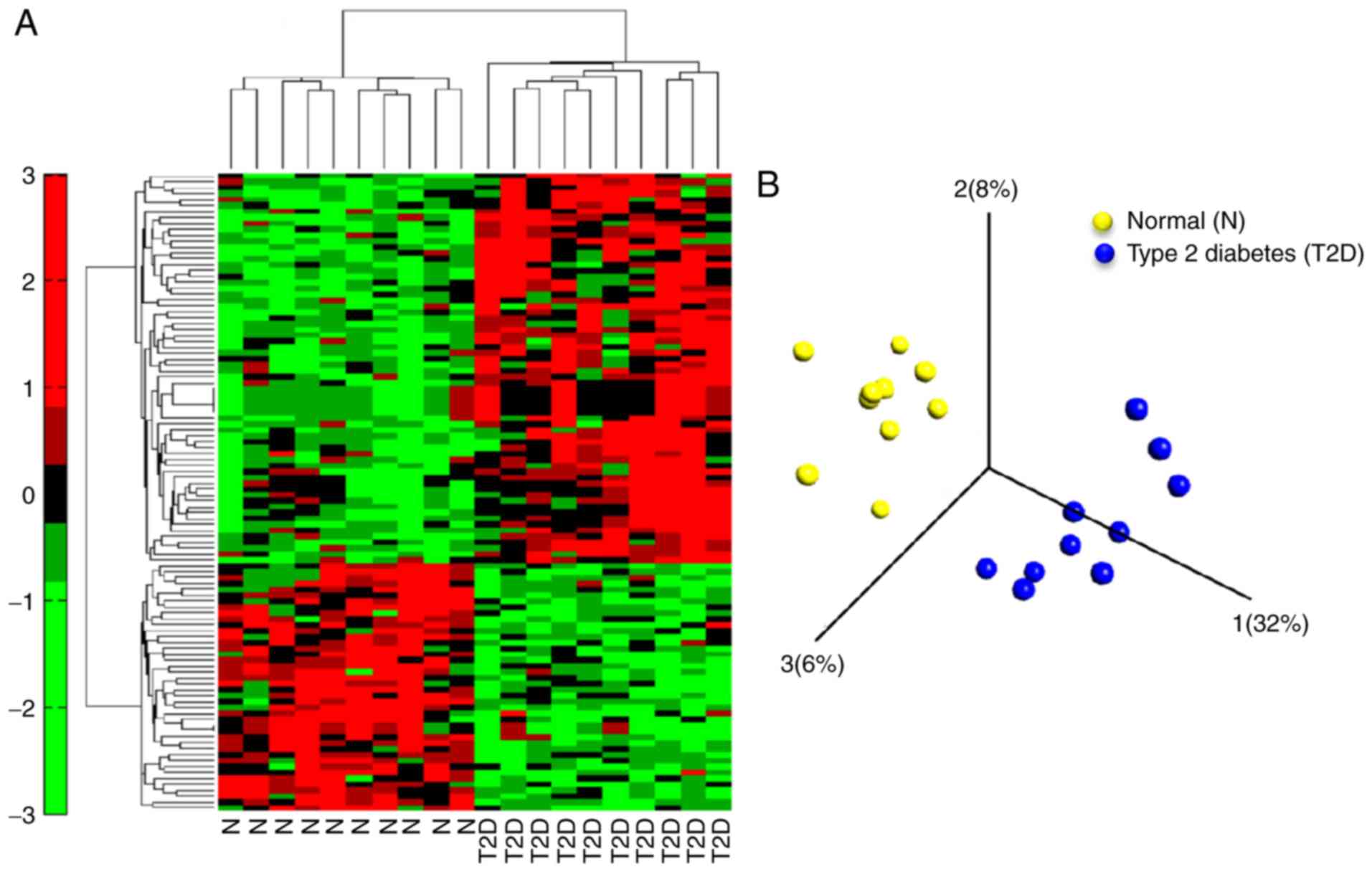
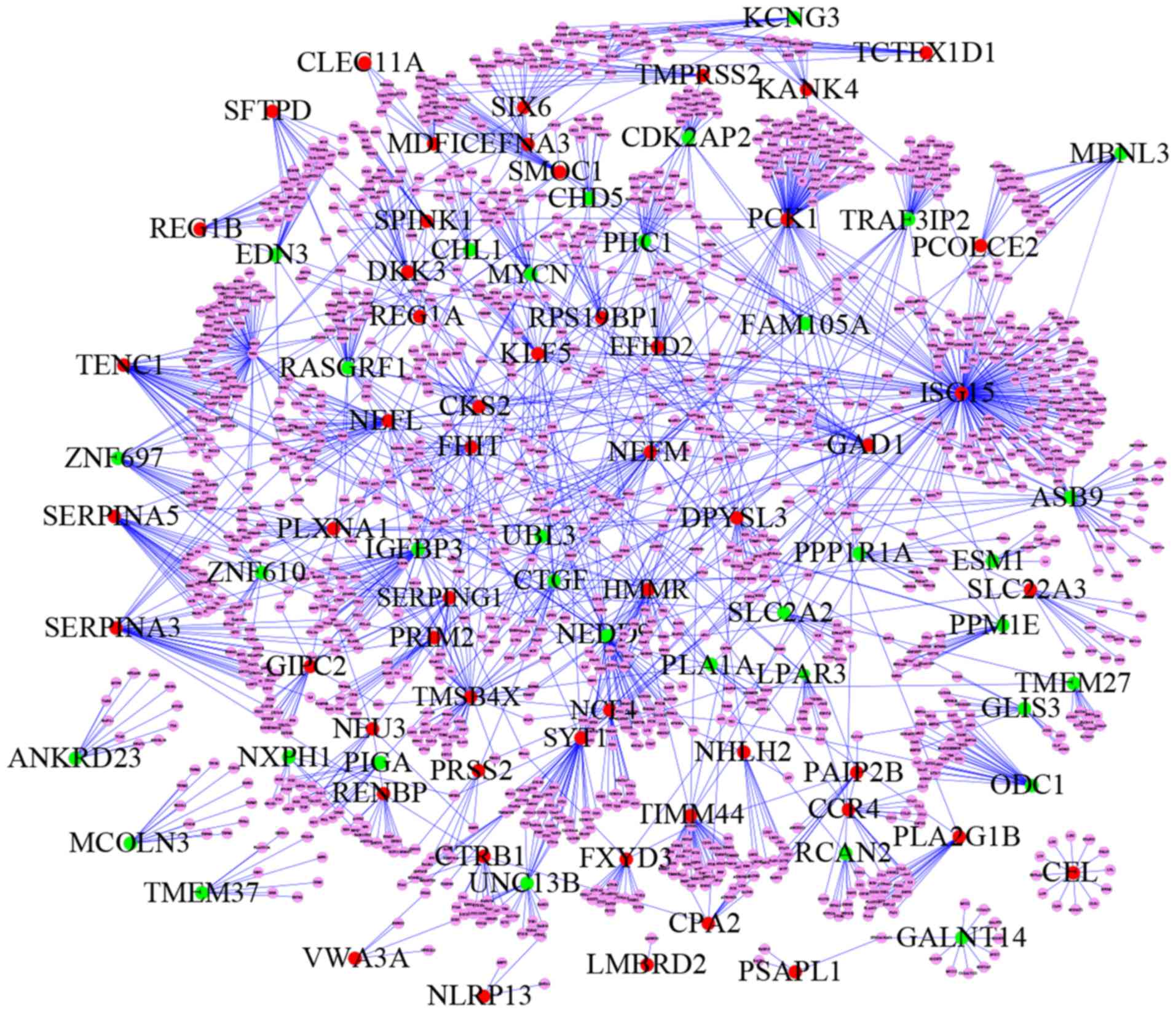
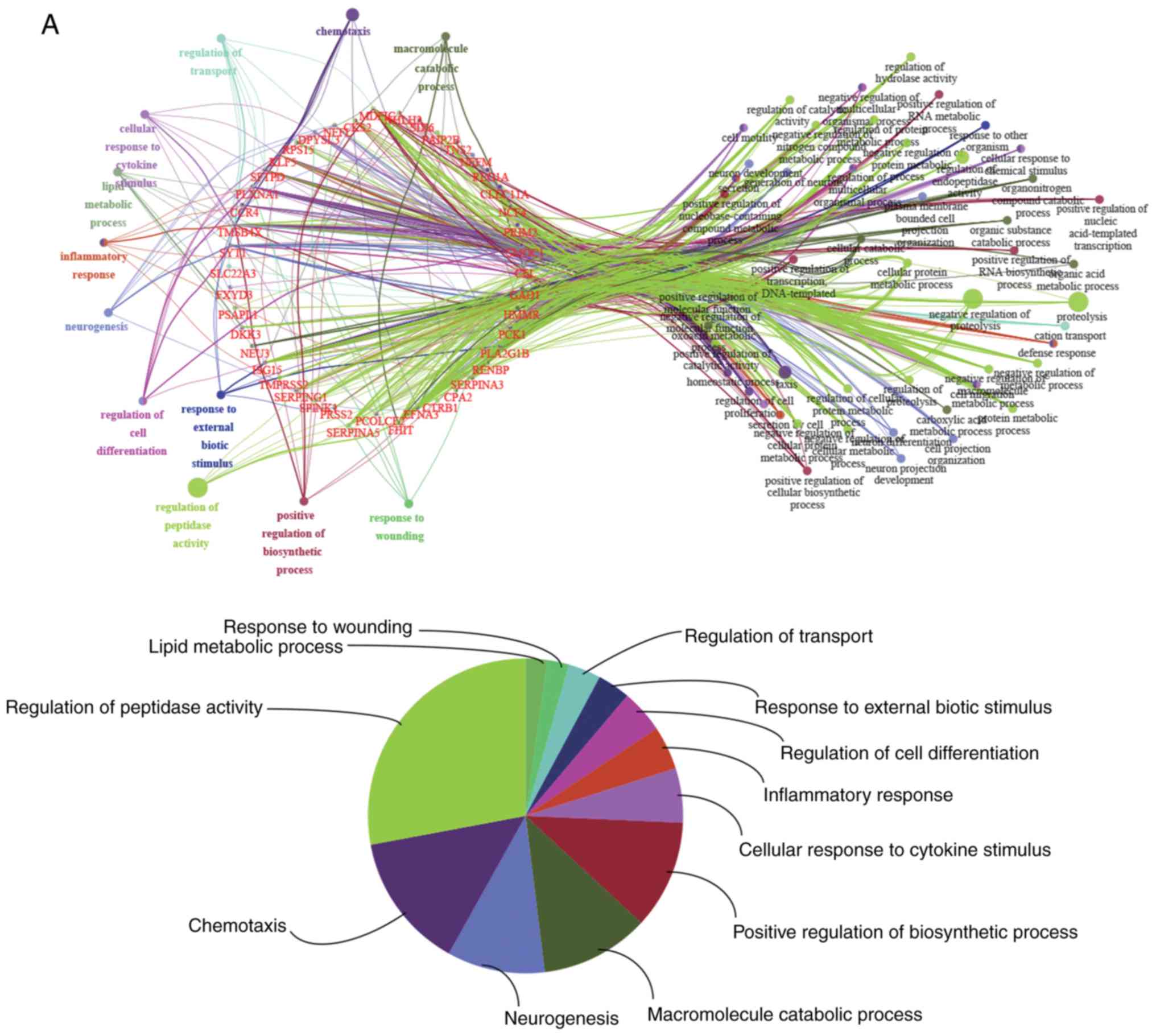
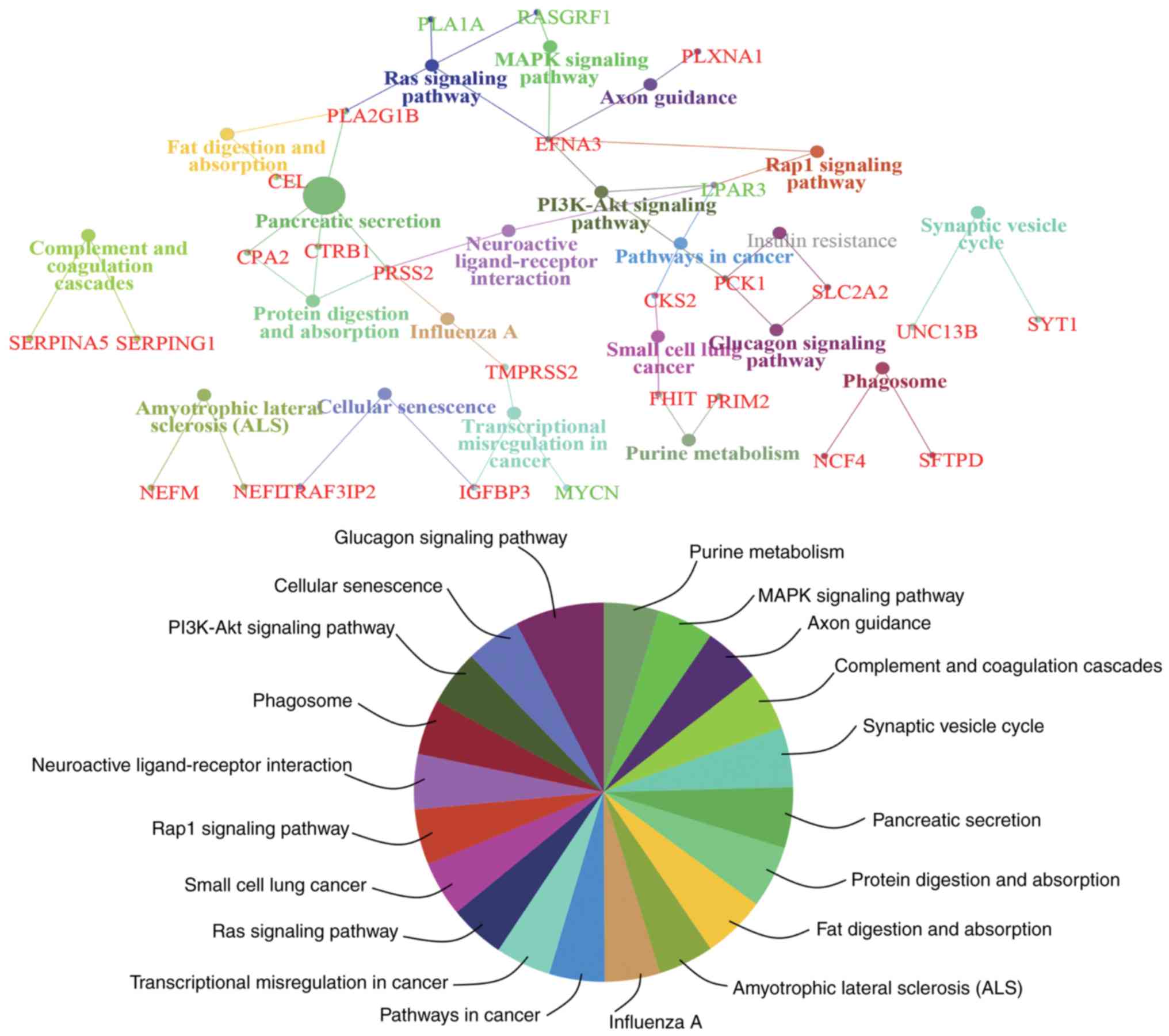
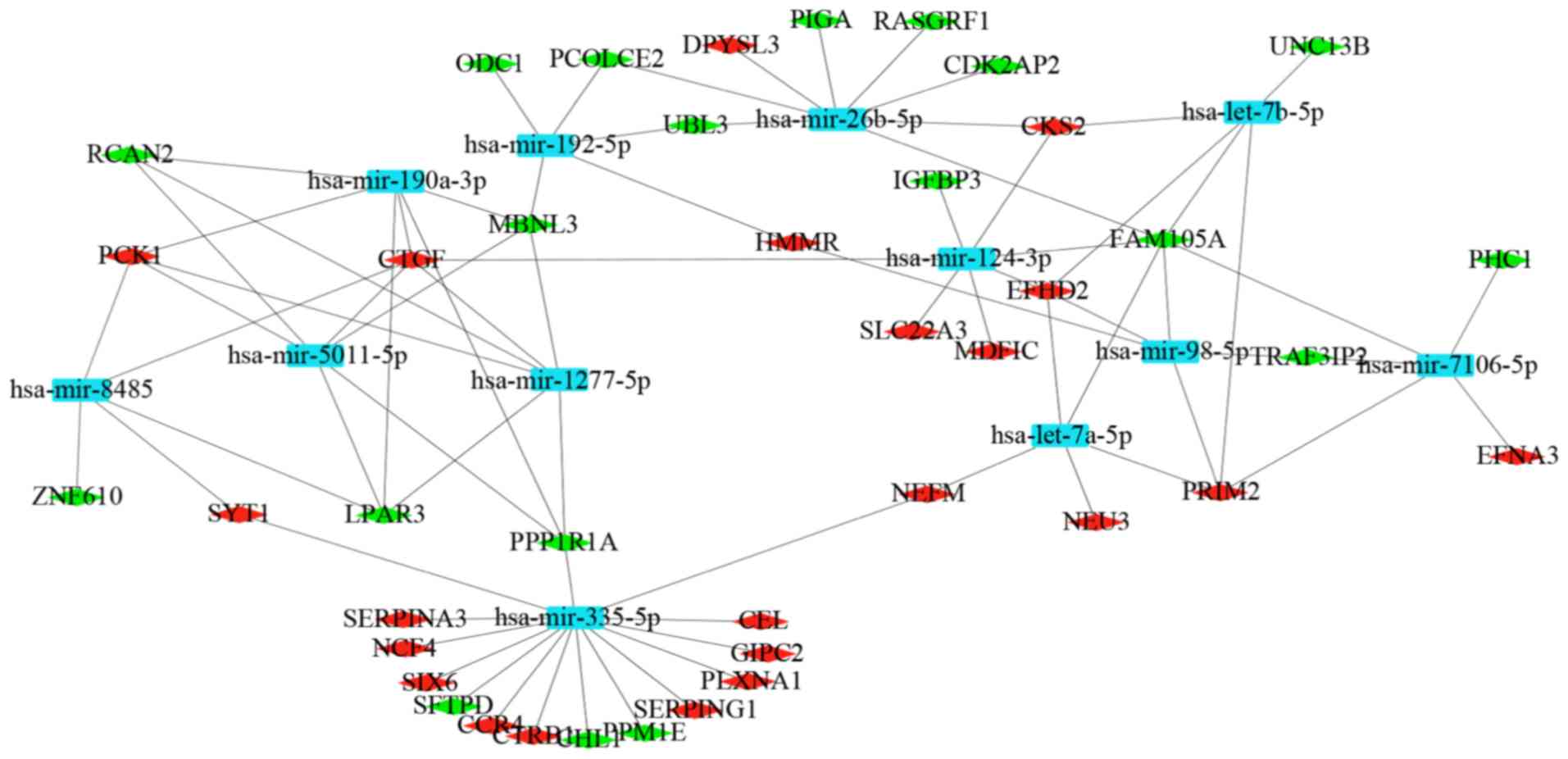
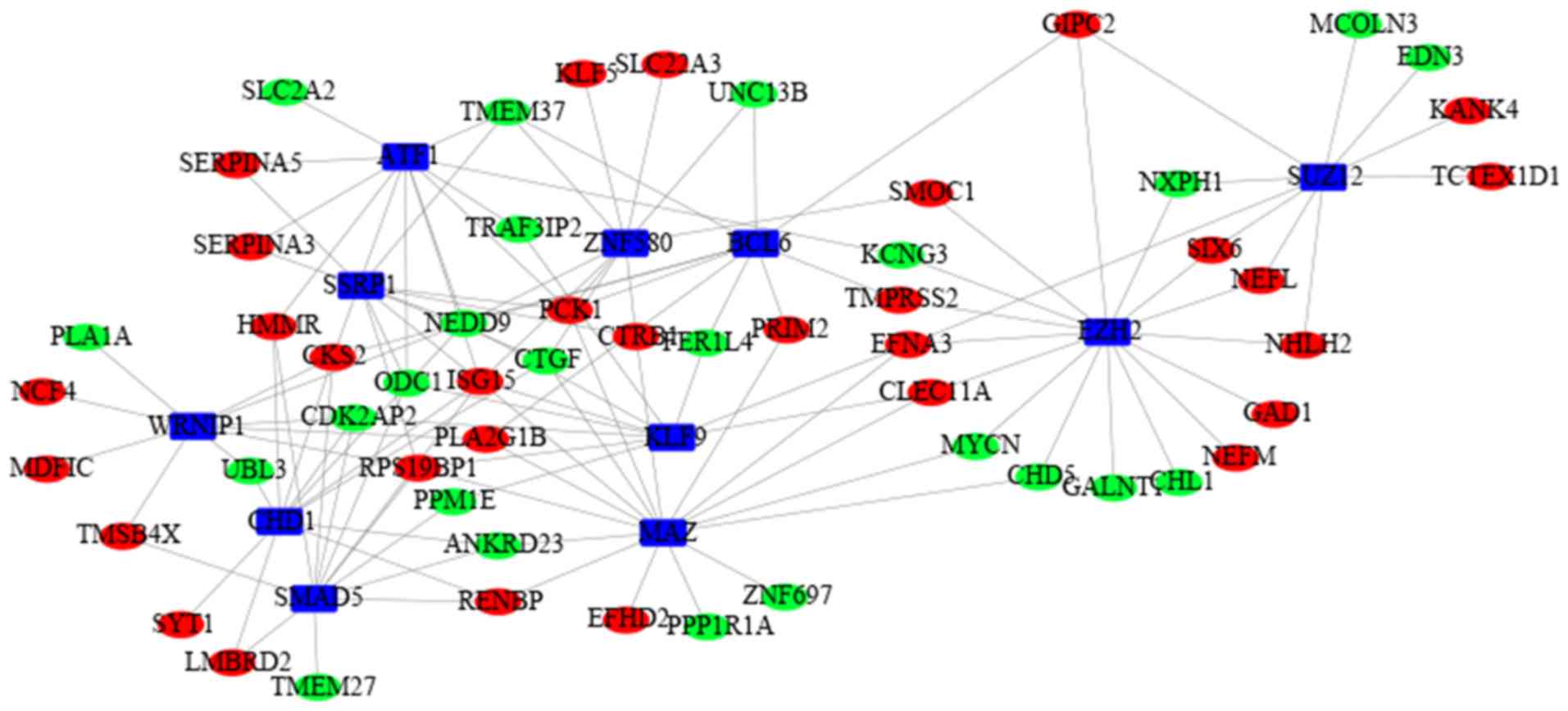
|
1
|
Zheng Y, Ley SH and Hu FB: Global
aetiology and epidemiology of type 2 diabetes mellitus and its
complications. Nat Rev Endocrinol. 14:88–98. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
International Diabetes Federation: IDF
Diabetes Atlas. (8th). IDF. (Brussels, Belgium). 2017.
|
|
3
|
Ohn JH, Kwak SH, Cho YM, Lim S, Jang HC,
Park KS and Cho NH: 10-year trajectory of β-cell function and
insulin sensitivity in the development of type 2 diabetes: A
community-based prospective cohort study. Lancet Diabetes
