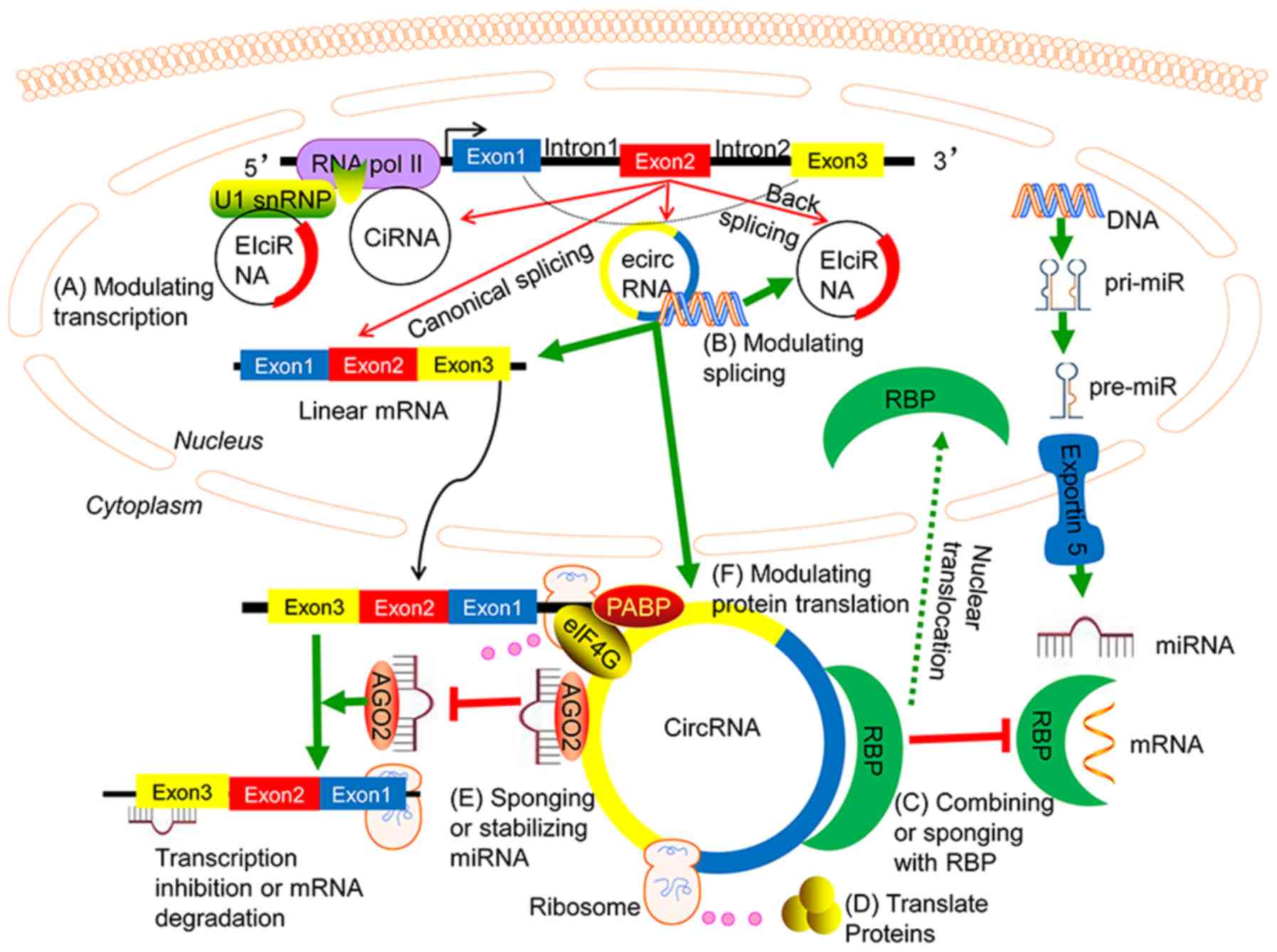
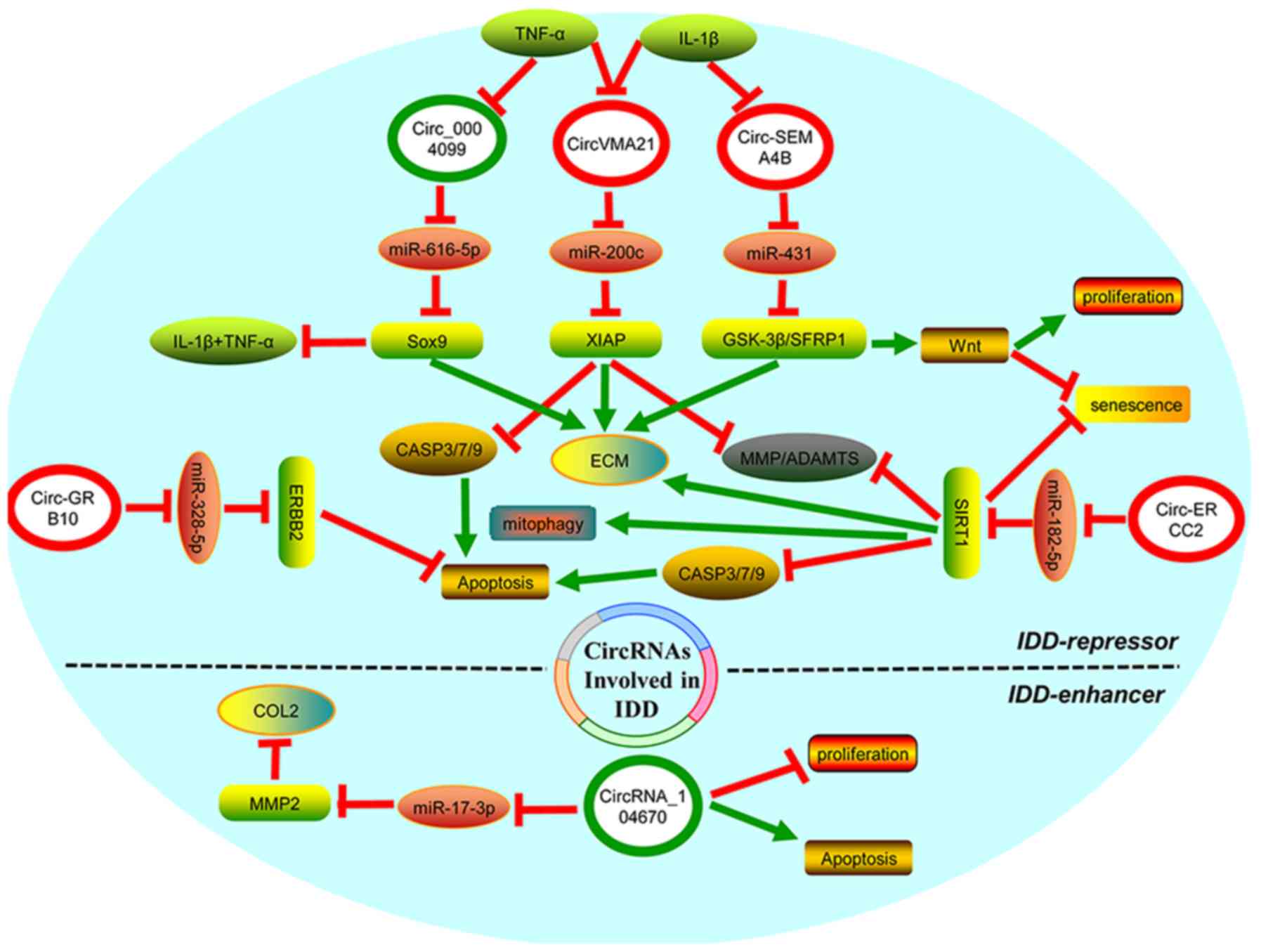
|
1
|
Vos T, Abajobir AA, Abate KH, Abbafati C,
Abbas KM, Abd-Allah F, Abdulkadre RS, Abdulle AM, Abebo TA, Abera
SF, et al: Global, regional, and national incidence, prevalence,
and years lived with disability for 328 diseases and injuries for
195 countries, 1990–2016: A systematic analysis for the global
burden of disease study 2016. Lancet. 390:3057–1259. 2017.
