Introduction
Esophageal cancer is the 8th most common
human cancer and 6th highest in mortality (1). There are obvious regional and
histological differences in the incidence of esophageal cancer.
China has one of the highest incidence rates in the world, with the
histological type of most patients being squamous cell carcinoma,
which exceeds 90% of the total number of cases (2). Esophageal cancer is one of the most
difficult gastrointestinal malignancies to treat and cure. Patients
often experience distant metastasis or local disease recurrence,
even after undergoing curative resection. Although multi-modality
approaches based on surgery combined with preoperative chemotherapy
and/or radiotherapy have been attempted, the efficacy of these
treatments is limited, and overall survival remains poor (3–6).
Therefore, novel strategies against esophageal cancer need to be
developed and established to improve the prognosis of patients.
SH3 domain-containing kinase-binding protein 1
(CIN85) was first identified in human cells as a Cbl-interacting 85
kDa protein. CIN85 contains three Src homology 3 (SH3) domains at
its N-terminus, followed by a proline-rich region and a C-terminal
coiled-coil region (7). In
association with casitas B-lineage lymphoma, an E3 ubiquitin
ligase, CIN85 controls the intracellular internalization,
trafficking and sorting of several activated receptor tyrosine
kinases, including the epidermal growth factor receptor (EGFR)
(8). Moreover, through the SH3
domains and the proline-rich region, CIN85 is implicated in a
number of protein-protein interactions and has been found to play
important roles in other processes, such as signal transduction,
vesicle-mediated transport, cytoskeleton remodeling, immunological
synapse, cell migration and invasion (9–11).
Previously, CIN85 was detected on lamellipodia and invadopodia,
which are involved in cell adhesion and migration, suggesting that
the overexpression of CIN85 could promote the invasiveness of
cancer cells (12,13).
However, the expression profiles and clinical
relevance of CIN85 in esophageal squamous cell carcinoma (ESCC)
remain unknown. The present study was designed to elucidate the
clinical significance of CIN85 in ESCC, as well as its in
vitro functions.
Materials and methods
Patient cohort and cell line
A total of 129 patients were included in the present
study, which was approved by the Institutional Ethics Committee of
The First Affiliated Hospital of China Medical University
(Shenyang, China). All patients were operated with curative intent
between January 2014 and January 2017 at the Department of Thoracic
Surgery, The First Affiliated Hospital of China Medical University.
All patients were included except those who had undergone
chemotherapy before the operation. The distance between cancer
tissue and the adjacent normal tissue was >5 cm. A summary of
the clinicopathological data is provided in Table I. No radiotherapy, chemotherapy or
other adjuvant therapy was performed prior to surgery. Tumor
staging in the present study was based on the 8th edition of the
World Health Organization Tumor Node Metastasis (TNM) staging
criteria for ESCC, published in 2016 (14).
 | Table I.Association between the expression of
CIN85 and the clinicopathological features of patients with
esophageal squamous cell carcinoma. |
Table I.
Association between the expression of
CIN85 and the clinicopathological features of patients with
esophageal squamous cell carcinoma.
|
|
| CIN85 expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Clinical
characteristic | Number of
cases | Negative, n
(%) | Positive, n
(%) | χ2 | P-value |
|---|
| Sex |
|
|
| 0.548 | 0.459 |
|
Male | 102 | 30 (29.4) | 72 (70.6) |
|
|
|
Female | 27 | 6 (22.2) | 21 (77.8) |
|
|
| Age, years |
|
|
| 0.568 | 0.451 |
|
<60 | 72 | 22 (30.6) | 50 (69.4) |
|
|
|
≥60 | 57 | 14 (24.6) | 43 (75.4) |
|
|
| Tumor invasion,
T |
|
|
| 0.520 | 0.471 |
|
T1-T2 | 87 | 26 (29.9) | 61 (70.1) |
|
|
|
T3-T4 | 42 | 10 (23.8) | 32 (76.2) |
|
|
| Lymph node
metastasis, N |
|
|
| 11.356 | 0.001 |
| N0 | 66 | 27 (40.9) | 39 (59.1) |
|
|
|
N1-N3 | 63 | 9 (14.3) | 54 (85.7) |
|
|
| Cell grading |
|
|
| 0.000 | 0.989 |
|
G1-G2 | 111 | 31 (27.9) | 80 (72.1) |
|
|
| G3 | 18 | 5 (27.8) | 13 (72.2) |
|
|
| TNM stage |
|
|
| 9.768 | 0.002 |
|
I–II | 72 | 28 (38.9) | 44 (61.1) |
|
|
|
III–IV | 57 | 8 (14.0) | 49 (86.0) |
|
|
A human ESCC TE1 cell line was obtained from The
Cell Bank of Type Culture Collection of the Chinese Academy of
Sciences. Cells were routinely cultured in RPMI-1640 medium (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine
serum (Cytiva) and 1% penicillin/streptomycin at 37°C in 5%
CO2.
Immunohistochemistry (IHC) and
scoring
IHC was performed on 4-µm thick paraffin sections
fixed in 100% formaldehyde for 24 h at room temperature, which were
then dewaxed and rehydrated in a descending ethanol series (100,
95, 85 and 75% for 5 min each at room temperature). For antigen
retrieval, slides were placed in 0.01 ml/l citrate buffer in a
pressure cooker on high pressure at 110–120°C for 10 min.
Endogenous peroxidase activity was blocked with 3%
H2O2 for 30 min at room temperature, and 5%
goat serum (abs933; Shanghai Universal Biotech Co., Ltd.) was used
for non-specific antibody blocking for 30 min at room temperature.
Rabbit anti-human CIN85 monoclonal antibody (1:50; cat. no.
ab151574; Abcam) was used overnight at 4°C. Next, an
anti-mouse/rabbit ready-to-use kit(cat. no. abs957; Shanghai
Universal Biotech Co., Ltd.) and DAB kit (cat. no. 8059S; Shanghai
Universal Biotech Co., Ltd.) were used according to the
manufacturer's instructions. Finally, the slides were
counterstained for 4 min with hematoxylin at room temperature.
