Introduction
Aortic valve stenosis (AS) involving aortic valve
calcification is the most common type of acquired valve diseases
worldwide (1,2). A key risk factor of AS in clinical
practice is bicuspid aortic valve (BAV), which occurs in 1–2% of
the general population (3,4). By definition, a BAV consists of two
functional leaflets, while a normal tricuspid aortic valve (TAV)
contains three (4). The
progression of valve calcification in adults with BAV is similar to
that observed in patients with TAV; however, calcification with BAV
is faster and often more severe (3). Thus, it remains critical to
investigate the differences between calcified BAVs and calcified
TAVs to identify key features that are associated with the
deterioration of valve calcification.
The progression of valve calcification is
predominantly regulated by valve interstitial cells (VICs) that
populate the interstitial matrix, and is accompanied by their
changes in the expression of different genes (5–8),
including bone morphogenetic protein 2 (BMP2), NOTCH 1, and the
SMADs, which result in differences in calcification-associated
protein expression and activities (9–12).
microRNAs (miRs/miRNAs) are well-known regulators of gene
expression at the post-transcriptional level (13,14),
and growing evidence suggested that several miRs are involved in
valve calcification via their target genes (13,15–17).
The present study performed miR profiling in calcified BAV and TAV
tissues, in order to compare their differentially expressed miRs.
The differentially expressed miRs were further analyzed, in which
miR-330-3p was demonstrated to be involved in the deterioration of
calcification of BAVs. The present study aimed to investigate the
potential signaling pathways associated with upregulated miR-330-3p
in BAVs and its calcification-associated target gene, CREB-binding
protein (CREBBP).
Materials and methods
Study population and aortic valve
collection
The present study was approved by the Ethics
Committee of the First Affiliated Hospital of Nanjing Medical
University, and written informed consent was obtained from all
patients prior to the study. Aortic leaflets were collected from
patients receiving aortic replacement between January 2016 and
December 2018 at Jiangsu Province Hospital. Echocardiography was
performed to confirm the morphology and function of the leaflets
prior to surgery. A total of 15 BAV samples were examined in the
present study, all of which presented type 1 BAV leaflets, with
fusion of the right and left coronary leaflets. A total of 15
degenerative TAVs were selected according to the echocardiographic
results and intra surgical inspections. All leaflets were sectioned
into two parts. The larger part was flash frozen in liquid nitrogen
and stored for RNA or protein extraction, while the smaller part
was fixed in 10% formalin and embedded in paraffin for subsequent
experimentation.
miRNA microarray
Agilent Human miRNA Microarray Slide (version 21.0;
8×60K; Design ID: 070156; Agilent Technologies, Inc.) was used to
profile miRNA expression in 8 aortic valve tissue samples from
patients with BAV or TAV. Total RNA was quantified using a NanoDrop
ND-2000 spectrophotometer (Thermo Fisher Scientific, Inc.) and the
RNA integrity was assessed using an Agilent Bioanalyzer 2100
(Agilent Technologies, Inc.). The sample labeling, microarray
hybridization and washing were performed based on the
manufacturer's standard protocol. Briefly, total RNA was
dephosphorylated, denaturated and labeled with Cyanine-3-CTP. After
purification the labeled RNAs were hybridized onto the microarray.
After washing, the arrays were scanned using the Agilent Scanner
G2505C (Agilent Technologies, Inc.).
Feature Extraction software (version 10.7.1.1;
Agilent Technologies, Inc.) was used to analyze array images to get
raw data. Subsequently, Genespring software (version 13.1; Agilent
Technologies, Inc.) was employed to finish the basic analysis with
the raw data. To begin with, the raw data was normalized with the
quantile algorithm. The probes that at least 100.0 percent of
samples in any 1 condition out of 2 conditions have flags in
‘Detected’ were chosen for further data analysis. Differentially
expressed miRNAs were then identified through fold change as well
as P-value calculated using t-test. The threshold set for up- and
down-regulated genes was a fold change>=1.5 and a
P-value<=0.05. Target genes of differentially expressed miRNAs
were the intersection predicted with 3 databases [(TargetScan
(www.targetscan.org), microRNAorg
(www.microrna.org) and PITA (www.pictar.org)].
Reverse transcription-quantitative PCR
(RT-qPCR)
miRs or mRNAs were extracted from the tissues
obtained from surgical operations or cultured VICs using the
miRNeasy Mini kit (Qiagen, Inc.) according to the manufacturer's
instructions, and reverse transcribed into cDNA using the Takara
PrimeScript™ RT master mix (Takara Bio, Inc.) according to the
manufacturer's instructions. qPCR was subsequently performed using
the 7900HT Fast Real-Time PCR System (Thermo Fisher Scientific,
Inc.) according to manufacturer's protocol. The primer sequences
used for qPCR were purchased from Realgene Bio-Technologies, Inc.
and are presented in Table I. The
following thermal cycling conditions were used for the qPCR:
Initial denaturation at 95°C for 10 min; then 40 cycles of 95°C for
15 sec and 60°C for 1 min. Relative expression levels were measured
using the 2−ΔΔCq method (18) and normalized to the internal
reference gene GAPDH. U6 small nuclear RNA was used as internal
control to normalize miRNA expression. All experiments were
performed in triplicate.
 | Table I.Primer sequences used for reverse
transcription-quantitative PCR. |
Table I.
Primer sequences used for reverse
transcription-quantitative PCR.
| Name | Primer sequence
(5′-3′) |
|---|
| hsa-CREBBP | F:
CAACCCCAAAAGAGCCAAACT |
|
| R:
CCTCGTAGAAGCTCCGACAGT |
| ssc-GAPDH | F:
TCGGAGTGAACGGATTTGGC |
|
| R:
TGACAAGCTTCCCGTTCTCC |
| hsa-GAPDH | F:
AGAAGGCTGGGGCTCATTTG |
|
| R:
AGGGGCCATCCACAGTCTTC |
| ssc-collagen I | F:
GAGGGCCAAGACGAAGACATC |
|
| R:
CAGATCACGTCATCGCACAAC |
| hsa-collagen I | F:
AGCCCTGGTGAAAATGGAGC |
|
| R:
CACCCTTAGCACCAACAGCA |
| ssc-Runx 2 | F:
AGCCACCTACCACAGAGCTA |
|
| R:
GGATGAGGAATGCGCCCTAA |
| ssc-BMP2 | F:
GAGCTAGCACTGAGCGACC |
|
| R:
GAAGTCTCCAGCCAAGTGCT |
| hsa-U6 | F:
GTGGGGAGAAGAGGACAGGA |
|
| R:
GTGGTACCCACTTTCGCACA |
Histological examination and
immunohistochemistry
The tissue samples were fixed with 10% formalin at
room temperature for 24 h, embedded in paraffin and cut into 20-µm
thick sections for Von Kossa and Sirius Red staining and
semi-quantitative analysis. Tissues were also used for
immunohistochemical staining for CREBBP, collagen I, Runt-related
transcription factor 2 (Runx 2), matrix metalloproteinase (MMP)-2
and MMP-9. In brief, sections were washed in Tris-buffered
saline/0.1% Tween-20 and subsequently blocked in 5% milk at room
temperature for 2 h. Tissue sections were incubated with the
primary antibodies (all 1:1,000) listed in Table II at 4°C overnight. Sections were
washed in PBS prior to incubation with biotinylated secondary
antibody (1:200; cat. no. 21537; Merck KGaA) at room temperature
for 30 min. The sections were subsequently treated with Vectastain
ABC reagent (Vector Laboratories, Inc.) and 3,3′-diaminobenzidine,
prior to counterstaining with hematoxylin at room temperature for 2
min. A light optical microscope was used for image capture
(magnification, ×10 or ×200) using visible light source without any
filters. Image-Pro plus software (version 6.0; Media Cybernetics,
Inc.) was used for signal quantification. In brief, the positive
area was automatically selected according to criteria that
contained a specific Hue-Saturation-Intensity (HIS) parameter
adjusted to the different staining method. Subsequently, the
optical density (OD) of the positive areas and the whole image were
calculated automatically. The data were extracted and the OD of per
unit area was calculated and presented as a semi-quantitative
indicator.
 | Table II.Antibody information. |
Table II.