Endocrinol. 4:27–34. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ashcroft FM, Rohm M, Clark A and Brereton
MF: Is type 2 diabetes a glycogen storage disease of pancreatic β
cells? Cell Metab. 26:17–23. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Almaça J, Weitz J, Rodriguez-Diaz R,
Pereira E and Caicedo A: The pericyte of the pancreatic islet
regulates capillary diameter and local blood flow. Cell Metab.
27:630–644.e4. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wei FJ, Cai CY, Yu P, Lv J, Ling C, Shi
WT, Jiao HX, Chang BC, Yang FH, Tian Y, et al: Quantitative
candidate gene association studies of metabolic traits in Han
Chinese type 2 diabetes patients. Genet Mol Res. 14:15471–15481.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen J, Meng Y, Zhou J, Zhuo M, Ling F,
Zhang Y, Du H and Wang X: Identifying candidate genes for type 2
diabetes mellitus and obesity through gene expression profiling in
multiple tissues or cells. J Diabetes Res. 2013:9704352013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lynch CJ and Adams SH: Branched-chain
amino acids in metabolic signalling and insulin resistance. Nat Rev
Endocrinol. 10:723–736. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xue A, Wu Y, Zhu Z, Zhang F, Kemper KE,
Zheng Z, Yengo L, Lloyd-Jones LR, Sidorenko J, Wu Y, et al: eQTLGen
Consortium: Genome-wide association analyses identify 143 risk
variants and putative regulatory mechanisms for type 2 diabetes.
Nat Commun. 9:29412018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lawlor N, Khetan S, Ucar D and Stitzel ML:
Genomics of Islet (Dys)function and Type 2 Diabetes. Trends Genet.
33:244–255. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee I, Blom UM, Wang PI, Shim JE and
Marcotte EM: Prioritizing candidate disease genes by network-based
boosting of genome-wide association data. Genome Res. 21:1109–1121.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lawlor N, George J, Bolisetty M, Kursawe
R, Sun L, Sivakamasundari V, Kycia I, Robson P and Stitzel ML:
Single-cell transcriptomes identify human islet cell signatures and
reveal cell-type-specific expression changes in type 2 diabetes.
Genome Res. 27:208–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Segerstolpe Å, Palasantza A, Eliasson P,
Andersson EM, Andréasson AC, Sun X, Picelli S, Sabirsh A, Clausen
M, Bjursell MK, et al: Single-cell transcriptome profiling of human
pancreatic islets in health and type 2 diabetes. Cell Metab.