View Article : Google Scholar
|
|
2
|
Deyo RA and Tsui-Wu YJ: Descriptive
epidemiology of low-back pain and its related medical care in the
United States. Spine (Phila Pa 1976). 12:264–268. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Risbud MV and Shapiro IM: Role of
cytokines in intervertebral disc degeneration: Pain and disc
content. Nat Rev Rheumatol. 10:44–56. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Battié MC, Videman T and Parent E: Lumbar
disc degeneration: Epidemiology and genetic influences. Spine
(Phila Pa 1976). 29:2679–2690. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Humzah MD and Soames RW: Human
intervertebral disc: Structure and function. Anat Rec. 220:337–356.
1988. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sakai D and Andersson GB: Stem cell
therapy for intervertebral disc regeneration: Obstacles and
solutions. Nat Rev Rheumatol. 11:243–256. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jacobs WC, van der Gaag NA, Kruyt MC,
Tuschel A, de Kleuver M, Peul WC, Verbout AJ and Oner FC: Total
disc replacement for chronic discogenic low back pain: A cochrane
review. Spine (Phila Pa 1976). 38:24–36. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Skvortsova K, Iovino N and Bogdanović O:
Functions and mechanisms of epigenetic inheritance in animals. Nat
Rev Mol Cell Biol. 19:774–790. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Frapin L, Clouet J, Delplace V, Fusellier
M, Guicheux J and Le Visage C: Lessons learned from intervertebral
disc pathophysiology to guide rational design of sequential
delivery systems for therapeutic biological factors. Advanced Drug
Delivery Reviews. 149-150:49–71. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fontana G, See E and Pandit A: Current
trends in biologics delivery to restore intervertebral disc
anabolism. Adv Drug Deliv Rev. 84:146–158. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vergroesen PP, Kingma I, Emanuel KS,
Hoogendoorn RJ, Welting TJ, van Royen BJ, van Dieën JH and Smit TH:
Mechanics and biology in intervertebral disc degeneration: A
vicious circle. Osteoarthritis Cartilage. 23:1057–1070. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cheng X, Zhang L, Zhang K, Zhang G, Hu Y,
Sun X, Zhao C, Li H, Li YM and Zhao J: Circular RNA VMA21 protects
against intervertebral disc degeneration through targeting miR-200c
and X linked inhibitor-of-apoptosis protein. Ann Rheum Dis.
77:770–779. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guo W, Zhang B, Mu K, Feng SQ, Dong ZY,
Ning GZ, Li HR, Liu S, Zhao L, Li Y, et al: Circular RNA GRB10 as a
competitive endogenous RNA regulating nucleus pulposus cells death
in degenerative intervertebral disk. Cell Death Dis. 9:3192018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang H, He P, Pan H, Long J, Wang J, Li Z,
Liu H, Jiang W and Zheng Z: Circular RNA circ-4099 is induced by
TNF-α and regulates ECM synthesis by blocking miR-616-5p inhibition
of Sox9 in intervertebral disc degeneration. Exp Mol Med.
50:272018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Song J, Wang HL, Song KH, Ding ZW, Wang
HL, Ma XS, Lu FZ, Xia XL, Wang YW, Fei-Zou and Jiang JY:
CircularRNA_104670 plays a critical role in intervertebral disc
degeneration by functioning as a ceRNA. Exp Mol Med. 50:942018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang X, Wang B, Zou M, Li J, Lü G, Zhang
Q, Liu F and Lu C: CircSEMA4B targets miR-431 modulating
IL-1β-induced degradative changes in nucleus pulposus cells in
intervertebral disc degeneration via Wnt pathway. Biochim Biophys
Acta Mol Basis Dis. 1864:3754–3768. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xie L, Huang W, Fang Z, Ding F, Zou F, Ma
X, Tao J, Guo J, Xia X, Wang H, et al: CircERCC2 ameliorated
intervertebral disc degeneration by regulating mitophagy and
apoptosis through miR-182-5p/SIRT1 axis. Cell Death Dis.
