Introduction
Diabetic cardiomyopathy (DCM) is a cardiovascular
complication of a major chronic metabolic disease, diabetes
mellitus (DM) (1), and affects ~12%
of patients with DM in Greece (2).
In diabetes, myocardial structure alterations, namely left
ventricular hypertrophy and functional changes in systole and
diastole, emerge and trigger extensive metabolic perturbation
(3). The resulting metabolic
perturbations manifest as hyperglycaemia, augmented lipid
metabolism, hyperlipidaemia and hyperinsulinaemia, impaired cardiac
contractility and increased cardiomyocyte dysfunction, injury and
cell death, which are all indicative of the development of DCM
(4). The physical change in
diabetic myocardium is characterized by cardiac hypertrophy,
primarily posed by increased left ventricular hypertrophy and is
accompanied by decreased systolic and diastolic function (5). Altered diastolic function is
considered to be an early sign of diabetic myocardium injury,
whereas systolic function alteration may appear in the later stage
of injury. Eventually, patients with diabetes develop a high risk
of heart failure, with clinical trails reporting a 19–26% incidence
rate worldwide (6,7).
Hyperglycaemia is considered to be the primary
pathogenic factor of DCM, although the driver of DCM is
multifactorial (8). High
concentration glucose (HG)-induced hyperglycaemia can cause
oxidative burst, which activates maladaptive signaling to further
disrupt cellular environmental homeostasis (8). Interactions between numerous molecular
mechanisms are implicated in the diabetic myocardium, such as
increased concentration of pro-inflammatory cytokines, increased
numbers of apoptotic and necrotic cells, fibrosis, mitochondrial
dysfunction, oxidative stress, autophagy and altered expression
level patterns of microRNAs (miRNAs or miRs) (4). Notably, miRNAs can impose a memory
effect related to their altered expressions, which are induced by
certain mechanisms in the diabetic heart, and fail to be normalized
by glycemic control (9). Therefore,
the interplay of miRNA expression levels and other molecular
mechanisms in the diabetic heart needs to be further
investigated.
Long non-coding RNAs (lncRNAs) are defined as ncRNAs
comprising >200 nucleotides in length, without protein-coding
capacity. They have been discovered to participate in multiple
physiological and pathological processes via modulating gene
transcription, sequestering RNA-binding proteins and sponging
miRNAs (10). Emerging evidence has
revealed that lncRNAs modulate autophagy by acting as a competing
endogenous RNA to sponge miRNA (11,12).
The regulatory role of lncRNAs in the pathogenesis of
cardiovascular disease has recently been highlighted (13). Hence, the present study aimed to
elucidate the mechanism of certain lncRNAs in the development of
DCM and its association with cardiac autophagy.
Growth-arrest specific transcript 5 (GAS5) is an
lncRNA in the 5′-terminal oligopyrimidine (TOP) class, with the
capacity to regulate cell growth, survival and proliferation
(14,15). GAS5 serves as the host gene of small
nucleolar RNAs (snoRNAs); encoded snoRNAs in the introns of GAS5
have been predicted to function in the 2′-O-methylation of
ribosomal RNA (16). The 5′ TOP
sequence involves protein synthesis and the mTOR pathway partly
controls translation of 5′-TOP RNAs (17) and influences the RNA expression
levels of GAS5 (16). The GAS5
expression levels in serum of patients with diabetes have been
demonstrated to decrease, with GAS5 values <10 ng/µl raising the
risk of diabetes twelve-fold (18).
The present study established a rat model of DM and generated a
HG-processed cardiomyocyte environment to compare the performance
of GAS5 in vitro and in vivo and investigated the
mechanism underlying GAS5 interactions with its targeted miRNA in
DCM to identify potential methods for cardiovascular risk
mitigation in DM.
Materials and methods
Ethics statement
All animal experiments were performed in accordance
with the guidelines for the Care and Use of Laboratory Animals
(19). The present study was
approved by the Committee of Experimental Animals of Yongchuan
Hospital of Chongqing Medical University (approval no.
YHC20190537). Every effort was made to minimize pain and discomfort
to the animals. The animal experiments were performed at Yongchuan
Hospital of Chongqing Medical University.
Cell culture
H9C2 cells [H9c2 (2–1); The
Cell Bank of Type Culture Collection of Chinese Academy of
Sciences] were cultured in DMEM [cat. no. 30-2002; American Type
Culture Collection (ATCC)] with 10% FBS (cat. no. 30-2020; ATCC) at
37°C and a pH of 7.2–7.4 in 5% CO2. A total of 1 ml
pancreatic enzyme (cat. no. P816199-50g; Casmart) was added to
1×106 cells for digestion; digestion was stopped by
adding cells to DMEM containing 10% FBS. Cells were centrifuged at
1,000 × g for 5 min at 37°C and maintained at 5% CO2 at
37°C for subsequent experiments.
293A cells (293A; BioVector NTCC, Inc.) were
cultured in DMEM with 10% FBS and Penicillin-Streptomycin
antibiotics (cat. no. P4333; Sigma-Aldrich; Merck KGaA) and
sub-cultured in HG DMEM in 5% CO2 at 37°C for virus
packaging and dual luciferase reporter assay.
Vector construction and
adeno-associated virus (AAV-9A) packaging
A certain segment of GAS5 nucleotide sequence
(lnc6160405085202-1-5; Guangzhou RiboBio Co., Ltd.) was subcloned
into pHBAd (Hanbio Biotechnology Co., Ltd.). Scrambled-small
interfering (si)RNA (siG141112151026-1-5; Guangzhou RiboBio Co.,
Ltd.) was used as a control. 293A cells were co-transfected with 50
ng pHBAd containing the segment of GAS5 nucleotide sequence and
pBHGlox (Δ) E1, 3Cre for 6 h using LipoFiter3 transfection reagent
(both Hanbio Biotechnology Co., Ltd.). Virus in the cell
supernatant, which was obtained via centrifugation at 3,000 × g at
37°C for 15 min, was precipitated using PEG8000 (cat. no. 07164;
Sigma Aldrich; Merck KGaA), harvested and purified via the
iodixanol method (20). Virus titer
was detected by quantitative (q)PCR (21). Vector construction and virus
(AAV-9A) packaging were performed by Hanbio Biotechnology Co., Ltd.
The acquired virus was used for rat myocardium injection and H9C2
transfection.
Establishment of diabetic rats
A total of 40 6-week-old male rats (Slac:SD; weight,
160–180 g; Shanghai SLAC Animal Laboratory Co., Ltd.) were kept at
21–22°C, 50–70% humidity and 12/12-h day/night cycle in a specific
pathogen-free environment, with free access to food and water.
Streptozotocin (STZ) used to induce diabetes was
procured from Sigma Aldrich (cat. no. 572201; Merck KGaA) and 60
mg/kg/day of STZ was dissolved in 10 mM citrate buffer (pH, 4.5;
cat. no. A55926-500ml; Casmart) to prepare STZ injection. The rats
were randomly distributed into the following four groups
(n=10/group): Control (rats with no injection), DM (rats injected
with STZ), DM + NC (rats injected with STZ and AAV-9 containing
scrambled-siRNA) and DM + GAS5 (rats injected with STZ and AAV-9
containing GAS5 nucleotide sequence). In the latter three groups,
rat myocardium was intraperitoneally injected with prepared STZ for
5 consecutive days, in the presence or absence of injection of 100
µl AAV-9 containing scrambled-RNA or GAS5 nucleotide sequence.
