Introduction
Colorectal cancer (CRC) is a common digestive tract
malignancy, which ranks third amongst cancer incidence rates and is
the second leading cause of cancer-related death worldwide. CRC
accounts for >6.1% of all malignant incidences, with ~1.8
million new CRC cases diagnosed worldwide in 2018 (1). Despite the significant progress made
in CRC treatment, including in chemotherapy and surgical
techniques, patients with distant metastases have a poor prognosis.
Indeed, the 5-year survival rate is <10% for patients with
distant metastases (2). Tumor
metastasis is the primary cause of poor prognosis in patients with
CRC (3). Thus, understanding the
underlying molecular mechanisms associated with CRC metastasis is
an urgent requirement.
Long non-coding RNAs (lncRNAs) are
non-protein-encoding RNAs >200 nucleotides in length (4). A previous study indicated that lncRNAs
participate in the regulation of gene expression and serve
functional roles both in physiological and pathological processes
(5). Furthermore, aberrant
expression of lncRNAs has been implicated in tumorigenesis and the
development of hepatocellular carcinoma and pancreatic ductal
adenocarcinoma (6,7). For example, lncRNA SNHG6 has been
reported to promote the migration, invasion and
epithelial-mesenchymal transition of CRC cells through the microRNA
(miRNA/miR)-26a/EZH2 axis, thereby acting as an oncogene (8). Zhan et al (9) reported that the overexpression of
lncRNA ROR promoted the invasion and metastasis of pancreatic
cancer by upregulating ZEB1 expression levels.
Kong et al (10) demonstrated that the recently
identified lncRNA ZFPM2-AS1, located on human chromosome 8, was
upregulated in gastric cancer tissue and that ZFPM2-AS1
overexpression promoted the proliferation and inhibited the
apoptosis of gastric cancer cells by inhibiting the p53 pathway.
Furthermore, Xue et al (11)
indicated that ZFPM2-AS1 played an oncogenic role in lung
adenocarcinoma progression through miR-18b-5p and VMA21. However,
whether ZFPM2-AS1 is expressed in CRC tissue or plays a biological
role in CRC progression remains unknown.
The aim of the present study was to elucidate the
function of ZFPM2-AS1 in the development of CRC. This study
demonstrated that ZFPM2-AS1 expression was upregulated in CRC
tissues compared with normal tissue, and that high ZFPM2-AS1
expression was associated with poor prognosis for patients with
CRC. Moreover, ZFPM2-AS1 knockdown inhibited the proliferation,
migration and invasion of CRC cells in vitro. Prediction of
target genes, luciferase reporter and rescue experiment assays
indicated that miR-137 was a target of ZFPM2-AS1, and tripartite
motif containing 24 (TRIM24) was identified as a target of miR-137.
Mechanistic studies further clarified that ZFPM2-AS1 served as a
competing endogenous RNA (ceRNA) that upregulates TRIM24 expression
by sponging miR-137 in CRC.
Materials and methods
Patient tissues
Human CRC tissue samples and the matched normal
tissue (n=58 pairs) were collected from The First Affiliated
Hospital of the University of South China between May 2016 and
March 2019. The inclusion criteria included: Patients who were
diagnosed with CRC by a pathologist and underwent radical surgery.
The exclusion criteria included the following: Patients had
received radiotherapy or chemotherapy prior to the operation.
Written informed consent was obtained from all patients. The
present study was approved by the Medical Ethics Committees of The
First Affiliated Hospital of University of South China.
Cell lines and culture
The human CRC cell lines (SW480, SW620, HT-29,
Caco-2 and HCT116) and the normal human colorectal mucosa cell line
FHC were purchased from The Cell Bank of Type Culture Collection of
the Chinese Academy of Sciences. All cells were cultured in
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.),
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.)
and 1% penicillin-streptomycin (Beijing Solarbio Science &
Technology Co., Ltd.) in an incubator at 37°C with 5%
CO2.
Cell transfection
To stably knockdown the expression of ZFPM2-AS1,
three short hairpin (sh) RNA targeting ZFPM2-AS1 (sh-ZFPM2-AS1-1,
5′-GCGACCTAGGATCGAACCGAT-3′; sh-ZFPM2-AS1-2,
5′-AGCTGAATTCGAGCCAATTGCGT-3′; sh-ZFPM2-AS1-3,
5′-GCCGGAACGGATTTCGGAGAC-3′) and sh-negative control (NC,
5′-TTCTCCGAACGTGTCACGT-3′) were synthesized by Shanghai GenePharma
Co., Ltd. These sequences were inserted into a pLKO.1 vector
(BioSettia, Inc.). miR-137 mimics (5′-GUACUUCCUAACCCAUUGUUCA-3′),
miR-137 inhibitors (5′-AUUCUUCAGAUUUCCAGGGACU-3′), mimics NC
(5′-CAGUACUUUUGUGUAGUACAA-3′) and inhibitor NC
(5′-CAGUACUUUUGUGUAGUACAA-3′) were also obtained from Shanghai
GenePharma Co., Ltd. To construct the TRIM24 expression vector, the
full-length sequence of TRIM24 were cloned into the pcDNA3.1 vector
(pcDNA3.1-TRIM24). Before transfection, cells were sub-cultured
into 6-well plates (Corning, Inc.) at ~50% confluence
(3×105). sh-RNAs (100 nM) or miR-137 mimics (50 nM) or
miR-137 inhibitor (150 nM) or plasmids (1.5 µg per well) were
transfected into SW480 and HCT116 cells. Cell transfection was
performed using Lipofectamine® LTX (Invitrogen; Thermo
Fisher Scientific, Inc.). After incubation for 48 h, cells were
collected for subsequent experiments.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA from CRC tissue, normal tissue, SW480 and
HCT116 cells were extracted using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). To determine mRNA and
lncRNA expression levels, the extracted RNA was reverse transcribed
into cDNA using a PrimeScript RT reagent kit (Takara Bio, Inc.).