24:593–607. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zeggini E, Scott LJ, Saxena R, Voight BF,
Marchini JL, Hu T, de Bakker PI, Abecasis GR, Almgren P, Andersen
G, et al Wellcome Trust Case Control Consortium, : Meta-analysis of
genome-wide association data and large-scale replication identifies
additional susceptibility loci for type 2 diabetes. Nat Genet.
40:638–645. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Maruthur NM, Gribble MO, Bennett WL, Bolen
S, Wilson LM, Balakrishnan P, Sahu A, Bass E, Kao WH and Clark JM:
The pharmacogenetics of type 2 diabetes: A systematic review.
Diabetes Care. 37:876–886. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
van de Bunt M, Manning Fox JE, Dai X,
Barrett A, Grey C, Li L, Bennett AJ, Johnson PR, Rajotte RV,
Gaulton KJ, et al: Transcript expression data from human islets
links regulatory signals from genome-wide association studies for
type 2 diabetes and glycemic traits to their downstream effectors.
PLoS Genet. 11:e10056942015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bonnefond A and Froguel P: Rare and common
genetic events in type 2 diabetes: What should biologists know?
Cell Metab. 21:357–368. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ndiaye FK, Ortalli A, Canouil M, Huyvaert
M, Salazar-Cardozo C, Lecoeur C, Verbanck M, Pawlowski V, Boutry R,
Durand E, et al: Expression and functional assessment of candidate
type 2 diabetes susceptibility genes identify four new genes
contributing to human insulin secretion. Mol Metab. 6:459–470.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pellegrino M, Sciambi A, Treusch S,
Durruthy-Durruthy R, Gokhale K, Jacob J, Chen TX, Geis JA, Oldham
W, Matthews J, et al: High-throughput single-cell DNA sequencing of
acute myeloid leukemia tumors with droplet microfluidics. Genome
Res. 28:1345–1352. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang P, Xia JH, Zhu J, Gao P, Tian YJ, Du
M, Guo YC, Suleman S, Zhang Q, Kohli M, et al: High-throughput
screening of prostate cancer risk loci by single nucleotide
polymorphisms sequencing. Nat Commun. 9:20222018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nepal C, O'Rourke CJ, Oliveira DVNP,
Taranta A, Shema S, Gautam P, Calderaro J, Barbour A, Raggi C,
Wennerberg K, et al: Genomic perturbations reveal distinct
regulatory networks in intrahepatic cholangiocarcinoma. Hepatology.
68:949–963. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fuchsberger C, Flannick J, Teslovich TM,
Mahajan A, Agarwala V, Gaulton KJ, Ma C, Fontanillas P, Moutsianas
L, McCarthy DJ, et al: The genetic architecture of type 2 diabetes.
Nature. 536:41–47. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Thul PJ, Åkesson L, Wiking M, Mahdessian
D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM,
et al: A subcellular map of the human proteome. Science.
356:8062017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu H, Bilgin M and Snyder M: Proteomics.
Annu Rev Biochem. 72:783–812. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Larance M and Lamond AI: Multidimensional
proteomics for cell biology. Nat Rev Mol Cell Biol. 16:269–280.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Stuart LM, Boulais J, Charriere GM,
Hennessy EJ, Brunet S, Jutras I, Goyette G, Rondeau C, Letarte S,
Huang H, et al: A systems biology analysis of the Drosophila
phagosome. Nature. 445:95–101. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhao L, Chen Y, Bajaj AO, Eblimit A, Xu M,
Soens ZT, Wang F, Ge Z, Jung SY, He F, et al: Integrative
subcellular proteomic analysis allows accurate prediction of human
disease-causing genes. Genome Res. 26:660–669. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hunt KK, Karakas C, Ha MJ, Biernacka A, Yi
M, Sahin AA, Adjapong O, Hortobagyi GN, Bondy M, Thompson P, et al:
Cytoplasmic Cyclin E Predicts Recurrence in Patients with Breast
Cancer. Clin Cancer Res. 23:2991–3002. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mezzanzanica D, Fabbi M, Bagnoli M,
Staurengo S, Losa M, Balladore E, Alberti P, Lusa L, Ditto A,
Ferrini S, et al: Subcellular localization of activated leukocyte
cell adhesion molecule is a molecular predictor of survival in
ovarian carcinoma patients. Clin Cancer Res. 14:1726–1733. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhu W, Yang L and Du Z: Layered functional
network analysis of gene expression in human heart failure. PLoS
One. 4:e62882009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Du ZP, Wu BL, Wang SH, Shen JH, Lin XH,
Zheng CP, Wu ZY, Qiu XY, Zhan XF, Xu LY, et al: Shortest path
analyses in the protein-protein interaction network of NGAL
(neutrophil gelatinase-associated lipocalin) overexpression in
esophageal squamous cell carcinoma. Asian Pac J Cancer Prev.
15:6899–6904. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Diao B, Liu Y, Zhang Y, Liu Q, Lu WJ and
Xu G: Functional network analysis with the subcellular location and
gene ontology information in human allergic asthma. Genet Test Mol
Biomarkers. 16:1287–1292. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee S, Zhang C, Kilicarslan M, Piening BD,
Bjornson E, Hallström BM, Groen AK, Ferrannini E, Laakso M, Snyder
M, et al: Integrated Network Analysis Reveals an Association
between Plasma Mannose Levels and Insulin Resistance. Cell Metab.
24:172–184. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Alvarez MJ, Shen Y, Giorgi FM, Lachmann A,
Ding BB, Ye BH and Califano A: Functional characterization of
somatic mutations in cancer using network-based inference of
protein activity. Nat Genet. 48:838–847. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Krell J, Stebbing J, Frampton AE,
Carissimi C, Harding V, De Giorgio A, Fulci V, Macino G, Colombo T
and Castellano L: The role of TP53 in miRNA loading onto AGO2 and
in remodelling the miRNA-mRNA interaction network. Lancet. 385
(Suppl 1):S152015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Marselli L, Thorne J, Dahiya S, Sgroi DC,
Sharma A, Bonner-Weir S, Marchetti P and Weir GC: Gene expression
profiles of Beta-cell enriched tissue obtained by laser capture
microdissection from subjects with type 2 diabetes. PLoS One.
5:e114992010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Keshava Prasad TS, Goel R, Kandasamy K,
Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R,
Shafreen B, Venugopal A, et al: Human Protein Reference
Database--2009 update. Nucleic Acids Res 37 (Database). D767–D772.
