Introduction
Hepatocellular carcinoma (HCC) is a primary liver
cancer, caused by cirrhosis resulting from infection with a
Hepatitis virus, typically Hepatitis B or C, or alcohol (1). In 2012, 782,000 individuals suffered
from liver cancer, and in 2015 HCC resulted in 810,500
cancer-associated deaths worldwide (2). The high incidence and mortality rates
indicate that it is imperative to identify new therapeutic methods
and molecular targets for HCC treatment.
Long non-coding (lnc)RNAs are a class of RNA
molecules, >200 nucleotides in length, which do not encode
proteins (3). Unusual expression of
lncRNAs often appears in different types of cancer (4–6),
including HCC (7,8). Accumulating data demonstrated that
lncRNAs played a central role in cancer initiation, progression and
growth in multiple tumors (9–11). The
lncRNA, nuclear-enriched abundant transcript 1 (NEAT1), is
specifically localized in nuclear paraspeckles (12). It was reported to be abnormally,
highly expressed in glioma and lung cancer, but was lowly expressed
in de novo acute promyelocytic leukemia (13–15).
Zhu et al (16) found that
NEAT1 was upregulated in HCC and promoted the malignant biological
properties of HCC. However, the mechanism of NEAT1 in regulating
HCC requires further investigation.
MicroRNAs (miRNAs/miRs) are highly conserved and
small non-coding RNAs (~22 nucleotides in length), which regulate
gene expression levels by specifically targeting the
3′-untranslated regions (3′-UTR) of mRNA at the
post-transcriptional level (17).
Research into different types of cancer have increased the
understanding of the role of miRNAs (18–20).
Several studies have confirmed that miR-503 could serve as a tumor
suppressor in HCC (21,22). However, the precise mechanism of
miR-503 has not been completely elucidated.
Smoothened, frizzled class receptor (SMO) is a class
of frizzled G protein-coupled receptor. Increasing evidence has
indicated that SMO participated in the regulation of numerous types
of cancer, including basal-cell carcinomas and breast cancer, and
it might function as an oncogene (23,24).
However, whether SMO could be an attractive cancer drug target
requires further investigation.
In the present study, the expression levels of NEAT1
were investigated in HCC tissues and cells. In addition, the
function of NEAT1 and the regulatory mechanism between NEAT1,
miR-503 and SMO in HCC progression were also further
investigated.
Materials and methods
Samples and cell culture
Hepatocellular carcinoma tissues and adjacent normal
tissues (>5 cm laterally from the edge of the cancerous region)
were collected from 31 patients with HCC (<43 age ≤67; 19 male
and 12 female patients) during surgeries between July 2016 and
January 2019 at Weifang People's Hospital (Shandong, China). The
included patients did not receive any chemotherapy or radiation
treatment prior to surgery. All patients provided written informed
consent and the present study was authorized by the Ethics
Committee of Weifang People's Hospital (Shandong, China).
All cell lines (the human normal hepatic cell line,
QZG, and the HCC cell lines, Huh-7 and Hep3B) were purchased from
The Cell Bank of Type Culture Collection of the Chinese Academy of
Sciences. Dulbecco's modified Eagle's medium, supplemented with 10%
fetal bovine serum (FBS) (both from Sigma-Aldrich; Merck KGaA), was
used to culture the cells at 37°C in a humidified incubator with 5%
CO2.