Reverse transcription protocol consisted of 37°C for 15 min,
followed by 85°C for 5 sec. To determine miRNA expression levels,
cDNA was synthesized using the qScript microRNA cDNA Synthesis kit
(Guangzhou RiboBio Co., Ltd.). The thermocycling conditions were as
follows: 94°C for 3 min, followed by 30 cycles of 94°C for 30 sec,
55°C for 30 sec and 72°C for 60 sec, and a final extension at 72°C
for 5 min. RT-qPCR was subsequently performed using SYBR-Green
(Takara Bio, Inc.), GAPDH and U6 were used as the internal loading
controls for lncRNA, mRNA and miRNA, respectively. The relative
expression of these genes was determined using the
2−ΔΔCq method (12). The
primers used for the RT-qPCR are presented in Table I.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Gene | Primer sequences
(5→3) |
|---|
| ZFPM2-AS1 | F:
TTTCCTACAATGAATCCACCAG |
|
| R:
TTTGAGCCACTCTTTGAGG |
| miR-137 | F:
UUAUUGCUUAAGAAUACGCGUAG |
|
| R:
GTCGTATCCAGTGCAGGGTCCGAGGT |
| U6 | F:
CTCGCTTCGGCAGCACA |
|
| R:
AACGCTTCACGAATTTGCGT |
| TRIM24 | F:
CGCCACCCAAGTTGGAGT |
|
| R:
GCTGGGAACCTCAGTAGTGTCCT |
| GAPDH | F:
GCACCGTCAAGGCTGAGAAC |
|
| R:
GCCTTCTCCATGGTGGTGAA |
Cell proliferation assay
The proliferative capacity of SW480 and HCT116 cells
were determined using MTS and colony formation assays. For the MTS
assay, 2×103 cells/well were seeded into 96-well plates
and cultured for 0, 24, 48, 72, 96 and 120 h. Following the
incubation, 10 µl MTS solution/well was added for 2 h, and then
dimethyl sulfoxide was used to dissolve formazan. Finally, the
absorbance at 450 nm was measured using a microplate reader
(Bio-Rad Laboratories, Inc.).
For the colony formation assay, 800 cells/well were
seeded into 6-well plates and incubated at 37°C in 5%
CO2 for 14 days. Following the incubation, the colonies
were fixed with methanol at room temperature for 10 min and stained
with 0.1% crystal violet at room temperature for 10 min. The number
of colonies (a single colony was defined as cell number >50) was
counted using an inverted microscope (magnification, ×200; CKX41;
Olympus Corporation).
Transwell migration and invasion
assays
Transwell assays were performed to analyze the
migratory and invasive abilities of SW480 and HCT116 cells. For
invasion assays, the Transwell membrane was coated with Matrigel™
(BD Biosciences) and 5×104 cells/well were suspended in
200 µl serum-free medium and plated into the upper chambers. For
migration assays, cells were plated in the same conditions, but the
Transwell membrane was left uncoated. A volume of 750 µl medium
containing 10% FBS was added to the lower chamber. Following 48 h
of incubation at 37°C with 5% CO2, the cells were fixed
with methanol at room temperature for 10 min and stained with 0.1%
crystal violet at room temperature for 10 min. The stained cells
were counted under an inverted microscope (magnification, ×200;
CKX41; Olympus Corporation).
Dual-luciferase reporter assay
The potential targets of ZFPM2-AS1 or miR-137 were
predicted using miRBase 21.0 (http://www.mirbase.org/) or TargetScan 7.2 (http://www.targetscan.org/vert_72/). The
sequences of the NC, wild-type (WT) ZFPM2-AS1 (ZFPM2-AS1-WT) and
mutant (MUT) ZFPM2-AS1 (ZFPM2-AS1-MUT) were inserted into the
pmirGLO reporter vector (Shanghai GenePharma Co., Ltd.).
Subsequently, pmirGLO-NC, pmirGLO-ZFPM2-AS1-WT and
pmirGLO-ZFPM2-AS1-MUT were co-transfected into SW480 and HCT116
cells along with the miR-137 mimics or miR-NC using LTX
(Invitrogen; Thermo Fisher Scientific, Inc.).
Similarly, the sequences of NC, TRIM24-WT and
TRIM24-MUT were inserted into the pmirGLO reporter vector.
pmirGLO-NC, pmirGLO-TRIM24-WT and pmirGLO-TRIM24-MUT were then
co-transfected into SW480 and HCT116 cells along with the miR-NC or
miR-137 mimics with Lipofectamine 2000. Following 48 h of
transfection, the relative luciferase activity was analyzed using
the Dual Luciferase Reporter Assay System (Promega Corporation).
The Renilla luciferase was selected as an internal
control.
RNA immunoprecipitation (RIP)
assay
A RIP assay was conducted to determine the
interaction between ZFPM2-AS1 and miR-137 using a Magna RIP kit
(EMD Millipore). Briefly, cells were lysed in RIP lysis buffer (EMD
Millipore) containing magnetic beads (2 µg) conjugated with human
anti-Argonaute 2 (Ago2) antibody (5 µg) (cat. no. 186733; Abcam).
Input and normal IgG (EMD Millipore) were used as internal
controls. Then, RT-qPCR was performed to analyze ZFPM2-AS1 and
miR-137 expression levels.
Western blotting
Western blotting was performed according to a
previously described study (13).
Total protein was extracted with a pre-cooled RIPA lysis (cat. no.
KGP703-100; Nanjing KeyGen Biotech Co., Ltd.). Protein
concentration was detected with a BCA assay (Thermo Fisher
Scientific, Inc.). Equal amounts (60 µg/lane) of protein aliquots
were loaded on 10% gel and separated via SDS-PAGE, following which,
separated proteins were transferred to PVDF membranes (EMD
Millipore). Membranes were blocked with 5% skimmed milk at room
temperature for 2 h, and then incubated with primary antibodies at
4°C overnight. The following primary antibodies were used:
Anti-TRIM24 (1:3,000; Abcam; cat. no. 70560) and anti-GAPDH
(1:2,500; Abcam; cat. no. 9485). Subsequently, membranes were
incubated with a HRP-conjugated rabbit anti-goat IgG antibody
(1:10,000; cat. no. BA1060; Wuhan Boster Biological Technology,
Ltd.) at room temperature for 1 h. Finally, the immunoblots were
visualized with LI-COR Odyssey CLX Two-colour infrared laser
imaging system (LI-COR Biosciences). ImageJ v1.8.0 software
(National Institutes of Health) was used for densitometric
analysis.
Statistical analysis
Statistical analysis was performed using SPSS 18.0
software (SPSS Inc.) and data are presented as the mean ± SD. A
paired t-test was performed to compare expression between tumor and
healthy tissues. For other comparisons between two groups, the
unpaired t-test was performed. For all comparisons among ≥3 groups
of data, Tukey's test was performed after one-way ANOVA analysis.