All tissue slides were examined by two independent
pathologists. The immunohistochemical results for the CIN85 protein
were recorded by analyzing the average cells in five randomly
selected high-power fields under a light microscope (magnification,
×400). Positive cells were considered those whose cytoplasm or cell
membrane was pale yellow to brown. The scoring was system was as
follows: i) 0 points, no positive cells; ii) 1 point, <25%
positive cells; iii) 2 points, 26–50% positive cells; iv) 3 points,
51–75% positive cells; and v) 4 points, >75% positive cells.
Coloring intensity was sored as follows: i) 0 points,
colorlessness; ii) 1 point, pale yellow; iii) 2 points, brown; and
iv) 3 points, sepia. The sum of the two scores gave the final
score, with scores 0–3 being negative expression and 4–7 being
positive expression.
Immunocytochemistry assay
200-µl cell suspension (1×105/ml) was
added on coverslips in a 24-well culture plate and fixed with 4%
paraformaldehyde for 15 min at room temperature. Fixed cells were
washed three times with 1X PBS, then 5% goat serum (abs933;
Univ,China) was used for non-specific antibody blocking for 30 min
at room temperature and followed by overnight incubation with 2
µg/ml rabbit anti-human CIN85 monoclonal antibody (1:50; cat. no.
ab151574; Abcam) at 4°C. The cells were rinsed three times with PBS
and incubated at room temperature with Alexa Fluor®
488-conjugated goat anti-rabbit IgG secondary polyclonal antibody
(1:1,500; cat. no. 150077; Abcam) for 1 h. The cells were rinsed
three times with PBS, then counterstained with Fluoroshield™
containing DAPI Staining Solution (cat. no. C1005; Beyotime
Institute of Biotechnology). The slides were examined under an
immunofluorescence microscope (Olympus FV-100) at ×200
magnification.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted using the miRNeasy Mini kit
(Qiagen AB), according to the manufacturer's instructions. cDNA was
synthesized using 1 µg extracted mRNA as the template with a
GoScript™ Reverse Transcription kit (Promega Corporation),
according to the manufacturer's instructions. Primer sequences for
CIN85 and GAPDH were as follows: CIN85 sense,
5′-ACGATCAGCGTGGGTGAAAT-3′ and antisense,
5′-CGCTCGCCTCTCTTATTGGT-3′; GAPDH sense,
5′-AAGAGCACAAGAGGAAGAGAGAGAC-3′ and antisense,
5′-GTCTACATGGCAACTGTGAGGAG-3′. mRNA expression levels were
quantified using the RT2 SYBR Green qPCR Master Mix
(Promega Corporation) and detected using the 7900HT Fast Real-Time
PCR system (Thermo Fisher Scientific, Inc.). The qPCR mix contained
0.4 µl of each primer, 10 µl RT2 SYBR Green qPCR Master
Mix and 2 µl cDNA. Nuclease-free water was added to achieve a final
reaction volume of 20 µl. The qPCR reaction condition was set to
95°C for 2 min, followed by 40 cycles of 95°C elongation for 15 sec
and 60°C for 1 min each. A melting curve was then calculated for
each PCR product to confirm synthesis specificity (15).
Protein extraction and western
blotting
Cells were harvested and lysed using lysis buffer
(Cell Signaling Technology, Inc.) with 1 mM PMSF (Beyotime
Institute of Biotechnology). The concentration of total protein was
measured using the Pierce™ BCA Protein Assay kit (Pierce; Thermo
Fisher Scientific, Inc.), according to the manufacturer's
instructions. Protein aliquots (20 µg) were loaded with SDS buffer
(Beyotime Institute of Biotechnology) and boiled at 95°C for 10
min. The denatured protein samples were then subjected to western
blotting. Same amounts of protein samples were isolated by 12%
SDS-PAGE gels, then transferred onto PVDF membranes. The membranes
were then blocked with TBS-T containing 5% non-fat milk powder for
2 h at room temperature and incubated with primary antibodies at
4°C overnight. The membranes were then incubated with secondary
antibody for 1 h at room temperature. Rabbit anti-human CIN85
monoclonal (1:200; cat. no. ab151574; Abcam) and anti-human GAPDH
monoclonal (1:500; cat. no. ab181602; Abcam) primary antibodies
were used. Then, membranes were incubated with a HRP-conjugated
goat anti-rabbit IgG polyclonal secondary antibody (1:500; cat. no.
7074; Cell Signaling Technology, Inc.). Antibodies were all diluted
according to the manufacturer's instructions. Western blotting was
performed as previously described (16). The grayscale values of the resulting
bands were measured using ImageJ (version 1.46r) software (National
Institutes of Health).
Lentivirus construction and cell
transduction
Lentiviral vectors were cloned and packaged with the
GV248 plasmid, which contained a target interfering short hairpin
RNA (shRNA), Hepler1.0 and Helper2.0. The shRNA targeting human
CIN85 (5′-AAGACCAGAAATGCTTCCAAA-3′) and the negative control shRNA
(5′-TTCTCCGAACGTGTCACGT-3′) were designed, synthesized and inserted
into the GV248 plasmid by Shanghai GeneChem Co. Ltd. The
second-generation system was used. 293T cells were purchased from
the American Type Culture Collection and were used as the interim
cell line. The mass of lentiviral plasmid used was 20 µg and the
ratio of the lentiviral plasmid, packaging vector and envelope
vector was 4:3:2. After 15 days of the lentivirus construction,
when the cells grew to 70–80% confluence, TE1 cells were infected
with different titers (5×108 U/ml, 6×108 U/ml
and 1×109 U/ml) of the virus in the presence of 5 µg/ml
Enhanced Infection Solution and polybrene (Shanghai GeneChem Co.,
Ltd.). The medium was changed 12 h later. After 48 h of cell
transduction, the most suitable multiplicity of infection (70%) was
determined by observing the minimum lentivirus and the relatively
largest number of fluorescent cells. Puromycin (Merck KGaA) was
used at 1 µg/ml to screen for TE1 cells that were successfully
transduced.