Antibody information.
| Name of
antibody | Catalog no. | Supplier | Type |
|---|
| Collagen I | WL0088 | Wanleibio Co.,
Ltd. | Polyclonal |
| BMP2 | ab14933 | Abcam | Polyclonal |
| Runx 2 | 12556 | Cell Signaling
Technology, Inc. | Monoclonal |
| GAPDH | ab8245 | Abcam | Monoclonal |
| CREBBP | ab2832 | Abcam | Polyclonal |
| MMP2 | sc-13594 | Santa Cruz
Biotechnology, Inc. | Monoclonal |
| MMP9 | sc-21736 | Santa Cruz
Biotechnology, Inc. | Monoclonal |
VIC culture and transient
transfection
All procedures involving the use of animals were
approved by the Institutional Animal Care and Use Committee of
Nanjing Medical University. Porcine aortic VICs were harvested from
porcine hearts, which were supplied by the Laboratory Animal Center
of Nanjing Medical University and isolated by collagenase digestion
as previously described (19). In
brief, pigs at 3–4 months old were sedated using a cocktail of
ketamine hydrochloride (20 mg/kg, intramuscular injection) and
xylazine (2 mg/kg, intramuscular injection). Following adequate
sedation (~15 min), pigs were euthanized by overdose of
pentobarbital (156 mg/kg body weight) administered intravenously
and their hearts were harvested immediately. Aortic valves were
excised with precision to exclude non-leaflet tissue. The excised
leaflets were carefully scraped on the aortic and ventricular
aspects to remove the endothelial layer. Tissues samples were
subsequently sectioned into ~2×2 mm explants, which were cultured
in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
1% penicillin-streptomycin (Beyotime Institute of Biotechnology),
and 15% fetal bovine serum (ScienCell Research Laboratories, Inc.),
in 5% CO2 at 37°C. Cells that reached ~100% confluence
were extracted from the explants for subculture and those of the
2nd or 3rd generations were used for further analyses.
An antisense-based strategy was adapted to block
miRNA function in cultured cells. Anti-miR-330 2′-O-methyl
oligoribonucleotides (Guangzhou RiboBio Co., Ltd.) were synthesized
to sequence-specifically inactivate miR-330 and thus to release the
suppression on target mRNAs without changing its expression level
(20). A total of
7.5×104 cells/cm2 VICs were transfected with
5 nmol/l miR-330-3p mimic or inhibitor (micrONTM miR
mimic/inhibitor/nc_Standard; cat. nos. miR10000751-1-5 and
miR20000751-1-5; Guangzhou RiboBio Co., Ltd.) using Lipofectamine™
3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The
transfected cells were incubated in DMEM supplemented with 15% FBS
for 6 h and subsequently were incubated in DMEM supplemented with
1% penicillin-streptomycin and 15% FBS. At 48 h post-transfection,
cells were used for subsequent experiments.
VICs were also transfected with miR-659 mimic,
miR-663 mimic, or miR-146 inhibitor (Guangzhou RiboBio Co., Ltd.)
using the aforementioned protocol.
Pro-osteogenic medium preparation and
Alizarin Red S staining for calcium deposits
To prepare the calcification medium, 1 mmol of
CaCl2 and 1 mmol of
Na5P3O10 were dissolved in 50 ml
of water to produce reagent one and reagent two, respectively.
Subsequently, 500 µl reagent one, 375 µl reagent two and 4 ml of 8%
FBS were added to 45 ml DMEM. VICs were cultured with or without
pro-osteogenic medium for 48 h post-transfection, and subsequently
stained with Alizarin Red S Staining (Beyotime Institute of
Biotechnology) according to the manufacturer's protocol. Briefly,
cells were washed with Ca2+-free PBS three times, fixed
with 4% paraformaldehyde for 10 min at room temperature and then
fixed with 95% ethanol for 20 min at room temperature.
Subsequently, cells were stained with 1% alizarin red solution (pH
4.2; cat. no. ST1078; Beyotime Institute of Biotechnology) for 1
min at room temperature to visualize matrix calcium deposition. The
solution was washed out with distilled water and the stained cells
were photographed using a digital camera. The images were then
processed and analyzed using Image-Pro Plus software (version 6.0,
Media Cybernetics, Inc.).
Western blotting
Transfected cells were harvested and used for
preparation of protein extracts. Cells were lysed using a cell
lysis kit (Nanjing KeyGen Biotech Co., Ltd.). The bicinchoninic
acid method was used for quantification of total protein of the
samples. Equal amounts of protein extracts (30 µg) were separated
by 10% SDS-PAGE and transferred to PVDF membranes. Following
blocking with TBS-Tween-20 solution containing 5% BSA (cat. no.
B600036; Sangon Biotech Co., Ltd.) for 2 h at room temperature, the
membranes were incubated with the following primary antibodies:
Rabbit polyclonal antibody against Runx 2 (1:1,000; cat. no. 12556;
Cell Signaling Technology, Inc.), rabbit polyclonal antibody
against BMP2 (1:1,000; cat. no. 14933; Abcam), rabbit polyclonal
antibody against CREBBP (1:1,000; cat. no. 2832; Abcam), rabbit
polyclonal antibody against collagen I (1:1,000; cat. no. WL0088;
Wanleibio Co., Ltd.), and mouse monoclonal antibody against GAPDH
(1:1,000; cat. no. 8245; Abcam) overnight at 4°C. The membranes
were then probed using either a goat anti-rabbit or a goat
anti-mouse horseradish peroxidase-conjugated secondary antibody
(1:5,000; cat. nos. ab150077 and ab205719; Abcam) for 2 h at room
temperature. The blots were developed with an Omni-ECL™ Enhanced
Pico Light Chemiluminescence kit (Epizyme Co.) and exposed on a
ChemiDoc MP imaging system (Bio-Rad Laboratories, Inc.). Band
density was quantified using ImageJ software (version 1.8.0;
National Institutes of Health) and normalized to GAPDH.
Dual-luciferase reporter assay
Specific culture medium was prepared by mixing DMEM,
10% FBS (ScienCell Research Laboratories, Inc.), 1% nonessential
amino acids, 1% L-glutamine and 1% penicillin with streptomycin.
293T cells (National Infrastructure of Cell Line Resource) were
seeded into 12-well plates (5×104 cells/cm2)
and cultured in specific medium at 37°C in 5% CO2.
The pmirGLO dual luciferase miR target expression
vector (pmirGLO), containing both firefly and Renilla
luciferase genes was purchased from Promega Corporation. Human
CREBBP 3′-untraslated region (3′-UTR), including the predicted
binding site of miR-330-3p, was amplified via RT-PCR and inserted
into the 3′-UTR downstream of the firefly luciferase gene in the
pmirGLO vector (pmirGLO-UTR) using XbaI and SacI
restriction sites. The QuikChange XL Site-Directed Mutagenesis kit
(Agilent Technologies, Inc.) was used to construct the mutant
miR-330-3p-binding site vector (pmirGLO-UTR-MUT) according to the
manufacturer's protocol. Restriction enzyme digestion and
sequencing was performed to validate the constructs. After cellular
transfection for 36 h, luciferase activity was assessed with
Lipofectamine™ 3000 reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) using a Dual-Glo luciferase assay system (Promega
Corporation). Firefly luciferase activity was normalized to
Renilla luciferase activity.
Engineered clustered regularly
interspaced shot palindromic repeats (CRISPR)/CRISPR associated
protein 9 (−Cas9) and DNA constructs
The design and use of engineered Cas9 complex
(pLenti-CMV-NLS-dCas9-VP64-2A-Puro; cat. no. H7281; Heyuan Kangning
Medical Company Biotechnology Limited Company) and efficient single
guide RNA (gRNA;
plenti-U6-gRNA-2×(wt+f6)MS2-CMV-MCP-P65-HSF1-IRES-blasticidin; cat.
no. H7284; Heyuan Kangning Medical Company Biotechnology Limited
Company) to induce CREBBP transcriptional activation was performed
according to previously published protocols (21,22).
CCTop, a CRISPR/Cas9 target online predictor
(cctop.cos.uni-heidelberg.de) was used to design the gRNAs used in
the present study. The sequences of both control and CREBBP gRNAs
are as follows: 5′-GCACTACCAGAGCTAACTCA-3′ (control gRNA) and
5′-CCACTTAATGAATTCGCTCG-3′ (CREBBP gRNA). The gRNA primers were
annealed and cloned into gRNA (MS2)-plasmids via the BbsI
sites. All CRISPR constructs were purchased from Heyuan Biotech
Company (cat. nos. 61422, 61423 and 61424). The CREBBP
promoter-luciferase construct was generated by synthesizing the DNA
fragment corresponding to the CREBBP promoter region from
GenScript, and sub-cloned into the pGL4.20 vector (Heyuan Biotech
Company).
Statistical analysis
Statistical analysis was performed using SPSS
software (version 19.0; IBM Corp.). Univariate analyses were
performed using the χ2 test for categorical variables,
and Student's t-test or one-way ANOVA followed by Dunnett's post
hoc were performed for continuous variables. Pearson's correlation
analysis was used to investigate the relationship between two
quantitative, continuous variables. Data are presented as the mean
± SD. P<0.05 was considered to indicate a statistically
significant difference.
