Introduction
Glioma, which is characterized at a high rate of
local recurrence, is located in the frontal lobe, temporal,
parietal, occipital lobe, or deeper structures and accounts for
~80% of malignant primary brain and CNS tumors in recent years
(1–3). Local recurrence and resistance to
therapies are the predictors for glioma prognosis, and approaches
aimed at eliminating cancer recurrence and alleviating therapeutic
resistance are potential strategies for glioma treatment (4). Cancer stem cells (CSCs) are regarded
as the main contributor to cancer recurrence, and are known to
survive following radiotherapy and chemotherapy of different types
of cancer, such as glioma, liver cancer and breast cancer (5–8).
Thus, targeting CSCs may be an effective therapy in improving the
prognosis of patients with glioma.
Long non-coding RNAs (lncRNAs) exert important
functions in biological processes, such as chromatin remodeling,
transcriptional activation and interference, RNA processing and
mRNA translation (9), thereby
modulating cell proliferation and development, cell cycle, cell
differentiation, stress responses and the occurrence of several
diseases in humans and animals, such as cancer, cardiovascular
disease and Alzheimer's disease (10). Previous studies have reported that
lncRNAs have crucial roles in modulating cancer stemness (11–15).
For example, the lncRNA, TP73-AS1 has been demonstrated to promote
tumor aggressiveness and temozolomide resistance in CSCs, which
confers a poor prognosis in glioblastoma multiforme (GBM) (16). Another lncRNA, Linc00152, has been
reported to enhance the malignant behavior of CSCs via the microRNA
(miRNA/miR)-103a-3p/FEZ family zinc finger 1/Cell Division Cycle
25A axis, thereby upregulating the PI3K/AKT signaling pathway
(17). Furthermore, nano complex
targeting metastasis associated lung adenocarcinoma transcript 1
may lower the stemness of GBM cells and improve the sensitivity of
GBM cells to the current first-line therapy of GBM (18). In addition, lncRNAs have also been
indicated to participate in the regulation of glioma
radioresistance (19). The
upregulation of the lncRNA HIF1A antisense RNA 2 and lncRNA small
nucleolar RNA host gene 18 was demonstrated to promote glioma cell
radioresistance (20,21). However, the function of the lncRNA
transmembrane phosphatase with tensin homology pseudogene 1
(TPTEP1) in glioma currently remains unknown, and any potential
influence of the lncRNA TPTEP1 in the regulation of stemness and
radioresistance of glioma is yet to be determined.
The present study aimed to investigate the impact of
lncRNA TPTEP1 on glioma and determine the underlying molecular
mechanism of TPTEP1 on the regulation of glioma progression. The
results demonstrated that TPTEP1 upregulated MAPK14 (P38) by
competitively interacting with miR-106a-5p, and inhibited glioma
cell stemness and radioresistance both in vitro and in
vivo. Notably, TPTEP1/miR-106a-5p was demonstrated to form a
reciprocal regulatory circuit to impede glioma development.
Furthermore, low TPTEP1 expression levels were detected in
high-grade glioma tissues compared with low-grade glioma tissues,
and were associated with poor overall survival time of patients
with glioma.
Materials and methods
Cell culture
Human low malignancy grade SHG44 cells and high
malignancy grade U-251MG cells were purchased from the Cell Bank of
Type Culture Collection of the Chinese Academy of Sciences. Cells
were maintained in DMEM medium supplemented with 10% fetal bovine
serum (HyClone; GE Healthcare) and 1% penicillin/streptomycin, at
37°C in a humidified incubator with 5% CO2. The cell
lines were verified to be free of mycoplasma contamination.
Cell transfection
Specific short hairpin (sh)RNAs against TPTEP1
(sh-TPTEP1), lncRNA TPTEP1 sequence (TPTEP1) and their
corresponding negative control (NC) sequences were purchased from
GeneChem, Inc. Lentivirus particles were subsequently constructed
and amplified, and polybrene reagent (GeneChem, Inc.) was used for
transfection (MOI: 10, 37°C for 12 h) in glioma cells
(1×104). Plasmids were purchased from Vigene Bioscience,
Inc. Mimics (5′-AAAAGUGCUUACAGUGCAGGUAG-3′), mimics control
(5′-UUUGUACUACACAAAAGUACUG-3′), inhibitors
(5′-CUACCUGCACUGUAAGCACUUUU-3′), inhibitors control
(5′-CAGUACUUUUGUGUAGUACAAA3′), small interfering (si)RNAs
(5′-ACAGCAUCGAAGUAAGAGAUCUCUUACUUCGAUGCUGU-3′) and siRNA control
(5′-UUUGUACUACACAAAAGUACUG-3′) were purchased from Guangzhou
RiboBio Co., Ltd. Lipofectamine™ 2000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was to transfect 4 µg plasmids (oe-TPTEP1;
GeneChem, Inc.), 50 nM siRNAs (si-TPTEP1), 100 nM inhibitors and/or
50 nM mimics into glioma cells (1×105). The cells were
subjected to subsequent experiments 48–72 h post-transfection.
Tumorspheres formation assay
Glioma cells (5×103) in the exponential
growth phase were seeded into ultra-low adherence 6-well plates,
and propagated in serum-free DMEM/Ham's F-12 medium supplemented
with 20 ng/ml epidermal growth factor, 20 ng/ml fibroblast growth
factor and 2% B27 supplement (for supporting neuronal cell
proliferation), all purchased from Invitrogen; Thermo Fisher
Scientific, Inc.. Following cell culture for 2–3 weeks at 37°C, the
resultant tumorspheres were imaged under a light microscope (×100
magnification) and subjected to further analysis.
MTT assay
Glioma cells (1×103) in the exponential
growth phase were seeded into 96-well plates and incubated with MTT
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 4 h.
Following the MTT incubation, the purple formazan crystals were
dissolved using dimethyl sulfoxide (Invitrogen; Thermo Fisher
Scientific, Inc.) and viability was subsequently analyzed at 570 nm
using a Multiskan™ FC (Thermo Fisher Scientific, Inc.).
Immunofluorescence
Cells (5×104) were seeded onto glass
slides and allowed to adhere. Cells were subsequently fixed with 4%
paraformaldehyde diluted with PBS for 15 min at room temperature,
permeabilized with 0.2% Triton X-100 diluted with PBS for 20 min at
room temperature and blocked with 10% goat serum diluted with PBS
(Fuzhou Maixin Biotech Co., Ltd.) for 30 min at 37°C. Cells were
incubated against the following antibodies: CD133 (1:500;
ProteinTech Group, Inc.; cat. no. 66666-1-Ig) or γ-histone H2AX
(H2AX; 1:500; Abcam; cat. no. ab2893) at 4°C for 16 h, and
PE-conjugated secondary antibody at 37°C for 1 h (1:200;
Invitrogen; Thermo Fisher Scientific, Inc.; cat. no. P-2771MP.)
diluted with PBS. Cells were subsequently co-stained with DAPI
(Sigma-Aldrich; Merck KGaG) for 10 min at room temperature and
observed under a fluorescence or confocal microscope (×400
magnification).
Ionizing radiation treatment
Cells (5×106) were cultured at with DMEM
in 10 cm plates and irradiated with a 6 MV linear accelerator
(Siemens AG). During irradiation for 1–2 min, cells were treated 50
cm from the source of radiation, with a dose of 4 Gy at room
temperature.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from glioma cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and was reverse-transcribed into cDNA using the reverse
transcriptase kit (cat. no. D350A, Takara Bio, Inc.), according to
the manufacturer's protocols. qPCR was subsequently performed using
the SYBR Premix Ex Taq™ II kit (cat. no. DRR081, Takara Bio, Inc.),
on an ABI ViiA 7 sequence detection system (Applied Biosystems;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocols. The following primer sequences were used for qPCR:
Hsa-miR-106a-5p primer, 5′-AAAAGTGCTTACAGTGCAGGTAG-3′; TPTEP1
forward, 5′-CTGGGAGAAGTGCCCTTGC-3′ and reverse,
5′-CACCTCATCAGTCATTTGCTCA-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′
and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; and GAPDH forward,
5′-GCAAATTCCATGGCACCGT-3′ and reverse, 5′-TCGCCCCACTTGATTTTGG-3′.
The following thermocycling conditions were used for qPCR: Initial
denaturation at 95°C for 10 sec, followed by 40 cycles of
denaturation at 94°C for 5 sec and annealing/extension at 60°C for
30 sec. Relative gene expression levels were calculated using the
2−ΔΔCq method (22) and
normalized to the internal reference genes U6 or GAPDH.