10:7512019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Grabowski PJ, Zaug AJ and Cech TR: The
intervening sequence of the ribosomal RNA precursor is converted to
a circular RNA in isolated nuclei of Tetrahymena. Cell. 23:467–476.
1981. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nigro JM, Cho KR, Fearon ER, Kern SE,
Ruppert JM, Oliner JD, Kinzler KW and Vogelstein B: Scrambled
exons. Cell. 64:607–613. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Capel B, Swain A, Nicolis S, Hacker A,
Walter M, Koopman P, Goodfellow P and Lovell-Badge R: Circular
transcripts of the testis-determining gene Sry in adult mouse
testis. Cell. 73:1019–1030. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z
and Sharpless NE: Expression of linear and novel circular forms of
an INK4/ARF associated non-coding RNA correlates with
atherosclerosis risk. PLoS Genet. 6:e10012332010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Morris KV and Mattick JS: The rise of
regulatory RNA. Nat Rev Genet. 15:423–437. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li X, Yang L and Chen LL: The biogenesis,
functions, and challenges of circular RNAs. Mol Cell. 71:428–442.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Boeckel JN, Jaé N, Heumüller AW, Chen W,
Boon RA, Stellos K, Zeiher AM, John D, Uchida S and Dimmeler S:
Identification and characterization of hypoxia-regulated
endothelial circular RNA. Circ Res. 117:884–890. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vicens Q and Westhof E: Biogenesis of
circular RNAs. Cell. 159:13–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Suzuki H, Zuo Y, Wang J, Zhang MQ,
Malhotra A and Mayeda A: Characterization of RNase R-digested
cellular RNA source that consists of lariat and circular RNAs from
pre-mRNA splicing. Nucleic Acids Res. 34:e632006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao
L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, et al: The
Landscape of circular RNA in cancer. Cell. 176:869–881.e13. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Patop IL, Wüst S and Kadener S: Past,
present, and future of circRNAs. EMBO J. 38:e1008362019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang
W, Wang G, Wu P, Wang H, Jiang L, et al: Exosomal circRNAs:
Biogenesis, effect and application in human diseases. Mol Cancer.
18:1162019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Schmidt CA, Giusto JD, Bao A, Hopper AK
and Matera AG: Molecular determinants of metazoan tricRNA
biogenesis. Nucleic Acids Res. 47:6452–6465. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Guarnerio J, Bezzi M, Jeong JC, Paffenholz
SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH and Pandolfi PP:
Oncogenic role of fusion-circRNAs derived from cancer-associated
chromosomal translocations. Cell. 165:289–302. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu X, Wang X, Li J, Hu S, Deng Y, Yin H,
Bao X, Zhang QC, Wang G, Wang B, et al: Identification of mecciRNAs
and their roles in mitochondrial entry of proteins. Sci China Life
Sci. Jan 21–2020.(Epub ahead of print).
|
|
38
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Graves P and Zeng Y: Biogenesis of
mammalian MicroRNAs: A global view. Genomics Proteomics
Bioinformatics. 10:239–245. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bartel DP: MiRNAs: Target recognition and
regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ye F, Gao G, Zou Y, Zheng S, Zhang L, Ou
X, Xie X and Tang H: circFBXW7 inhibits malignant progression by
sponging miR-197-3p and encoding a 185-aa protein in
triple-negative breast cancer. Mol Ther Nucleic Acids. 18:88–98.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bai N, Peng E, Qiu X, Lyu N, Zhang Z, Tao
Y, Li X and Wang Z: circFBLIM1 act as a ceRNA to promote
hepatocellular cancer progression by sponging miR-346. J Exp Clin
Cancer Res. 37:1722018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang PF, Wei CY, Huang XY, Peng R, Yang
X, Lu JC, Zhang C, Gao C, Cai JB, Gao PT, et al: Circular RNA
circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress
hepatocellular carcinoma progression. Mol Cancer. 18:1052019.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen S, Huang V, Xu X, Livingstone J,
Soares F, Jeon J, Zeng Y, Hua JT, Petricca J, Guo H, et al:
Widespread and functional RNA circularization in localized prostate
cancer. Cell. 176:831–843.e22. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Piwecka M, Glazar P, Hernandez-Miranda LR,
Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda
Jara CA, Fenske P, et al: Loss of a mammalian circular RNA locus
causes miRNA deregulation and affects brain function. Science.