Results
Clinical characteristics of patients
with BAV and TAV
Clinical characteristics of the patients involved in
the present study are listed in Table III. Patients with BAV were
significantly younger compared with patients with TAV. No
significant differences were observed between the two groups in sex
and other risk factors concerning cardiovascular calcification.
Both groups presented similar preoperative echocardiography
results, which measured the left ventricular ejection fraction,
aortic valve area and mean trans valvular gradient. Based on the
echocardiography results, patients with a mean aortic valve area
<0.75 cm2 were diagnosed with severe AS.
 | Table III.Patient demographics. |
Table III.
Patient demographics.
| Variable | TAV (n=15) | BAV (n=15) | P-value |
|---|
| Age (years) | 61.40±8.91 | 49.20±10.72 | 0.00 |
| Females, % (n) | 46.67 (7) | 53.33 (8) | 0.72 |
| Risk factors |
|
Hypertension, % (n) | 37.50 (3) | 25.00 (2) | 1.00 |
|
Hypercholesterolemia, %
(n) | 50.00 (4) | 62.50 (5) | 1.00 |
|
Diabetes, % (n) | 37.50 (3) | 50.00 (4) | 1.00 |
| Smoking
history, % (n) | 25.00 (2) | 37.50 (3) | 1.00 |
| Chronic
kidney disease, % (n) | 0.00 (0) | 0.00 (0) | N/A |
| Echocardiographic
parameters |
| Left
ventricular ejection fraction, % | 63.25±4.61 | 62.99±5.71 | 0.89 |
| Flow
velocity maximum of aortic valve, m/s | 4.56±0.88 | 4.20±0.86 | 0.48 |
| Mean
gradient, mmHg | 70.27±26.33 | 65.33±34.21 | 0.70 |
|
Ascending aorta maximal
diameter, mm | 40.23±4.42 | 44.83±5.92 | 0.07 |
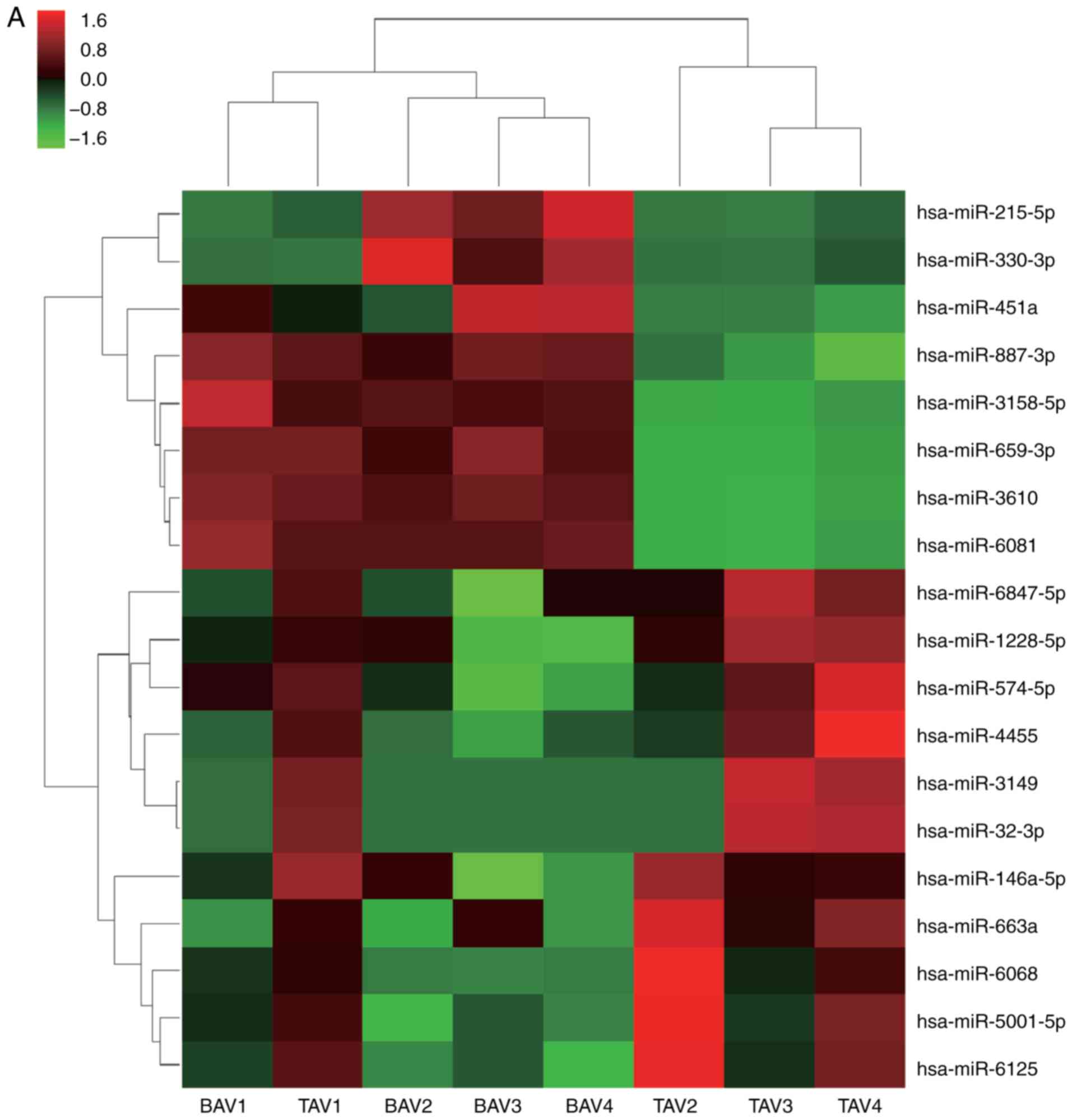
Differences in miR expression profiles
of valve tissue from AS patients with BAV and TAV
Valve tissues from patients with AS, with either BAV
(n=4) or TAV (n=4) were used for miR profiling. miRs that were
commonly upregulated or downregulated in patients with BAV compared
with patients with TAV were selected, including 11 upregulated and
eight downregulated genes, respectively (Table IV). The heat map of the
differentially expressed miRs are presented in Fig. 1A.
 | Table IV.Differentially expressed microRNAs of
aortic valves from patients with BAV compared with patients with
TAV. |
Table IV.
Differentially expressed microRNAs of
aortic valves from patients with BAV compared with patients with
TAV.
| A, Downregulated
differentially expressed genes |
|---|
|
|---|
| microRNA | P-value (TAV vs.
BAV) | Fold change |
|---|
|
hsa-miR-1228-5p | 0.038 | 3.195 |
|
hsa-miR-146a-5p | 0.036 | 3.104 |
| hsa-miR-3149 | 0.027 | 15.946 |
| hsa-miR-32-3p | 0.026 | 13.893 |
| hsa-miR-4455 | 0.022 | 1.676 |
|
hsa-miR-5001-5p | 0.031 | 1.645 |
| hsa-miR-574-5p | 0.045 | 1.763 |
| hsa-miR-6068 | 0.044 | 1.537 |
| hsa-miR-6125 | 0.015 | 1.597 |
| hsa-miR-663a | 0.020 | 1.518 |
|
hsa-miR-6847-5p | 0.035 | 1.575 |
|
| B, Upregulated
differentially expressed genes |
|
|
microRNA | P-value (TAV vs.
BAV) |
Fold-change |
|
| hsa-miR-215-5p | 0.036 | 6.212 |
|
hsa-miR-3158-5p | 0.017 | 11.207 |
| hsa-miR-330-3p | 0.041 | 4.462 |
| hsa-miR-3610 | 0.026 | 21.066 |
| hsa-miR-451a | 0.038 | 6.534 |
| hsa-miR-6081 | 0.017 | 9.466 |
| hsa-miR-659-3p | 0.045 | 12.583 |
| hsa-miR-887-3p | 0.030 | 12.512 |
miR sequencing results of the patients' valve
tissues were validated via RT-qPCR analysis. miR-330-3p, miR-659
and miR-663 expression levels in the BAV leaflets were
significantly increased compared with the degenerative TAV leaflets
by 3.72-fold, 5.04-fold and 4.22-fold, respectively (Fig. 1B-D); however, miR-146 expression
was significantly decreased by −11.56-fold (Fig. 1E). No significant differences were
observed in the other 15 miRs (Fig.
S1).