Western blotting
Glioma cells in the exponential growth phase were
lysed and collected using RIPA lysis buffer (Beyotime Institute of
Biotechnology). Total protein was quantified using the
bicinchoninic acid assay protein kit (cat. no. 23227, Pierce;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. A total of 30–50 µg proteins were loaded to each well and
separated by 10% SDS-PAGE gel. The separated proteins were
transferred onto polyvinylidene difluoride membranes (Thermo Fisher
Scientific, Inc.) and blocked with 3% bovine serum albumin
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 1 h. The
membranes were incubated with primary antibodies against: p38 MAPK
(1:2,000; ProteinTech Group, Inc.; cat. no. 66234-1-Ig),
phosphorylated (p-)p38 MAPK (1:1,000; Abcam; cat. no. ab195049),
aldehyde dehydrogenase 1 (ALDH1; 1:1,000; ProteinTech Group,
Inc.; cat. no. 60171-1-Ig), octamer binding protein 4 (OCT4;
1:1,000; ProteinTech Group, Inc.; cat. no. 60242-1-Ig), γ-H2AX
(1:500; Abcam; cat. no. ab2893), H2AX (1:1,000; Abcam; cat. no.
ab124781) and β-actin (1:5,000; ProteinTech Group, Inc.; cat. no.
60008-1-Ig). Following the primary incubation at 4°C for 16 h,
membranes were incubated with HRP-conjugated anti-Rabbit IgG
(1:5,000; ProteinTech Group, Inc.; cat. no. SA00001-2) and
anti-Mouse IgG (1:5,000; ProteinTech Group, Inc.; cat. no.
SA00001-1) secondary antibodies at 37°C for 1 h. Protein bands were
visualized using the ECL Plus kit (cat. no. PI80196X3, Thermo
Fisher Scientific, Inc.), according to the manufacturer's
protocol.
In vivo tumor xenograft study
Male BALB/c nude mice aged 4–5 weeks were adopted
for the construction of a subcutaneous transplantation tumor model
(n=5/group). The dorsal flank of the mice were inoculated with
glioma cells (5×106/100 µl). Following tumor
development, tumor diameters (<1 cm) were detected using digital
calipers and tumor volumes were calculated using the following
formula: Length × width2 ×0.5, as previously described
(23). Animal health and behavior,
and tumor volume were monitored every 2 days. Tumors were
irradiated with 2 Gy/day for 5 consecutive days when they reached
~50–100 mm3, and tumor growth was recorded during the
observation. All mice were euthanized 60 days after cell
inoculation via intraperitoneal injection of 200 mg/kg
pentobarbital sodium. All animal experiment were approved by The
Institutional Animal Ethical Committee of Fujian Medical University
(Fuzhou, China; approval no. 2018102). The welfare of animals in
the present study was protected, and standard protocols were
performed to minimize suffering.
Dual-luciferase reporter assay
The synthesized TPTEP1 or MAPK14 3′-untranslated
region (UTR) wild type (WT)/mutant (Mut) sequence that harbors
binding sites was subcloned into the pmirGLO dual-luciferase vector
(Promega Corporation) to form pmirGLOTPTEP1/MAPK14 3′-UTR-WT or
pmirGLO-TPTEP1/MAPK14 3′-UTR-Mut, which was subsequently used for
co-transfection in glioma cells with mimics, inhibitor or the
control sequence using Lipofectamine™ 2000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). After cell transfection (48 h),
Firefly and Renilla luciferase activities were detected
using a Dual-Luciferase Reporter Assay System (Promega
Corporation), according to the manufacturer's protocol. Firefly
luciferase activity was normalized to Renilla luciferase.
RNA immunoprecipitation (RIP)
assay
The Magna RNA-Binding Protein Immunoprecipitation
kit (cat. no. 03-110, Millipore; Merck KGaA) was used to perform
RIP assays according to the manufacturer's protocol. Cells
(2×107) were initially scraped in a cell culture dish,
followed by lysis with RIP lysis buffer. The magnetic beads
(Invitrogen; Thermo Fisher Scientific, Inc.) were applied for
incubation of cell extracts and conjugated with 5 ug anti-IgG
antibody (cat. no. ab172730; Abcam) or anti- Agonaute 2 (Ago2)
antibody (1:200; cat. no. 03-110; Millipore; Merck KGaA) supplied
with the kit at 4°C for 16 h. Samples were subsequently treated
with Proteinase K supplied with the kit and assayed via
RT-qPCR.
Bioinformatics analysis
Bioinformatics analysis was performed on the
extracted mRNA-Seq data of brain lower grade glioma (LGG) and
glioblastoma multiforme (GBM). The glioma expression profiles were
assessed using data from The Cancer Genome Atlas (TCGA) database
(https://www.cancer.gov/tcga).
Accordingly, patients with lncRNA expression values lower than the
median value were classified into the low expression group (n=257),
while the residual patients were classified into the high
expression group (n=257). The Gene Expression Profiling Interactive
Analysis database (http://gepia.cancer-pku.cn) (24) was used to assess the association
between TPTEP1 expression and overall survival time of the
patients. The impact of TPTEP1 on signaling pathways was assessed
via gene set enrichment analyses (GSEA). Bioinformatics tools
(miRcode: http://www.mircode.org/ and RNAhybrid:
http://bibiserv.cebitec.uni-bielefeld.de/rnahybrid?id=rnahybrid_view_submission)
(25,26) were applied to identify the miRNA
that potentially interacts with TPTEP1.
Patient tissues
A total of 177 paraffin-embedded glioma samples were
collected from departments of Neurology, The Second Affiliated
Hospital of Fujian Medical University, (Quzhou, China). The medical
records of these patients were used for collecting clinical data,
including age, sex, World Health Organization (WHO) grade,
recurrence status, and survival time. There are 111 male and 66
female patients, and the median age is 41 years old. All specimens
were pathologically diagnosed as glioma from 2008.2–2011.10. The
follow-up period ranged from 6–9.4 years. All experiments were
strictly performed according to the guidelines approved by the
Institutional Review Board (approval no. 2019-075), and written
informed consent was provided by all patients prior to the study
start.
In situ hybridization (ISH)
The paraffin-embedded samples were subjected to ISH
analysis. Briefly, sections were deparaffinized, rehydrated and
subsequently treated with proteinase K. Following incubation with
proteinase K at 37°C for 15 min, the sections were washed with
glycine once and PBS twice, and prehybridized with TPTEP1 or
miR-106-5p digoxygenin-labeled probes constructed by BersinBio
Corporation (http://www.bersinbio.com/). Samples were rinsed and
incubated with anti-digoxygenin-HRP Fab fragments (1:200; cat. no.
200-032-156; Jackson ImmunoResearch Laboratories, Inc.) for 1 h at
room temperature, prior to addition of DAB substrate (cat. no.
DAB-0031; Fuzhou Maixin Biotech Co., Ltd.) for signal detection.
Staining intensity was assessed, as previously described (7).
Immunohistochemistry (IHC)
Tissues were fixed in 10% neutral formalin overnight
at room temperature and dehydrated to make wax blocks. Paraffin
sections (3 µm) were deparaffinized, dehydrated and subsequently
subjected to antigen retrieval using citrate buffer. Deparaffinized
sections were blocked with 10% goat serum (Fuzhou Maixin Biotech
Co., Ltd.) for 15 min at room temperature, and incubated with 3%
H2O2 for 15 min at room temperature to
inhibit endogenous peroxidase activity and non-specific antigens.
Sections were incubated with anti-p38 MAPK (1:250; ProteinTech
Group, Inc.; cat. no. 66234-1-Ig) at 4°C for 16 h and
HRP-conjugated secondary antibody (1:200; ProteinTech Group, Inc.;
cat. no. SA00001-1) for 30 min at room temperature. DAB substrate
was applied to assess signal detection and observed under a light
microscope (×400 magnification). Staining intensity was assessed,
as previously described (27).
Statistical analysis
Statistical analysis was performed using SPSS 21.0
software (IBM Corp.) and data are presented as the mean ± standard
deviation. Unpaired student's t-test was used to assess differences
between two groups, while one-way ANOVA followed by
Student-Newman-Keuls post hoc test was used to assess differences
between multiple groups. Spearman's rank correlation test was used
to measure the correlation between genes expression. The general
linear model was applied for repeated measures variance analysis in
assessing differences between tumor growth. The χ2 test
was used to assess the association between gene expression and
clinical characteristics. Survival analyses were performed using
the Kaplan-Meier method. The Cox regression model was applied for
univariate and multivariate analyses. P<0.05 was considered to
indicate a statistically significant difference.