MTS assay
An MTS assay (Promega Corporation) was performed to
assess the cell viability of TE1 cells, according to the
manufacturer's instructions. In brief, logarithmic growth phase
cells were suspended and diluted to 1×104/ml with medium
and inoculated at 200 µl per well in 96-well plates. Cell viability
was then measured by detecting the absorbance at 490 nm on days 0,
1, 2, 3, 4 and 5. Each experiment was conducted in triplicate.
Clone formation assay
A total of 200 cells per well were inoculated into
6-well plates and allowed to grow under conditions of 37°C with 5%
CO2 for 2 weeks. The medium was changed every other day.
The clones (≥50 cells) were counted under a light microscope
(magnification, ×400) after fixation with 4% paraformaldehyde for
15 min and staining with crystal violet solution for 5 min at room
temperature.
Wound healing assay
When cells grew to 80–90% confluence
(~1×106 cells per well) in 6-well plates, sterilized
1-ml tips were used to generate wounding across the cell monolayer,
and the debris was washed with PBS. The medium was then replaced
with serum-free medium. Cells were observed and photographed in
three fields randomly selected from each well under an inverted
microscope (magnification, ×200) at 0, 12 and 24 h. Wound areas
were measured at 0 and 24 h using ImageJ (version1.46r; National
Institutes of Health). The migration rate was then calculated as
(wound area at 24 h migration/wound area at 0 h. Each experiment
was conducted in triplicate.
Transwell invasion assay
Cell suspension was prepared at a density of
1×105/ml in a serum-free medium. A 200-µl cell
suspension was added to the upper Transwell chamber (Corning, Inc.)
with an insert coated with Matrigel™ (1:8; BD Biosciences) for 30
min at 37°C for the invasion assay. Complete medium (500 µl)
containing 10% fetal bovine serum was added to the lower chamber.
After 24 h of cultivation at 37°C, the Matrigel and cells on the
upper chamber were gently wiped off with a cotton swab. The
Transwell membranes were cut off using a surgical blade and stained
with crystal violate dye for 5 min at room temperature. For
quantification, the integral optical density (IOD) of the two
groups was measured under a light microscope under low
magnification (magnification, ×4).