2009. View Article : Google Scholar
|
|
38
|
Chatr-Aryamontri A, Breitkreutz BJ,
Oughtred R, Boucher L, Heinicke S, Chen D, Stark C, Breitkreutz A,
Kolas N, O'Donnell L, et al: The BioGRID interaction database: 2015
update. Nucleic Acids Res. 43(D1): D470–D478. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kerrien S, Aranda B, Breuza L, Bridge A,
Broackes-Carter F, Chen C, Duesbury M, Dumousseau M, Feuermann M,
Hinz U, et al: The IntAct molecular interaction database in 2012.
Nucleic Acids Res. 40(D1): D841–D846. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43(D1): D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pontén F, Jirström K and Uhlen M: The
Human Protein Atlas--a tool for pathology. J Pathol. 216:387–393.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Barsky A, Gardy JL, Hancock RE and Munzner
T: Cerebral: A Cytoscape plugin for layout of and interaction with
biological networks using subcellular localization annotation.
Bioinformatics. 23:1040–1042. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bindea G, Galon J and Mlecnik B: CluePedia
Cytoscape plugin: Pathway insights using integrated experimental
and in silico data. Bioinformatics. 29:661–663. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bindea G, Mlecnik B, Hackl H, Charoentong
P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z and
Galon J: ClueGO: A Cytoscape plug-in to decipher functionally
grouped gene ontology and pathway annotation networks.
Bioinformatics. 25:1091–1093. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gene Ontology Consortium: Gene Ontology
Consortium: Going forward. Nucleic Acids Res. 43(D1): D1049–D1056.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45(D1): D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fan Y, Siklenka K, Arora SK, Ribeiro P,
Kimmins S and Xia J: miRNet - dissecting miRNA-target interactions
and functional associations through network-based visual analysis.
Nucleic Acids Res 44 (W1). W135–41. 2016. View Article : Google Scholar
|
|
49
|
Chou CH, Chang NW, Shrestha S, Hsu SD, Lin
YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, et al: miRTarBase
2016: Updates to the experimentally validated miRNA-target
interactions database. Nucleic Acids Res. 44(D1): D239–D247. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Xiao F, Zuo Z, Cai G, Kang S, Gao X and Li
T: miRecords: An integrated resource for microRNA-target
interactions. Nucleic Acids Res 37 (Database). D105–D110. 2009.
View Article : Google Scholar
|
|
51
|
Xia J, Gill EE and Hancock RE:
NetworkAnalyst for statistical, visual and network-based
meta-analysis of gene expression data. Nat Protoc. 10:823–844.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang S, Sun H, Ma J, Zang C, Wang C, Wang
J, Tang Q, Meyer CA, Zhang Y and Liu XS: Target analysis by
integration of transcriptome and ChIP-seq data with BETA. Nat
Protoc. 8:2502–2515. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yoshikawa A, Imagawa A, Nakata S, Fukui K,
Kuroda Y, Miyata Y, Sato Y, Hanafusa T, Matsuoka TA, Kaneto H, et
al: Interferon stimulated gene 15 has an anti-apoptotic effect on
MIN6 cells. Endocr J. 61:883–890. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Rees SD, Britten AC, Bellary S, O'Hare JP,
Kumar S, Barnett AH and Kelly MA: The promoter polymorphism −232C/G
of the PCK1 gene is associated with type 2 diabetes in a
UK-resident South Asian population. BMC Med Genet. 10:832009.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Gustavsson N and Han W: Calcium-sensing
beyond neurotransmitters: Functions of synaptotagmins in
neuroendocrine and endocrine secretion. Biosci Rep. 29:245–259.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Marshall C, Hitman GA, Partridge CJ, Clark
A, Ma H, Shearer TR and Turner MD: Evidence that an isoform of
calpain-10 is a regulator of exocytosis in pancreatic beta-cells.
Mol Endocrinol. 19:213–224. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Rajpathak SN, He M, Sun Q, Kaplan RC,
Muzumdar R, Rohan TE, Gunter MJ, Pollak M, Kim M, Pessin JE, et al:
Insulin-like growth factor axis and risk of type 2 diabetes in
women. Diabetes. 61:2248–2254. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Drogan D, Schulze MB, Boeing H and Pischon
T: Insulin-like growth factor 1 and insulin-like growth
factor-binding protein 3 in relation to the risk of type 2 diabetes
mellitus: Results from the EPIC-Potsdam study. Am J Epidemiol.
183:553–560. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang Y, Katayama A, Terami T, Han X,
Nunoue T, Zhang D, Teshigawara S, Eguchi J, Nakatsuka A, Murakami
K, et al: Translocase of inner mitochondrial membrane 44 alters the
mitochondrial fusion and fission dynamics and protects from type 2
diabetes. Metabolism. 64:677–688. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Venkatesan B, Valente AJ, Das NA,
Carpenter AJ, Yoshida T, Delafontaine JL, Siebenlist U and
Chandrasekar B: CIKS (Act1 or TRAF3IP2) mediates high
glucose-induced endothelial dysfunction. Cell Signal. 25:359–371.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Bergholdt R, Brorsson C, Palleja A,
Berchtold LA, Fløyel T, Bang-Berthelsen CH, Frederiksen KS, Jensen
LJ, Størling J and Pociot F: Identification of novel type 1
diabetes candidate genes by integrating genome-wide association
data, protein-protein interactions, and human pancreatic islet gene
expression. Diabetes. 61:954–962. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kokkola T, Suuronen T, Molnár F, Määttä J,
Salminen A, Jarho EM and Lahtela-Kakkonen M: AROS has a
context-dependent effect on SIRT1. FEBS Lett. 588:1523–1528. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Haigis MC and Sinclair DA: Mammalian
sirtuins: Biological insights and disease relevance. Annu Rev
Pathol. 5:253–295. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhou CH, Zhang MX, Zhou SS, Li H, Gao J,
Du L and Yin XX: SIRT1 attenuates neuropathic pain by epigenetic
regulation of mGluR1/5 expressions in type 2 diabetic rats. Pain.
158:130–139. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Ortega FJ, Pueyo N, Moreno-Navarrete JM,
Sabater M, Rodriguez-Hermosa JI, Ricart W, Tinahones FJ and
Fernandez-Real JM: The lung innate immune gene surfactant protein-D
is expressed in adipose tissue and linked to obesity status. Int J
Obes (Lond) (2005). 37:1532–1538. 2013. View Article : Google Scholar
|
|
66
|
Crook M: Type 2 diabetes mellitus: A
disease of the innate immune system? An update. Diabet Med.