Cell transfection
Small interfering (si) RNA targeting NEAT1
(si-NEAT1), miR-503 mimic (miR-503) and its inhibitor
(anti-miR-503), as well as the corresponding negative controls,
were purchased from Shanghai GenePharma Co., Ltd. The sequences for
siRNAs and miRNAs were as follows: si-NEAT1,
5′-UGGUAAUGGUGGAGGAAGAUU-3′; si-NC, 5′-UUCUCCGAACGUGUCACGU-3′;
miR-503 mimic, 5′-UAGCAGCGGGAACAGUUCUGCAG-3′; miR-NC mimic,
5′-UCACAACCUCCUAGAAAGAGUAGA-3′; anti-miR-503,
5′-UAGCAGCGGGAACAGUUCUGCAG-3′; and anti-miR-NC,
5′-UUGUACUACACAAAAGUACUG-3′. The full-length NEAT1 cDNA and the
3′UTR of SMO were synthesized by Guangzhou RiboBio Co., Ltd., and
cloned into the vector, pcDNA3.1 (Invitrogen; Thermo Fisher
Scientific, Inc.). These were subsequently verified using
sequencing (conducted by Guangzhou RiboBio Co., Ltd.), named as
pcDNA-NEAT1 and pcDNA-SMO, and the empty vector was used as a
negative control (pcDNA-control). When cells were cultured to ~70%
confluence, the siRNAs (final concentration, 20 nM), miRNAs (at
final concentration of 20 nM for miRNA mimic, and 25 nM for miRNA
inhibitor) and vectors (at final concentration of 1 µg/ml) were
transfected into the cells using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Then, the cells were incubated in a
humidified incubator with 5% CO2 at 37°C for 24 or 48 h
upon transfection. Cells were collected for further research after
this incubation.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA from the tissues or cells were isolated
using a RNAiso Plus kit (Takara Biotechnology Co., Ltd.) according
to the manufacturer's instructions. After the concentration and
purity of RNA were detected using a NanoDrop 2000/2000c (Thermo
Fisher Scientific, Inc.), 1 µg RNA was reverse transcribed into
cDNA using the PrimeScript™ RT Master Mix kit (Takara Biotechnology
Co., Ltd.) at 37°C for 15 min and for a further 5 sec at 85°C. The
RT-qPCR was conducted using a Fast SYBR® Green Master
Mix kit (Thermo Fisher Scientific, Inc.) using the following
thermocycling conditions: Initial denaturation at 95°C for 10 min
followed by 35 cycles of 95°C for 15 sec and 60°C for 1 min. The
data were analyzed using the 2−ΔΔCq method (25). β-actin and U6 were used as the
endogenous controls. The primer sequences used are shown in
Table I.
 | Table I.Primer sequences used in RT-qPCR. |
Table I.
Primer sequences used in RT-qPCR.
| Gene name | Forward
(5′-3′) | Reverse
(5′-3′) |
|---|
| NEAT1 |
TGGCTAGCTCAGGGCTTCAG |
TCTCCTTGCCAAGCTTCCTTC |
| miR-503 |
CCTATTTCCCATGATTCCTTCATA |
GTAATACGGTTATCCACGCG |
| SMO |
TGCTCATCGTGGGAGGCTACTT |
ATCTTGCTGGCAGCCTTCTCAC |
| β-actin |
GCACCACACCTTCTACAATG |
TGCTTGCTGATCCACATCTG |
| U6 |
CTTCGGCAGCACATATACT |
AAAATATGGAACGCTTCACG |
MTT assay
MTT (Sigma-Aldrich; Merck KGaA) was used to
investigate cell viability. Transfected cells were seeded in
96-well plates (5×103 cells per well) for 0, 24, 48 or
72 h. A total of 20 µl MTT solution was added to the wells and the
cells were incubated at 37°C for another 4 h. Following which, the
media was aspirated off and 200 µl dimethyl sulfoxide (Thermo
Fisher Scientific, Inc.) was added. Optical density values were
examined at 490 nm using a microplate reader (Bio-Rad Laboratories,
Inc.) and were analyzed with Microplate Manager Software v. 6.0
(Bio-Rad Laboratories, Inc.).
Cell migration and invasion
assays
Transwell chambers, that were precoated with or
without Matrigel at room temperature for 1 h (Corning Inc.) prior
to use, were utilized to detect the invasion or migration abilities
of the cells. Transfected cells in serum-free media were added to
the upper chamber, and the lower chamber contained the media with
10% FBS. After incubation at 37°C for 24 h, the cells on the upper
surface of the membrane were wiped off with a cotton swab and the
cells in lower surface were analyzed using an inverted microscope
(Leica DMi1; Leica Microsystems, Inc.) at ×40 magnification,
following staining with crystal violet for 10 min at 37°C. The
upper chambers, coated with Matrigel (Corning Inc.), were used to
determine cell invasion with the same method.