Following injection, the rats were maintained in the aforementioned
conditions. On the 7th day after the final injection, the caudal
vein blood glucose concentration and body weight of rats were
measured; blood glucose level >16.7 mmol/l was considered to
indicate a diabetic rat.
Reverse transcription-qPCR
mRNA was extracted using a TRIzol Plus RNA
Purification kit (cat. no. 12183555; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. A total of
50–100 ml myocardium from one rat was lysed using TRIzol reagent (1
ml) to retain the lysate of mRNA. Then, 0.2 ml chloroform (cat. no.
48520-U; Sigma Aldrich; Merck KGaA) was used to extract the lysate.
The lysate was centrifuged (12,000 × g) for 15 min at 4°C, then an
equal volume of 70% ethanol (cat. no. 459836; Sigma-Aldrich; Merck
KGaA) was added and mixed well by vortex. miRNA was lysed and
extracted using an RNAmisi microRNA kit (cat. no. 110501; Beijing
BLKW Biotechnology Co., Ltd.). Following isolation of the
precipitate from the supernatant, 75% ethanol was used to resuspend
the precipitate. The precipitate underwent 7,500 × g centrifugation
for 10 min at 4°C and was then dissolved in 20 µl diethyl
pyrocarbonate (cat. no. rats (Slac:SD). cDNA for mRNA was produced
using a First Strand cDNA Synthesis kit (cat. no. K1621, Thermo
Fisher, Inc.) and the following temperature protocol: 65°C for 5
min, 42°C for 60 min and 70°C for 5 min, according to the
manufacturer's instructions. cDNA for miRNA was produced using a
TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The cDNA was transferred to an
Applied Biosystems 7500 FAST real-time PCR machine (Applied
Biosystems; Thermo Fisher Scientific, Inc.) for PCR using IV
One-Step RT-PCR System with ezDNase (cat. no. 12595025; Thermo
Fisher Scientific, Inc.). The following thermocycling conditions
were used for qPCR: 95°C for 10 min; and 35 cycles of 95°C for 15
sec and 60°C for 60 sec. The sense and antisense primers are listed
in Table I. Fold change in the
expression levels of mRNA and miRNA was determined using the
2−ΔΔCq method (22).
 | Table I.Primers for quantitative PCR
detection of genes. |
Table I.
Primers for quantitative PCR
detection of genes.
| Gene | Sense (5′→3′) | Antisense
(5′→3′) |
|---|
| Growth arrest
specific transcript 5 |
AGCCAGAAAATGGGATGGTGG |
ACTGCACTGTCCACTTGTCA |
| GAPDH |
CAATGACCCCTTCATTGACC |
GACAAGCTTCCCGTTCTCAG |
| p62 |
TCAGTTAAAGCCCCGGAAGA |
AACAACCTCTAGCTCCACCC |
| Collagen I |
GCTCCTCTTAGGGGCCACT |
CCACGTCTCACCATTGGGG |
| Collagen III |
CTGTAACATGGAAACTGGGGAAA |
CCATAGCTGAACTGAAAACCACC |
| TGF-β |
CTCCCGTGGCTTCTAGTGC |
GCCTTAGTTTGGACAGGATCTG |
| Connective tissue
growth factor |
GGGCCTCTTCTGCGATTTC |
ATCCAGGCAAGTGCATTGGTA |
| LC3B I/II |
ATGGTAGTCTGTGGTGGTGG |
CATACCTTGAATCCCAGCGC |
|
Rno-microRNA-221-3p |
A(24)CACTGTCATGCCGTTACGTAGCGTATCGTTGACAGCTAACCAA |
GCTGTCAACGATACGCTACGTAACG |
| U6 |
CTCGCTTCGGCAGCACA |
AACGCTTCACGAATTTGCGT |
Hematoxylin and eosin (H&E)
staining
In order to observe rat myocardium and calculate
cardiomyocyte cross-sectional area (CSA), rat myocardium
(thickness, 0.3 cm) was fixed in 10% formalin at 37°C for 24 h
(cat. no. HT5011; Sigma-Aldrich; Merck KGaA), dehydrated by
ethanol, blocked by paraffin (cat. no. 1.07150; Sigma-Aldrich;
Merck KGaA) and sliced (5-µm thick). Later, the sliced rat
myocardium was dewaxed by soaking in xylene (cat. no. X749400-250g;
Casmart) twice (5 min each). Rehydration was performed via soaking
in gradient ethanol (100, 95, 85 and 75%; 2 min each) and rinsing
with water. The cells were stained with 5% hematoxylin (cat. no.
H9627-100G; Casmart) at 37°C for 15 min, then soaked in 5% acetic
acid (cat. no. A465500; Casmart) to be differentiated and soaked in
distilled water for 20 min to develop blue. Then, the cells were
stained by 0.5% eosin (cat. no. 170; Casmart) at 37°C for 10 min,
then rinsed with distilled water at 37°C for 2 sec. Dehydration was
performed with graded ethanol (75%, 1 min; 85%, 1 min; 90%, 1 min;
100%, 2 min; and 100%, 4 min). Cells were soaked in xylene twice (5
min each), then sealed by neutral resin (cat. no. XY-23474-1;
Casmart) at 37°C for 2 h for observation using a light microscope
(CX43; Olympus Corporation).
Masson staining
Rat myocardium was stained using a Masson staining
kit (cat. no. G1340; Beijing Solarbio Science & Technology,
Co., Ltd.) according to the manufacturer's instructions for
collagen volume fraction calculation. Rat myocardium was dewaxed by
soaking in xylene twice (10 min each). Weigert dye solutions A and
B were mixed at a ratio of 1:1 to prepare Weigert Iron hematoxylin
dye solution. Myocardium was stained with Weigert Iron hematoxylin
solution at 37°C for 10 min and differentiated by acidic ethanol
for 10 sec. Masson solution was used to develop blue on the
myocardium. Then, 10% porcelain Red Magenta solution was used to
stain the myocardium at 37°C for 10 min. The myocardium underwent
sequential weak and phosphomolybdic acid washes. Myocardium was
immersed in aniline blue solution at 37°C for 2 min, then
dehydrated with anhydrous ethanol and soaked in xylene three times.
Finally, the myocardium was blow-dried and sealed and tissue
observation was performed using a light microscope (CX43; Olympus
Corporation).
Western blot
A total of 250 mg rat myocardium was homogenized
using 1 ml ice cold RIPA buffer (cat. no. P0013B; Beyotime
Institute of Biotechnology). Following homogenization, total
protein was extracted. Measurement of protein concentration was
conducted using a BCA Protein Assay kit (cat. no. 23225; Thermo
Fisher Scientific, Inc.). The obtained protein (20 µl per lane) and
protein marker (10–170 kD) were subjected to gel electrophoresis
with 12% SDS-PAGE (cat. no. P0012A; Beyotime Institute of
Biotechnology). Following electrophoresis, the protein was
transferred to nitrocellulose membranes (cat. no. N8395; Sigma
Aldrich; Merck KGaA). Following blocking in TBS with 1% Tween-20
(TBST; cat. no. TA-125-TT; Thermo Fisher Scientific, Inc.) at 37°C
for 1 h, the membrane was incubated with primary antibody at 4°C
overnight and subsequently incubated with HRP-conjugated secondary
antibody for 1 h at room temperature. The protein was transferred
to an ECL system (Amersham Pharmaci; Cytiva) with ECL reagent (EMD
Millipore). ImageJ software 1.50 (National Institutes of Health)
was utilized to analyze the optical density of protein bands. The
following primary antibodies (all Abcam) were used: Rabbit Anti-p27
KIP 1 (27 kD; 1:5,000; product code ab32034), Mouse Anti-GAPDH (36
kD; 1:1,000; product code ab8245), Mouse Anti-SQSTM1/p62 antibody
(62 kD; 1:1,000; product code ab56416) and Rabbit Anti-LC3B
I/LC3BII antibody (19 and 17 kD, respectively; 1:1,000; product
code ab48394). The following secondary antibodies were used: Goat
Anti-Rabbit IgG H&L (HRP) and Goat Anti-Mouse IgG H&L (HRP)
(both 1:5,000; product codes ab205718 and ab205719, respectively;
both Abcam).