The association between the clinical parameters of patients with
CRC and ZFPM2-AS1 expression levels in the CRC cohort were analyzed
using a χ2 test. Pearson's correlation was used to
analyze the expression levels of ZFPM2-AS1, miR137 and TRIM24. All
experiments were performed in triplicate. P<0.05 was considered
to indicate a statistically significant difference.
Results
ZFPM2-AS1 expression levels are
upregulated in CRC tissue and predict an unfavorable prognosis for
patients with CRC
The expression levels of ZFPM2-AS1 in 58 CRC tissue
samples and matched normal tissue were analyzed by RT-qPCR.
ZFPM2-AS1 expression levels were significantly upregulated in CRC
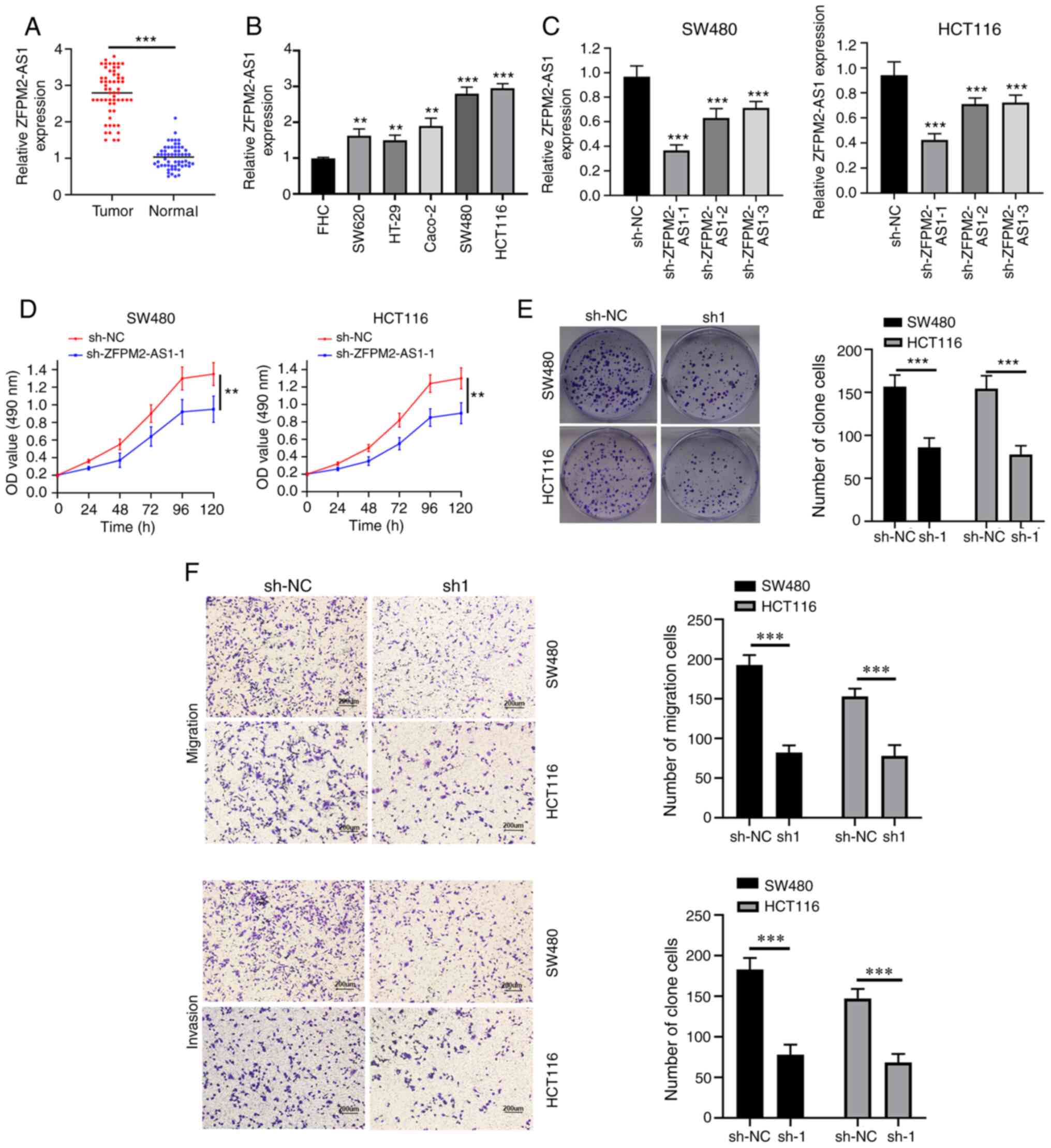
tissue, compared with healthy tissue (Fig. 1A). The expression levels of
ZFPM2-AS1 in the SW480, SW620, HT-29, Caco-2 and HCT116 human CRC
cell lines, were also significantly upregulated, compared with FHC
cells (Fig. 1B).
Subsequently, to further determine the clinical
significance of ZFPM2-AS1 expression levels in CRC, patients with
CRC were divided into a high expression group (n=29) and a low
expression group (n=29), according to the median expression levels
of ZFPM2-AS1 across all CRC tissue samples. The expression levels
of ZFPM2-AS1 were positively associated with tumor size (P=0.004),
histological differentiation (P=0.012), lymph node metastasis
(P=0.019) and TNM stage (P=0.031) (Table II). Taken together, these results
suggested that ZFPM2-AS1 could function as an oncogenic lncRNA in
CRC.
 | Table II.Association between ZFPM2-AS1
expression and clinicopathological characteristics of patients with
colorectal cancer. |
Table II.
Association between ZFPM2-AS1
expression and clinicopathological characteristics of patients with
colorectal cancer.
|
|
| ZFPM2-AS1
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Clinicopathological
characteristics | Cases, n | Low, n | High, n | χ2
value | P-value |
|---|
| Age, years |
|
<60 | 23 | 11 | 12 | 0.232 | 0.630 |
| ≥60 | 35 | 19 | 16 |
|
|
| Sex |
| Male | 31 | 15 | 16 | 0.297 | 0.586 |
|
Female | 27 | 15 | 12 |
|
|
| Tumor size, cm |
|
<5 | 28 | 20 | 8 | 8.417 | 0.004 |
| ≥5 | 30 | 10 | 20 |
|
|
|
Differentiation |
|
Good/moderate | 22 | 16 | 6 | 6.262 | 0.012 |
|
Poor | 36 | 14 | 22 |
|
|
| Lymph node
metastasis |
| No | 19 | 14 | 5 | 5.457 | 0.019 |
|
Yes | 39 | 16 | 23 |
|
|
| TNM stage |
| I +
II | 25 | 17 | 8 | 6.661 | 0.031 |
| III +
IV | 33 | 13 | 20 |
|
|
ZFPM2-AS1 knockdown inhibits the
proliferation, migration and invasion of CRC cells
Loss-of-function experiments were conducted to
investigate the function of ZFPM2-AS1 in CRC progression. SW480 and
HCT116 cells were transfected with sh-ZFPM2-AS1-1, −2 or −3
downregulate ZFPM2-AS1 expression. sh-ZFPM2-AS1-1 used in
subsequent experiments, due to its superior knockdown efficiency
(Fig. 1C), compared with the other
two shRNA candidates.