Statistical analysis
Statistical analysis was performed with SPSS
software 24.0 (IBM Corp.) and GraphPad Prism 6.0 software (GraphPad
Software, Inc.). The differential expression of CIN85 and its
association with clinicopathological factors was analyzed by
χ2 and Fisher's exact tests. Quantitative data are
expressed as the mean ± SD and were analyzed by Student's t-test.
The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/) was screened using the
‘esophageal carcinoma’ and ‘CIN85/SH3KBP1’ search terms. The
patients with upper 25% CIN85 expression were considered as the
high-expression group. Kaplan-Meier survival curves were
constructed using The Biomedical Informatics Institute OSescc tool
(bioinfo.henu.edu.cn/DBList.jsp) (17) and hazard ratio (HR) and log-rank
P-value were calculated. P<0.05 was considered to indicate a
statistically significant difference.
Results
CIN85 is differentially expressed in
ESCC and non-neoplastic esophageal tissue
A total of 129 patients with ESCC were included in
the present study. In 54 of the adjacent normal tissue specimens,
CIN85 was found to be positively expressed in the basal cells of
the esophageal epithelium in the adjacent normal tissue specimens.
The remaining 75 normal samples showed negative expression of
CIN85. Among the cancer specimens, 93 were positive and 36 negative
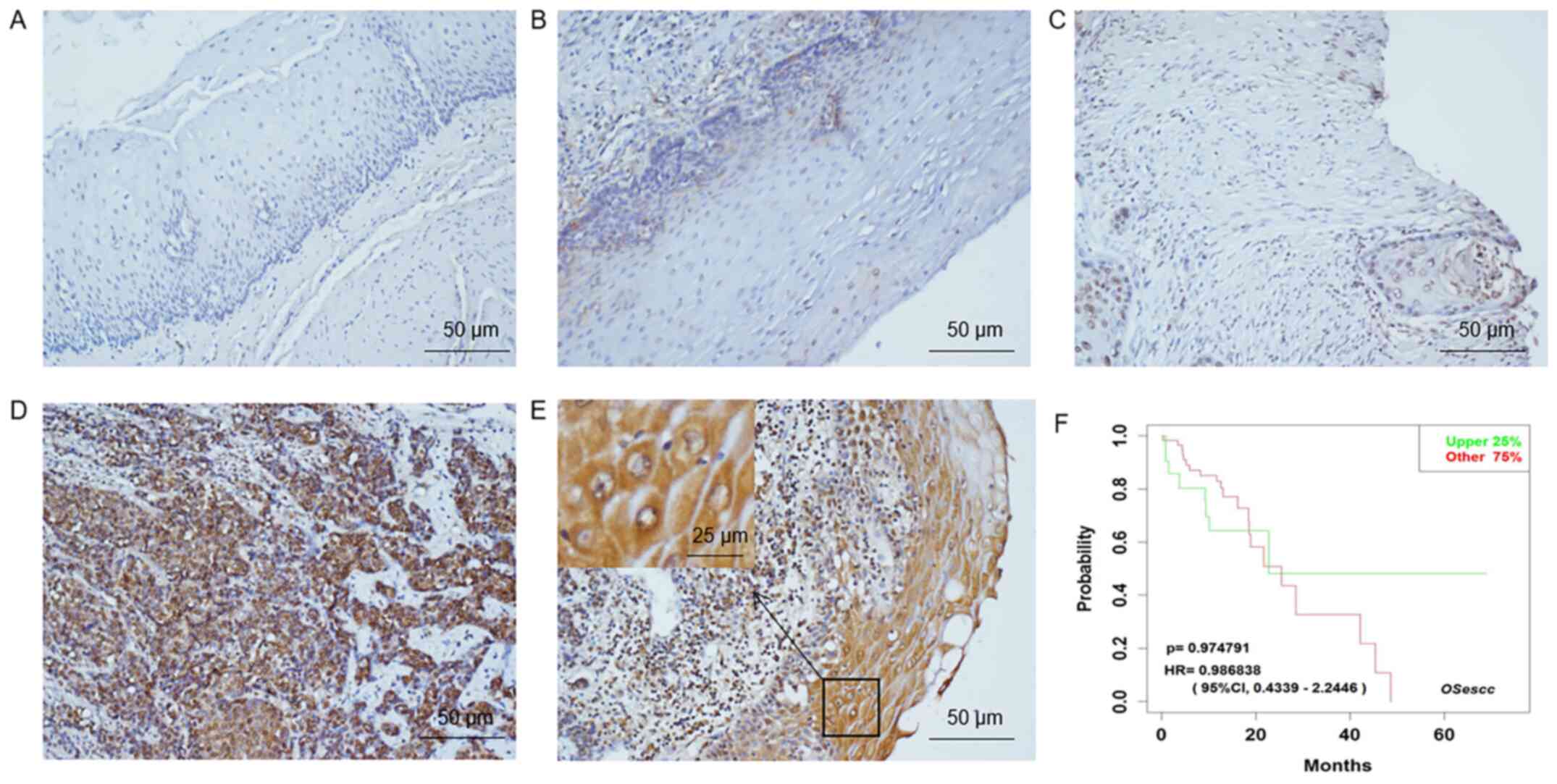
(Fig. 1A-E), corresponding to a
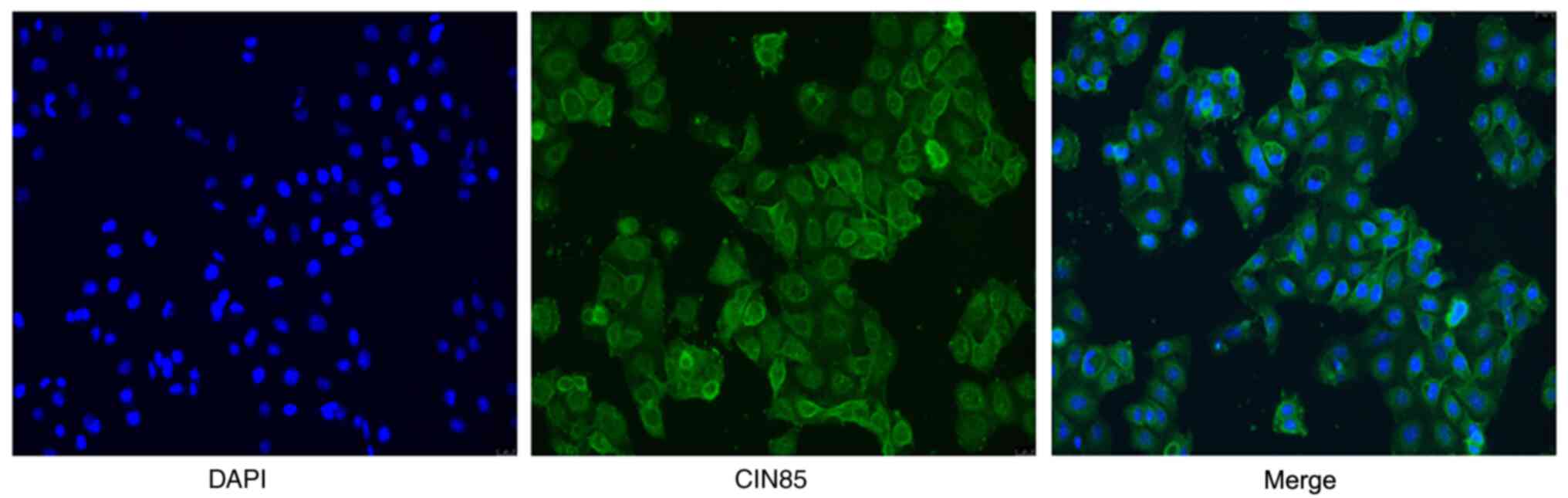
positive rate of 72.1%. CIN85 was primarily expressed in the cell
membrane and cytoplasm (Fig.
2).
Increase in CIN85 expression is
associated with advanced tumor stage and metastatic disease
To evaluate the potential role of CIN85 in ESCC,
TCGA was searched, and a total of 29 patients were included. The
OSescc tool for ESCC prognosis analysis was used to plot a
Kaplan-Meier survival curve (Fig.
1F). There was no significant difference in the overall
survival rate between patients with low and high CIN85 expression.
Table I summarizes the association
between the expression of CIN85 and the clinicopathological
features of patients with ESCC. It was found that CIN85 was highly
expressed in patients with advanced TNM stage (P=0.002) and those
with lymph node metastasis (P=0.001) (Table I).