Calcification progress is accelerated
by miR-330-3p in sus scrofa VICs
VICs were successfully transfected with miR-659 or
miR-663 mimics or miR-146 inhibitor as the expression of these
miRNAs were significantly increased (miR-659 and miR-663) or
decreased (miR-146) following transfection (Fig. S2A, E and I). However, levels of
calcification-associated factors Runx 2 and BMP2 and the
fibrosis-associated factor collagen I in miR-659 mimic- and miR-663
mimic-transfected cells did not show any significant difference
compared with controls (Fig. S2B-D,
F-H). In addition, mRNA expression levels of Runx 2 and BMP2
were significantly decreased in miR-146 inhibitor-transfected cells
compared with controls (Fig. S2J and
K). As the simulation of tissue expression alterations to the
three microRNAs in cultured VICs failed to demonstrate similar
calcification-associated protein alterations to those observed in
BAV tissue samples, further experimentation on miR-659, miR-663 and
miR-146 was ceased.
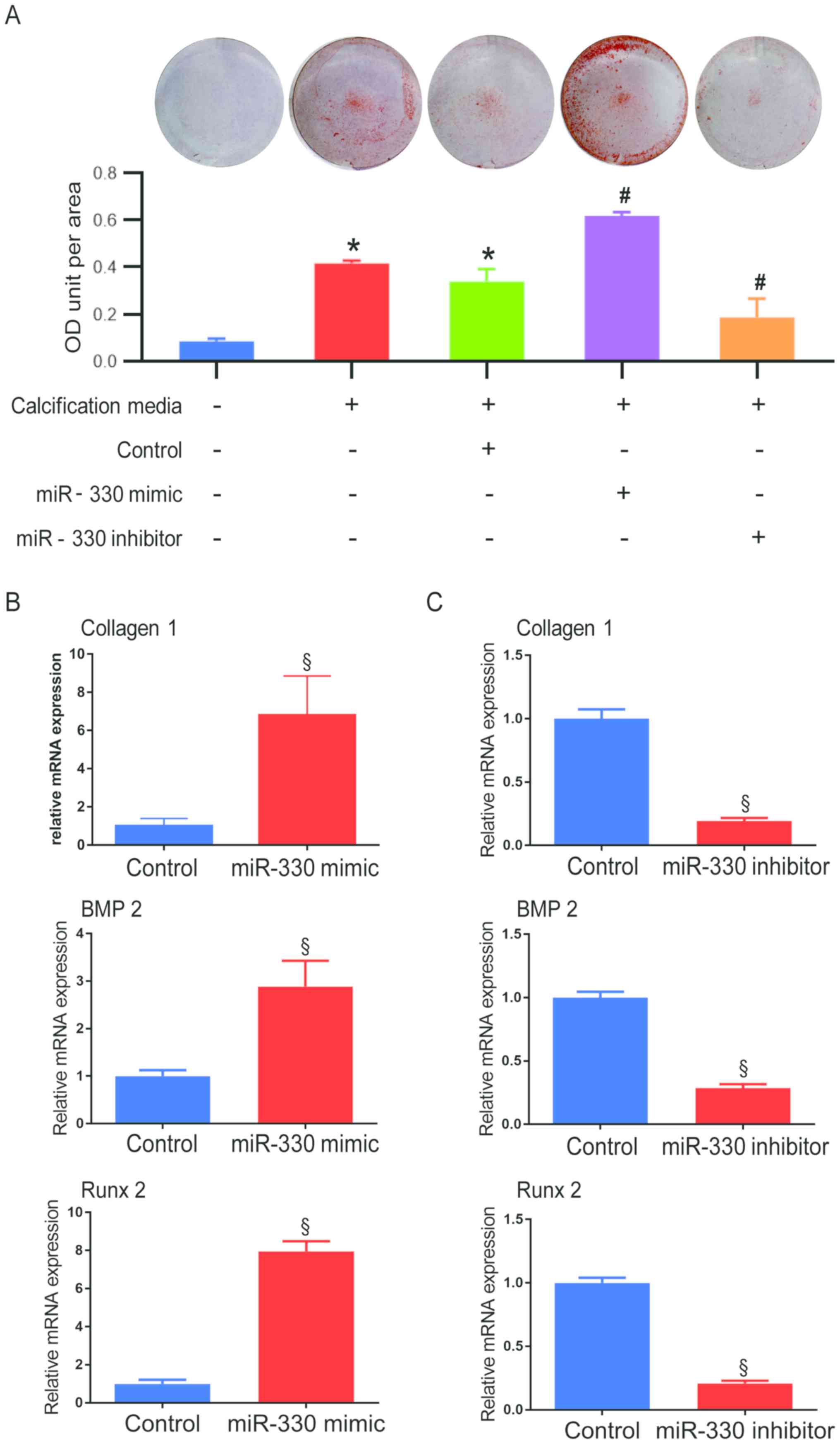
To investigate whether miR-330-3p plays a role in
cellular calcification, VICs transfected with miR-330 mimic or
inhibitor were incubated under pro-osteogenic conditions (Fig. 2A). After 48 h, both under
pro-osteogenic conditions, a significantly higher degree of
calcification was observed in the miR-330-3p mimic group compared
with the miR-330-3p negative control group, while cells transfected
with miR-330 inhibitor demonstrated a significant decrease of
calcification compared with the miR-330-3p negative control group
(Fig. 2A). Higher collagen I mRNA
expression was observed in VICs treated with miR-330-3p mimic
compared with the control group (Fig.
2B), indicating that the fibrosis progress was accelerated by
miR-330, thus fibrosis is considered a pro-calcification progress.
Concurrently, calcification-associated pathways were investigated
at the mRNA level. VICs exhibited a 2.88-fold increase in BMP2 mRNA
expression in the miR-330 mimic group compared with the control
group (Fig. 2B). Similar
alteration patterns were demonstrated in the expression of the
downstream pro-osteogenic transcription factor, Runx 2 (7.95-fold
increase vs. control; Fig. 2B).
Alternatively, VICs transfected with miR-330 inhibitor demonstrated
a significant decrease in mRNA expression of collagen I, BMP2 and
Runx 2 compared with their respective control groups (Fig. 2C).
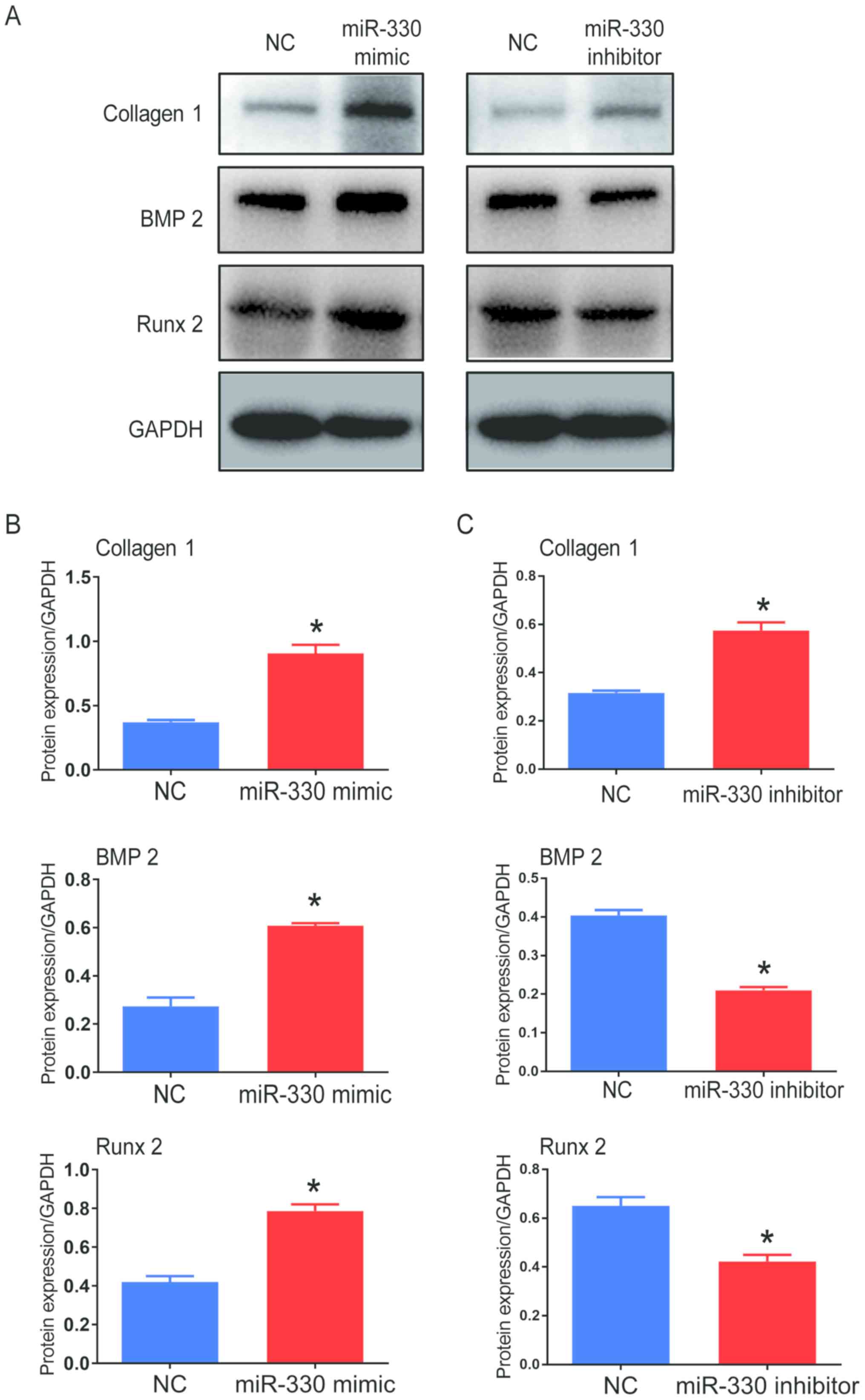
Furthermore, the protein expression of collagen I,
BMP2 and Runx 2 significantly increased in cells transfected with
miR-330-3p mimic compared with the control groups following
pro-calcification culture, indicating that the calcification
progress was accelerated in these cells (Fig. 3A and B). VICs transfected with
miR-330-3p inhibitor showed decreased protein expression levels of
BMP2 and Runx 2 compared with the control group; however, protein
expression level of collagen 1 increased (Fig. 3A and C). Western blots from
different experiments are shown in Fig. S3A and B. Taken together, these
results suggested that miR-330-3p might facilitate the
calcification progress in cultured VICs.