21:203–207. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Yang W, Li Y, Wang JY, Han R and Wang L:
Circulating levels of adipose tissue-derived inflammatory factors
in elderly diabetes patients with carotid atherosclerosis: A
retrospective study. Cardiovasc Diabetol. 17:752018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Prattichizzo F, De Nigris V, Spiga R,
Mancuso E, La Sala L, Antonicelli R, Testa R, Procopio AD, Olivieri
F and Ceriello A: Inflammageing and metaflammation: The yin and
yang of type 2 diabetes. Ageing Res Rev. 41:1–17. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Naidoo V, Naidoo M and Ghai M: Cell- and
tissue-specific epigenetic changes associated with chronic
inflammation in insulin resistance and type 2 diabetes mellitus.
Scand J Immunol. 88:e127232018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Ehses JA, Perren A, Eppler E, Ribaux P,
Pospisilik JA, Maor-Cahn R, Gueripel X, Ellingsgaard H, Schneider
MK, Biollaz G, et al: Increased number of islet-associated
macrophages in type 2 diabetes. Diabetes. 56:2356–2370. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Böni-Schnetzler M, Thorne J, Parnaud G,
Marselli L, Ehses JA, Kerr-Conte J, Pattou F, Halban PA, Weir GC
and Donath MY: Increased interleukin (IL)-1beta messenger
ribonucleic acid expression in beta -cells of individuals with type
2 diabetes and regulation of IL-1beta in human islets by glucose
and autostimulation. J Clin Endocrinol Metab. 93:4065–4074. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Donath MY, Böni-Schnetzler M, Ellingsgaard
H and Ehses JA: Islet inflammation impairs the pancreatic beta-cell
in type 2 diabetes. Physiology (Bethesda). 24:325–331.
2009.PubMed/NCBI
|
|
73
|
Marchetti P, Lupi R, Del Guerra S,
Bugliani M, D'Aleo V, Occhipinti M, Boggi U, Marselli L and Masini
M: Goals of treatment for type 2 diabetes: Beta-cell preservation
for glycemic control. Diabetes Care. 32 (Suppl 2):S178–S183. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Cnop M: Fatty acids and glucolipotoxicity
in the pathogenesis of Type 2 diabetes. Biochem Soc Trans.
36:348–352. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Saltiel AR and Kahn CR: Insulin signalling
and the regulation of glucose and lipid metabolism. Nature.
414:799–806. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wajchenberg BL: beta-cell failure in
diabetes and preservation by clinical treatment. Endocr Rev.
28:187–218. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Takahashi E, Unoki-Kubota H, Shimizu Y,
Okamura T, Iwata W, Kajio H, Yamamoto-Honda R, Shiga T, Yamashita
S, Tobe K, et al: Proteomic analysis of serum biomarkers for
prediabetes using the Long-Evans Agouti rat, a spontaneous animal
model of type 2 diabetes mellitus. J Diabetes Investig. 8:661–671.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Hart LM, Fritsche A, Nijpels G, van
Leeuwen N, Donnelly LA, Dekker JM, Alssema M, Fadista J, Carlotti
F, Gjesing AP, et al: The CTRB1/2 locus affects diabetes
susceptibility and treatment via the incretin pathway. Diabetes.
62:3275–3281. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Xue M and Jackson CJ: Activated protein C
and its potential applications in prevention of islet β-cell damage
and diabetes. Vitam Horm. 95:323–363. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Matsumoto K, Yano Y, Gabazza EC, Araki R,
Bruno NE, Suematsu M, Akatsuka H, Katsuki A, Taguchi O, Adachi Y,
et al: Inverse correlation between activated protein C generation
and carotid atherosclerosis in Type 2 diabetic patients. Diabet
Med. 24:1322–1328. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Wang JY, Lu Q, Tao Y, Jiang YR and Jonas
JB: Intraocular expression of thymosin β4 in proliferative diabetic
retinopathy. Acta Ophthalmol. 89:e396–e403. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Sidarala V and Kowluru A: The regulatory
roles of mitogen-activated protein kinase (MAPK) pathways in health
and diabetes: Lessons learned from the pancreatic β-cell. Recent
Pat Endocr Metab Immune Drug Discov. 10:76–84. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Pang Y, Zhu H, Xu J, Yang L, Liu L and Li
J: β-arrestin-2 is involved in irisin induced glucose metabolism in
type 2 diabetes via p38 MAPK signaling. Exp Cell Res. 360:199–204.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Stewart AF, Hussain MA, García-Ocaña A,
Vasavada RC, Bhushan A, Bernal-Mizrachi E and Kulkarni RN: Human
β-cell proliferation and intracellular signaling: Part 3. Diabetes.
64:1872–1885. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Fraenkel M, Ketzinel-Gilad M, Ariav Y,
Pappo O, Karaca M, Castel J, Berthault MF, Magnan C, Cerasi E,
Kaiser N, et al: mTOR inhibition by rapamycin prevents beta-cell
adaptation to hyperglycemia and exacerbates the metabolic state in
type 2 diabetes. Diabetes. 57:945–957. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Baeder WL, Sredy J, Sehgal SN, Chang JY
and Adams LM: Rapamycin prevents the onset of insulin-dependent
diabetes mellitus (IDDM) in NOD mice. Clin Exp Immunol. 89:174–178.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zhang CL, Katoh M, Shibasaki T, Minami K,
Sunaga Y, Takahashi H, Yokoi N, Iwasaki M, Miki T and Seino S: The
cAMP sensor Epac2 is a direct target of antidiabetic sulfonylurea
drugs. Science. 325:607–610. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Sabbatini ME, Chen X, Ernst SA and
Williams JA: Rap1 activation plays a regulatory role in pancreatic
amylase secretion. J Biol Chem. 283:23884–23894. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Manyes L, Arribas M, Gomez C, Calzada N,
Fernandez-Medarde A and Santos E: Transcriptional profiling reveals
functional links between RasGrf1 and Pttg1 in pancreatic beta
cells. BMC Genomics. 15:10192014. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Hoffmann A and Spengler D: Transient
neonatal diabetes mellitus gene Zac1 impairs insulin secretion in
mice through Rasgrf1. Mol Cell Biol. 32:2549–2560. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Yang Y, Fu Q, Wang X, Liu Y, Zeng Q, Li Y,
Gao S, Bao L, Liu S, Gao D, et al: Comparative transcriptome
analysis of the swimbladder reveals expression signatures in
response to low oxygen stress in channel catfish, Ictalurus
punctatus. Physiol Genomics. 50:636–647. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Dong Q, Ginsberg HN and Erlanger BF:
Overexpression of the A1 adenosine receptor in adipose tissue
protects mice from obesity-related insulin resistance. Diabetes
Obes Metab. 3:360–366. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Rampersaud E, Damcott CM, Fu M, Shen H,
McArdle P, Shi X, Shelton J, Yin J, Chang YP, Ott SH, et al:
Identification of novel candidate genes for type 2 diabetes from a
genome-wide association scan in the Old Order Amish: Evidence for
replication from diabetes-related quantitative traits and from
independent populations. Diabetes. 56:3053–3062. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Straub SG and Sharp GW: Glucose-stimulated
signaling pathways in biphasic insulin secretion. Diabetes Metab
Res Rev. 18:451–463. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Ma X, Lu C, Lv C, Wu C and Wang Q: The
expression of miR-192 and its significance in diabetic nephropathy
patients with different urine albumin creatinine ratio. J Diabetes
Res. 2016:67894022016. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Wu C, Chen X, Shu J and Lee CT:
Whole-genome expression analyses of type 2 diabetes in human skin
reveal altered immune function and burden of infection. Oncotarget.