Transfection and verification of
CIN85-specific interfering RNA
In order to study the effect of CIN85 on the
biological behavior of ESCC cells, the TE1 cell line was infected
with a virus containing a specific shRNA and a negative control
virus, respectively designated as TE1-KD and TE1-C cells. Fig. S1 demonstrates that a high frequency
of green fluorescent ESCC cells was observed 48 h after
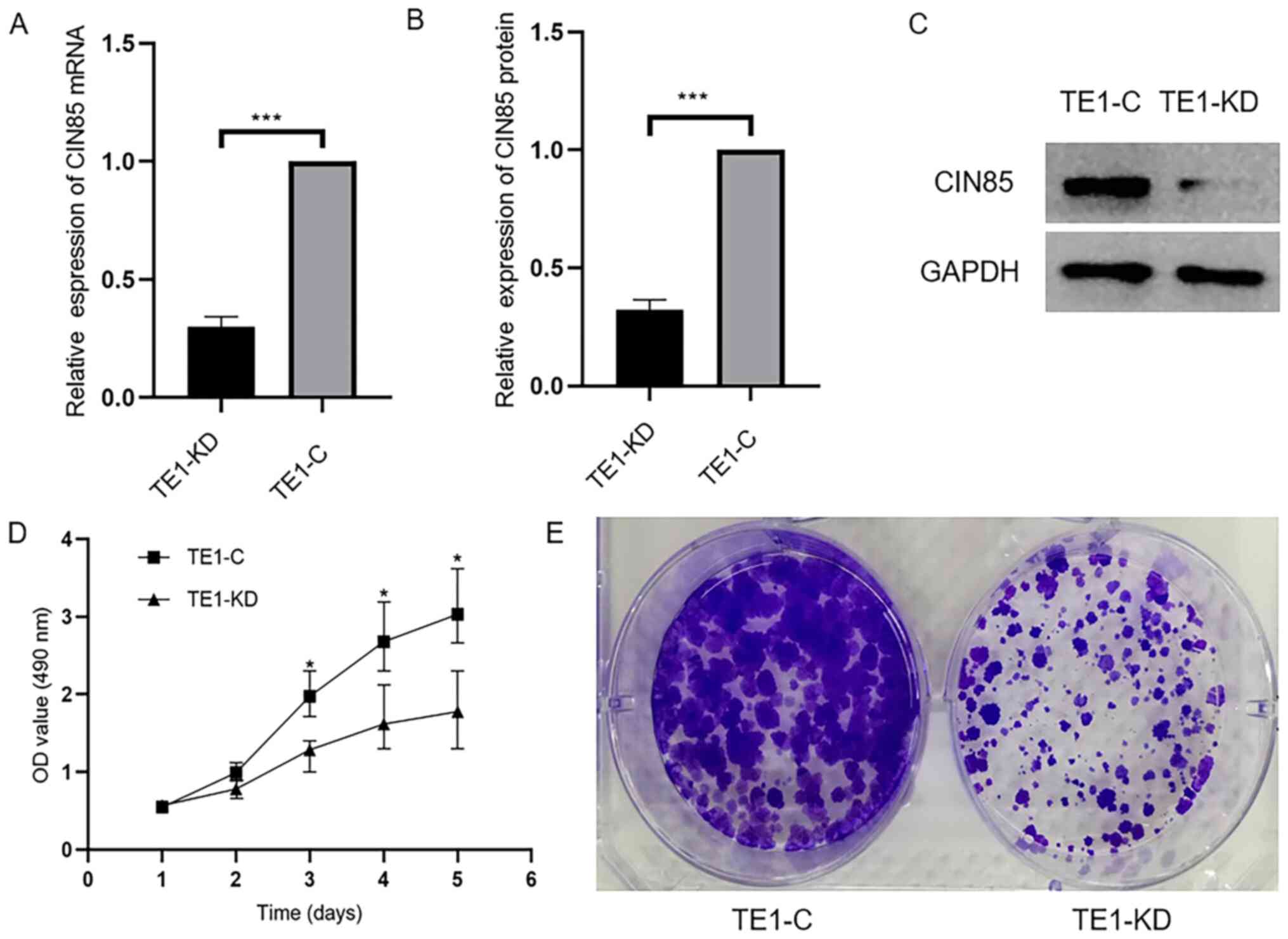
transfection, suggesting successful transfection. Next, RT-qPCR and
western blotting were performed to detect transfection efficiency.
The mRNA and protein expression levels of CIN85 were decreased
significantly in TE1-KD cells, as compared with the controls
(Fig. 3A-C).
Downregulation of CIN85 can inhibit
cell proliferation
After the successful construction of the TE1-KD cell
and TE1-C cell group, MTS and monoclonal formation assays were
conducted. It was found that the proliferation of ESCC cells was
significantly decreased following the downregulation of CIN85
(Fig. 3D and E).