Results
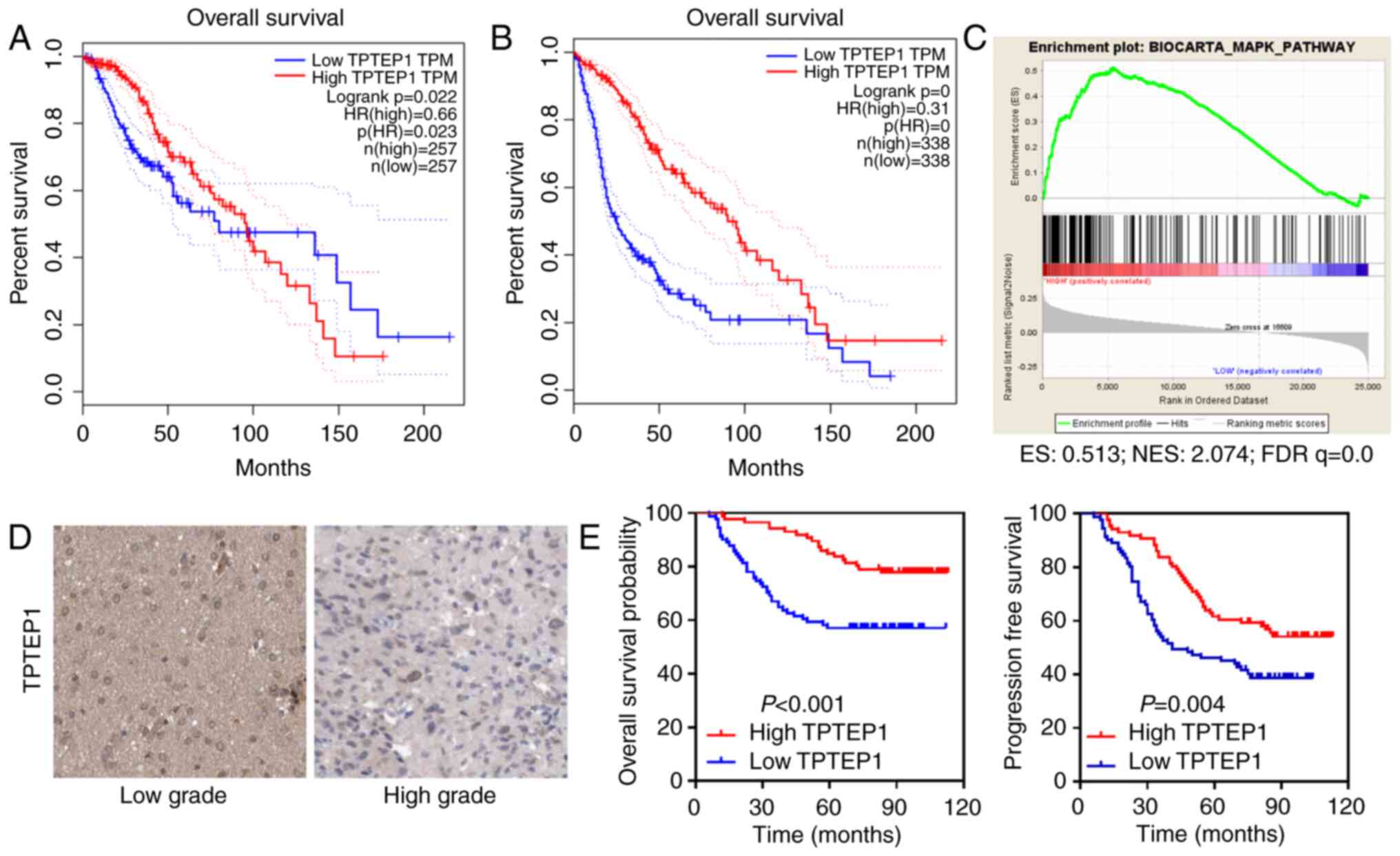
TPTEP1 is downregulated in glioma and
is associated with good patient survival
TCGA data analyses were performed to determine the
function of lncRNAs in glioma. TPTEP1 was identified as a potential
tumor suppressor that conferred good patient prognosis (Fig. 1A and B). Relying on data from TCGA
datasets, GSEA suggested that TPTEP1 participated in the modulation
of the MAPK signaling pathway (Fig.
1C).
ISH was performed to detect TPTEP1 levels in 177
patients with glioma. TPTEP1 expression levels were downregulated
in high-grade glioma tissues compared with low-grade glioma tissues
(Fig. 1D). The clinicopathological
characteristic of patients with glioma are listed in Table I. Association analysis revealed
that low TPTEP1 expression levels were associated with advanced WHO
grade and cancer recurrence, but not with other parameters
(Table I). Consistently,
Kaplan-Meier analysis revealed that patients with glioma who had
high TPTEP1 expression levels had a prolonged overall survival time
compared with those who had low TPTEP1 expression levels
(P<0.001, log-rank test). In addition, patients with glioma with
high TPTEP1 expression levels had a prolonged progression-free
survival time compared with those with low TPTEP1 expression levels
(P=0.004; Fig. 1E, log-rank test).
Univariate analysis was performed to elucidate the association
between clinical characteristics and overall patient survival in
glioma. Low TPTEP1 expression, older patients and high WHO grades
were all associated with shortened overall survival time of
patients with glioma. Multivariate analysis was subsequently
performed to determine the independent characteristics. The results
demonstrated that TPTEP1 expression and WHO grade were independent
prognostic indicators for patients with glioma (Table II).
 | Table I.Association between TPTEP1 expression
and clinicopathological characteristics of patients with glioma
(n=117). |
Table I.
Association between TPTEP1 expression
and clinicopathological characteristics of patients with glioma
(n=117).
|
|
| TPTEP1
expression |
|
|---|
|
|
|
|
|
|---|
| Characteristic | Patient number,
n | High, % | Low, % | P-value |
|---|
| Age, years |
|
|
|
|
|
≤Median | 90 | 41 (45.6) | 49 (54.4) | 0.955 |
|
>Median | 87 | 40 (46.0) | 47 (54.0) |
|
| Sex |
|
|
|
|
|
Male | 111 | 49 (44.1) | 62 (55.9) | 0.575 |
|
Female | 66 | 32 (48.5) | 34 (51.5) |
|
| WHO grade |
|
|
|
|
|
I–II | 105 | 55 (52.4) | 50 (47.6) | 0.033 |
|
III–IV | 72 | 26 (36.1) | 46 (63.9) |
|
| Recurrence |
|
|
|
|
| No | 83 | 46 (55.4) | 37 (44.6) | 0.015 |
|
Yes | 94 | 35 (37.2) | 59 (62.8) |
|
 | Table II.Univariate and multivariate Cox
regression survival analyses of the prognostic factors of
glioma. |
Table II.
Univariate and multivariate Cox
regression survival analyses of the prognostic factors of
glioma.
|
| Overall
survival |
|---|
|
|
|
|---|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Clinical
parameter | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| TPTEP1 expression
(low vs. high) | 2.572 | (1.516–4.366) | <0.001 | 2.143 | (1.062–3.554) | 0.031 |
| Age, years (≤median
vs. >median) | 0.481 | (0.287–0.808) | 0.007 | 0.590 | (0.331–1.054) | 0.075 |
| Sex (female vs.
male) | 0.660 | (0.389–1.119) | 0.118 | 0.736 | (0.407–1.330) | 0.310 |
| WHO grade (I–II vs.
III–IV) | 0.073 | (0.041–0.128) | <0.001 | 0.427 | (0.238–0.821) | 0.010 |
| Recurrence (no vs.
yes) | 0.095 | (0.056–0.161) | <0.001 | 0.485 | (0.269–0.974) | 0.043 |
TPTEP1 suppresses glioma cell stemness
and radioresistance both in vitro and in vivo
To determine the impact of TPTEP1 on glioma cell
stemness and radioresistance, TPTEP1 expression levels were
detected in SHG44 and U251 cells, and stable/transient
TPTEP1-silenced SHG44 cells and TPTEP1-overexpressed U251 cells
were established using lentiviral vectors (Fig. 2A and B). Both in vitro and
in vivo experiments were performed to assess the stemness
and radioresistance of the cell lines. TPTEP1 knockdown augmented
cells stemness and radioresistance in SHG44 (Fig. 2C-F). In addition, TPTEP1 knockdown
promoted cells stemness and expression of
radioresistance-associated genes, including OCT4, ALDH1 and γ-H2AX
(Fig. 2G). To further verify the
effect of TPTEP1 on glioma cell radioresistance, mice were
implanted with TPTEP1-silenced SHG44 cells and the corresponding
control cells. After developing to the indicated volumes, the
tumors were irradiated with 2 Gy/day for 5 consecutive days. The
radiation treatment inhibited the growth of control tumors and
inhibition of TPTEP1 led to resistance to radiation treatment
(Fig. 2H). However, TPTEP1
overexpression induced the opposite effects in U251 cells (Fig. 2C-G).