Apoptosis assay
Apoptosis was investigated using an Apoptosis
Detection kit (BD Biosciences) according to the manufacturer's
instructions. Cells were centrifuged at 200 × g for 5 min at room
temperature, resuspended using the provided binding buffer. Then, 5
µl Annexin V-fluorescein isothiocyanate (Annexin V-FITC) and 5 µl
propidium iodide (PI) were added to the buffer in the dark for a
further 5-min incubation at 4°C. Stained cells were analyzed using
an Attune NxT flow cytometer and Attune NxT Software version 3.2.1
(Thermo Fisher Scientific, Inc.).
Luciferase reporter assay
The target sites were analyzed using StarBase online
software version 3.0 (between NEAT1 and miR-503) (26) or TargetScanHuman 7.1 (between SMO
and miR-503) (27). The sequences
of wild-type NEAT1 (WT-NEAT1), mutant NEAT1 (MUT-NEAT1), WT SMO
3′-UTR (WT-SMO) and MUT SMO 3′-UTR (MUT-SMO) containing the
putative binding sites of miR-503 were amplified and ligated into
the pGL3 luciferase reporter vector (Promega Corporation). The
reporter vector and miR-503 or miR-control were cotransfected into
the Huh-7 and Hep3B cells using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) as aforementioned.
After 24 h of transfection, the luciferase activity was measured
using the Dual-Glo® Luciferase Assay system (Promega
Corporation). The firefly luciferase activity was normalized to the
Renilla luciferase activity.
Western blot analysis
Total protein was extracted using the RIPA buffer
(Thermo Fisher Scientific, Inc.) from tissues and cells. The
protein concentration was determined using a Pierce™ Rapid Gold BCA
Protein Assay kit (Thermo Fisher Scientific, Inc.). Equal amounts
of total protein (30 µg/lane) was separated using SDS-PAGE (10%
gels) then, transferred onto PVD membranes (Merck KGaA).
Subsequently, the membranes were blocked with 5% skimmed milk for 3
h at 4°C. After being washed three times with PBS, the membranes
were incubated with the primary antibodies against SMO (1:1,000;
cat. no. ab235183) or β-actin (1:2,500; cat. no. ab8226) (both from
Abcam) overnight at 4°C. The secondary antibody Goat Anti-Rabbit
IgG H&L [horseradish peroxidase (HRP)] (1:3,000; cat. no.
ab205718; Abcam) and Goat Anti-Mouse IgG H&L (HRP) (1:10,000;
cat. no. ab205718; Abcam) were added for incubation for another 2 h
at room temperature after being washed three times with PBS at room
temperature. The protein bands were visualized using Super ECL
Detection Reagent (Vazyme Biotech Co., Ltd.) and the ChemiDoc™ MP
Imaging System (Bio-Rad Laboratories, Inc.). ImageJ software
V1.8.0_172 (National Institutes of Health) was used for
semi-quantification of western blotting results.