357:eaam85262017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P
and Yang BB: Foxo3 circular RNA retards cell cycle progression via
forming ternary complexes with p21 and CDK2. Nucleic Acids Res.
44:2846–2858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhu YJ, Zheng B, Luo GJ, Ma XK, Lu XY, Lin
XM, Yang S, Zhao Q, Wu T, Li ZX, et al: Circular RNAs negatively
regulate cancer stem cells by physically binding FMRP against CCAR1
complex in hepatocellular carcinoma. Theranostics. 9:3526–3540.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Du WW, Yang W, Chen Y, Wu ZK, Foster FS,
Yang Z, Li X and Yang BB: Foxo3 circular RNA promotes cardiac
senescence by modulating multiple factors associated with stress
and senescence responses. Eur Heart J. 38:1402–1412.
2017.PubMed/NCBI
|
|
52
|
Du WW, Fang L, Yang W, Wu N, Awan FM, Yang
Z and Yang BB: Induction of tumor apoptosis through a circular RNA
enhancing Foxo3 activity. Cell Death Differ. 24:357–370. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liang WC, Wong CW, Liang PP, Shi M, Cao Y,
Rao ST, Tsui SK, Waye MM, Zhang Q, Fu WM and Zhang JF: Translation
of the circular RNA circβ-catenin promotes liver cancer cell growth
through activation of the Wnt pathway. Genome Biol. 20:842019.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z,
Xu B, Wu C, Zhou Q, Hu W, Wu C and Jiang J: A novel protein encoded
by a circular RNA circPPP1R12A promotes tumor pathogenesis and
metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer.
18:472019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao
F, Huang N, Yang X, Zhao K, Zhou H, et al: Novel role of FBXW7
circular RNA in repressing glioma tumorigenesis. J Natl Cancer
Inst. 110:304–315. 2018. View Article : Google Scholar
|
|
56
|
Wang L, Long H, Zheng Q, Bo X, Xiao X and
Li B: Circular RNA circRHOT1 promotes hepatocellular carcinoma
progression by initiation of NR2F6 expression. Mol Cancer.
18:1192019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wu N, Yuan Z, Du KY, Fang L, Lyu J, Zhang
C, He A, Eshaghi E, Zeng K, Ma J, et al: Translation of
yes-associated protein (YAP) was antagonized by its circular RNA
via suppressing the assembly of the translation initiation
machinery. Cell Death Differ. 26:2758–2773. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Sun YM, Wang WT, Zeng ZC, Chen TQ, Han C,
Pan Q, Huang W, Fang K, Sun LY, Zhou YF, et al: CircMYBL2, a
circRNA from, MYBL2 regulates FLT3 translation by recruiting PTBP1
to promote FLP3-ITD AML progression. Blood. 134:1533–1546. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang BG, Li JS, Liu YF and Xu Q:
MicroRNA-200b suppresses the invasion and migration of
hepatocellular carcinoma by downregulating RhoA and circRNA_000839.