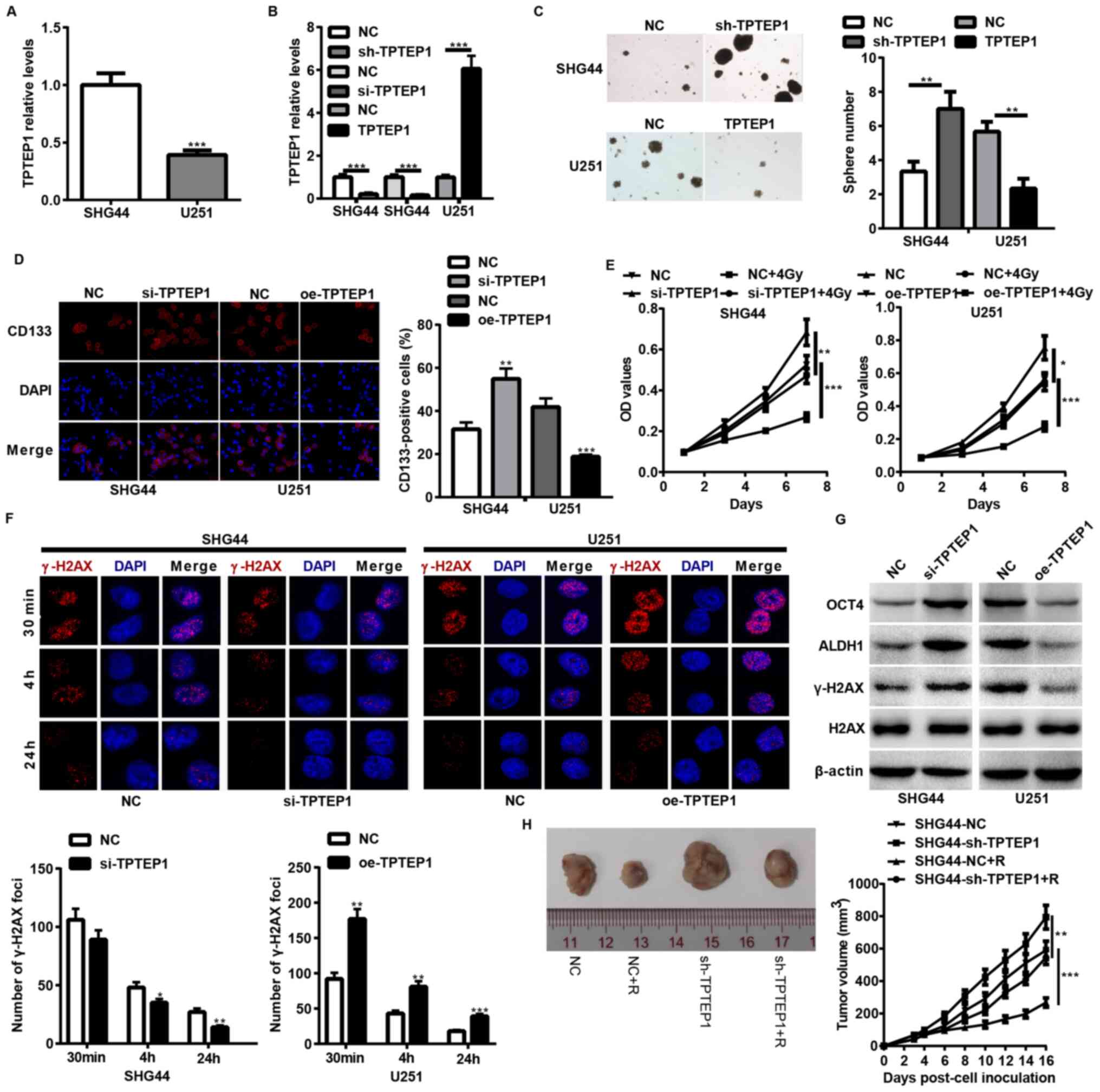 | Figure 2.TPTEP1 suppresses glioma cell
stemness and radioresistance in vitro and in vivo.
RT-qPCR analysis was performed to detect TPTEP1 expression levels
in (A) SHG44 and U251 cells and in (B) stable/transient
TPTEP1-depleted SHG44, stable TPTEP1-overexpressed U251 cells and
the control cells. (C) Tumorspheres formation assay was performed
in stable TPTEP1-depleted SHG44 cells, stable TPTEP1-overexpressed
U251 cells and the control cells (×100 magnification). (D)
Immunofluorescence analysis was performed to detect the expression
and location of CD133 in transient TPTEP1-depleted SHG44 cells,
transient TPTEP1-overexpressed U251 cells and their corresponding
controls (×400 magnification). (E) Cell proliferation and
radiosensitivity assays were performed in transient TPTEP1-depleted
SHG44 cells, transient TPTEP1-overexpressed U251 cells and their
corresponding controls. (F) Immunofluorescence analysis was
performed to evaluate γ-H2AX foci formation in transient
TPTEP1-silenced SHG44 cells, transient TPTEP1-overexpressed U251
cells and their corresponding controls. (G) Western blot analysis
was performed to detect OCT4, ALDH1, γ-H2AX and H2AX expression
levels in transient TPTEP1-silenced SHG44 cells, transient
TPTEP1-overexpressed U251 cells and their corresponding controls.
(H) A subcutaneous transplantation tumor model was applied to
assess glioma cell proliferation regulated by TPTEP1 and R.
*P<0.05, **P<0.01, ***P<0.001. TPTEP1, transmembrane
phosphatase with tensin homology pseudogene 1; RT-qPCR, reverse
transcription-quantitative PCR; OCT4, Octamer binding protein 4;
ALDH1, aldehyde dehydrogenase 1; γ-H2AX, Histone H2AX; R,
radiation. |
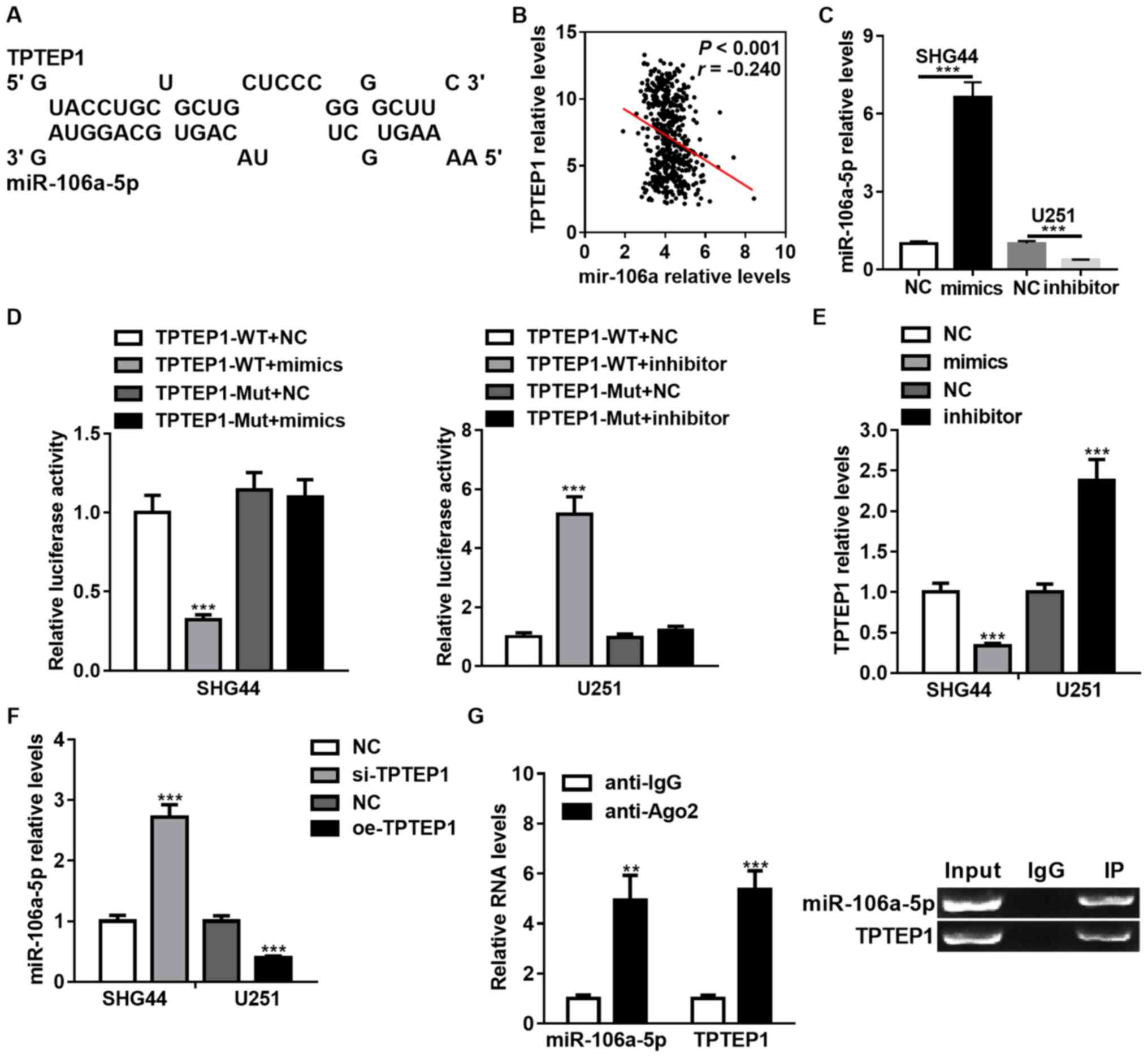
TPTEP1 interacts with miR-106a-5p in
glioma cells
Previous studies have verified the function of
lncRNAs as competing endogenous RNAs (ceRNAs) in protecting
miRNAs-targeted mRNAs (28,29).