MTS and colony formation assays demonstrated that
ZFPM2-AS1-1 knockdown significantly inhibited the proliferative
ability of CRC cells (Fig. 1D and
E), compared with sh-NC. In addition, Transwell assays
indicated that the migratory and invasive abilities of CRC cells
transfected with sh-ZFPM2-AS1 were markedly decreased (Fig. 1F). Thus, ZFPM2-AS1-1 knockdown may
lead to significant inhibition of the proliferation, migration and
invasion of CRC cells.
ZFPM2-AS1 serves as a sponge for
miR-137
To further identify the underlying mechanisms of
ZFPM2-AS1 in CRC progression, miRBase was used to predict potential
target miRNAs for ZFPM2-AS1. Thus, miR-137 was identified as a
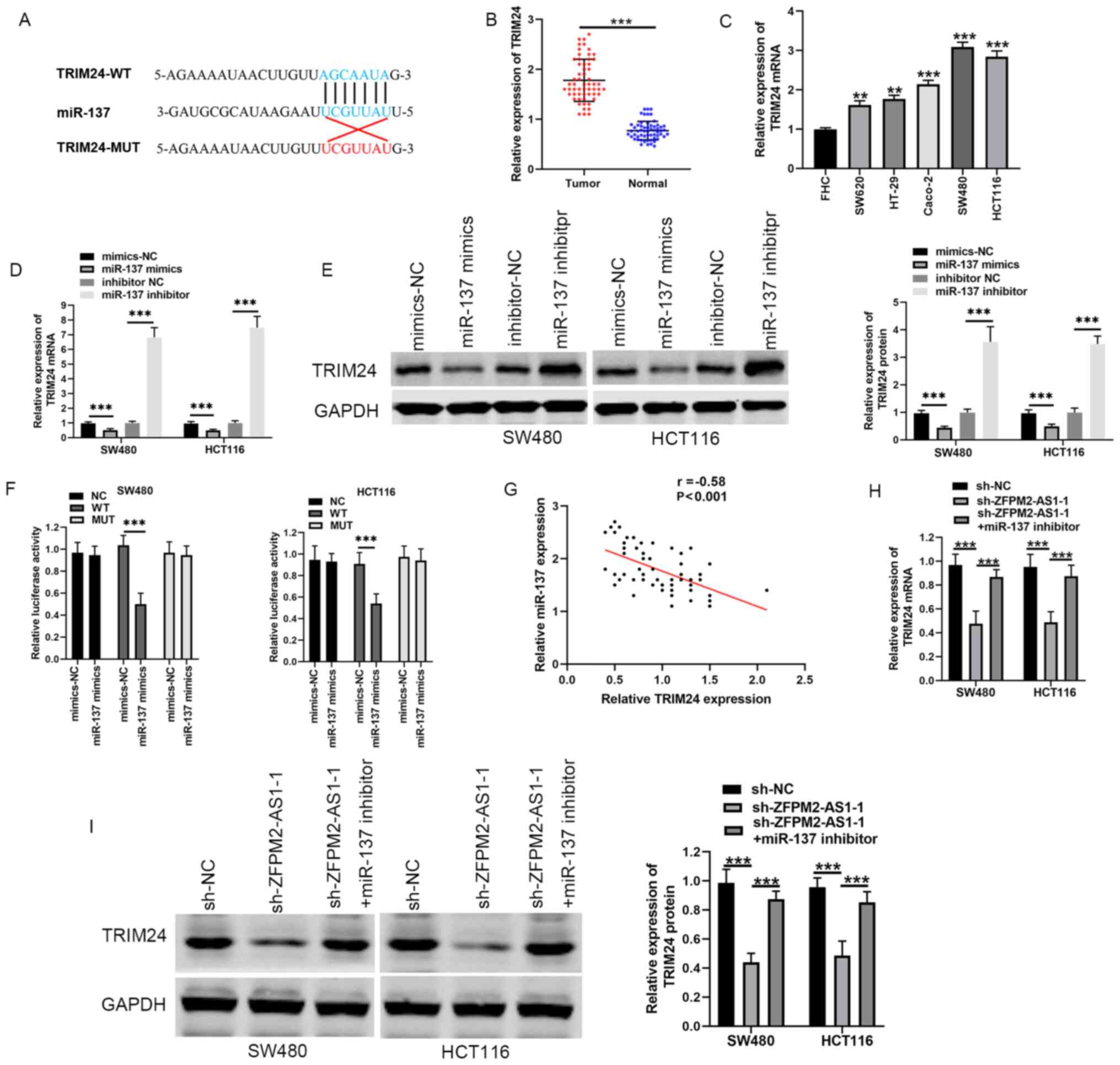
candidate target of ZFPM2-AS1 (Fig.
2A). Following ZFPM2-AS1 knockdown, miR-137 expression levels
were significantly upregulated, compared with sh-NC (Fig. 2B). miR-137 mimics and inhibitor, as
well as their respective NC were then transfected into SW480 and
HCT116 cell lines. The expression of miR-137 was significantly
upregulated in miR-137 mimics-transfected cells, whereas miR-137
expression was significantly decreased in miR-137
inhibitor-transfected cells, compared with their NC (Fig. 2C). The miR mimics and mimics NC were
then used in dual-luciferase reporter assays. Co-transfection with
miR-137 mimics significantly decreased the relative luciferase
activity of ZFPM2-AS1-WT, but not MUT, compared with mimics-NC
(Fig. 2D).