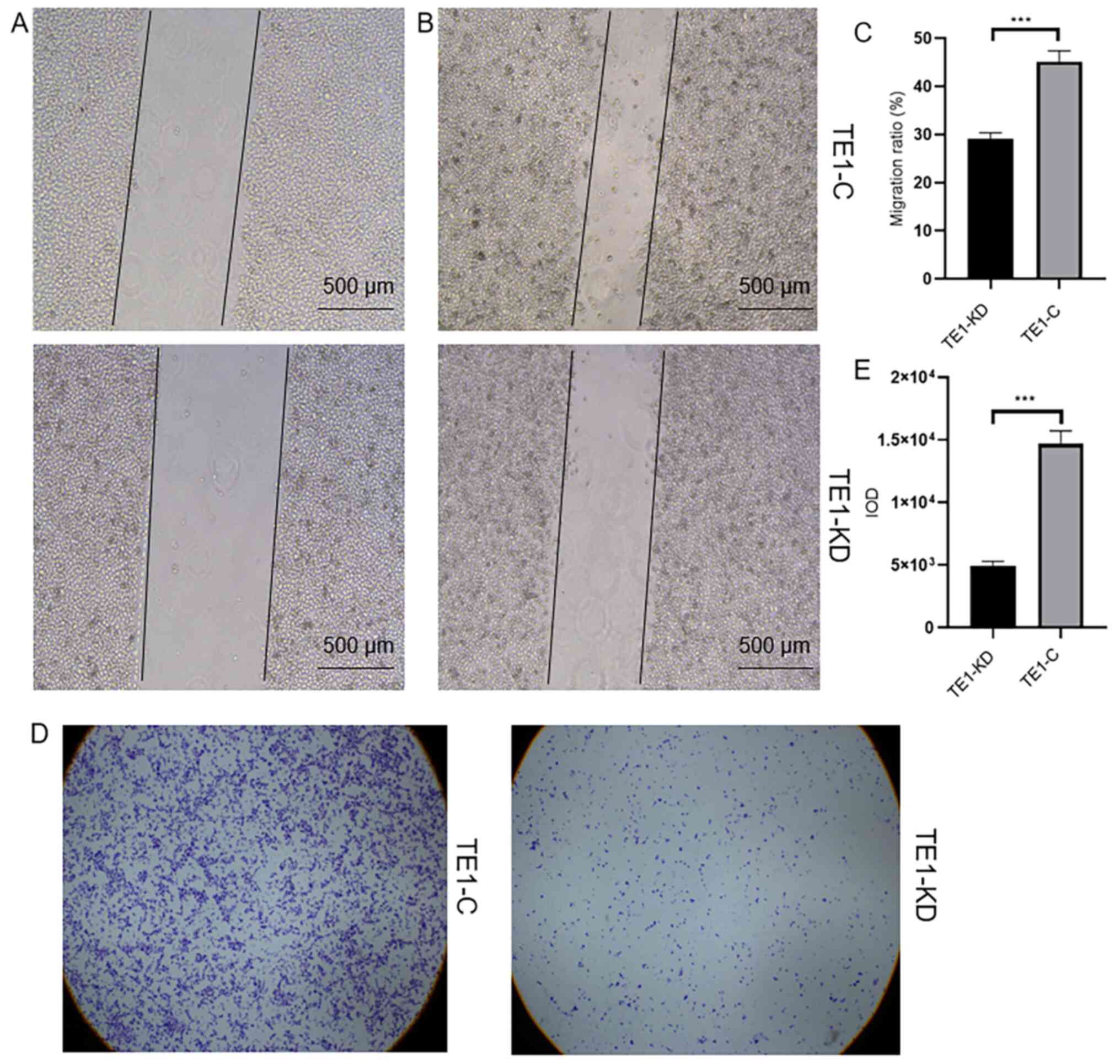
Downregulation of CIN85 can inhibit
cell invasion
In an attempt to explore the effect of CIN85 on TE1
cell invasion, the migration ratio and IOD were assessed by wound
healing and Transwell assays, respectively. Migration ratio and IOD
in the TE1-KD group was significantly decreased, as compared with
that in the control group (TE1-KD 29.06±1.315 vs. TE1-C
45.14±2.275, P<0.001; TE1-KD 4,866±320.8 vs. TE1-C 14,671±2,039,
P<0.001; Fig. 4), suggesting
that the downregulation of CIN85 can inhibit cell migration and
invasion.
Discussion
Esophageal cancer is one of the most common
malignant tumors of the digestive tract. ESCC is the main
pathological type of esophageal cancer in China (2). At present, the primary treatment of
esophageal cancer is a comprehensive treatment that is based on
surgical treatment, combined with chemotherapy, radiotherapy,
chemoradiotherapy or immunotherapy (3). Although the cure rate of early
esophageal cancer through endoscopic surgery has reached >90%,
early esophageal cancer was diagnosed less frequently (4). Additionally, the poor 5-year survival
rate and high recurrence of advanced esophageal cancer remains a
problem (18). With the
introduction and development of individualized treatment, further
studies are required to identify effective therapeutic targets for
esophageal cancer. EGFR has been found to be highly expressed in
42.5–85.7% of ESCC cases and is closely associated with the
recurrence and poor prognosis of esophageal cancer (19). Tyrosine kinase receptor inhibitors
have been used in combination with chemotherapeutic drugs, and
inhibitors of vascular EGFR in combination with chemotherapy for
the treatment of esophageal cancer; however, the two aforementioned
agents have not achieved outstanding results (20). The mechanism of resistance among
them is not yet clear, so knowledge of the precise expression
pattern and characterization of esophageal cancer is required to
further explore novel therapeutic targets. The CIN85 gene is
located on the distal end of the human X chromosome and was first
discovered in human gliomas (21).
Previous studies have found that there are >100 proteins that
interact with the CIN85 adaptor protein, of which the tyrosine
receptor kinase is a major class (22–24).
CIN85 accomplishes downstream signal transduction by mediating
tyrosine receptor kinase endocytosis and the trafficking of its
vesicles. Abnormal endocytosis and transport cause pathological
changes in cells (25). Other
studies have demonstrated that CIN85 is involved in the formation
of cell membranes and the cytoskeleton, and is associated with the
remodeling of the cytoskeleton, thereby promoting the invasion and
metastasis of tumor cells (26,27).
Nam et al (28) demonstrated
that the CIN85 complex is a component of the invasive machinery of
pseudotumor breast cancer cells, and is directly linked to
malignant behavior. Another study revealed that the CIN85 adaptor
protein could be directly associated with the proto-oncogene H-ras
(29).
A number of studies have found that CIN85 is highly
expressed in colon and breast cancer, oral squamous cell carcinoma,
glioma, melanoma, and other cancer tissues (11–13,30),
but at present no study has investigated the role of CIN85 in ESCC.
The present study found that CIN85 was highly expressed in patients
with advanced TNM stage and those with lymph node metastasis,
suggesting a poor prognosis, which was consistent with other
previous studies. On this basis, the present study further
constructed CIN85-knockdown cell lines and found that the
proliferation, migration and invasion of ESCC cells were
significantly inhibited in the CIN85 knockdown group. This
indicated that CIN85 could promote the proliferation and metastasis
of ESCC cells, and was directly associated with malignant
behaviors, such as tumor recurrence, and as a consequence, affects
the prognosis of patients.