Luciferase reporter assay
Online databases starBase (version 2.0; starbase.sysu.edu.cn/agoClipRNA.php?source=lncRNA) and
Targetscan7.2 (targetscan.org/vert_72/) were used to predict
potential binding sites of miR-221-3p on GAS5 and p27. The
3′-untranslated region (3′-UTR) of GAS5
(5′-AUUCUGCAUUCCCAUGUAGC-3′) and p27 wild-type (WT)
(5′-CUCUAAAAGCGUUGGAUGUAGCA-3′) containing the potential miR-221-3p
binding sites were cloned into pRL-CMV luciferase reporter plasmid
(cat. no. E2261; Promega Corporation). The mutant (MUT) sequence
(GAS5, 5′-CUUAUCGUUUCGCCCACGUA-3′; p27,
5′-CUCUAAACCAGUUGGCAUAGAUA-3′) was used as the MUT control. The
293A cells were co-transfected with the luciferase reporter plasmid
and miR-221-3p mimic (miR10000278-1-5; Guangzhou RiboBio Co., Ltd.)
or mimic control using Lipofectamine® 2000 (cat. no.
11668019; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. At 48 h post-transfection, firefly
luciferase activity was normalized to Renilla luciferase
activity, and detected using Promega Dual Luciferase Reporter Assay
System (Promega Corporation), according to manufacturer's
instructions.
Transfection of sip27
sip27 (sense, 5′-UGGAUUUGUACCAUUCUUCUG-3′ and
antisense, 5′-GAAGAAUGGUACAAAUCCAAG-3′) (cat. no. 12324; Cell
Signaling Technology, Inc.), miR-221-3p mimic (miR10000890-1-5;
Guangzhou RiboBio Co., Ltd.) and mimic control were transfected
into H9C2 cells using Lipofectamine® 2000 Transfection
Reagent (cat. no. 11668019; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. A total of
1×104 H9C2 cells were seeded to reach 70–90% confluence
per well in a 96-well plate (cat. no. 14-245-101; Thermo Fisher
Scientific, Inc.). Lipofectamine reagent (1 µl) was diluted in
Opti-MEM (25 µl) (cat. no. 31985062; Thermo Fisher Scientific,
Inc.). Then, 2.5 µg (0.5 µg/µl) RNA of interest was diluted in 125
µl Opti-MEM. The diluted RNA was mixed 1:1 with diluted
Lipofectamine and incubated at room temperature for 5 min.
Following incubation, 10 µl RNA-lipid complex, 100 ng final RNA and
0.5 µl Lipofectamine were added per well and again incubated at
37°C for 3 days.
Statistical analysis
Data are presented as the mean ± SEM of three
independent repeats. One-way ANOVA was used to compare multiple
groups, followed by post hoc Tukey's test. An unpaired Student's
t-test was used to analyze the differences between two groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
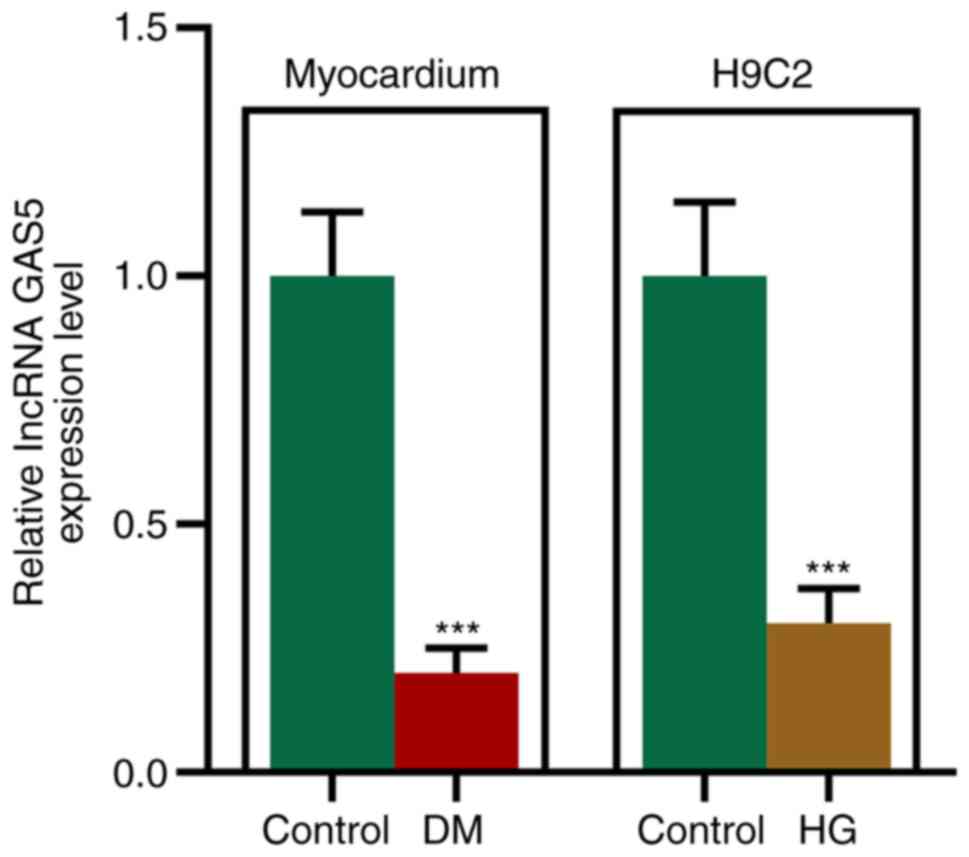
GAS5 expression levels are decreased
in the myocardium of STZ-induced diabetic rats and HG-processed
H9C2 cells
At the start of the experiment, there was no notable
association between rat body weight and blood glucose levels
(Table II). However, on day 7
after STZ injection, the body weight in the DM, DM + GAS5 and DM +
NC groups exhibited a significant decrease, compared with the
control group (Table II).
Moreover, the blood glucose concentration in these three groups
increased to >16.7 mM 7 days after STZ injection (Table II). It was confirmed that STZ
successfully established a DM model in rats, as the blood glucose
increased and body weight decreased following STZ injection
(Table II). GAS5 was significantly
downregulated in the myocardium of diabetic rats and HG-processed
H9C2 cells. GAS5 expression levels in STZ-induced diabetic rats
were decreased, which was similar to that in HG-processed
cardiomyocytes (Fig. 1).
 | Table II.Change in body weight and blood
glucose of STZ-induced diabetic rats. |
Table II.