Statistical analysis
Experimental data was analyzed using GraphPad Prism
v7 software (GraphPad Software, Inc.) and are presented using the
mean ± standard deviation (SD). A total of 2 independent groups
were compared using a paired or unpaired Student's t-test, while
>2 groups were analyzed using the one-way ANOVA followed by
Tukey's post hoc test. Every experiment was repeated three times,
independently. The correlation between the expression levels of
miR-503 and NEAT1 or SMO was analyzed using Pearson correlation
analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
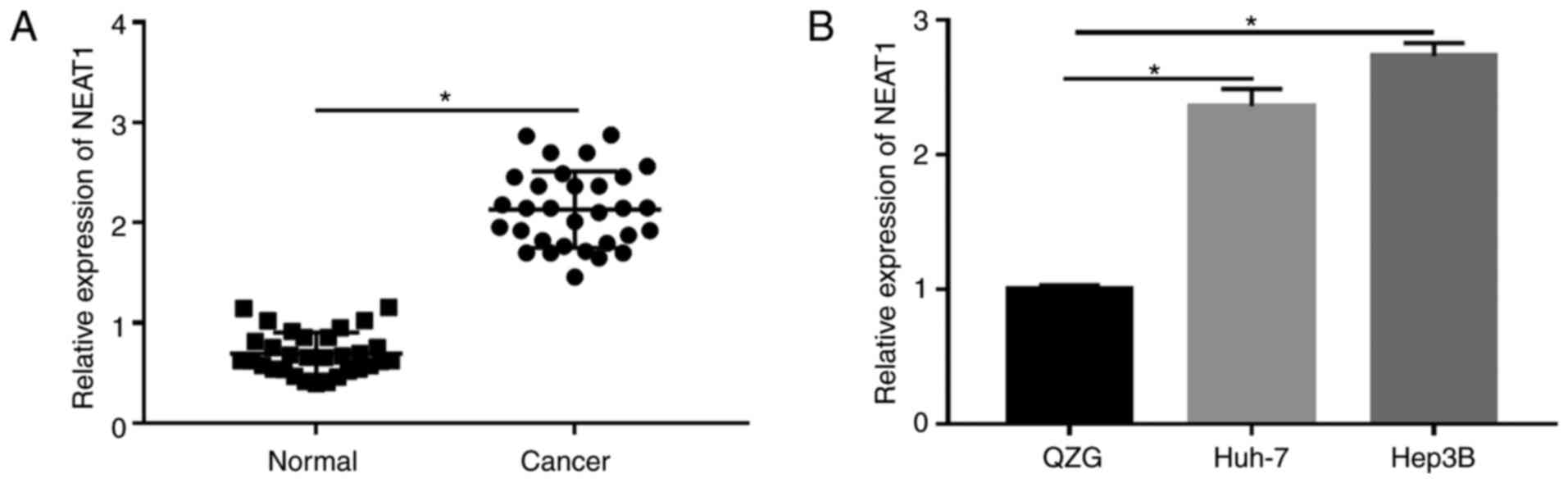
NEAT1 lncRNA was significantly
increased in HCC tissues and cells
To investigate the role of NEAT1 in HCC, the mRNA
expression level was first measured using RT-qPCR in HCC tissues.
The data showed that NEAT1 was significantly upregulated in HCC
tissues compared with that in the corresponding normal tissues
(Fig. 1A). Subsequently, the mRNA
expression levels of NEAT1 in the HCC cell lines Huh-7 and Hep3B
were determined and the results revealed that NEAT1 expression was
also highly elevated (Fig. 1B).
These results supported the hypothesis that NEAT1 might function as
an oncogene in the progression of HCC.
NEAT1 silencing induces apoptosis and
inhibits the viability, migration and invasion of HCC cells
To further investigate the function of NEAT1 in HCC,
the Huh-7 and Hep3B cell lines were transfected with si-NEAT1 and
the transfection efficiency was determined using RT-qPCR (Fig. 2A). MTT assays revealed that the cell
viability of the Huh-7 and Hep3B cell lines were both significantly
suppressed in the si-NEAT1 group compared with that in the
si-control group (Fig. 2B). Flow
cytometry analyses of apoptosis demonstrated that knockdown of
NEAT1 caused a significant increase in the rate of apoptosis in the
two cell lines (Fig. 2C).
Furthermore, Transwell assay, with or without Matrigel, indicated
that downregulation of NEAT1 weakened the ability of HCC cells to
invade or migrate (Fig. 2D and E).
Taken together, these data demonstrated that NEAT1 silencing
promoted cell apoptosis and suppressed the viability, migration and
invasion of HCC cells in vitro.