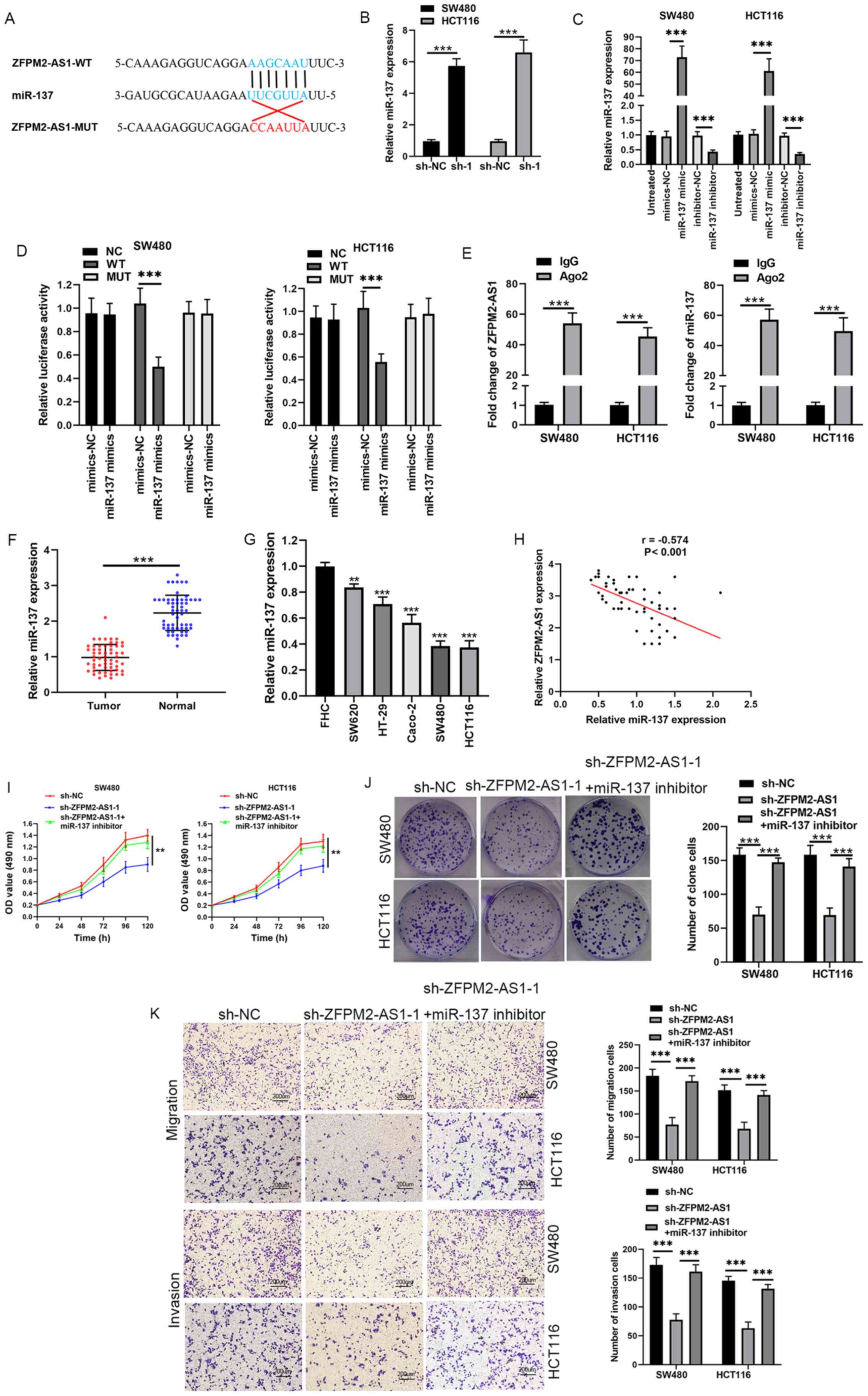 | Figure 2.miR-137 is a direct target of
ZFPM2-AS1. (A) Predicted binding site of miR-137 in ZFPM2-AS1. (B)
miR-137 expression in SW480 and HCT116 cells transfected with
sh-ZFPM2-AS1. (C) miR-137 expression in SW480 and HCT116 cells
following transfection with miR-137 mimics or miR-137 inhibitor.
(D) Effects of miR-137 mimics on luciferase activity WT or MUT
ZFPM2-AS1 gene constructs. (E) Radioimmunoprecipitation and reverse
transcription-quantitative PCR analysis of the binding between
ZFPM2-AS1 and miR-137. (F) miR-137 expression in CRC tumor tissues
and in adjacent normal tissues. (G) miR-137 expression in CRC cell
lines. (H) Correlation between ZFPM2-AS1 and miR-137 expressions in
CRC tissues. (I and J) Effects of sh-ZFPM2-AS1 and miR-137
inhibitor on proliferation of SW480 and HCT116 cells. (K) Effects
of sh-ZFPM2-AS1 and miR-137 inhibitor on cell migration and
invasion of SW480 and HCT116 cells (scale bar, 200 µm).
**P<0.01, ***P<0.001 vs. control group. miR, microRNA; CRC,
colorectal cancer; sh, short hairpin; NC, negative control; OD,
optical density; WT, wild-type; MUT, mutant; Ig, immunoglobulin;
Ago2, Argonaute 2. |
In a RIP assay, both ZFPM2-AS1 and miR-137 were
significantly enriched in Ago2-containing beads, compared with the
IgG-containing beads (Fig. 2E).
Furthermore, the relative expression levels of miR-137 were
significantly downregulated in CRC tissue compared with matched
normal tissue (Fig. 2F). The
expression levels of miR-137 were significantly downregulated in
CRC cell lines (SW620, HT-29, Caco-2, SW480 and HCT116) compared
with FHC cells (Fig. 2G). In
addition, miR-137 expression levels were negatively correlated with
ZFPM2-AS1 expression in CRC tissue (Fig. 2H). Notably, co-transfection of SW480
and HCT116 cells with miR-137 inhibitor and sh-ZFPM2-AS1-1
significantly increased cell proliferation, migration and invasion
abilities, compared with shRNA alone (Fig. 2I-K). These results suggested that
ZFPM2-AS1 may promote CRC progression by sponging miR-137.
miR-137 targets TRIM24 in CRC
TargetScan7 was used to predict the downstream
targets of miR-137, which identified TRIM24 as a potential
candidate gene (Fig. 3A). TRIM24
mRNA expression levels were significantly upregulated in CRC
tissues compared with the adjacent normal tissues (Fig. 3B). Moreover, TRIM24 mRNA levels were
significantly upregulated in CRC cell lines (SW620, HT-29, Caco-2,
SW480 and HCT116), compared with FHC cells (Fig. 3C). The effects of miR-137 mimics and
miR-137 inhibitor on TRIM24 mRNA and protein expression in CRC
cells were then evaluated. Compared with mimics NC, TRIM24
expression was significantly decreased both at the mRNA and protein
levels in CRC cells transfected with miR-137 mimics. By contrast,
the miR-137 inhibitor increased the expression levels of TRIM24
(Fig. 3D and E). In a
dual-luciferase reporter assay, miR-137 mimics significantly
inhibited the relative luciferase activity of TRIM24-WT, but not
TRIM24-MUT (Fig. 3F). Pearson's
correlation analysis also indicated that miR-137 expression levels
were negatively correlated with TRIM24 expression levels in CRC
tissues (Fig. 3G).