Change in body weight and blood
glucose of STZ-induced diabetic rats.
|
| Body weight, g | Blood glucose,
mmol/l |
|---|
|
|
|
|
|---|
| Group | Pre-STZ | Day 7 post-STZ | Pre-STZ | Day 7 post-STZ |
|---|
| Control | 142.36±3.62 |
181.60±5.07a | 5.63±0.52 | 5.71±0.42 |
| DM | 144.28±3.96 |
153.42±5.42a | 5.66±0.42 |
23.67±2.92a |
| DM + negative
control | 140.66±4.32 |
152.71±4.66a | 5.68±0.45 |
23.74±3.07a |
| DM + growth arrest
specific transcript 5 | 146.33±5.02 |
154.55±5.02a | 5.73±0.58 |
20.45±2.82a |
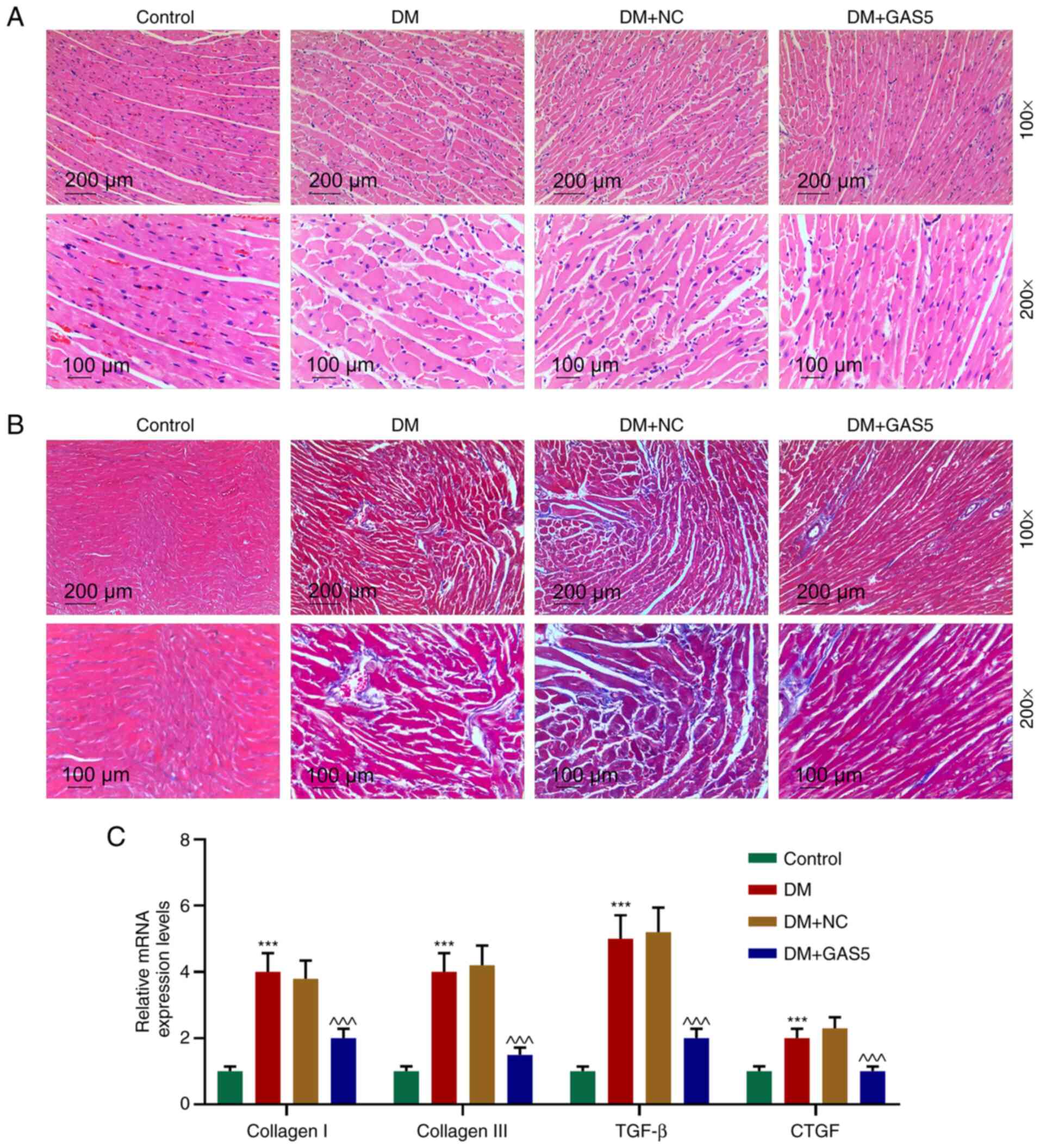
GAS5 improves histological
abnormalities in diabetic rats
H&E staining was utilized to measure the CSA of
cardiomyocytes in diabetic rats. The results demonstrated typical
histological abnormalities (23),
including cardiomyocyte hypertrophy, myocardial fiber breakage and
increased intercellular space in diabetic rats, whereas GAS5
ameliorated these abnormalities (Fig.
2A). As fibrosis is a biological feature of DCM, which
manifests in collagen accumulation in myocardium (24), the extent of fibrosis was determined
based on collagen volume observation via Masson staining,
calculation of collagen volume fraction and fibrosis marker
detection by qPCR (Fig. 2B and C).
Myocardial fibrosis was increased in diabetic rats, but notably
relieved by GAS5 (Fig. 2B).
Expression levels of fibrosis markers (collagen I, collagen III,
TGF-β and connective tissue growth factor) were significantly
increased in the myocardium in DM, indicating synthesis of collagen
and significant growth of connective tissue, whereas the expression
levels were inhibited by GAS5 (Fig. 2B
and C). The results indicated that GAS5 alleviated
diabetes-induced myocardial histological abnormalities and fibrosis
(Fig. 2).