Tumour Biol. 39:10104283177195772017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Dong W, Dai ZH, Liu FC, Guo XG, Ge CM,
Ding J, Liu H and Yang F: The RNA-binding protein RBM3 promotes
cell proliferation in hepatocellular carcinoma by regulating
circular RNA SCD-circRNA 2 production. EBioMedicine. 45:155–167.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhang N, Li G, Li X, Xu L and Chen M:
Circ5379-6, a circular form of tumor suppressor PPARα, participates
in the inhibition of hepatocellular carcinoma tumorigenesis and
metastasis. Am J Transl Res. 10:3493–3503. 2018.PubMed/NCBI
|
|
62
|
Yao Z, Luo J, Hu K, Lin J, Huang H, Wang
Q, Zhang P, Xiong Z, He C, Huang Z, et al: ZKSCAN1 gene and its
related circular RNA (circZKSCAN1) both inhibit hepatocellular
carcinoma cell growth, migration, and invasion but through
different signaling pathways. Mol Oncol. 11:422–437. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Shen S, Wu Y, Chen J, Xie Z, Huang K, Wang
G, Yang Y, Ni W, Chen Z, Shi P, et al: CircSERPINE2 protects
against osteoarthritis by targeting miR-1271 and ETS-related gene.
Ann Rheum Dis. 78:826–836. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Shang Q, Yang Z, Jia R and Ge S: The novel
roles of circRNAs in human cancer. Mol Cancer. 18:62019. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Aufiero S, Reckman YJ, Pinto YM and
Creemers EE: Circular RNAs open a new chapter in cardiovascular
biology. Nat Rev Cardiol. 16:503–514. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Akhter R: Circular RNA and Alzheimer's
disease. Adv Exp Med Biol. 1087:239–243. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Floris G, Zhang L, Follesa P and Sun T:
Regulatory role of circular RNAs and neurological disorders. Mol
Neurobiol. 54:5156–5165. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Chen X, Yang T, Wang W, Xi W, Zhang T, Li
Q, Yang A and Wang T: Circular RNAs in immune responses and immune
diseases. Theranostics. 9:588–607. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Yang L, Fu J and Zhou Y: Circular RNAs and
their emerging roles in immune regulation. Front Immunol.
9:29772018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zou F, Ding Z, Jiang J, Lu F, Xia X and Ma
X: Confirmation and preliminary analysis of circRNAs potentially
involved in human intervertebral disc degeneration. Mol Med Rep.
16:9173–9180. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wang S, Sun J, Yang H, Zou W, Zheng B,
Chen Y, Guo Y and Shi J: Profiling and bioinformatics analysis of
differentially expressed circular RNAs in human intervertebral disc
degeneration. Acta Biochim Biophys Sin (Shanghai). 51:571–579.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zhu J, Zhang X, Gao W, Hu H, Wang X and
Hao D: lncRNA/circRNA-miRNA-mRNA ceRNA network in lumbar
intervertebral disc degeneration. Mol Med Rep. 20:3160–3174.
2019.PubMed/NCBI
|
|
73
|
Collison J: Degenerative disc disease:
Circular RNA reduces cell death in IVD disease. Nat Rev Rheumatol.
14:1232018. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Pfirrmann CW, Metzdorf A, Zanetti M,
Hodler J and Boos N: Magnetic resonance classification of lumbar
intervertebral disc degeneration. Spine (Phila PA 1976).
26:1873–1878. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Lan PH, Liu ZH, Pei YJ, Wu ZG, Yu Y, Yang
YF, Liu X, Che L, Ma CJ, Xie YK, et al: Landscape of RNAs in human
lumbar disc degeneration. Oncotarget. 7:63166–63176. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Sekiya I, Tsuji K, Koopman P, Watanabe H,
Yamada Y, Shinomiya K, Nifuji A and Noda M: SOX9 enhances aggrecan
gene promoter/enhancer activity and is up-regulated by retinoic
acid in a cartilage-derived cell line, TC6. J Biol Chem.
275:10738–10744. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Lefebvre V, Huang W, Harley VR, Goodfellow
PN and de Crombrugghe B: SOX9 is a potent activator of the
chondrocyte-specific enhancer of the pro alpha1(II) collagen gene.
