8055 confers acquired resistance in neuroblastoma.
Biochem Biophys Res Commun. 499:425–432. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rodrik-Outmezguine VS, Chandarlapaty S,
Pagano NC, Poulikakos PI, Scaltriti M, Moskatel E, Baselga J,
Guichard S and Rosen N: mTOR kinase inhibition causes
feedback-dependent biphasic regulation of AKT signaling. Cancer
Discov. 1:248–259. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen H, Shi D, Niu P, Zhu Y and Zhou J:
Anti-inflammatory effects of cardamonin in ovarian cancer cells are
mediated via mTOR suppression. Planta Med. 84:1183–1190. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Niu PG, Zhang YX, Shi DH, Liu Y, Chen YY
and Deng J: Cardamonin inhibits metastasis of lewis lung carcinoma
cells by decreasing mTOR activity. PLoS One. 10:e01277782015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tang Y, Fang Q, Shi D, Niu P, Chen Y and
Deng J: mTOR inhibition of cardamonin on antiproliferation of A549
cells is involved in a FKBP12 independent fashion. Life Sci.
99:44–51. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Break MKB, Hossan MS, Khoo Y, Qazzaz ME,
Al-Hayali MZK, Chow SC, Wiart C, Bradshaw TD, Collins H and Khoo
TJ: Discovery of a highly active anticancer analogue of cardamonin
that acts as an inducer of caspase-dependent apoptosis and
modulator of the mTOR pathway. Fitoterapia. 125:161–173. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou X, Zhou R, Li Q, Jie X, Hong J, Zong
Y, Dong X, Zhang S, Li Z and Wu G: Cardamonin inhibits the
proliferation and metastasis of non-small-cell lung cancer cells by
suppressing the PI3K/Akt/mTOR pathway. Anticancer Drugs.
30:241–250. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jin J, Qiu S, Wang P, Liang X, Huang F, Wu
H, Zhang B, Zhang W, Tian X, Xu R, et al: Cardamonin inhibits
breast cancer growth by repressing HIF-1α-dependent metabolic
reprogramming. J Exp Clin Cancer Res. 38:3772019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shi D, Niu P, Heng X, Chen L, Zhu Y and
Zhou J: Autophagy induced by cardamonin is associated with mTORC1
inhibition in SKOV3 cells. Pharmacol Rep. 70:908–916. 2018.
View Article : Google Scholar
|
|
25
|
Shi D, Zhu Y, Niu P, Zhou J and Chen H:
Raptor mediates the antiproliferation of cardamonin by mTORC1
inhibition in SKOV3 cells. Onco Targets Ther. 11:757–767. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
You W, Wu Z, Ye F and Wu X: Cardamonin
protects against adverse cardiac remodeling through mTORC1
inhibition in mice with myocardial infarction. Pharmazie.
73:508–512. 2018.PubMed/NCBI
|
|
27
|
Du L, Li X, Zhen L, Chen W, Mu L, Zhang Y
and Song A: Everolimus inhibits breast cancer cell growth through
PI3K/AKT/mTOR signaling pathway. Mol Med Rep. 17:7163–7169.
2018.PubMed/NCBI
|
|
28
|
Guerrero-Zotano A, Mayer IA and Arteaga
CL: PI3K/AKT/mTOR: Role in breast cancer progression, drug
resistance, and treatment. Cancer Metastasis Rev. 35:515–524. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dey N, De P and Leyland-Jones B:
PI3K-AKT-mTOR inhibitors in breast cancers: From tumor cell
signaling to clinical trials. Pharmacol Ther. 175:91–106. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tian T, Li X and Zhang J: mTOR signaling
in cancer and mTOR inhibitors in solid tumor targeting therapy. Int
J Mol Sci. 20(pii): E7552019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kapoor A and Figlin RA: Targeted
inhibition of mammalian target of rapamycin for the treatment of
advanced renal cell carcinoma. Cancer. 115:3618–3630. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hudes G, Carducci M, Tomczak P, Dutcher J,
Figlin R, Kapoor A, Staroslawska E, Sosman J, McDermott D, Bodrogi
I, et al: Temsirolimus, interferon alfa, or both for advanced
renal-cell carcinoma. N Engl J Med. 356:2271–2281. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Koh KX, Tan GH, Hui Low SH, Mohd Omar MF,
Han MJ, Iacopetta B, Soo R, Beloueche-Babari M, Bhattacharya B and
Soong R: Acquired resistance to PI3K/mTOR inhibition is associated
with mitochondrial DNA mutation and glycolysis. Oncotarget.
8:110133–110144. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hassan B, Akcakanat A, Sangai T, Evans KW,
Adkins F, Eterovic AK, Zhao H, Chen K, Chen H, Do KA, et al:
Catalytic mTOR inhibitors can overcome intrinsic and acquired
resistance to allosteric mTOR inhibitors. Oncotarget. 5:8544–8557.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rodrik-Outmezguine VS, Okaniwa M, Yao Z,
Novotny CJ, McWhirter C, Banaji A, Won H, Wong W, Berger M, de
Stanchina E, et al: Overcoming mTOR resistance mutations with a
new-generation mTOR inhibitor. Nature. 534:272–276. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Santoni M, Pantano F, Amantini C, Nabissi
M, Conti A, Burattini L, Zoccoli A, Berardi R, Santoni G, Tonini G,
et al: Emerging strategies to overcome the resistance to current
mTOR inhibitors in renal cell carcinoma. Biochim Biophys Acta.
1845:221–231. 2014.PubMed/NCBI
|
|
37
|
Weisman R and Choder M: The fission yeast
TOR homolog, tor1+, is required for the response to starvation and
other stresses via a conserved serine. J Biol Chem. 276:7027–7032.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Grabiner BC, Nardi V, Birsoy K, Possemato
R, Shen K, Sinha S, Jordan A, Beck AH and Sabatini DM: A diverse
array of cancer-associated MTOR mutations are hyperactivating and
can predict rapamycin sensitivity. Cancer Discov. 4:554–563. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wagle N, Grabiner BC, Van Allen EM,
Amin-Mansour A, Taylor-Weiner A, Rosenberg M, Gray N, Barletta JA,
Guo Y, Swanson SJ, et al: Response and acquired resistance to
everolimus in anaplastic thyroid cancer. N Engl J Med.
371:1426–1433. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Earwaker P, Anderson C, Willenbrock F,
Harris AL, Protheroe AS and Macaulay VM: RAPTOR up-regulation
contributes to resistance of renal cancer cells to PI3K-mTOR
inhibition. PLoS One. 13:e01918902018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mateo F, Arenas EJ, Aguilar H,
Serra-Musach J, de Garibay GR, Boni J, Maicas M, Du S, Iorio F,
Herranz-Ors C, et al: Stem cell-like transcriptional reprogramming
mediates metastatic resistance to mTOR inhibition. Oncogene.
36:2737–2749. 2017. View Article : Google Scholar : PubMed/NCBI
|