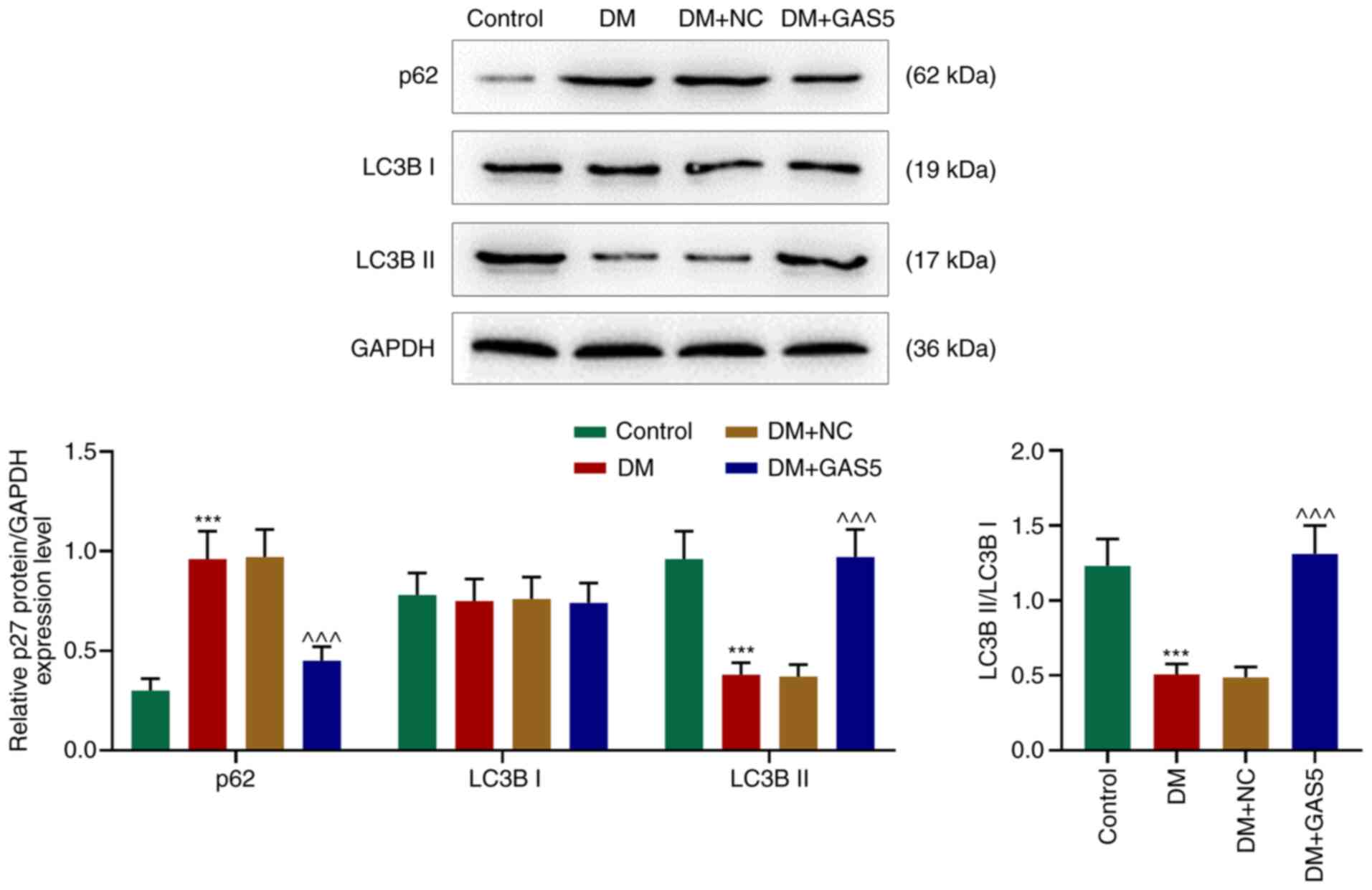
GAS5 reverses inhibition of autophagy
in the myocardium of diabetic rats
In order to investigate the role of GAS5 in cardiac
autophagy in diabetic rats, LC3B I, LC3B II and p62 expression
levels were measured using western blotting. p62 levels were
increased in the myocardium of diabetic rats, however LC3B I
remained stable, LC3B II and the ratio of LC3B II/LC3B I were
decreased, demonstrating a reduction in the formation of
autophagosomes and suppressed degradation of autophagosomes, which
further indicated inhibited cardiac autophagy in diabetic rats
(Fig. 3). GAS5 counteracted
DM-induced autophagy inhibition, which was demonstrated by an
increased ratio of LC3B II/LC3B I and decreased p62 expression
levels, suggesting that GAS5 reversed the inhibitory effect of DM
on cardiac autophagy (Fig. 3).
GAS5 competitively binds miR-221-3p,
which is upregulated in the myocardium of diabetic rats and
HG-processed H9C2 cells, to increase p27 in HG-processed H9C2
cells
In order to determine the internal mechanism by
which GAS5 promotes autophagy, starBase was employed to predict the
potential binding site between miR-221-3p and GAS5 as well as
miR-221-3p and p27. GAS5-WT had binding sites with miR-221-3p and
mutual binding sites between miR-221-3p and p27 were identified at
position 201–208 of the p27-WT 3′UTR (Fig. 4A and D). Regulatory associations
between mutual binding sites were assessed via luciferase reporter
assay. The expression levels of GATS5-WT and p27-WT were negatively
regulated by miR-221-3p mimic, however the MUT expression levels
were unaffected (Fig. 4B and
E).
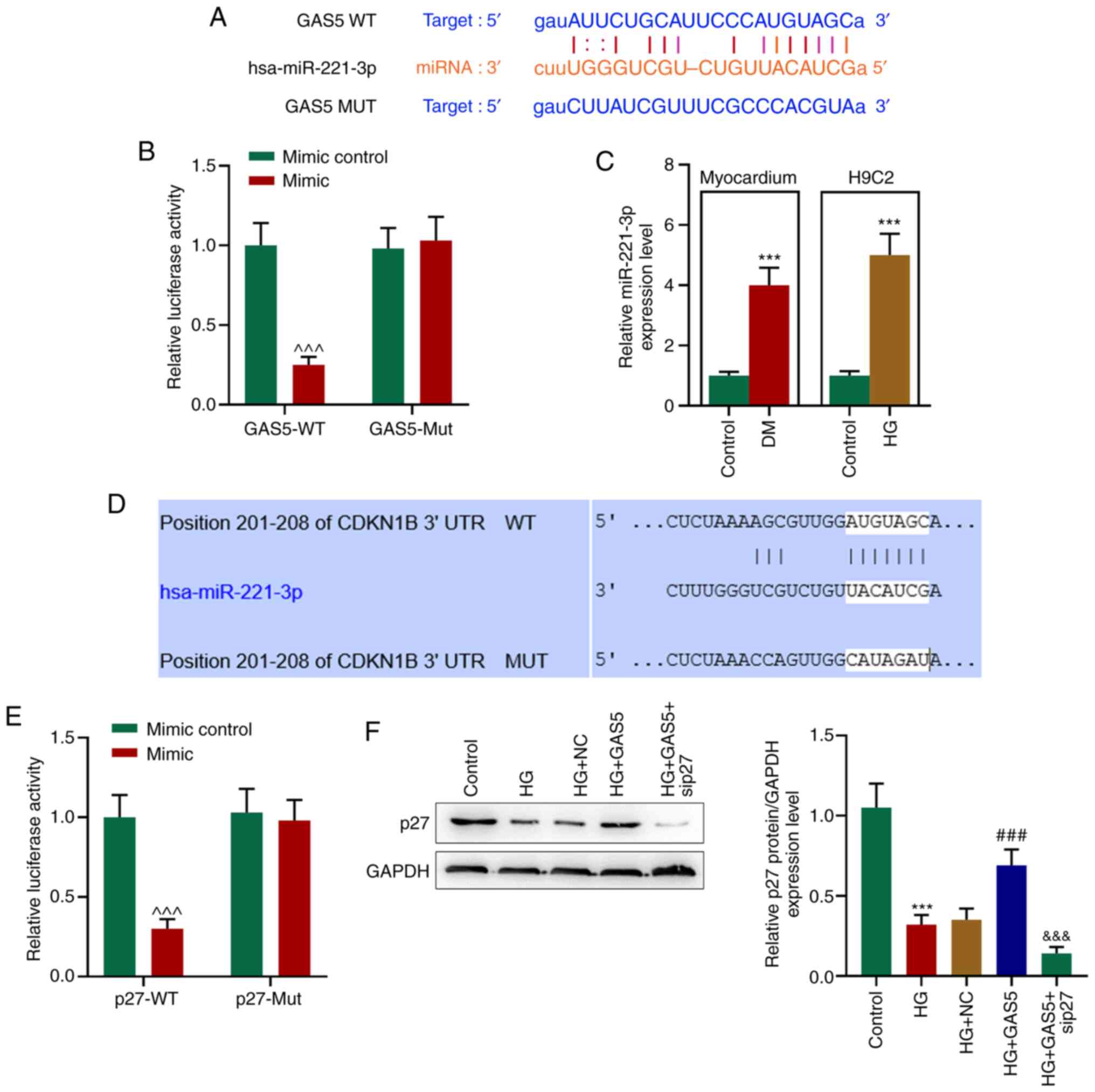 | Figure 4.GAS5 competitively binds miR-221-3p,
which is increased in the myocardium of diabetic rats and
HG-processed H9C2 cells, to increase p27 in HG-processed H9C2
cells. (A) starBase predicted binding sites between GAS5 and
miR-221-3p. (B) Dual luciferase reporter assay assessed the effect
of miR-221-3p mimic on the activity of GAS5. (C) miR-221-3p
expression levels in the myocardium of diabetic rats and
HG-processed H9C2 cells were assessed via quantitative PCR. U6 was
used as a reference gene. (D) Targetscan7.2 predicted binding sites
between miR-221-3p and p27. (E) Dual luciferase reporter assayed
the effect of miR-221-3p mimic on the activity of p27. (F) Western
blotting examined the protein expression levels of p27 in
HG-processed H9C2 cells. GAPDH was used as a reference gene.