Thus, bioinformatics tools were applied to identify the miRNA that
potentially interacts with TPTEP1. Considering base-pair
conservation, expression relevance from TCGA database and reported
functions of miRNAs in the regulation of CSC properties,
miR-106a-5p was the focus for further study (Fig. 3A and B). Furthermore, the
expression levels of TPTEP1 and miR-106a were comparable in TCGA
database (Fig. 3B). The
miR-106a-5p-overexpressed SHG44 cells and miR-106a-5p-silenced U251
cells were established using miR-106a-5p mimics and inhibitor,
respectively (Fig. 3C). A
dual-luciferase reporter assay was performed to determine whether
TPTEP1 is targeted by miR-106a-5p, and luciferase activities were
downregulated by miR-106a-5p overexpression and upregulated by
miR-106a-5p knockdown in SHG44 and U251 cells treated with WT
TPTEP1. In glioma cells transfected with Mut TPTEP1 harboring
mutations in the miR-106a-5p binding sequence inside TPTEP1, these
impacts on luciferase activities were abrogated (Fig. 3D).
RT-qPCR analysis demonstrated that depletion of
miR-106a-5p promoted the expression levels of TPTEP1, while
upregulation of miR-106a-5p inhibited the expression levels of
TPTEP1 (Fig. 3E). The impact of
TPTEP1 on the expression levels of miR-106a-5p was subsequently
investigated. TPTEP1 knockdown elevated miR-106a-5p expression
levels, and TPTEP1 overexpression induced the opposite effect
(Fig. 3F). Furthermore, TPTEP1 and
miR-106a-5p were demonstrated to interact with RNA-induced
silencing complexes (RISC)-related Ago2 by RNA immunoprecipitation
assays (Fig. 3G). Collectively,
these results suggest that TPTEP1 interacts with miR-106a-5p, and
both are assembled to RISC, forming a reciprocal regulatory
circuit.
TPTEP1 promotes MAPK14 expression by
antagonizing miR-106a-5p binding and activates the P38 MAPK
signaling pathway
A previous study identified MAPK14 as the target of
miR-106a-5p in the modulation of cell differentiation (30). To validate the interaction between
the miR-106-5p binding sequence inside TPTEP1 and the MAPK14 3′UTR
(Fig. 4A), western blot analysis
and dual-luciferase reporter assay were performed. The results
demonstrated that overexpression of miR-106-5p decreased MAPK14 and
phosphorylated-MAPK14 expression levels, and miR-106-5p knockdown
elevated MAPK14 expression levels (Fig. 4B). In addition, the luciferase
activities were downregulated by miR-106-5p overexpression and were
upregulated by miR-106-5p knockdown in SHG44 and U251 cells treated
with WT MAPK14 3′UTR reporter vector (Fig. 4C). Notably, these impacts were
abrogated in glioma cells transfected with Mut MAPK14 reporter
vector (Fig. 4C). Taken together,
these results indicate that the miR-106a-5p binding sequence within
TPTEP1 interacts with the MAPK14 3′UTR, and TPTEP1 may
competitively interplay with miR-106a-5p to prevent MAPK14 from
repressing by miR-106a-5p.
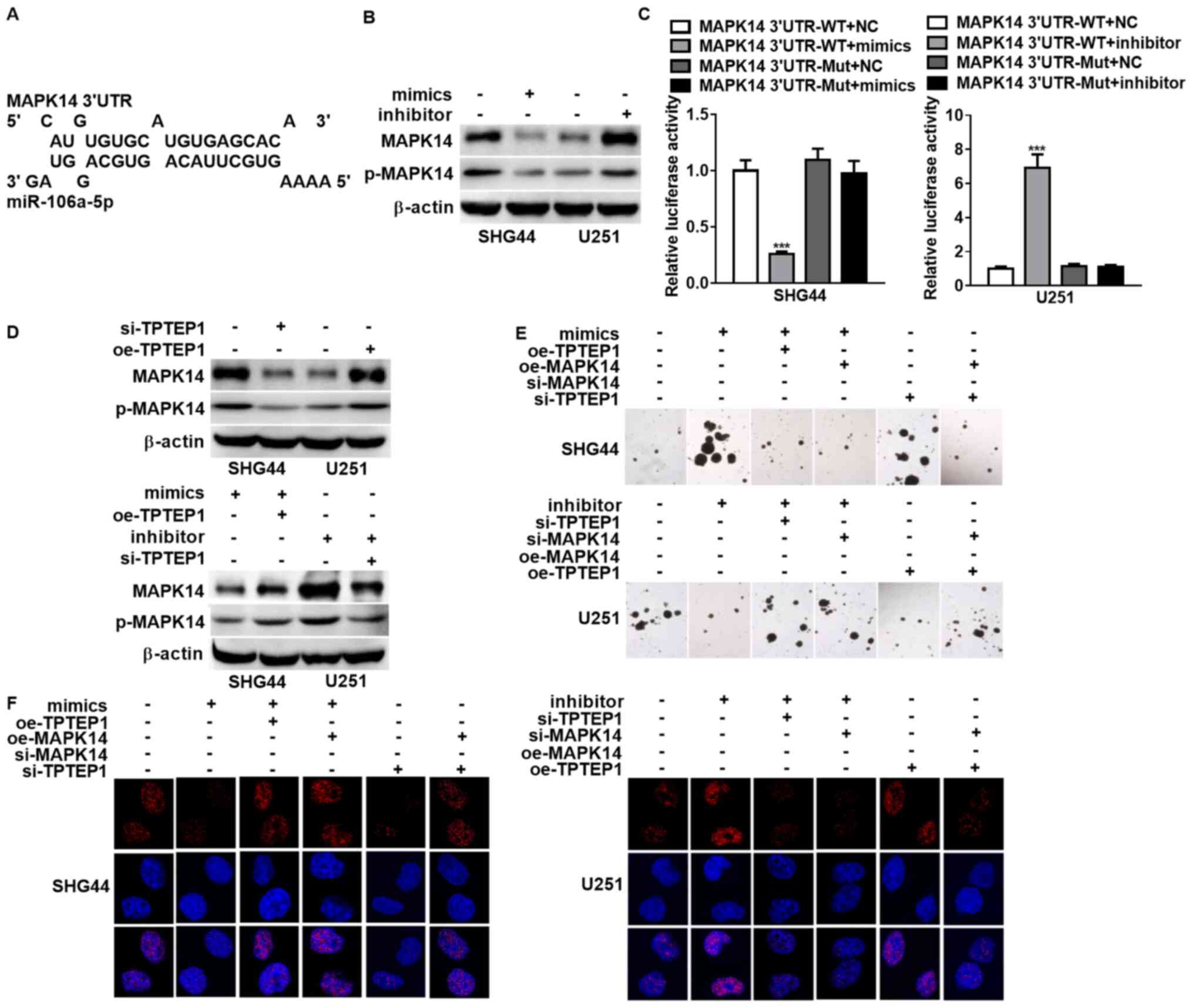 | Figure 4.TPTEP1 upregulates MAPK14 by
antagonizing miR-106a-5p binding and activates the P38 MAPK
signaling pathway. (A) Binding sites of miR-106a-5p inside MAPK14
3′-UTR. (B) Western blot analysis was performed to detect MAPK14
and phosphorylated-MAPK14 protein expression levels in
miR-106a-5p-overexpressed SHG44 cells, miR-106a-5p-silenced U251
cells and the control cells. (C) Dual-luciferase reporter assay was
performed to determine the interplay between MAPK14 3′UTR and
miR-106a-5p in SHG44 and U251 cells. (D) Western blot analysis was
performed to detect MAPK14 and phosphorylated-MAPK14 protein
expression levels in TPTEP1-depleted SHG44 cells,
TPTEP1-overexpressed U251 cells, miR-106a-5p-overexpressed SHG44
cells with TPTEP1 overexpression, miR-106a-5p-silenced U251 cells
with TPTEP1 knockdown and the control cells. (E) Tumorspheres
formation assay was performed to assess glioma cell stemness
regulated by miR-106a-5p, TPTEP1 and MAPK14. (F) Immunofluorescence
analysis were performed to assess γ-H2AX foci formation regulated
by miR-106a-5p, TPTEP1 and MAPK14. Sale bar=5 µm. ***P<0.001.