CREBBP is a direct target of
miR-330-3p
Based on online bioinformatics analysis using
TargetScan software (www.targetscan.org, version 7.0) and relevant studies
investigating vascular calcification (23,24),
CREBBP was identified as a potential target gene of miR-330-3p.
Immunohistochemical staining of CREBBP was performed in human
aortic valve tissue samples, which demonstrated significantly
increased CREBBP protein expression in BAV leaflet tissues compared
with TAV leaflets (Fig. 4A and B).
CREBBP mRNA expression in human leaflets was also measured, and the
results demonstrated a −2.08-fold decrease of CREBBP mRNA
expression in BAV leaflets compared with TAV tissues (Fig. 4C). Pearson's correlation analysis
on these human tissue samples indicated that the CREBBP protein
expression levels negatively correlated with miR-330-3p expression
(r=−0.897; P<0.05; Fig.
4D).
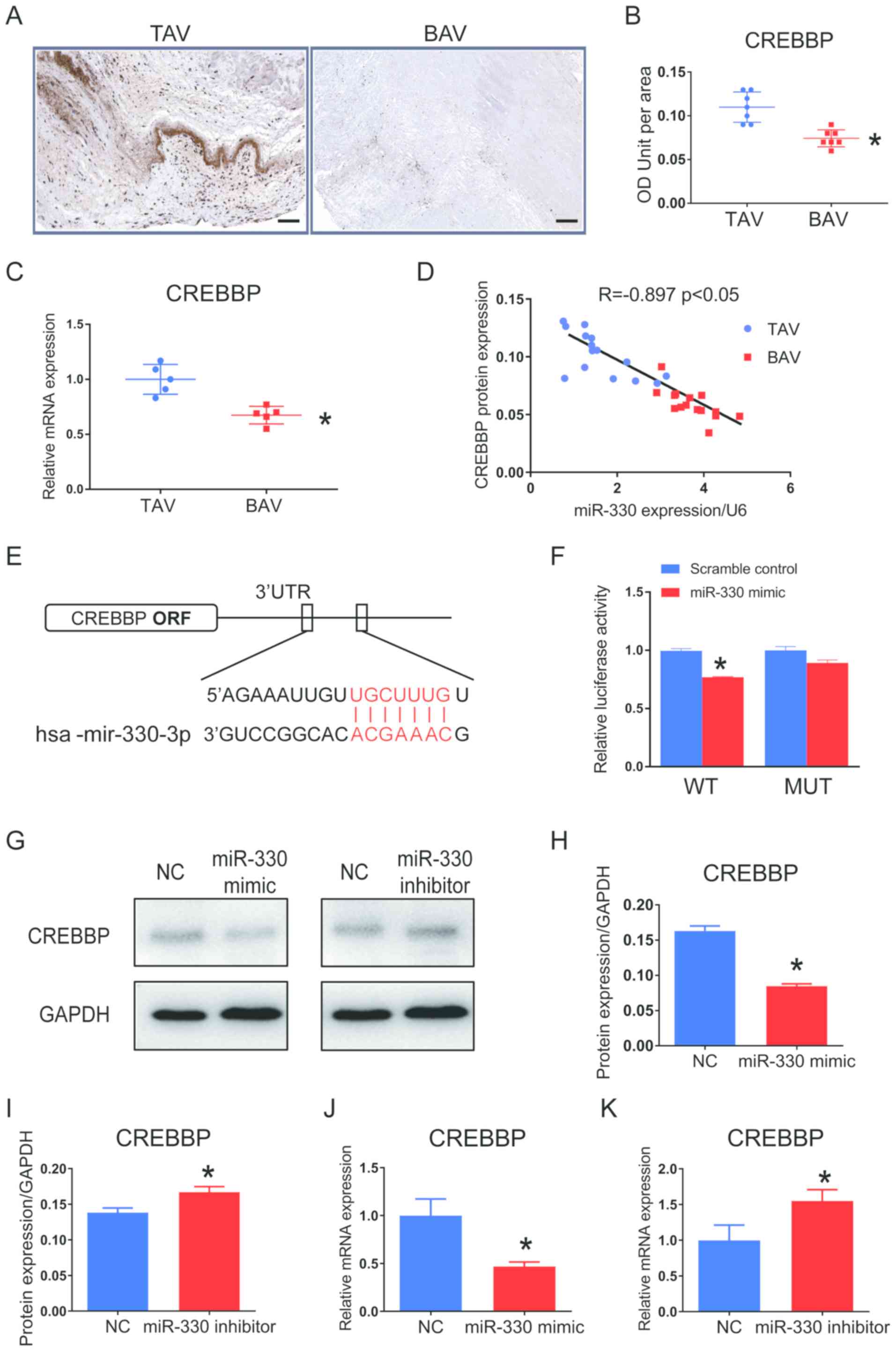 | Figure 4.CREBBP is a direct target of miR-330
in both human valve tissues and in cultured VICs. (A)
Representative images of human aortic valve tissue slices assessed
via immunohistochemistry for CREBBP. Scale bar, 50 µm. CREBBP
expression was lower in the human BAV group compared with the TAV
group at both the (B) protein and (C) mRNA levels. *P<0.05 vs.
TAV. n=6 in the TAV group; n=8 in the BAV group. (D) Pearson's
correlation analysis demonstrated that CREBBP protein levels
negatively correlated with miR-330 expression. n=15 per group;
R=−0.897; P<0.05. (E) Predicted binding sites of miR-330 with
CREBBP. (F) miR-330 mimic significantly suppressed luciferase
activity in 3′-UTR WT clones but not in 3′-UTR mutant clones.
*P<0.05 vs. scramble control. (G) Western blots of CREBBP
protein expression levels in VICs transfected with miR-330 mimic or
miR-330 inhibitor. Quantification of CREBBP protein expression
levels in VICs transfected with (H) miR-330 mimic or (I) miR-330
inhibitor. *P<0.05 vs. TAV. CREBBP mRNA expression in VICs
transfected with (J) miR-330 mimic or (K) miR-330 inhibitor.
*P<0.05 vs. TAV. Student's t-test was performed. Data are
presented as the mean ± standard deviation. miR, microRNA; VICs,
valvular interstitial cells; BAV, bicuspid aortic valve; TAV,
tricuspid aortic valve; UTR, untranslated region; OD, optical
density; ORF, open reading frame; WT, wild-type; MUT, mutant; NC,
negative control; CREBBP, CREB-binding protein. |
Computational analysis demonstrated that the seed
sequence of hsa-miR-330-3p was complementary to the 56–62 nt of the
3′-UTR in CREBBP mRNA, which is a potential binding site for
miR-330-3p directly targeting CREBBP (Fig. 4E). Dual-luciferase reporter assay
was subsequently performed using 293T cells transfected with
wild-type or point mutation vectors of the 3′-UTR region of CREBBP
at the predicted binding site. Luciferase activity was
significantly decreased in cells treated with miR-330-3p mimic in
the wild type group compared with the negative control group, while
there was no significant difference in luciferase activity between
the mutated groups (Fig. 4F),
indicating that miR-330-3p directly binds to the CREBBP 3′-UTR.