***P<0.001 vs. Control; ^^^P <0.001 vs. mimic
control; ###P<0.001 vs. HG + NC;
&&&P<0.001 vs. HG + GAS5. GAS5, growth
arrest specific transcript; miR, microRNA; HG, high concentration
glucose; NC, negative control; WT, wild-type; MUT, mutant; UTR,
untranslated region; si, small interfering. |
The expression levels of miR-221-3p were
significantly increased in the myocardium of diabetic rats as well
as in HG-processed H9C2 cells, whereas the protein expression
levels of p27 were decreased in a HG environment (Fig. 4C and F). Notably, GAS5 reversed this
inhibition of p27 protein expression levels, which may be due to
GAS5 competitively binding to miR-221-3p, preventing p27 binding to
miR-221-3p and thus increasing p27 protein expression levels
(Fig. 4F). Moreover, transfection
of sip27 significantly counteracted the effect of GAS5, resulting
in low protein expression levels of p27 (Fig. 4F) in a HG environment.
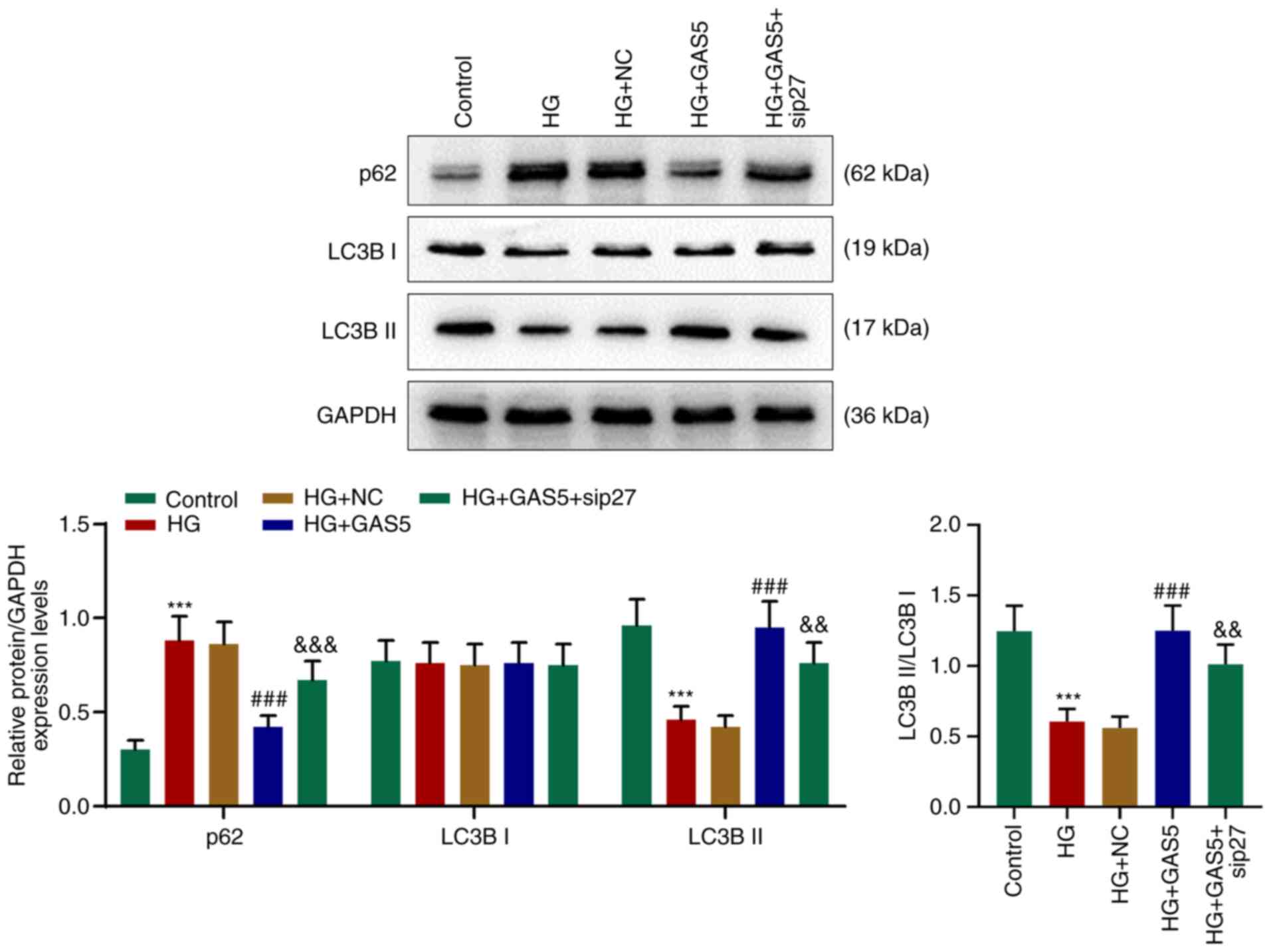
Sip27 reverses the facilitating effect
of GAS5 on HG-inhibited autophagy in HG-processed H9C2 cells
Similar to the impact of GAS5 on the myocardium of
diabetic rats, the significantly upregulated p62 expression levels
in HG-processed H9C2 cells was downregulated by GAS5, whereas LC3B
II expression levels were decreased in HG-processed H9C2 cells but
increased following exposure to GAS5 (Fig. 5). Similarly, the ratio of LC3B
II/LC3B I was decreased in HG-processed H9C2 cells, whereas this
effect was reversed by GAS5 (Fig.
5). The addition of sip27 counteracted the effects of GAS5 on
the expression levels of p62 and LC3B II, ratio of LC3B II/LC3B I
and cardiomyocyte autophagy.