RT-qPCR and western blot analysis were then carried
out to determine whether ZFPM2-AS1 could regulate TRIM24 expression
levels. TRIM24 expression levels were significantly downregulated
following ZFPM2-AS1 knockdown. However, transfection with the
miR-137 inhibitor and sh-ZFPM2-AS1-1 increased the expression
levels of TRIM24, compared with shRNA alone (Fig. 3H and I). Thus, ZFPM2-AS1 may promote
TRIM24 expression by sponging miR-137 in CRC.
ZFPM2-AS1 promotes CRC progression
through TRIM24
To determine whether ZFPM2-AS1 promoted CRC
progression through TRIM24, RT-qPCR and western blotting analyses
were carried out following TRIM24 overexpression. TRIM24 expression
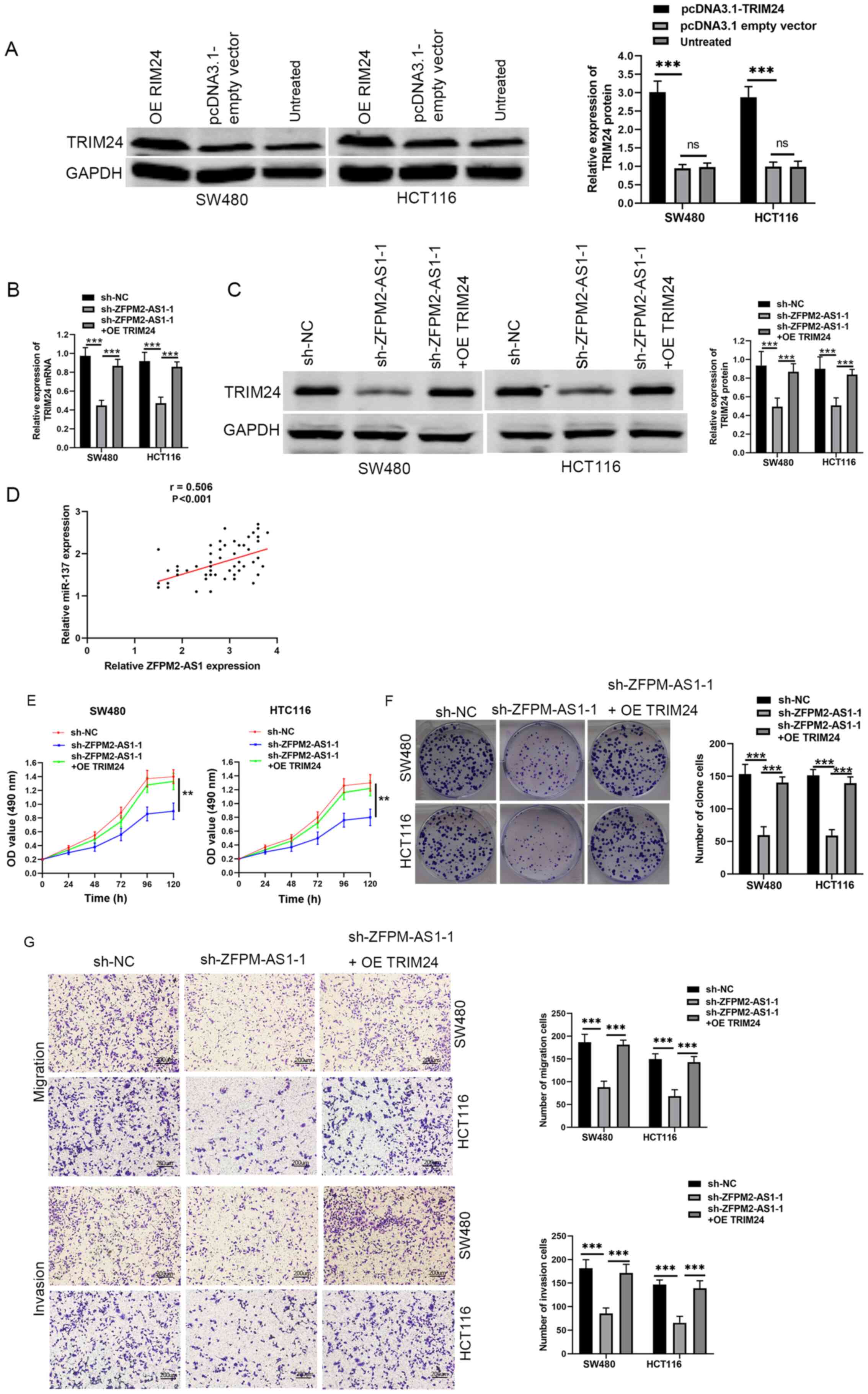
was significantly increased after transfection with pcDNA3.1-TRIM24
(Fig. 4A). Compared with sh-NC and
TRIM24 overexpression, ZFPM2-AS1 knockdown significantly
downregulated TRIM24 expression levels, both at the mRNA and
protein level (Fig. 4B and C).
Pearson's correlation analysis indicated that TRIM24 expression
levels were positively correlated with ZFPM2-AS1 expression levels
in CRC tissue (Fig. 4D). Moreover,
proliferation, migration and invasion of CRC cell lines were
significantly inhibited by ZFPM2-AS1 silencing, which was partly
reversed following TRIM24 overexpression (Fig. 4E-G). Taken together, these findings
suggested that ZFPM2-AS1 may serve an oncogenic role in CRC through
a miR-137/TRIM24 axis.
Discussion
Accumulating evidence suggests that lncRNAs exert
pivotal functions in the development and progression of CRC. For
example, Gao et al (14)
reported that the upregulation of lncRNA CACS15 contributed to
oxaliplatin resistance of CRC by regulating the miR-145/ABCC1 axis.