Mol Cell Biol. 17:2336–2346. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Hiyama A, Sakai D, Risbud MV, Tanaka M,
Arai F, Abe K and Mochida J: Enhancement of intervertebral disc
cell senescence by WNT/β-catenin signaling induced matrix
metalloproteinase expression. Arthritis Rheum. 62:3036–3047. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Hiyama A, Sakai D, Tanaka M, Arai F,
Nakajima D, Abe K and Mochida J: The relationship between the
Wnt/β-catenin and TGF-β/BMP signals in the intervertebral disc
cell. J Cell Physiol. 226:1139–1148. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Guezguez B, Almakadi M, Benoit YD,
Shapovalova Z, Rahmig S, Fiebig-Comyn A, Casado FL, Tanasijevic B,
Bresolin S, Masetti R, et al: GSK3 deficiencies in hematopoietic
stem cells initiate pre-neoplastic state that is predictive of
clinical outcomes of human acute leukemia. Cancer Cell. 29:61–74.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Tao J, Abudoukelimu M, Ma YT, Yang YN, Li
XM, Chen BD, Liu F, He CH and Li HY: Secreted frizzled related
protein 1 protects H9C2 cells from hypoxia/re-oxygenation injury by
blocking the Wnt signaling pathway. Lipids Health Dis. 15:722016.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Guo J, Shao M, Lu F, Jiang J and Xia X:
Role of Sirt1 plays in nucleus pulposus cells and intervertebral
disc degeneration. Spine (Phila Pa 1976). 42:E757–E766. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Wang D, Hu Z, Hao J, He B, Gan Q, Zhong X,
Zhang X, Shen J, Fang J and Jiang W: SIRT1 inhibits apoptosis of
degenerative human disc nucleus pulposus cells through activation
of Akt pathway. Age (Dordr). 35:1741–1753. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Yao ZQ, Zhang X, Zhen Y, He XY, Zhao S, Li
XF, Yang B, Gao F, Guo FY, Fu L, et al: A novel small-molecule
activator of Sirtuin-1 induces autophagic cell death/mitophagy as a
potential therapeutic strategy in glioblastoma. Cell Death Dis.
9:7672018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Wei Y, Chen X, Liang C, Ling Y, Yang X, Ye
X, Zhang H, Yang P, Cui X, Ren Y, et al: A noncoding regulatory
RNAs network driven by Circ-CDYL acts specifically in the early
stages hepatocellular carcinoma. Hepatology. 71:130–147. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Ren S, Liu J, Feng Y, Li Z, He L, Li L,
Cao X, Wang Z and Zhang Y: Knockdown of circDENND4C inhibits
glycolysis, migration and invasion by up-regulating miR-200b/c in
breast cancer under hypoxia. J Exp Clin Cancer Res. 38:3882019.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Ren GL, Zhu J, Li J and Meng XM: Noncoding
RNAs in acute kidney injury. J Cell Physiol. 234:2266–2276. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Chaichian S, Shafabakhsh R, Mirhashemi SM,
Moazzami B and Asemi Z: Circular RNAs: A novel biomarker for
cervical cancer. J Cell Physiol. 235:718–724. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Braicu C, Tomuleasa C, Monroig P, Cucuianu
A, Berindan-Neagoe I and Calin GA: Exosomes as divine messengers:
Are they the hermes of modern molecular oncology? Cell Death
Differ. 22:34–45. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Engelman JA, Luo J and Cantley LC: The
evolution of phosphatidylinositol 3-kinases as regulators of growth
and metabolism. Nat Rev Genet. 7:606–619. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Liu Y, Hou J, Zhang M, Seleh-Zo E, Wang J,
Cao B and An X: circ-016910 sponges miR-574-5p to regulate cell
physiology and milk synthesis via MAPK and PI3K/AKT-mTOR pathways
in GMECs. J Cell Physiol. 235:4198–4216. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Chen T, Yu Q, Xin L and Guo L: Circular
RNA circC3P1 restrains kidney cancer cell activity by regulating
miR-21/PTEN axis and inactivating PI3K/AKT and NF-kB pathways. J
Cell Physiol. 235:4001–4010. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Xu G, Liu C, Jiang J, Liang T, Yu C, Qin
Z, Zhang Z, Lu Z and Zhan X: A novel mechanism of intervertebral
disc degeneration: Imbalance between autophagy and apoptosis.
Epigenomics. Apr 14–2020.(Epub ahead of print). View Article : Google Scholar
|