8:34601–34609. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Zhu Z, Yin J, Li DC and Mao ZQ: Role of
microRNAs in the treatment of type 2 diabetes mellitus with
Roux-en-Y gastric bypass. Braz J Med Biol Res. 50:e58172017.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Pivovarova O, Fisher E, Dudziak K,
Ilkavets I, Dooley S, Slominsky P, Limborska S, Weickert MO,
Spranger J, Fritsche A, et al: A polymorphism within the connective
tissue growth factor (CTGF) gene has no effect on non-invasive
markers of beta-cell area and risk of type 2 diabetes. Dis Markers.
31:241–246. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Salunkhe VA, Ofori JK, Gandasi NR, Salo
SA, Hansson S, Andersson ME, Wendt A, Barg S, Esguerra JLS and
Eliasson L: MiR-335 overexpression impairs insulin secretion
through defective priming of insulin vesicles. Physiol Rep. 52017.
PubMed/NCBI
|
|
100
|
Taneera J, Fadista J, Ahlqvist E, Atac D,
Ottosson-Laakso E, Wollheim CB and Groop L: Identification of novel
genes for glucose metabolism based upon expression pattern in human
islets and effect on insulin secretion and glycemia. Hum Mol Genet.
24:1945–1955. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Aso Y, Matsuura H, Momobayashi A, Inukai
Y, Sugawara N, Nakano T, Yamamoto R, Wakabayashi S, Takebayashi K
and Inukai T: Profound reduction in T-helper (Th) 1 lymphocytes in
peripheral blood from patients with concurrent type 1 diabetes and
Graves' disease. Endocr J. 53:377–385. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Voss M, Paterson J, Kelsall IR,
Martín-Granados C, Hastie CJ, Peggie MW and Cohen PT: Ppm1E is an
in cellulo AMP-activated protein kinase phosphatase. Cell Signal.
23:114–124. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Torsvik J, Johansson S, Johansen A, Ek J,
Minton J, Raeder H, Ellard S, Hattersley A, Pedersen O, Hansen T,
et al: Mutations in the VNTR of the carboxyl-ester lipase gene
(CEL) are a rare cause of monogenic diabetes. Hum Genet. 127:55–64.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Taneera J, Lang S, Sharma A, Fadista J,
Zhou Y, Ahlqvist E, Jonsson A, Lyssenko V, Vikman P, Hansson O, et
al: A systems genetics approach identifies genes and pathways for
type 2 diabetes in human islets. Cell Metab. 16:122–134. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Anhê FF, Lellis-Santos C, Leite AR,
Hirabara SM, Boschero AC, Curi R, Anhê GF and Bordin S: Smad5
regulates Akt2 expression and insulin-induced glucose uptake in L6
myotubes. Mol Cell Endocrinol. 319:30–38. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Glauser DA and Schlegel W: The
FoxO/Bcl-6/cyclin D2 pathway mediates metabolic and growth factor
stimulation of proliferation in Min6 pancreatic beta-cells. J
Recept Signal Transduct Res. 29:293–298. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Rechsteiner MP, Floros X, Boehm BO,
Marselli L, Marchetti P, Stoffel M, Moch H and Spinas GA: Automated
assessment of β-cell area and density per islet and patient using
TMEM27 and BACE2 immunofluorescence staining in human pancreatic
β-cells. PLoS One. 9:e989322014. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Ikeda K, Emoto N, Matsuo M and Yokoyama M:
Molecular identification and characterization of a novel nuclear
protein whose expression is up-regulated in insulin-resistant
animals. J Biol Chem. 278:3514–3520. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Solimena M, Schulte AM, Marselli L,
Ehehalt F, Richter D, Kleeberg M, Mziaut H, Knoch KP, Parnis J,
Bugliani M, et al: Systems biology of the IMIDIA biobank from organ
donors and pancreatectomised patients defines a novel
transcriptomic signature of islets from individuals with type 2
diabetes. Diabetologia. 61:641–657. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Scott EM, Carter AM and Findlay JB: The
application of proteomics to diabetes. Diab Vasc Dis Res. 2:54–60.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Khan AR and Awan FR: Mining of protein
based biomarkers for type 2 diabetes mellitus. Pak J Pharm Sci.
25:889–901. 2012.PubMed/NCBI
|
|
112
|
Okada S, List EO, Sankaran S and Kopchick
JJ: Plasma protein biomarkers correlated with the development of
diet-induced Type 2 diabetes in mice. Clin Proteomics. 6:6–17.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Bustin SA and Mueller R: Real-time reverse
transcription PCR (qRT-PCR) and its potential use in clinical
diagnosis. Clin Sci (Lond). 109:365–379. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Guénin S, Mauriat M, Pelloux J, Van
Wuytswinkel O, Bellini C and Gutierrez L: Normalization of qRT-PCR
data: The necessity of adopting a systematic, experimental
conditions-specific, validation of references. J Exp Bot.
60:487–493. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Kim B: Western Blot Techniques. Methods
Mol Biol. 1606:133–139. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Ule J, Hwang HW and Darnell RB: The future
of cross-linking and immunoprecipitation (CLIP). Cold Spring Harb
Perspect Biol. 10:a0322432018. View Article : Google Scholar : PubMed/NCBI
|