Furthermore, cultured VICs treated with miR-330
mimic for 36 h exhibited significantly lower CREBBP protein
expression by −1.88-fold compared with the control group, while
treatment with miR-330-3p inhibitor significantly increased CREBBP
protein expression by 1.55-fold (Fig.
4G-I). Western blots from repeat experiments are shown in
Fig. S3A and B. CREBBP mRNA
expression in cultured VICs showed the same patterns to protein
expression (Fig. 4J and K).
Promotion of calcification induced by
miR-330-3p is suppressed by CREBBP upregulation
It was further investigated whether the
calcification promotion effect of miR-330-3p depends on the
downregulation of CREBBP. Since the coding sequence of CREBBP gene
is too long, CREBBP protein cannot be overexpressed by the
conventional construction of plasmids containing its cDNA sequence.
Hence, endogenous overexpression was performed using the
transcriptional activation method. CREBBP transcriptional
activation was induced by CRISPR activation (CRISPRa) strategy,
which exploits the targeting mechanism of an sgRNA to direct the
complex to a promoter or enhancer of a gene. In the present study,
CRISPRa targeted the CREBBP promoter region and significantly
increased the expression of endogenous CREBBP in VICs compared with
the control group (Fig. 5A and B).
CREBBP mRNA expression was also significantly increased in the
CREBBP sgRNA group compared with sgRNA negative control group in
the presence of miR-330-3p mimic (Fig.
5C). mRNA (Fig. 5D) and
protein (Fig. 5E and F; S3C and D)
expression levels of Runx 2 and BMP2 significantly decreased
following overexpression of CREBBP in the presence of miR-330-3p
mimic, indicating that the presence of CREBBP may alleviate the
degree of calcification in the high miR-330-3p environment.
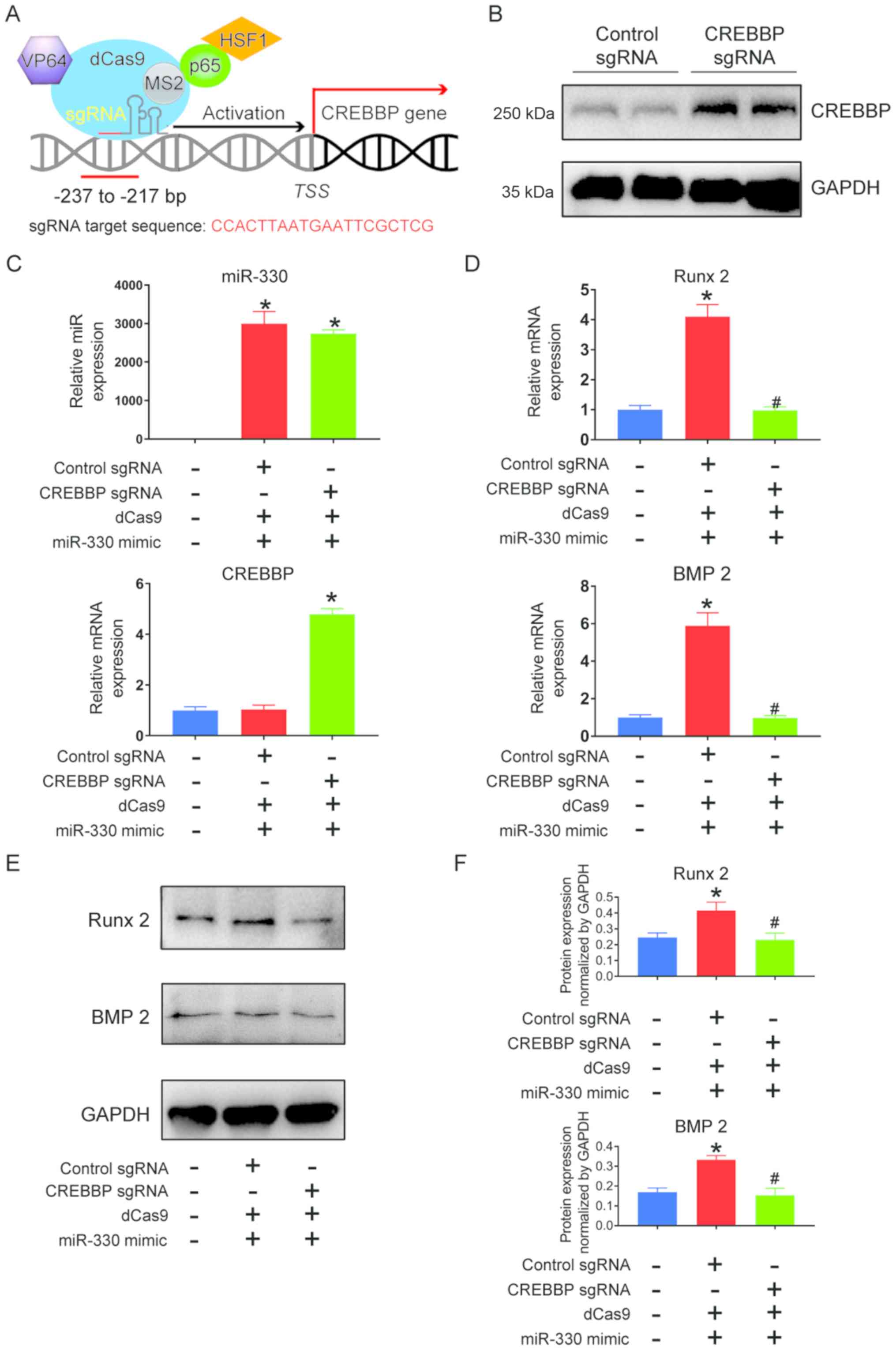 | Figure 5.Promotion of calcification via
miR-330 is suppressed by upregulating endogenous CREBBP in cultured
VICs. (A) Schematic of CRISPRa strategy to activate CREBBP gene
expression. (B) CRISPRa induction of CREBBP increased CREBBP
protein expression, as shown by Western blotting. (C) VICs were
transfected with either a negative control or sgRNA targeting the
proximal CREBBP promoter region in the presence of miR-330 mimic.
mRNA levels of miR-330 and CREBBP were determined via reverse
transcription-quantitative PCR analysis. (D) mRNA expression levels
of BMP2 and Runx 2 in VICs transfected with gRNA targeting CREBBP,
in the presence of miR-330 mimic. (E) Western blots and (F)
quantification of BMP2 and Runx 2 protein expression levels in VICs
transfected with gRNA targeting CREBBP in the presence of miR-330
mimic. One-way ANOVA with Dunnett's post hoc test was performed.
Data are presented as the mean ± standard deviation. *P<0.05 vs.