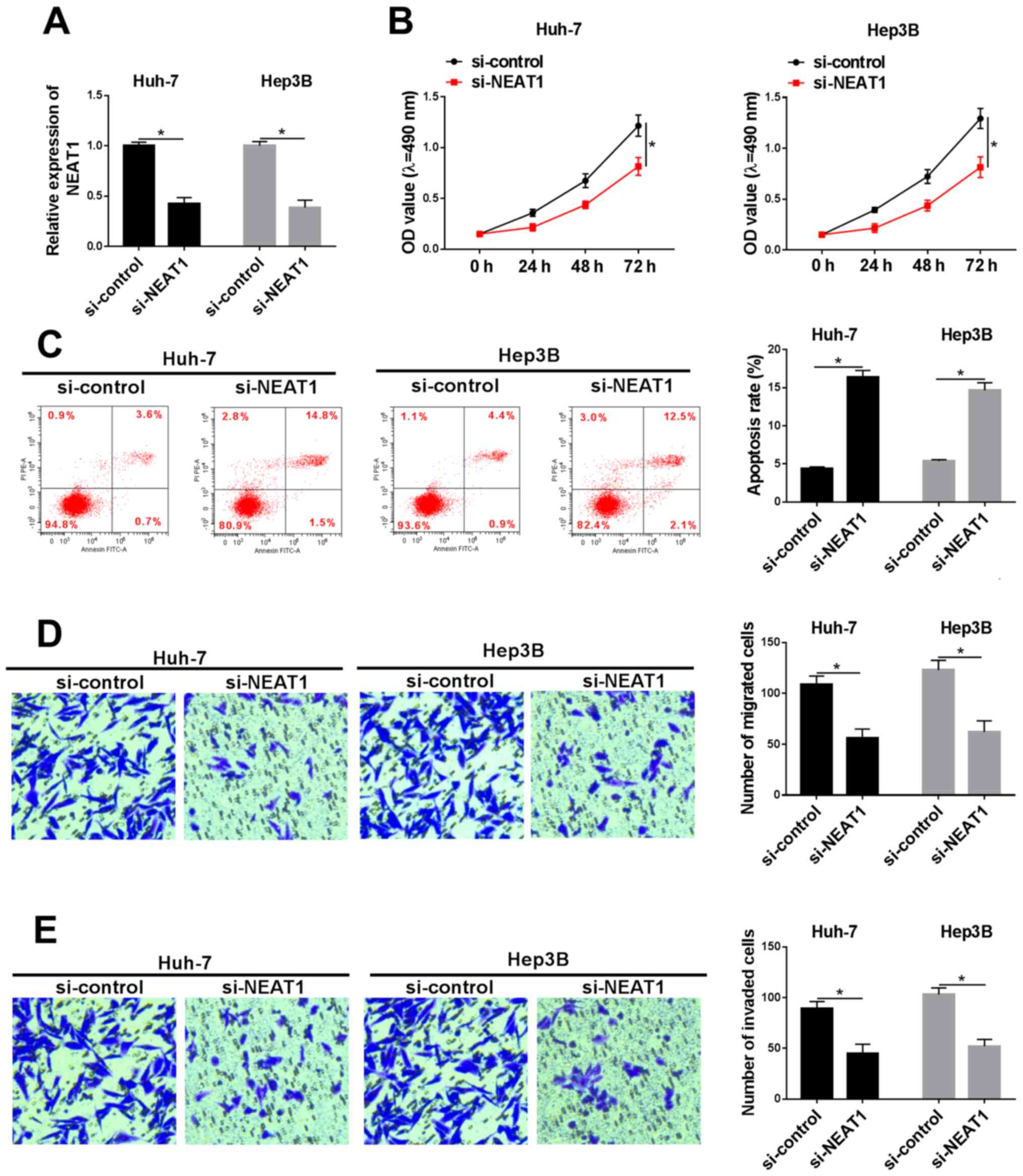 | Figure 2.Silencing of NEAT1 inhibits cell
viability, migration and invasion, and promoted apoptosis in
hepatocellular carcinoma cells. Following transfection with
si-NEAT1 or si-control, (A) the expression level of NEAT1, (B) cell
viability and (C) apoptosis was investigated in the Huh-7 and Hep3B
cell lines using reverse transcription-quantitative PCR, MTT assay
and flow cytometry, respectively. Transwell (D) migration and (E)
invasion assays were performed, and the numbers of migrated or
invaded cells were calculated, following transfection with si-NEAT1
or si-control (magnification, ×100). *P<0.05. NEAT1,
nuclear-enriched abundant transcript 1; si, small interfering RNA;
OD, optical density. |
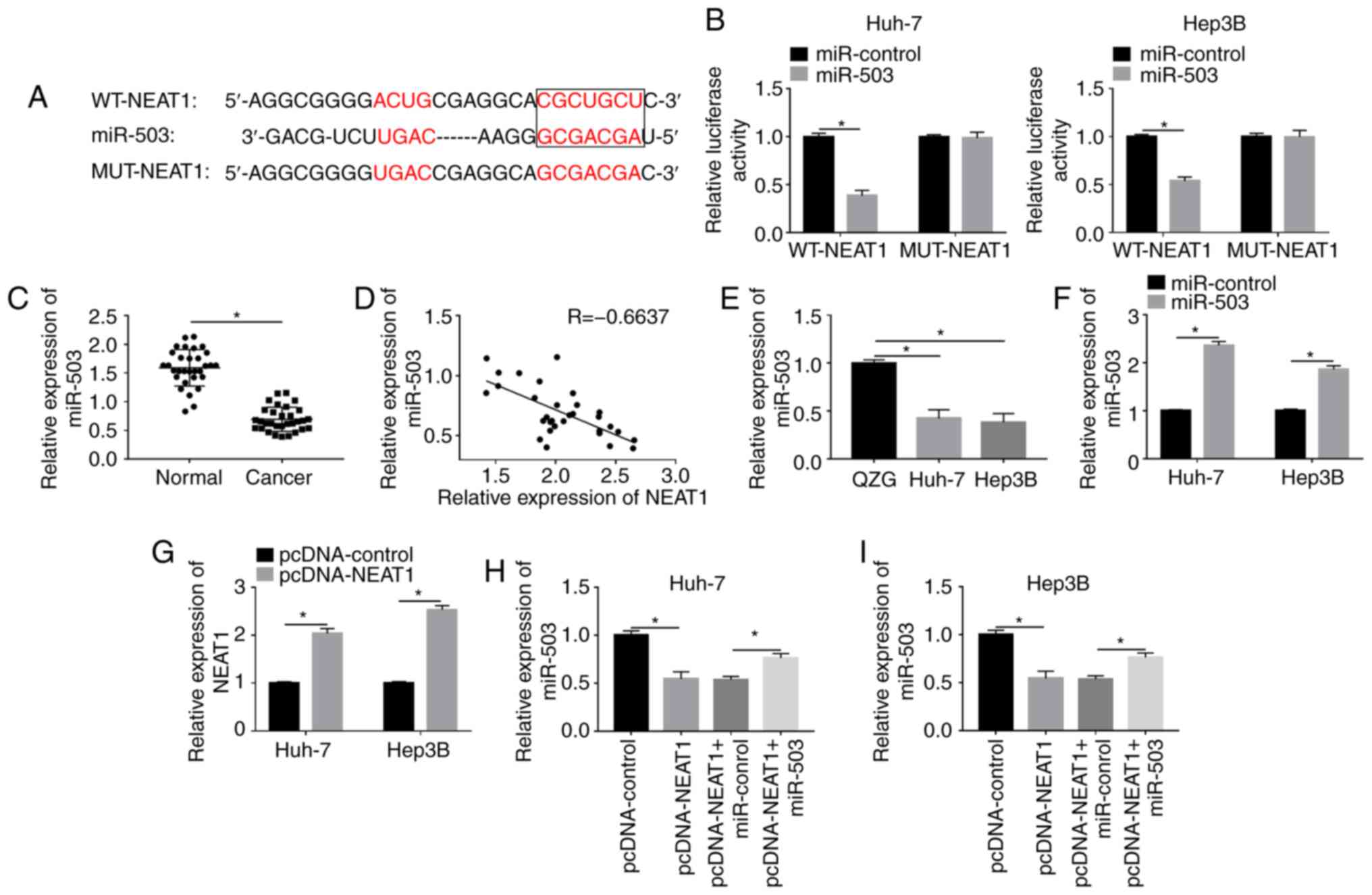
NEAT1 directly interacts with miR-503
and negatively regulates the expression level of miR-503
Previous studies have confirmed that the interaction
between lncRNA and miRNA has an essential role in the regulation of