Discussion
The key role of lncRNAs in the regulation of diverse
types of cardiovascular physio-pathology processes, including
atherosclerosis, coronary disease, cardiac remodeling and heart
failure, has been recorded (25–28)
and its involvement in the pathogenesis of DCM, an important
cardiovascular complication of diabetes, has previously been
investigated (29). lncRNA KCNQ1
opposite strand/antisense transcript 1 has been revealed to mediate
pyroptosis in DCM via affecting miR-214-3p expression levels
(30). lncRNAs H19 and myocardial
infarction-associated transcript have been revealed to influence
cardiomyocyte apoptosis via regulating their target miRNAs in DCM
(31,32). lncRNA HOX transcript antisense RNA
(HOTAIR) improved cardiac function, decreased oxidative stress and
inflammation and protected myocytes from death in mice with DCM
(33).
lncRNA GAS5 has been revealed to be aberrantly
increased in peripheral blood mononuclear cells (PBMCs) from
patients with type 2 diabetes and changes in its expression levels
are positively associated with poor glycemic control, insulin
resistance and transcriptional markers of senescence and
inflammation (34). Thus, it was
hypothesized that GAS5 expression levels serve a vital role in the
development of the diabetic heart. Furthermore, a previous study
demonstrated an association between decreased GAS5 expression
levels and diabetes; individuals with absolute GAS5 <10 ng/µl
were 12 times more likely to have diabetes (18). Therefore, in order to clarify the
mechanism of GAS5 in DCM, STZ-induced diabetic rats and
HG-processed H9C2 cells were established. Similar to the results
obtained by Tao et al (35),
STZ injection to rat myocardium resulted in decreased body weight
and increased blood glucose concentration, indicating successful
establishment of diabetic rats. Contrary to the increased
expression levels of GAS5 in PBMCs from patients with type 2
diabetes, the present results revealed decreased expression levels
of GAS5 both in the rat myocardium and HG-processed H9C2 cells,
suggesting that GAS5 expression patterns differ according to the
cell types in patients with DM.
Diabetes induces cardiac metabolic dysregulation
that is linked with oxidative stress and autophagy disturbance,
which induce cell death, fibrotic ‘backfill’ (increased fibrosis)
and cardiac dysfunction (36). A
previous study reported diabetes-induced cardiovascular
histological changes, such as increased left ventricular and wall
thickness, decreased volume of the left ventricular systolic
chamber and increased arterial stiffness (5). The present study further observed
enlarged CSA and aggravated fibrosis in the myocardium of diabetic
rats, which was consistent with results reported by Gao et
al (33). Moreover, Gao et
al also demonstrated that pathological changes (including
increased CSA and fibrosis) were reversed by HOTAIR overexpression.
The present study demonstrated that GAS5 serves a similar role to
HOTAIR in the myocardium of diabetic rats.
Prior studies have demonstrated inhibited cardiac
autophagy in diabetic OVE26 mice, as evidenced by decreased LC3-II
expression levels, and have observed that enhancement of cardiac
autophagy is concomitant with improved cardiac function in DCM
(37,38). Lower p62 expression levels are also
a sign of inhibited cardiac autophagy in DCM (39). The present study detected decreased
LC3-II expression levels and increased p62 expression levels, which
reflected inhibition of autophagosome formation in the myocardium
of diabetic rats. Moreover, GAS5 in the diabetic myocardium
resulted in cardiomyocyte autophagy rebounding, which indicated
that GAS5 ameliorates cardiac function of patients with DM patients
via promoting autophagy.
miRNA expression levels in diabetic mice have been
demonstrated to be significantly altered and do not recover with
subsequent normoglycaemia, which may explain why diabetic
cardiomyopathy progresses even under normal blood glucose levels;
this indicates that glycemic control is not a solution to prevent
DCM (9). A previous study
demonstrated that lncRNAs function as miRNA sponges to decrease
their regulatory effect on mRNAs (40). As lncRNAs also serve a regulatory
role, it was hypothesized that GAS5 sponges its direct target miRNA
to affect indirectly associated mRNAs and that this may be a
solution to control the development of DCM. Previous research
indicated that miR-221-3p can bind to the 3′UTR of p27 and the
overexpression of miR-221-3p in non-small cell lung cancer was
revealed to lower the expression levels of p27, promoting cell
cycle progression (41). In the
present study, overexpression of miR-221-3p was detected both in
the myocardium of diabetic rats and HG-processed H9C2 cells. As
anticipated, the expression levels of p27 decreased in HG-processed
H9C2 cells. Additionally, an axis of miR-221-3p and p27 was
implicated in the regulation of GAS5 in the present study. In order
to substantiate the regulatory effect of GAS5 on the miR-221-3p/p27
axis, the present study demonstrated that GAS5-mediated
upregulation of p27 and sip27 expression levels offset the effect
of GAS5 in HG-processed H9C2. Similarly to the in vivo
results, autophagy was inhibited in HG-processed H9C2 cells. GAS5
increased autophagy in HG-processed H9C2 cells, but sip27
counteracted this effect. The present results indicated that GAS5
may upregulate p27 via sponging miR-221-3p to promote cardiomyocyte
autophagy.
In conclusion, GAS5 reversed the histopathological
changes induced by DCM and enhanced cardiomyocyte autophagy to
ameliorate myocardial function. The mechanism underlying the effect
of GAS5 in the diabetic myocardium may be attributed to a
GAS5/miR-221-3p/p27 competing endogenous network and cardiomyocyte
autophagy. Thus, GAS5 may promote cardiomyocyte autophagy via
regulating the miR-221-3p/p27 axis to protect myocardial function
in DCM.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DC designed the study and wrote the manuscript. MZ
performed the research. MZ analyzed the data. All authors
contributed to editorial changes in the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Committee of
Experimental Animals of Yongchuan Hospital of Chongqing Medical
University (approval no. YHC20190537).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
lncRNA
|
long non-coding RNA
|
|
DM
|
diabetes mellitus
|
|
STZ
|
streptozotocin
|
|
HG
|
high concentration glucose
|
References
|
1
|
Boudina S and Abel ED: Diabetic
cardiomyopathy, causes and effects. Rev Endocr Metab Disord.
11:31–39. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Trachanas K, Sideris S, Aggeli C,
Poulidakis E, Gatzoulis K, Tousoulis D and Kallikazaros I: Diabetic
cardiomyopathy: From pathophysiology to treatment. Hellenic J
Cardiol. 55:411–421. 2014.PubMed/NCBI
|
|
3
|
Hayat SA, Patel B, Khattar RS and Malik
RA: Diabetic cardiomyopathy: Mechanisms, diagnosis and treatment.
Clin Sci (Lond). 107:539–557. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bugger H and Abel ED: Molecular mechanisms
of diabetic cardiomyopathy. Diabetologia. 57:660–671. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Devereux RB, Roman MJ, Paranicas M,
O'Grady MJ, Lee ET, Welty TK, Fabsitz RR, Robbins D, Rhoades ER and
Howard BV: Impact of diabetes on cardiac structure and function:
The strong heart study. Circulation. 101:2271–2276. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lüscher TF: Heart failure and
comorbidities: Renal failure, diabetes, atrial fibrillation, and
inflammation. Eur Heart J. 36:1415–1417. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jia G, Hill MA and Sowers JR: Diabetic
cardiomyopathy: An update of mechanisms contributing to this
clinical entity. Circ Res. 122:624–638. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Boudina S and Abel ED: Diabetic
cardiomyopathy revisited. Circulation. 115:3213–3223. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Costantino S, Paneni F, Lüscher TF and
Cosentino F: MicroRNA profiling unveils hyperglycaemic memory in
the diabetic heart. Eur Heart J. 37:572–576. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Moran VA, Perera RJ and Khalil AM:
Emerging functional and mechanistic paradigms of mammalian long
non-coding RNAs. Nucleic Acids Res. 40:6391–6400. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu K, Hou Y, Liu Y and Zheng J: LncRNA
SNHG15 contributes to proliferation, invasion and autophagy in
osteosarcoma cells by sponging miR-141. J Biomed Sci. 24:462017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang K, Liu CY, Zhou LY, Wang JX, Wang M,
Zhao B, Zhao WK, Xu SJ, Fan LH, Zhang XJ, et al: APF lncRNA
regulates autophagy and myocardial infarction by targeting
miR-188-3p. Nat Commun. 6:67792015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sallam T, Sandhu J and Tontonoz P: Long
noncoding RNA discovery in cardiovascular disease: Decoding form to
function. Circ Res. 122:155–166. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lander ES, Linton LM, Birren B, Nusbaum C,
Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al:
Initial sequencing and analysis of the human genome. Nature.