Xu et al (15) demonstrated
that lncRNA MALAT1 promoted CRC cell proliferation, invasion and
migration by upregulating SOX9 expression. Xu et al
(16) reported that the expression
levels of the lncRNA SATB2-AS1 were downregulated in CRC; clinical
analysis indicated that low SATB2-AS1 expression levels were
associated with poor prognosis. Functional analysis also
demonstrated that SATB2-AS1 knockdown enhanced migration and
invasion of CRC cells (16).
In the present study, the expression levels of
ZFPM2-AS1 were upregulated in human CRC tissues. In addition, there
was an association between ZFPM2-AS1 expression levels and clinical
characteristics. Indeed, high expression levels of ZFPM2-AS1 were
significantly associated with tumor size, histological
differentiation, lymph node metastasis and TNM stage. Thus, it was
hypothesized that ZFPM2-AS1 may serve as oncogenic role in CRC.
ZFPM2-AS1 expression levels were also markedly upregulated in CRC
cell lines, compared with a colorectal mucosa cell line.
Furthermore, loss-of-function experiments suggested that ZFPM2-AS1
knockdown decreased SW480 and HCT116 cell proliferation, migration
and invasion in vitro.
Previous studies have demonstrated that lncRNAs
function as ceRNA networks that sponge miRNAs and modulate the
expression of target mRNA molecules, resulting in changes in the
progression of multiple types of cancer, such as lung
adenocarcinoma and pancreatic ductal adenocarcinoma (17,18).
Thus, the potential miRNAs that can bind with ZFPM2-AS1 were
examined in the present study, and miR-137 was identified as a
potential target. miR-137 has previously been identified as a tumor
suppressor in numerous types of cancer. For example, Chen et
al (19) reported that miR-137
expression levels were downregulated in glioblastoma, and that the
overexpression of miR-137 markedly suppressed the proliferation and
invasion of glioma cell lines. In addition, Li et al
(20) demonstrated that miR-137
induced the apoptosis of ovarian cancer cells by regulating
X-linked inhibitor of apoptosis protein. miR-137 also inhibits the
malignant phenotype of malignant melanoma (21), tongue squamous carcinoma (22), cervical cancer (23) and CRC (24). In the present study, miR-137 could
directly bind ZFPM2-AS1. Moreover, miR-137 expression levels were
upregulated following ZFPM2-AS1 knockdown, and ZFPM2-AS1 expression
was inversely correlated with that of miR-137 in patients with CRC.
Moreover, transfection with a miR-137 inhibitor reversed the
effects of ZFPM2-AS1 knockdown in CRC cells.
The downstream targets of this ZFPM2-AS1/miR-137
axis were also examined. TRIM24 was predicted as a potential target
of miR-137. In a dual-luciferase reporter assay, miR-137 could
directly bind to TRIM24. Moreover, TRIM24 expression levels were
downregulated following the transfection with miR-137 mimics.
Several previous studies have suggested that TRIM24 may serve as an
oncogene in the development of several types of cancer. For
instance, Zhang et al (25)
reported that TRIM24 promoted glioma progression by enhancing the
PI3K/AKT signaling pathway. Lv et al (26) also demonstrated that TRIM24
contributed to human glioblastoma development. In addition, Zhu
et al (27) indicated that
TRIM24 was highly expressed in human hepatocellular carcinoma (HCC)
and that TRIM24 knockdown suppressed the proliferative and
migratory abilities of HCC cells.
Notably, a previous study also suggested that TRIM24
expression levels were significantly upregulated in CRC tissues,
compared with normal tissues, and that TRIM24 upregulation was
associated with poor prognosis in patients (28). Additionally, Wang et al
(29) reported that TRIM24
silencing inhibited the proliferation and promoted the apoptosis of
CRC cells. These previous studies suggested that TRIM24 may serve
as an oncogene in human CRC. Thus, it was investigated whether
TRIM24 participated in the miR-137/TRIM24 axis in CRC cells. It was
identified that ZFPM2-AS1 silencing downregulated TRIM24 expression
levels by binding to miR-137. In addition, TRIM24 overexpression
could rescue the effects of ZFPM2-AS1 knockdown on CRC cell
proliferation, migration and invasion.
In conclusion, the findings of the present study
suggested an important role for ZFPM2-AS1 in the proliferation,
migration and invasion of CRC cells. These results also identified
a novel mechanism through which the ZFPM2-AS1/miR-137/TRIM24 axis
may regulate the progression of CRC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MX and ZY carried out the study design and drafted
the manuscript. MX and ZL performed the experiments. ZY conceived
the study and revised the manuscript. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Medical Ethics
Committee of The First Affiliated Hospital of University of South
China (approval no. LL20170326021). Written informed consent was
obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar
|
|
2
|
Zhang J, Jiang Y, Zhu J, Wu T, Ma J, Du C,
Chen S, Li T, Han J and Wang X: Overexpression of long non-coding
RNA colon cancer-associated transcript 2 is associated with
advanced tumor progression and poor prognosis in patients with
colorectal cancer. Oncol Lett. 14:6907–6914. 2017.
|
|
3
|
Simon K: Colorectal cancer development and
advances in screening. Clin Interv Aging. 11:967–976. 2016.
View Article : Google Scholar
|
|
4
|
Bach DH and Lee SK: Long noncoding RNAs in
cancer cells. Cancer Lett. 419:152–166. 2018. View Article : Google Scholar
|
|
5
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar
|
|
6
|
Huang JL, Cao SW, Ou QS, Yang B, Zheng SH,
Tang J, Chen J, Hu YW, Zheng L and Wang Q: The long non-coding RNA
PTTG3P promotes cell growth and metastasis via up-regulating PTTG1
and activating PI3K/AKT signaling in hepatocellular carcinoma. Mol
Cancer. 17:932018. View Article : Google Scholar
|
|
7
|
Zheng S, Chen H, Wang Y, Gao W, Fu Z, Zhou
Q, Jiang Y, Lin Q, Tan L, Ye H, et al: Long non-coding RNA
LOC389641 promotes progression of pancreatic ductal adenocarcinoma
and increases cell invasion by regulating E-cadherin in a
TNFRSF10A-related manner. Cancer Lett. 371:354–365. 2016.
View Article : Google Scholar
|
|
8
|
Zhang M, Duan W and Sun W: LncRNA SNHG6
promotes the migration, invasion, and epithelial-mesenchymal
transition of colorectal cancer cells by miR-26a/EZH2 axis.