negative control; #P<0.05 vs. negative control in the
presence of miR-330 mimic. miR, microRNA; VICs, valvular
interstitial cells; CRISPR, clustered regularly interspaced short
palindromic repeats; sgRNA, single guide RNA; BMP2, bone
morphogenic protein 2; Runx 2, Runt-related transcription factor 2;
CRISPRa, CRISPR activation; CREBBP, CREB-binding protein. Cas9,
CRISPR-associated protein 9; HSF1, Heat shock transcription factor
1. |
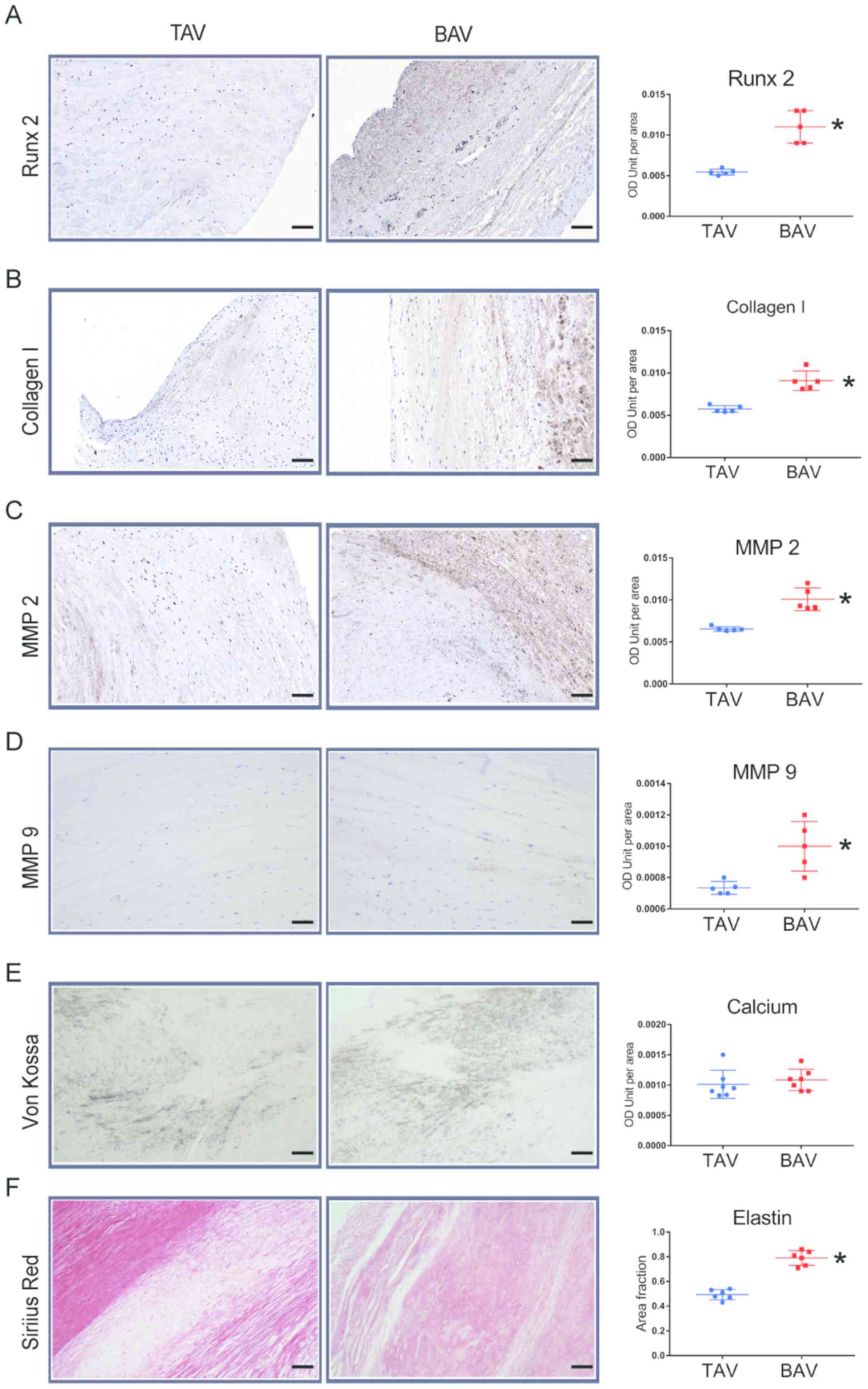
BAV leaflets present more
calcification-associated alterations similar to cellular
alterations
To validate that upregulated microRNA-330-3p
promotes calcification in patients with BAV, the positive areas of
pro-calcification proteins in aortic valve samples from patients
with BAV and TAV were examined. Von Kossa staining demonstrated
similar calcium deposits between TAV and BAV leaflet slices
(Fig. 6E). Furthermore,
immunohistochemical staining of Runx 2, collagen 1, MMP 2 and MMP 9
demonstrated significantly higher positive areas in BAV samples
compared with TAV samples (Fig.
6A-D). These alterations suggest a poor disorder of the
interstitial function of VICs in patients with BAV compared with
patients with TAV. Furthermore, Sirius Red staining demonstrated a
higher concentration of elastin in the BAV group compared with the
TAV group, which is prone to calcification in a number of disease
processes (Fig. 6F).
Discussion
The present study identified a total of 19
differentially expressed miRs between BAVs and TAVs via microarray
analysis. Of the 19, only four miRs (miR-146, miR-330-3p, miR-659
and miR-663) demonstrated a significant difference between the two
groups when compared with the leaflets obtained from patients with
AS. The alteration in the calcification progress was indicated in
sus scrofa VICs when these miRs were overexpressed or knocked down.
Of the four miRs, only miR-330-3p was demonstrated to influence the
calcification by remarkably accelerating its calcification
progress. Thus, miR-330-3p was selected for further
experimentation. Both CREBBP mRNA and protein expression levels
were demonstrated to increase in BAV leaflets; however, CREBBP
protein expression in human aortic valves was negatively correlated
to miR-330-3p expression. Furthermore, increased miR-330-3p
expression in VICs was demonstrated to decrease CREBBP mRNA and
protein expression, while miR-330-3p knockdown increased CREBBP
mRNA and protein expression levels. In addition, BAV leaflets
presented calcification-associated alterations similar to the
cellular alterations. miR-330-3p expression was upregulated in
human BAV leaflets, which suggested that it may play a role in
early calcification by regulating CREBBP, which, to the best of our
knowledge, has not been previously reported.
CBP/p300 interacting transactivator 1 (CiTED1) was
reported to be an inhibitor for the early calcification of BAVs
(25,26). Lin et al (26) reported that the translocation of
CiTED1 from the cytoplasm to the nucleus was promoted by the
parathyroid hormone in the mouse osteoblastic MC3T3-E1 subclone 14
cell line, which impaired the calcification progress. They also
found that serine-to-alanine mutations at position 79 in the 63–84
domain of CiTED1 affect Runx 2 expression, which is a key protein
relevant to calcification (26).
CREBBP is known as a key co-activator of CiTED1 (25). Therefore, the aberrant expression
of CREBBP may have impact on calcification which is consistent with
the results of the present study. Furthermore, it was reported that
the translocation of CiTED1 is enhanced by the protein kinase C
activator (27,28). A number of studies have set out to
determine the role of miR-330-3p in different types of tumors, most
of which have reported miR-330-3p-3p as a tumor suppressor gene.
For example, miR-330-3p plays a critical role in glioma, as well as
in breast (29), liver, esophageal
(30), lung, gastric, colon and
prostate cancers (31–35). Although its role in cardiovascular
calcification remains unclear, Liu et al (28) has proposed that miR-330-3p directs
the proliferation of several types of cells, and demonstrated its
negative influence of protein kinase C on the endothelial
monocyte-activating polypeptide-II in the immortalized human brain
endothelial cell line, hCMEC/D3. These findings indicate that
miR-330-3p may be involved in the process of calcification in
cardiovascular system, which was consistent with the results of the
present study.
A number of classic alteration patterns are involved
in calcification, including receptor translocation mediated by
serine mutation and epigenetic modulation regulated by histone
acetyltransferase (19,26). Previous studies reported the
association between CREBBP and aortic calcification, and suggested
that CREBBP ultimately promotes calcification via its translocation
from the cytoplasm to the nucleus (27). Taken together, the results of the
present study and findings from previous studies indicated that the
alteration of CREBBP via miR-330-3p might aggravate susceptibility
toward early calcification observed in patients with BAV.
The functional differences between BAVs and TAVs
were assessed in the leaflets of patients. MMP-2 protein, MMP-9
protein and collagen 1 expression were higher in BAV leaflets,
suggesting a disorder in the matrix function. This further
validated the hypothesis that upregulation of miR-330-3p in BAV
leaflets inhibited CREBBP signaling and promoted fibrosis in the
extracellular matrix, which ultimately causes valvular
calcification.
In the present study, VICs were incubated in
pro-osteogenetic medium for 48 h to promote cellular calcification.
This medium contains a high dose of phosphate and calcium ions,
which simulates an environment often observed in acute renal
failure models characterized by nephric calcification (36). It was shown that the imbalance of
phosphate metabolism and high blood calcium levels are the
pathogenic factors in the process of vascular calcification in
patients with chronic kidney disease (37). This method was successfully applied
to develop calcification in valve interstitial cells in 48 h
(19). Compared with other
methods, which usually uses dexamethasone and vitamin D in the
medium and takes >20 days to develop calcification in cells, the
present approach was more advantageous in terms of lower incubation
time and higher cytoactiveness.
However, there are limitations in the present study.
The experimental setting involved a number of models, which may not
be particularly relevant for BAV as it is a type of heart valve
deformation at the organ level. It is difficult to find a cellular
model that mimics BAV at a cellular level. However, the in
vitro model of cell calcification in the present study is well
established and the results are reliable in terms of the mechanisms
of promoting cellular calcification. These results need further
confirmation on BAV animal models, which will be investigated in
the future.
The results of the present study demonstrated that
microRNA-330-3p was upregulated in the stenotic aortic leaflets
from patients with BAV compared with patients with TAV.
Upregulation of miR-330-3p was associated with valvular
calcification via CREBBP targeting, which involves remodeling of
the extracellular matrix. Taken together, these results introduced
a novel perspective concerning the molecular mechanism underlying
accelerated valvular calcification in BAV, which may contribute to
the prevention of this disease.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural
Science Foundation of China (grant no. 81974033).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YS and JD conceived and designed the study. RZ, HL,
JG, BN, HS, YG, CS and KH collected the data. RZ, HL, JG, BN, HS,
YG, CS and KH performed the experiments, acquired the data and
prepared the diagrams. RZ, HL, JD and YS drafted the manuscript.
RZ, JD and YS reviewed and edited the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
This study was granted ethical approval by the First
Affiliated Hospital of Nanjing Medical University Ethics Committee.