However, the major limitation of this research was
that the effects of CIN85 on the proliferative and migratory
activities of ESCC cells were only confirmed in one ESCC cell line.
Cell experiments were actually conducted with three cell lines,
Kyse 30, Kyse 350 and TE 1, and two knockdown sequences were
designed for each cell type. However, the knockdown of CIN85
expression failed in the Kyse 30 (Fig.
S2) and Kyse 350 (data not shown) cell lines, so only one cell
line was used in the subsequent studies. It was speculated that the
different cell types caused the failure. Although the three cell
lines are all ESCC cell lines, TE1 is a low differentiated ESCC
cell line, whereas Kyse 350 is medium differentiated and Kyse 30 is
high differentiated. To the best of our knowledge, this is the
first report to investigate the abnormal expression of CIN85 in
ESCC, so further research in this field are encouraged to
demonstrate that the present results are not accidental.
CIN85 plays a role in multiple tumors, mainly
through its N-terminal SH3 domain interacting with other proteins,
so the SH3 domain may be the most promising research target
(31). At present, there have been
numerous signs of progress in preventing the development of tumor
cells by focusing on the SH3 domain. In particular, a previous
study by Hashimoto et al (32), used peptide ligands targeting SH3
not only in vitro but also in vivo to successfully
reduce the invasion and metastasis of breast cancer without
significant adverse events. Sato et al (33) also demonstrated that the inhibition
of the SH3 domain of CIN85 using a lysyl oxidase precursor peptide
could reduce the degradation of the surrounding matrix and decrease
the invasive and metastatic ability of breast cancer cells. These
studies indicated that the use of certain molecules to block the
SH3 domain of CIN85 can, in principle, serve as a basis for the
study of novel antitumor drugs.
In conclusion, the present study provided possible
target genes for basic and clinical studies of ESCC. CIN85 is
closely associated with the growth and migration of ESCC and may be
an effective target for the treatment of esophageal cancer.
However, the occurrence and development of tumors is a multi-factor
and multi-stage process. Therefore, the specific underlying
mechanism of CIN85 involved in the occurrence and development of
esophageal cancer requires further study.
Supplementary Material
Supporting Data
Acknowledgements
The authors gratefully acknowledge the contribution
of Dr Si-Yuan Dong (Department of Thoracic Surgery, The First
Hospital of China Medical University) in data extraction and
software input.
Funding
The present research was supported by grants from
the Natural Science Foundation of China (grant no. 81201890) and
Research Foundation of Education Bureau of Liaoning Province, China
(grant no. LK201614).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SGZ and XYH designed the research. XYH, XXB and XC
performed the research and analyzed the results. SGZ and XYH wrote
the paper. XXB and XC edited the manuscript and provided critical
comments. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Ethics Committee of The First Affiliated Hospital of China Medical
University (Shenyang, China). Patients provided written informed
consent.
Patient consent for publication
All patients provided written informed consent for
the publication of any associated data and accompanying images.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Smyth EC, Lagergren J, Fitzgerald RC,
Lordick F, Shah MA, Lagergren P and Cunningham D: Oesophageal
cancer. Nat Rev Dis Prim. 3:170482017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Murphy G, McCormack V, Abedi-Ardekani B,
Arnold M, Camargo MC, Dar NA, Dawsey SM, Etemadi A, Fitzgerald RC,
Fleischer DE, et al: International cancer seminars: A focus on
esophageal squamous cell carcinoma. Ann Oncol. 28:2086–2093. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Borggreve AS, Kingma BF, Domrachev SA,
Koshkin MA, Ruurda JP, van Hillegersberg R, Takeda FR and Goense L:
Surgical treatment of esophageal cancer in the era of multimodality
management. Ann N Y Acad Sci. 1434:192–209. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Saeki H, Nakashima Y, Zaitsu Y, Tsuda Y,
Kasagi Y, Ando K, Imamura Y, Ohgaki K, Ito S, Kimura Y, et al:
Current status of and perspectives regarding neoadjuvant
chemoradiotherapy for locally advanced esophageal squamous cell
carcinoma. Surg Today. 46:261–267. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
van Workum F, Berkelmans GH, Klarenbeek
BR, Nieuwenhuijzen GAP, Luyer MDP and Rosman C: McKeown or Ivor
Lewis totally minimally invasive esophagectomy for cancer of the
esophagus and gastroesophageal junction: Systematic review and
meta-analysis. J Thorac Dis. 9 (Suppl 8):S826–S833. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yibulayin W, Abulizi S, Lv H and Sun W:
Minimally invasive oesophagectomy versus open esophagectomy for
resectable esophageal cancer: A meta-analysis. World J Surg Oncol.
14:3042016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kurakin AV, Wu S and Bredesen DE: Atypical
recognition consensus of CIN85/SETA/Ruk SH3 domains revealed by
target-assisted iterative screening. J Biol Chem. 278:34102–34109.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kowanetz K, Husnjak K, Höller D, Kowanetz
M, Soubeyran P, Hirsch D, Schmidt MHH, Pavelic K, De Camilli P,
Randazzo PA, et al: CIN85 associates with multiple effectors
controlling intracellular trafficking of epidermal growth factor
receptors. Mol Biol Cell. 15:3155–3166. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Havrylov S, Redowicz MJ and Buchman VL:
Emerging roles of Ruk/CIN85 in vesicle-mediated transport,
adhesion, migration and malignancy. Traffic. 11:721–731. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gout I, Middleton G, Adu J, Ninkina NN,
Drobot LB, Filonenko V, Matsuka G, Davies AM, Waterfield M and
Buchman VL: Negative regulation of PI 3-kinase by Ruk, a novel
adaptor protein. EMBO J. 19:4015–4025. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cascio S and Finn OJ: Complex of MUC1,
CIN85 and Cbl in colon cancer progression and metastasis. Cancers
(Basel). 7:342–352. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wakasaki T, Masuda M, Niiro H,
Jabbarzadeh-Tabrizi S, Noda K, Taniyama T, Komune S and Akashi K: A
critical role of c-Cbl-interacting protein of 85 kDa in the
development and progression of head and neck squamous cell
carcinomas through the ras-ERK pathway. Neoplasia. 12:789–796.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cascio S, Farkas AM, Hughey RP and Finn
OJ: Altered glycosylation of MUC1 influences its association with
CIN85: The role of this novel complex in cancer cell invasion and
migration. Oncotarget. 4:1686–1697. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rice TW, Ishwaran H, Hofstetter WL, Kelsen
DP, Apperson-Hansen C and Blackstone EH; Worldwide Esophageal
Cancer Collaboration Investigators, : Recommendations for
pathologic staging (pTNM) of cancer of the esophagus and
esophagogastric junction for the 8th edition AJCC/UICC staging
manuals. Dis Esophagus. 29:897–905. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Camacho Londoño J and Philipp SE: A
reliable method for quantification of splice variants using
RT-qPCR. BMC Mol Biol. 17:82016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hnasko TS and Hnasko RM: The Western Blot.