TPTEP1, transmembrane phosphatase with tensin homology pseudogene
1; miR, microRNA; 3′-URT, 3′-untranslated region; γ-H2AX, Histone
H2AX. |
Accordingly, the protein expression levels of MAPK14
and phosphorylated-MAPK14 were decreased in TPTEP1-silenced SHG44
cells and were increased in TPTEP1-upregulated U251 cells compared
with the corresponding controls. Consistently, TPTEP1
overexpression upregulated MAPK14 and phosphorylated-MAPK14
expression levels in miR-106a-5p-overexpressed SHG44 cells, while
TPTEP1 knockdown downregulated MAPK14 and phosphorylated-MAPK14
expression levels in miR-106a-5p-depleted U251 cells, which
suggests that TPTEP1 restores MAPK14 expression regulated by
miR-106a-5p (Fig. 4D). Notably,
the depletion of miR-106a-5p inhibited glioma cell stemness and
radioresistance, while downregulation of TPTEP1 or MAPK14 enhanced
glioma cell stemness and radioresistance in miR-106a-5p-silenced
U251. However, the upregulation of TPTEP1 or MAPK14 reversed glioma
cell stemness and radioresistance modulated by miR-106a-5p
overexpression in SHG44, which indicates that TPTEP1 or MAPK14 may
attenuate the stimulated impacts of miR-106a-5p on glioma cell
stemness and radioresistance. Furthermore, overexpression of TPTEP1
was demonstrated to suppress cells stemness and radioresistance,
and depletion of MAPK14 in TPTEP1-overexpressed U251 cells
augmented cell stemness and radioresistance. However,
overexpression of MAPK14 reversed the impacts of TPTEP1 depletion
on cells stemness and radioresistance in SHG44, which suggests that
MAPK14 may reverse glioma cell stemness and radioresistance
modulated by TPTEP1 (Fig. 4E and
F).
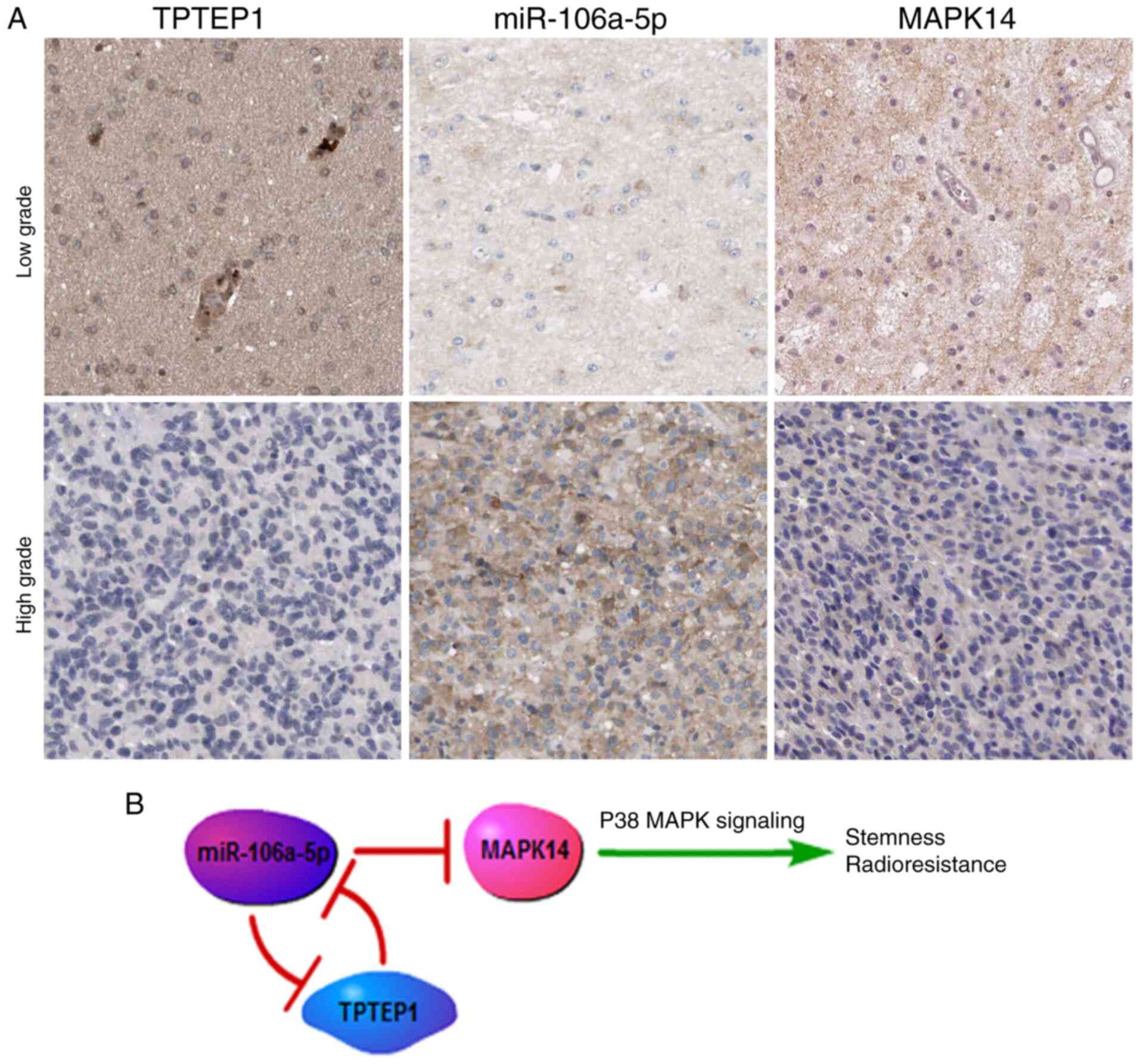
Associations of TPTEP1, miR-106a-5p
and MAPK14 in glioma tissues
ISH and IHC analyses were performed to detect the
expression levels of miR-106a-5p and MAPK14 in the 177 glioma
samples. TPTEP1 expression levels were demonstrated to be
negatively associated with miR-106a-5p expression levels (P=0.013,
κ=−0.185) and were positively associated with MAPK14
expression levels (P=0.032, κ=0.161) in glioma tissues.
miR-106a-5p expression levels were also demonstrated to be
negatively associated with MAPK14 expression levels in glioma
tissues (P=0.001, κ=−0.253) (Fig. 5A). Thus, the
TPTEP1/miR-106a-5p/MAPK14 axis may be responsible for regulating
the suppressive effects of TPTEP1 on glioma progression (Fig. 5B).