409:860–921. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Amaral PP, Clark MB, Gascoigne DK, Dinger
ME and Mattick JS: lncRNAdb: A reference database for long
noncoding RNAs. Nucleic Acids Res. 39:D146–D151. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Smith CM and Steitz JA: Classification of
gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member
of the 5′-terminal oligopyrimidine gene family reveals common
features of snoRNA host genes. Mol Cell Biol. 18:6897–6909. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Meyuhas O: Synthesis of the translational
apparatus is regulated at the translational level. Eur J Biochem.
267:6321–6330. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Carter G, Miladinovic B, Patel AA, Deland
L, Mastorides S and Patel NA: Circulating long noncoding RNA GAS5
levels are correlated to prevalence of type 2 diabetes mellitus.
BBA Clin. 4:102–107. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Clark JD, Baldwin RL, Bayne KA, Brown MJ,
Gebhart GF and Gonder JC: Guide for the care and use of laboratory
animals. Institute of Laboratory Animal Resources, Institute of
Laboratory Animal Resources Commission on Life Sciences, National
Research Council, National Academy Press; Washington, D.C.:
1996
|
|
20
|
Zolotukhin S, Byrne BJ, Mason E,
Zolotukhin I, Potter M, Chesnut K, Summerford C, Samulski RJ and
Muzyczka N: Recombinant adeno-associated virus purification using
novel methods improves infectious titer and yield. Gene Ther.
6:973–985. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Puglia AL, Rezende AG, Jorge SA, Wagner R,
Pereira CA and Astray RM: Quantitative RT-PCR for titration of
replication-defective recombinant Semliki forest virus. J Virol
Methods. 193:647–652. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pfaffl MW: A new mathematical model for
relative quantification in real-time RT-PCR. Nucleic Acids Res.
29:e452001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhong P, Wu L, Qian Y, Fang Q, Liang D,
Wang J, Zeng C, Wang Y and Liang G: Blockage of ROS and
NF-κB-mediated inflammation by a new chalcone L6H9 protects
cardiomyocytes from hyperglycemia-induced injuries. Biochim Biophys
Acta. 1852:1230–1241. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Y, Zhang YY, Li TT, Wang J, Jiang Y,
Zhao Y, Jin XX, Xue GL, Yang Y, Zhang XF, et al: Ablation of
interleukin-17 alleviated cardiac interstitial fibrosis and
improved cardiac function via inhibiting long non-coding
RNA-AK081284 in diabetic mice. J Mol Cell Cardiol. 115:64–72. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Uchida S and Dimmeler S: Long noncoding
RNAs in cardiovascular diseases. Circ Res. 116:737–750. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shen S, Jiang H, Bei Y, Xiao J and Li X:
Long non-coding RNAs in cardiac remodeling. Cell Physiol Biochem.
41:1830–1837. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Greco S, Zaccagnini G, Perfetti A, Fuschi
P, Valaperta R, Voellenkle C, Castelvecchio S, Gaetano C, Finato N,
Beltrami AP, et al: Long noncoding RNA dysregulation in ischemic
heart failure. J Transl Med. 14:1832016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yin Q, Wu A and Liu M: Plasma long
non-coding RNA (lncRNA) GAS5 is a new biomarker for coronary artery
disease. Med Sci Monit. 23:6042–6048. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lee WS and Kim J: Diabetic cardiomyopathy:
Where we are and where we are going. Korean J Intern Med.
32:404–421. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang F, Qin Y, Wang Y, Li A, Lv J, Sun X,
Che H, Han T, Meng S, Bai Y and Wang L: LncRNA KCNQ1OT1 mediates
pyroptosis in diabetic cardiomyopathy. Cell Physiol Biochem.
50:1230–1244. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li X, Wang H, Yao B, Xu W, Chen J and Zhou
X: lncRNA H19/miR-675 axis regulates cardiomyocyte apoptosis by
targeting VDAC1 in diabetic cardiomyopathy. Sci Rep. 6:363402016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhou X, Zhang W, Jin M, Chen J, Xu W and
Kong X: lncRNA MIAT functions as a competing endogenous RNA to
upregulate DAPK2 by sponging miR-22-3p in diabetic cardiomyopathy.
Cell Death Dis. 8:e29292017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gao L, Wang X, Guo S, Xiao L, Liang C,
Wang Z, Li Y, Liu Y, Yao R, Liu Y and Zhang Y: LncRNA HOTAIR
functions as a competing endogenous RNA to upregulate SIRT1 by
sponging miR-34a in diabetic cardiomyopathy. J Cell Physiol.
234:4944–4958. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sathishkumar C, Prabu P, Mohan V and
Balasubramanyam M: Linking a role of lncRNAs (long non-coding RNAs)
with insulin resistance, accelerated senescence, and inflammation
in patients with type 2 diabetes. Hum Genomics. 12:412018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tao S, Chen L, Song J, Zhu N, Song X, Shi
R, Ge G and Zhang Y: Tanshinone IIA ameliorates diabetic
cardiomyopathy by inhibiting Grp78 and CHOP expression in
STZ-induced diabetes rats. Exp Ther Med. 18:729–734.
2019.PubMed/NCBI
|
|
36
|
Varma U, Koutsifeli P, Benson VL, Mellor
KM and Delbridge LMD: Molecular mechanisms of cardiac pathology in
diabetes-Experimental insights. Biochim Biophys Acta Mol Basis Dis.
1864:1949–1959. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xie Z, Lau K, Eby B, Lozano P, He C,
Pennington B, Li H, Rathi S, Dong Y, Tian R, et al: Improvement of
cardiac functions by chronic metformin treatment is associated with
enhanced cardiac autophagy in diabetic OVE26 mice. Diabetes.
60:1770–1778. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yamaguchi O: Autophagy in the heart. Circ
J. 83:697–704. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang M, Lin J, Wang S, Cheng Z, Hu J,
Wang T, Man W, Yin T, Guo W, Gao E, et al: Melatonin protects
against diabetic cardiomyopathy through Mst1/Sirt3 signaling. J
Pineal Res. 63:2017. View Article : Google Scholar
|
|
40
|
Paraskevopoulou MD and Hatzigeorgiou AG:
Analyzing MiRNA-LncRNA interactions. Methods Mol Biol.
1402:271–286. 2016. View Article : Google Scholar
|
|
41
|
Yin G, Zhang B and Li J: miR2213p promotes
the cell growth of nonsmall cell lung cancer by targeting p27. Mol
Med Rep. 20:604–612. 2019.PubMed/NCBI
|