OncoTargets Ther. 12:3349–3360. 2019. View Article : Google Scholar
|
|
9
|
Zhan HX, Wang Y, Li C, Xu JW, Zhou B, Zhu
JK, Han HF, Wang L, Wang YS and Hu SY: LincRNA-ROR promotes
invasion, metastasis and tumor growth in pancreatic cancer through
activating ZEB1 pathway. Cancer Lett. 374:261–271. 2016. View Article : Google Scholar
|
|
10
|
Kong F, Deng X, Kong X, Du Y, Li L, Zhu H,
Wang Y, Xie D, Guha S, Li Z, et al: ZFPM2-AS1, a novel lncRNA,
attenuates the p53 pathway and promotes gastric carcinogenesis by
stabilizing MIF. Oncogene. 37:5982–5996. 2018. View Article : Google Scholar
|
|
11
|
Xue M, Tao W, Yu S, Yan Z, Peng Q, Jiang F
and Gao X: lncRNA ZFPM2-AS1 promotes proliferation via
miR-18b-5p/VMA21 axis in lung adenocarcinoma. J Cell Biochem.
121:313–321. 2020. View Article : Google Scholar
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
13
|
Duan X, Kong Z, Liu Y, Zeng Z, Li S, Wu W,
Ji W, Yang B, Zhao Z and Zeng G: β-Arrestin2 contributes to cell
viability and proliferation via the down-regulation of FOXO1 in
castration-resistant prostate cancer. J Cell Physiol.
230:2371–2381. 2015. View Article : Google Scholar
|
|
14
|
Gao R, Fang C, Xu J, Tan H, Li P and Ma L:
LncRNA CACS15 contributes to oxaliplatin resistance in colorectal
cancer by positively regulating ABCC1 through sponging miR-145.
Arch Biochem Biophys. 663:183–191. 2019. View Article : Google Scholar
|
|
15
|
Xu Y, Zhang X, Hu X, Zhou W, Zhang P,
Zhang J, Yang S and Liu Y: The effects of lncRNA MALAT1 on
proliferation, invasion and migration in colorectal cancer through
regulating SOX9. Mol Med. 24:522018. View Article : Google Scholar
|
|
16
|
Xu M, Xu X, Pan B, Chen X, Lin K, Zeng K,
Liu X, Xu T, Sun L, Qin J, et al: LncRNA SATB2-AS1 inhibits tumor
metastasis and affects the tumor immune cell microenvironment in
colorectal cancer by regulating SATB2. Mol Cancer. 18:1352019.
View Article : Google Scholar
|
|
17
|
Ge X, Li GY, Jiang L, Jia L, Zhang Z, Li
X, Wang R, Zhou M, Zhou Y, Zeng Z, et al: Long noncoding RNA CAR10
promotes lung adenocarcinoma metastasis via miR-203/30/SNAI axis.
Oncogene. 38:3061–3076. 2019. View Article : Google Scholar
|
|
18
|
Yang H, Liu P, Zhang J, Peng X, Lu Z, Yu
S, Meng Y, Tong WM and Chen J: Long noncoding RNA MIR31HG exhibits
oncogenic property in pancreatic ductal adenocarcinoma and is
negatively regulated by miR-193b. Oncogene. 35:3647–3657. 2016.
View Article : Google Scholar
|
|
19
|
Chen L, Wang X, Wang H, Li Y, Yan W, Han
L, Zhang K, Zhang J, Wang Y, Feng Y, et al: miR-137 is frequently
down-regulated in glioblastoma and is a negative regulator of
Cox-2. Eur J Cancer. 48:3104–3111. 2012. View Article : Google Scholar
|
|
20
|
Li X, Chen W, Zeng W, Wan C, Duan S and
Jiang S: microRNA-137 promotes apoptosis in ovarian cancer cells
via the regulation of XIAP. Br J Cancer. 116:66–76. 2017.
View Article : Google Scholar
|
|
21
|
Peres J, Kwesi-Maliepaard EM, Rambow F,
Larue L and Prince S: The tumour suppressor, miR-137, inhibits
malignant melanoma migration by targetting the TBX3 transcription
factor. Cancer Lett. 405:111–119. 2017. View Article : Google Scholar
|
|
22
|
Sun L, Liang J, Wang Q, Li Z, Du Y and Xu
X: MicroRNA-137 suppresses tongue squamous carcinoma cell
proliferation, migration and invasion. Cell Prolif. 49:628–635.
2016. View Article : Google Scholar
|
|
23
|
Miao H, Wang N, Shi LX, Wang Z and Song
WB: Overexpression of mircoRNA-137 inhibits cervical cancer cell
invasion, migration and epithelial-mesenchymal transition by
suppressing the TGF-β/smad pathway via binding to GREM1. Cancer
Cell Int. 19:1472019. View Article : Google Scholar
|
|
24
|
Balaguer F, Link A, Lozano JJ, Cuatrecasas
M, Nagasaka T, Boland CR and Goel A: Epigenetic silencing of
miR-137 is an early event in colorectal carcinogenesis. Cancer Res.
70:6609–6618. 2010. View Article : Google Scholar
|
|
25
|
Zhang LH, Yin AA, Cheng JX, Huang HY, Li
XM, Zhang YQ, Han N and Zhang X: TRIM24 promotes glioma progression
and enhances chemoresistance through activation of the PI3K/Akt
signaling pathway. Oncogene. 34:600–610. 2015. View Article : Google Scholar
|
|
26
|
Lv D, Li Y, Zhang W, Alvarez AA, Song L,
Tang J, Gao WQ, Hu B, Cheng SY and Feng H: TRIM24 is an oncogenic
transcriptional co-activator of STAT3 in glioblastoma. Nat Commun.
8:14542017. View Article : Google Scholar
|
|
27
|
Zhu Y, Zhao L, Shi K, Huang Z and Chen B:
TRIM24 promotes hepatocellular carcinoma progression via AMPK
signaling. Exp Cell Res. 367:274–281. 2018. View Article : Google Scholar
|
|
28
|
Wang FQ, Han Y, Yao W and Yu J: Prognostic
relevance of tripartite motif containing 24 expression in
colorectal cancer. Pathol Res Pract. 213:1271–1275. 2017.
View Article : Google Scholar
|
|
29
|
Wang J, Zhu J, Dong M, Yu H, Dai X and Li
K: Knockdown of tripartite motif containing 24 by lentivirus
suppresses cell growth and induces apoptosis in human colorectal
cancer cells. Oncol Res. 22:39–45. 2014. View Article : Google Scholar
|