Discussion
Increasing evidence have verified the
tumor-suppressive and -promoting (oncogenic) functions of lncRNAs
in different types of cancer, including lung cancer, breast cancer
and brain tumor (31,32). However, the function and
corresponding molecular mechanism of lncRNA TPTEP1 in glioma are
yet to be investigated. The results of the present study identified
TPTEP1 as a significant prognostic indicator for predicting
favorable prognosis in patients with glioma. Furthermore, TPTEP1
was demonstrated to suppress stemness and radioresistance of
glioma. Thus, the results of the present study suggest that
mechanistically, TPTEP1 exerts its tumor-suppressive effects by
triggering the P38 MAPK signaling pathway. TPTEP1 upregulates
MAPK14 by antagonizing miR-106a-5p binding, and TPTEP1 interlays
with miR-106a-5p to form a reciprocal regulatory circuit.
TPTEP1 acts as a tumor suppressor in hepatocellular
carcinoma (33,34) and lung cancer (35), and has been reported to participate
in the modulation of several biological functions, including cell
proliferation, invasion, chemoresistance and inflammation (33–35).
In addition, bioinformatic analysis suggests that TPTEP1 may serve
as a tumor suppressor in the development of GBM (36). The present study aimed to determine
the properties of CSCs and investigate radioresistance modulated by
lncRNA TPTEP1 in glioma, and also elucidate the tumor-suppressive
role of TPTEP1 in glioma. The results demonstrated that TPTEP1
inhibited glioma cell stemness and radioresistance, thereby
impeding cancer progression. TPTEP1 expression levels were
downregulated in high-grade glioma tissues compared with low-grade
glioma tissues. Furthermore, TPTEP1 was negatively associated with
high WHO grade, recurrence status and poor patient survival
time.
lncRNAs modulate gene expression through
multidimensional mechanisms (37)
and function as ceRNAs to prevent miRNAs from interacting with
shared mRNAs (38), thus forming a
lncRNA-miRNA-mRNA regulatory network. The present study
hypothesized that the direct interaction between TPTEP1 and
miR-106a-5p mutually represses their expression at the
post-transcription level. The inverse association between miR-106a
and TPTEP1 was validated using clinical samples. miR-106a-5p, also
referred to as miR-106a, exerts an oncogenic role in different
types of human cancer, including glioma (39,40).
In addition, miR-106a facilitates radioresistance of prostate
cancer (41) and promotes cell
stemness of colorectal cancer (42). The results of the present study
confirmed that miRNA-106a-5p serves as an oncogene by augmenting
TPTEP1-mediated stemness and radioresistance of glioma. Previous
studies have demonstrated the stimulating effects of TPTEP1 and the
inhibitory effect of miRNA-106a-5p on cancer cell apoptosis
(33,35,43).
Thus, prospective studies will aim to elucidate the effect of
TPTEP1 and miR-106-5p on glioma cell apoptosis.
P38 MAPK family members have dual roles in a cell
type-specific and cell context-specific manner, and transduce
signals that modulate cell differentiation, proliferation,
migration and survival (44).
Upregulating p38-MAPK signaling primes resistant GBM cells to
apoptosis, and activation of p38 MAPK in GSCs leads to decreased
neurosphere formation capacity and stem cell marker expression,
which suggests that decreased stemness and radioresistance is
attributed to activation of the P38 MAPK signaling pathway in most
cases of glioma (45–49). The results of the present study
demonstrated that MAPK14, namely P38, serves as a tumor suppressor
in the development of glioma. MAPK14 exerted its inhibitory
activity by antagonizing stemness and radioresistance of glioma. In
addition, MAPK14 was verified as a target gene of miR-106a-5p, and
cooperated with miR-106a-5p and TPTEP1 to regulate glioma
progression. The rescue experiments and association analysis in
tissues further confirmed the modulation of the
TPTEP1/miR-106a-5p/MAPK14 axis in glioma progression. Notably, this
study also has some limitations. For example, the effect of TPTEP1
and miR-106-5p on glioma cell apoptosis needs to be further
studied. Additionally, the impact of TPTEP1 on tumorigenesis needs
to be investigated further since this study focused on glioma
progression.
Taken together, the results of the present study
demonstrated that TPTEP1 functioned as a tumor suppressor that
antagonized stemness and radioresistance of glioma by competitively
inhibiting the interaction between miR-106a-5p and MAPK14. TPTEP1
and miR-106a-5p formed a reciprocal regulatory circuit to
facilitate P38 MAPK signaling activation in glioma. Collectively,
the results suggest that TPTEP1 may serve as a diagnostic and
prognostic indicator for glioma, and be used as an anti-cancer
target for glioma treatment.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Talent Training
Project of the Fujian Provincial Health and Family Planning
Commission (grant no. 2017-2-29) and the Quanzhou Science and
Technology Project [grant no. Z(2014)0106].
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
TT acquired, analyzed and interpreted the data, and
drafted the initial manuscript. RMZ designed the present study,
performed the literature review, and revised and reviewed the
manuscript. LXW performed the experiments and clinical studies, and
MLY performed statistical analysis. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
All animal experiments were approved by The
Institutional Animal Ethical Committee of Fujian Medical University
(Fuzhou, China; approval no. 2018102). All experiments performed
using human samples were approved by The institutional Ethics
Committee of The Second Affiliated Hospital of Fujian Medical
University (Quanzhou, China; approval no. 2019-075), and written
informed consent was provided by all patients prior to the study
start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ceRNAs
|
competing endogenous RNAs
|
|
CSCs
|
cancer stem cells
|
|
GBM
|
glioblastoma multiforme
|
|
GSEA
|
gene set enrichment analysis
|
|
LGG
|
brain lower grade glioma
|
|
lncRNAs
|
long non-coding RNAs
|
|
ISH
|
in situ hybridization
|
|
RIP
|
RNA immunoprecipitation
|
|
RISC
|
RNA-induced silencing complexes
|
References
|
1
|
Liu YZ, Du XX, Zhao QQ, Jiao Q and Jiang
H: The expression change of OTUD3-PTEN signaling axis in glioma
cells. Ann Transl Med. 8:4902020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Larjavaara S, Mäntylä R, Salminen T,
Haapasalo H, Raitanen J, Jääskeläinen J and Auvinen A: Incidence of
gliomas by anatomic location. Neuro Oncol. 9:319–325. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bauchet L and Ostrom QT: Epidemiology and
molecular epidemiology. Neurosurg Clin N Am. 30:1–16. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang H, Xu T, Jiang Y, Xu H, Yan Y, Fu D
and Chen J: The challenges and the promise of molecular targeted
therapy in malignant gliomas. Neoplasia. 17:239–255. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Balca-Silva J, Matias D, Carmo AD,
Sarmento-Ribeiro AB, Lopes MC and Moura-Neto V: Cellular and
molecular mechanisms of glioblastoma malignancy: Implications in
resistance and therapeutic strategies. Semin Cancer Biol.
58:130–141. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Visvanathan A, Patil V, Arora A, Hegde AS,
Arivazhagan A, Santosh V and Somasundaram K: Essential role of
METTL3-mediated m6A modification in glioma stem-like
cells maintenance and radioresistance. Oncogene. 37:522–533. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lin X, Zuo S, Luo R, Li Y, Yu G, Zou Y,
Zhou Y, Liu Z, Liu Y, Hu Y, et al: HBX-induced miR-5188 impairs
FOXO1 to stimulate β-catenin nuclear translocation and promotes
tumor stemness in hepatocellular carcinoma. Theranostics.
9:7583–7598. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zou Y, Lin X, Bu J, Lin Z, Chen Y, Qiu Y,
Mo H, Tang Y, Fang W and Wu Z: Timeless-stimulated
miR-5188-FOXO1/β-catenin-c-Jun feedback loop promotes stemness via
ubiquitination of β-catenin in breast cancer. Mol Ther. 28:313–327.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Camacho CV, Choudhari R and Gadad SS: Long
noncoding RNAs and cancer, an overview. Steroids. 133:93–95. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang X, Wang W, Zhu W, Dong J, Cheng Y,
Yin Z and Shen F: Mechanisms and functions of long non-coding RNAs
at multiple regulatory levels. Int J Mol Sci. 20:55732019.
View Article : Google Scholar
|
|
11
|
Peng Z, Liu C and Wu M: New insights into
long noncoding RNAs and their roles in glioma. Mol Cancer.