Written informed consent was obtained from the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kurabayashi M: Molecular mechanism of
vascular calcification. Clin Calcium. 29:157–163. 2019.(In
Japanese). PubMed/NCBI
|
|
2
|
Chen HY, Engert JC and Thanassoulis G:
Risk factors for valvular calcification. Curr Opin Endocrinol
Diabetes Obes. 26:96–102. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Girdauskas E and Borger MA: Bicuspid
aortic valve and associated aortopathy: An update. Semin Thorac
Cardiovasc Surg. 25:310–316. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siu SC and Silversides CK: Bicuspid aortic
valve disease. J Am Coll Cardiol. 55:2789–2800. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cirka HA, Kural MH and Billiar KL:
Mechanoregulation of aortic valvular interstitial cell life and
death. J Long Term Eff Med Implants. 25:3–16. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gu X and Masters KS: Role of the MAPK/ERK
pathway in valvular interstitial cell calcification. Am J Physiol
Heart Circ Physiol. 296:H1748–H1757. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gu X and Masters KS: Role of the Rho
pathway in regulating valvular interstitial cell phenotype and
nodule formation. Am J Physiol Heart Circ Physiol. 300:H448–H458.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hjortnaes J, Shapero K, Goettsch C,
Hutcheson JD, Keegan J, Kluin J, Mayer JE, Bischoff J and Aikawa E:
Valvular interstitial cells suppress calcification of valvular
endothelial cells. Atherosclerosis. 242:251–260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mohler ER, Gannon F, Reynolds C, Zimmerman
R, Keane MG and Kaplan FS: Bone formation and inflammation in
cardiac valves. Circulation. 103:1522–1528. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kaden JJ, Bickelhaupt S, Grobholz R, Vahl
CF, Hagl S, Brueckmann M, Haase KK, Dempfle CE and Borggrefe M:
Expression of bone sialoprotein and bone morphogenetic protein-2 in
calcific aortic stenosis. J Heart Valve Dis. 13:560–566.
2004.PubMed/NCBI
|
|
11
|
Wang Y, Wu B, Farrar E, Lui W, Lu P, Zhang
D, Alfieri CM, Mao K, Chu M, Yang D, et al: Notch-Tnf signalling is
required for development and homeostasis of arterial valves. Eur
Heart J. 38:675–686. 2017.PubMed/NCBI
|
|
12
|
Wu M, Chen G and Li YP: TGF-β and BMP
signaling in osteoblast, skeletal development, and bone formation,
homeostasis and disease. Bone Res. 4:160092016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Leopold JA: MicroRNAs regulate vascular
medial calcification. Cells. 3:963–980. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nigam V, Sievers HH, Jensen BC, Sier HA,
Simpson PC, Srivastava D and Mohamed SA: Altered microRNAs in
bicuspid aortic valve: A comparison between stenotic and
insufficient valves. J Heart Valve Dis. 19:459–465. 2010.PubMed/NCBI
|
|
15
|
Rathan S, Ankeny CJ, Arjunon S, Ferdous Z,
Kumar S, Fernandez EJ, Heath JM, Nerem RM, Yoganathan AP and Jo H:
Identification of side- and shear-dependent microRNAs regulating
porcine aortic valve pathogenesis. Sci Rep. 6:253972016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Patel V, Carrion K, Hollands A, Hinton A,
Gallegos T, Dyo J, Sasik R, Leire E, Hardiman G, Mohamed SA, et al:
The stretch responsive microRNA miR-148a-3p is a novel repressor of
IKBKB, NF-κB signaling, and inflammatory gene expression in human
aortic valve cells. FASEB J. 29:1859–1868. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Holliday CJ, Ankeny RF, Jo H and Nerem RM:
Discovery of shear- and side-specific mRNAs and miRNAs in human
aortic valvular endothelial cells. Am J Physiol Heart Circ Physiol.
301:H856–H867. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gu J, Lu Y, Deng M, Qiu M, Tian Y, Ji Y,
Zong P, Shao Y, Zheng R, Zhou B, et al: Inhibition of acetylation
of histones 3 and 4 attenuates aortic valve calcification. Exp Mol
Med. 51:792019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Meister G, Landthaler M, Dorsett Y and
Tuschl T: Sequence-specific inhibition of microRNA- and
siRNA-induced RNA silencing. RNA. 10:544–550. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nishimasu H, Shi X, Ishiguro S, Gao L,
Hirano S, Okazaki S, Noda T, Abudayyeh OO, Gootenberg JS, Mori H,
et al: Engineered CRISPR-Cas9 nuclease with expanded targeting
space. Science. 361:1259–1262. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Trevino AE and Zhang F: Genome editing
using Cas9 nickases. Methods Enzymol. 546:161–174. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gomez RA, Belyea B, Medrano S, Pentz ES
and Sequeira-Lopez ML: Fate and plasticity of renin precursors in
development and disease. Pediatr Nephrol. 29:721–726. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Devan J, Janikova A and Mraz M: New
concepts in follicular lymphoma biology: From BCL2 to epigenetic
regulators and non-coding RNAs. Semin Oncol. 45:291–302. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang D, Guo J, Divieti P, Shioda T and
Bringhurst FR: CBP/p300-interacting protein CITED1 modulates
parathyroid hormone regulation of osteoblastic differentiation.
Endocrinology. 149:1728–1735. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lin Z, Feng R, Li J, Meng Y, Yuan L, Fu Z,
Guo J, Bringhurst FR and Yang D: Nuclear translocation of
CBP/p300-interacting protein CITED1 induced by parathyroid hormone
requires serine phosphorylation at position 79 in its 63–84 domain.
Cell Signal. 26:2436–2445. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li SJ, Kao YH, Chung CC, Chen WY, Cheng WL
and Chen YJ: Activated p300 acetyltransferase activity modulates
aortic valvular calcification with osteogenic transdifferentiation
and downregulation of Klotho. Int J Cardiol. 232:271–279. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu J, Liu L, Chao S, Liu Y, Liu X, Zheng
J, Chen J, Gong W, Teng H, Li Z, et al: The Role of
miR-330-3p/PKC-α signaling pathway in Low-dose endothelial-monocyte
activating polypeptide-ii increasing the permeability of
blood-tumor barrier. Front Cell Neurosci. 11:3582017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang H, Chen SH, Kong P, Zhang LY, Zhang
LL, Zhang NQ and Gu H: Increased expression of miR-330-3p: A novel
independent indicator of poor prognosis in human breast cancer. Eur
Rev Med Pharmacol Sci. 22:1726–1730. 2018.PubMed/NCBI
|
|
30
|
Meng H, Wang K, Chen X, Guan X, Hu L,
Xiong G, Li J and Bai Y: MicroRNA-330-3p functions as an oncogene
in human esophageal cancer by targeting programmed cell death 4. Am
J Cancer Res. 5:1062–1075. 2015.PubMed/NCBI
|
|
31
|
Min M, Peng LH, Sun G, Guo MZ, Qiu ZW and
Yang YS: Aquaporin 8 expression is reduced and regulated by
microRNAs in patients with ulcerative colitis. Chin Med J (Engl).
126:1532–1537. 2013.PubMed/NCBI
|
|
32
|
Feng L, Ma J, Ji H, Liu Y and Hu W:
miR-330-5p suppresses glioblastoma cell proliferation and
invasiveness through targeting ITGA5. Biosci Rep.
37:BSR201700192017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen T, Yang Z, Liu C, Wang L, Yang J,
Chen L and Li W: Circ_0078767 suppresses non-small-cell lung cancer
by protecting RASSF1A expression via sponging miR-330-3p. Cell
Prolif. 52:e125482019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Guan A, Wang H, Li X, Xie H, Wang R, Zhu Y
and Li R: MiR-330-3p inhibits gastric cancer progression through
targeting MSI1. Am J Transl Res. 8:4802–4811. 2016.PubMed/NCBI
|
|
35
|
Lee KH, Chen YL, Yeh SD, Hsiao M, Lin JT,
Goan YG and Lu PJ: MicroRNA-330 acts as tumor suppressor and
induces apoptosis of prostate cancer cells through E2F1-mediated
suppression of Akt phosphorylation. Oncogene. 28:3360–3370. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pirklbauer M and Mayer G: The exchangeable
calcium pool: Physiology and pathophysiology in chronic kidney
disease. Nephrol Dial Transplant. 26:2438–2444. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Goodman WG, Goldin J, Kuizon BD, Yoon C,
Gales B, Sider D, Wang Y, Chung J, Emerick A, Greaser L, et al:
Coronary-artery calcification in young adults with end-stage renal
disease who are undergoing dialysis. N Engl J Med. 342:1478–1483.
2000. View Article : Google Scholar : PubMed/NCBI
|