Methods Mol Biol. 1318:87–96. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jeong DY, Lee KS, Choi JY, Chung MJ, Min
YW, Kim HK, Zo JI, Shim YM and Sun JM: Surgically Resected
Esophageal Squamous Cell Carcinoma: Patient Survival and
Clinicopathological Prognostic Factors. Sci Rep. 10:50772020.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Q, Wang F, Lv J, Xin J, Xie L, Zhu W,
Tang Y, Li Y, Zhao X, Wang Y, et al: Interactive online consensus
survival tool for esophageal squamous cell carcinoma prognosis
analysis. Oncol Lett. 18:1199–1206. 2019.PubMed/NCBI
|
|
19
|
Jiang D, Li X, Wang H, Shi Y, Xu C, Lu S,
Huang J, Xu Y, Zeng H, Su J, Hou Y and Tan L: The prognostic value
of EGFR overexpression and amplification in Esophageal squamous
cell Carcinoma. BMC Cancer. 15:3772015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Luo N, Zhao LC, Shi QQ, Feng ZQ, Chen DL
and Li J: Induction of Apoptosis in Human Leukemic Cell Lines by
Diallyl Disulfide via Modulation of EGFR/ERK/PKM2 Signaling
Pathways. Asian Pac J Cancer Prev. 16:3509–3515. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Take H, Watanabe S, Takeda K, Yu ZX, Iwata
N and Kajigaya S: Cloning and characterization of a novel adaptor
protein, CIN85, that interacts with c-Cbl. Biochem Biophys Res
Commun. 268:321–328. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Watanabe S, Take H, Takeda K, Yu ZX, Iwata
N and Kajigaya S: Characterization of the CIN85 adaptor protein and
identification of components involved in CIN85 complexes. Biochem
Biophys Res Commun. 278:167–174. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Niiro H, Jabbarzadeh-Tabrizi S, Kikushige
Y, Shima T, Noda K, Ota S, Tsuzuki H, Inoue Y, Arinobu Y, Iwasaki
H, et al: CIN85 is required for Cbl-mediated regulation of antigen
receptor signaling in human B cells. Blood. 119:2263–2273. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Brett TJ1, Traub LM and Fremont DH:
Accessory protein recruitment motifs in clathrin-mediated
endocytosis. Structure. 10:797–809. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Legendre-Guillemin V, Wasiak S, Hussain
NK, Angers A and McPherson PS: ENTH/ANTH proteins and
clathrin-mediated membrane budding. J Cell Sci. 117:9–18. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Donaldson JG and Jackson CL: ARF family G
proteins and their regulators: Roles in membrane transport,
development and disease. Nat Rev Mol Cell Biol. 12:362–375. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lanier MH, McConnell P and Cooper JA: Cell
Migration and Invadopodia Formation Require a Membrane-binding
Domain of CARMIL2. J Biol Chem. 291:1076–1091. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nam JM, Onodera Y, Mazaki Y, Miyoshi H,
Hashimoto S and Sabe H: CIN85, a Cbl-interacting protein, is a
component of AMAP1-mediated breast cancer invasion machinery. EMBO
J. 26:647–656. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lito P, Mets BD, Kleff S, O'Reilly S,
Maher VM and McCormick JJ: Evidence that sprouty 2 is necessary for
sarcoma formation by H-Ras oncogene-transformed human fibroblasts.
J Biol Chem. 283:2002–2009. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Samoylenko A, Vynnytska-Myronovska B, Byts
N, Kozlova N, Basaraba O, Pasichnyk G, Palyvoda K, Bobak Y, Barska
M, Mayevska O, et al: Increased levels of the HER1 adaptor protein
Rukl/CIN85 contribute to breast cancer malignancy. Carcinogenesis.
33:1976–1984. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bögler O, Furnari FB, Kindler-Roehrborn A,
Sykes VW, Yung R, Huang HJ and Cavenee WK: SETA: A novel SH3
domain-containing adapter molecule associated with malignancy in
astrocytes. Neuro-oncol. 2:6–15. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hashimoto S, Hirose M, Hashimoto A,
Morishige M, Yamada A, Hosaka H, Akagi K, Ogawa E, Oneyama C,
Agatsuma T, et al: Targeting AMAP1 and cortactin binding bearing an
atypical src homology 3/proline interface for prevention of breast
cancer invasion and metastasis. Proc Natl Acad Sci USA.
103:7036–7041. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sato S, Zhao Y, Imai M, Simister PC,
Feller SM, Trackman PC, Kirsch KH and Sonenshein GE: Inhibition of
CIN85-mediated invasion by a novel SH3 domain binding motif in the
lysyl oxidase propeptide. PLoS One. 8:772882013. View Article : Google Scholar
|