17:612018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kiang KM, Zhang XQ and Leung GK: Long
non-coding RNAs: The key players in glioma pathogenesis. Cancers
(Basel). 7:1406–1424. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang X, Xiao R, Pan S, Yang X, Yuan W, Tu
Z, Xu M, Zhu Y, Yin Q, Wu Y, et al: Uncovering the roles of long
non-coding RNAs in cancer stem cells. J Hematol Oncol. 10:622017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen
S, Wang Y, Wang T and Hou Y: Carcinoma-associated fibroblasts
promote the stemness and chemoresistance of colorectal cancer by
transferring exosomal lncRNA H19. Theranostics. 8:3932–3948. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhuang J, Shen L, Yang L, Huang X, Lu Q,
Cui Y, Zheng X, Zhao X, Zhang D, Huang R, et al: TGFβ1 promotes
gemcitabine resistance through regulating the
LncRNA-LET/NF90/miR-145 signaling axis in bladder cancer.
Theranostics. 7:3053–3067. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mazor G, Levin L, Picard D, Ahmadov U,
Carén H, Borkhardt A, Reifenberger G, Leprivier G, Remke M and
Rotblat B: The lncRNA TP73-AS1 is linked to aggressiveness in
glioblastoma and promotes temozolomide resistance in glioblastoma
cancer stem cells. Cell Death Dis. 10:2462019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu M, Xue Y, Zheng J, Liu X, Yu H, Liu L,
Li Z and Liu Y: Linc00152 promotes malignant progression of glioma
stem cells by regulating miR-103a-3p/FEZF1/CDC25A pathway. Mol
Cancer. 16:1102017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim SS, Harford JB, Moghe M, Rait A,
Pirollo KF and Chang EH: Targeted nanocomplex carrying siRNA
against MALAT1 sensitizes glioblastoma to temozolomide. Nucleic
Acids Res. 46:1424–1440. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dai X, Liao K, Zhuang Z, Chen B, Zhou Z,
Zhou S, Lin G, Zhang F, Lin Y, Miao Y, et al: AHIF promotes
glioblastoma progression and radioresistance via exosomes. Int J
Oncol. 54:261–270. 2019.PubMed/NCBI
|
|
20
|
Liao K, Ma X, Chen B, Lu X, Hu Y, Lin Y,
Huang R and Qiu Y: Upregulated AHIF-mediated radioresistance in
glioblastoma. Biochem Biophys Res Commun. 509:617–623. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zheng R, Yao Q, Ren C, Liu Y, Yang H, Xie
G, Du S, Yang K and Yuan Y: Upregulation of long noncoding RNA
small nucleolar RNA host gene 18 promotes radioresistance of glioma
by repressing semaphorin 5A. Int J Radiat Oncol Biol Phys.
96:877–887. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li P, Wang Q and Wang H: MicroRNA-204
inhibits the proliferation, migration and invasion of human lung
cancer cells by targeting PCNA-1 and inhibits tumor growth in
vivo. Int J Mol Med. 43:1149–1156. 2019.PubMed/NCBI
|
|
24
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45((W1)):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jeggari A, Marks DS and Larsson E:
miRcode: A map of putative microRNA target sites in the long
non-coding transcriptome. Bioinformatics. 28:2062–2063. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rehmsmeier M, Steffen P, Hochsmann M and
Giegerich R: Fast and effective prediction of microRNA/target
duplexes. RNA. 10:1507–1517. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lin X, Li AM, Li YH, Luo RC, Zou YJ, Liu
YY, Liu C, Xie YY, Zuo S, Liu Z, et al: Silencing MYH9 blocks
HBx-induced GSK3β ubiquitination and degradation to inhibit tumor
stemness in hepatocellular carcinoma. Signal Transduct Target Ther.
5:132020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Karreth FA and Pandolfi PP: ceRNA
cross-talk in cancer: When ce-bling rivalries go awry. Cancer
Discov. 3:1113–1121. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Thomson DW and Dinger ME: Endogenous
microRNA sponges: Evidence and controversy. Nat Rev Genet.
17:272–283. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Naka-Kaneda H, Nakamura S, Igarashi M, Aoi
H, Kanki H, Tsuyama J, Tsutsumi S, Aburatani H, Shimazaki T and
Okano H: The miR-17/106-p38 axis is a key regulator of the
neurogenic-to-gliogenic transition in developing neural
stem/progenitor cells. Proc Natl Acad Sci USA. 111:1604–1609. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang Y and Tang L: The application of
lncRNAs in cancer treatment and diagnosis. Recent Pat Anticancer
Drug Discov. 13:292–301. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ding H, Liu J, Zou R, Cheng P and Su Y:
Long non-coding RNA TPTEP1 inhibits hepatocellular carcinoma
progression by suppressing STAT3 phosphorylation. J Exp Clin Cancer
Res. 38:1892019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ding H, Zhang X, Su Y, Jia C and Dai C:
GNAS promotes inflammation-related hepatocellular carcinoma
progression by promoting STAT3 activation. Cell Mol Biol Lett.
25:82020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cao F, Wang Z, Feng Y, Zhu H, Yang M,
Zhang S and Wang X: lncRNA TPTEP1 competitively sponges miR-328-5p
to inhibit the proliferation of non-small cell lung cancer cells.
Oncol Rep. 43:1606–1618. 2020.PubMed/NCBI
|
|
36
|
Zheng J, Su Z, Kong Y, Lin Q, Liu H, Wang
Y and Wang J: lncRNAs predicted to interfere with the gene
regulation activity of miR-637 and miR-196a-5p in GBM. Front Oncol.
10:3032020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Marchese FP, Raimondi I and Huarte M: The
multidimensional mechanisms of long noncoding RNA function. Genome
Biol. 18:2062017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang Z, Wang B, Shi Y, Xu C, Xiao HL, Ma
LN, Xu SL, Yang L, Wang QL, Dang WQ, et al: Oncogenic miR-20a and
miR-106a enhance the invasiveness of human glioma stem cells by
directly targeting TIMP-2. Oncogene. 34:1407–1419. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhao H, Shen J, Hodges TR, Song R, Fuller
GN and Heimberger AB: Serum microRNA profiling in patients with
glioblastoma: A survival analysis. Mol Cancer. 16:592017.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hoey C, Ray J, Jeon J, Huang X, Taeb S,
Ylanko J, Andrews DW, Boutros PC and Liu SK: miRNA-106a and
prostate cancer radioresistance: A novel role for LITAF in ATM
regulation. Mol Oncol. 12:1324–1341. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Qin Y, Chen X, Liu Z, Tian X and Huo Z:
miR-106a reduces 5-fluorouracil (5-FU) sensitivity of colorectal
cancer by targeting dual-specificity phosphatases 2 (DUSP2). Med
Sci Monit. 24:4944–4951. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Dong S, Zhang X and Liu D: Overexpression
of long noncoding RNA GAS5 suppresses tumorigenesis and development
of gastric cancer by sponging miR-106a-5p through the Akt/mTOR
pathway. Biol Open. 8:bio0413432019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wagner EF and Nebreda AR: Signal
integration by JNK and p38 MAPK pathways in cancer development. Nat
Rev Cancer. 9:537–549. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sato A, Okada M, Shibuya K, Watanabe E,
Seino S, Narita Y, Shibui S, Kayama T and Kitanaka C: Pivotal role
for ROS activation of p38 MAPK in the control of differentiation
and tumor-initiating capacity of glioma-initiating cells. Stem Cell
Res. 12:119–131. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu J, Zhang H, Xu Y, Zhang J, Zhu W, Zhang
Y, Chen L, Hua W and Mao Y: Juglone induces apoptosis of tumor
stem-like cells through ROS-p38 pathway in glioblastoma. BMC
Neurol. 17:702017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu P, Brown S, Goktug T, Channathodiyil
P, Kannappan V, Hugnot JP, Guichet PO, Bian X, Armesilla AL,
Darling JL and Wang W: Cytotoxic effect of disulfiram/copper on
human glioblastoma cell lines and ALDH-positive cancer-stem-like
cells. Br J Cancer. 107:1488–1497. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tamura K, Wakimoto H, Agarwal AS, Rabkin
SD, Bhere D, Martuza RL, Kuroda T, Kasmieh R and Shah K:
Multimechanistic tumor targeted oncolytic virus overcomes
resistance in brain tumors. Mol Ther. 21:68–77. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yang W, Shen Y, Wei J and Liu F:
MicroRNA-153/Nrf-2/GPx1 pathway regulates radiosensitivity and
stemness of glioma stem cells via reactive oxygen species.
Oncotarget. 6:22006–22027. 2015. View Article : Google Scholar : PubMed/NCBI
|