cancer in general (28,29). Using the online database StarBase,
NEAT1 was found to contain binding sites for miR-503 (Fig. 3A), and dual-luciferase reporter
assays were conducted to validate the prediction. The results
showed that the luciferase activity of WT-NEAT1 was significantly
decreased by miR-503 in the Huh-7 and Hep3B cell lines, whereas the
activity of MUT-NEAT1 was not changed (Fig. 3B). In addition, the expression level
of miR-503 was significantly downregulated in tumor tissues
(Fig. 3C) and the mRNA expression
level of NEAT1 was negatively correlated with the expression of
miR-503 in 31 HCC tissues (Fig.
3D). Furthermore, miR-503 expression was also found to be
significantly decreased in the Huh-7 and Hep3B cell lines compared
with that in the normal liver cell line (Fig. 3E). Next, miR-503 mimic and
pcDNA-NEAT1 were used to overexpress miR-503 and NEAT1,
respectively, and the expression levels of miR-503 and NEAT1 in the
transfected cells were measured using RT-qPCR (Fig. 3F and G). The results revealed that
upregulation of NEAT1 reduced the expression of miR-503, which
could be rescued by enhancing miR-503 expression in HCC cells
(Fig. 3H and I). The results
illustrated that NEAT1 could interact with miR-503 and negatively
modulated the expression level of miR-503 in the HCC cell
lines.
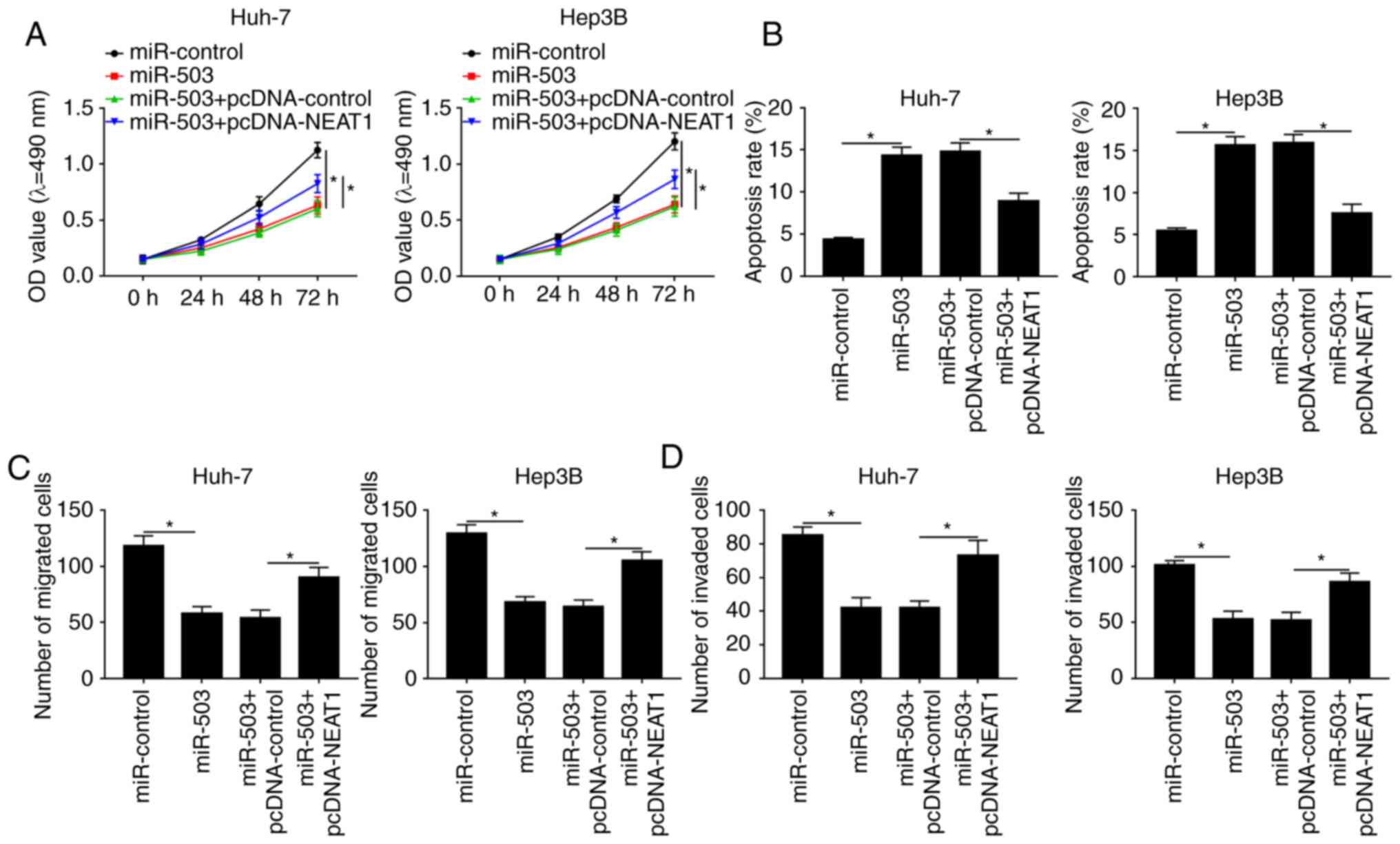
NEAT1 overexpression reverses the
miR-503-mediated effects on the viability, apoptosis, migration and
invasion of the HCC cells
To further investigate the functions of the
interaction between NEAT1 and miR-503 in the HCC cell lines, MTT,
Transwell and apoptosis assays were performed. MTT assays indicated
that overexpression of miR-503 reduced the viability of HCC cells
compared with the miR-control group, whereas elevating the
expression level of NEAT1 recovered cell viability and rescued the
effect of miR-503-mediated inhibition on cell viability (Fig. 4A). Flow cytometry analysis of
apoptosis demonstrated that NEAT1 overexpression reversed the
effect of miR-503-mediated promotion on apoptosis (Figs. 4B; S1A
and B). Similarly, Transwell assays revealed that miR-503
suppressed the migration and invasion of the HCC cell lines;
however, the effects could be abrogated by NEAT1 overexpression
(Figs. 4C and D; S1C-F). Collectively, these results
suggested that upregulation of NEAT1 abolished the effects of
miR-503-mediated inhibition on the viability, migration, invasion
and promoted apoptosis of the HCC cell lines.
miR-503 targets and negatively
regulates SMO expression level in vitro
To determine the underlying mechanism of miR-503 in
HCCs, the TargetScan online tool was utilized to identify the
possible target genes of miR-503. The result indicated that miR-503
had a potential binding site on the 3′-UTR of SMO (Fig. 5A). To validate the prediction,
luciferase reporter plasmids harboring miR-503 targeting region
(WT-SMO) and its mutant (MUT-SMO) were constructed. The results
showed that miR-503 could reduce the luciferase activity of WT-SMO
in the Huh and Hep3B cell lines, while it had little effect on the
luciferase activity of MUT-SMO (Fig.
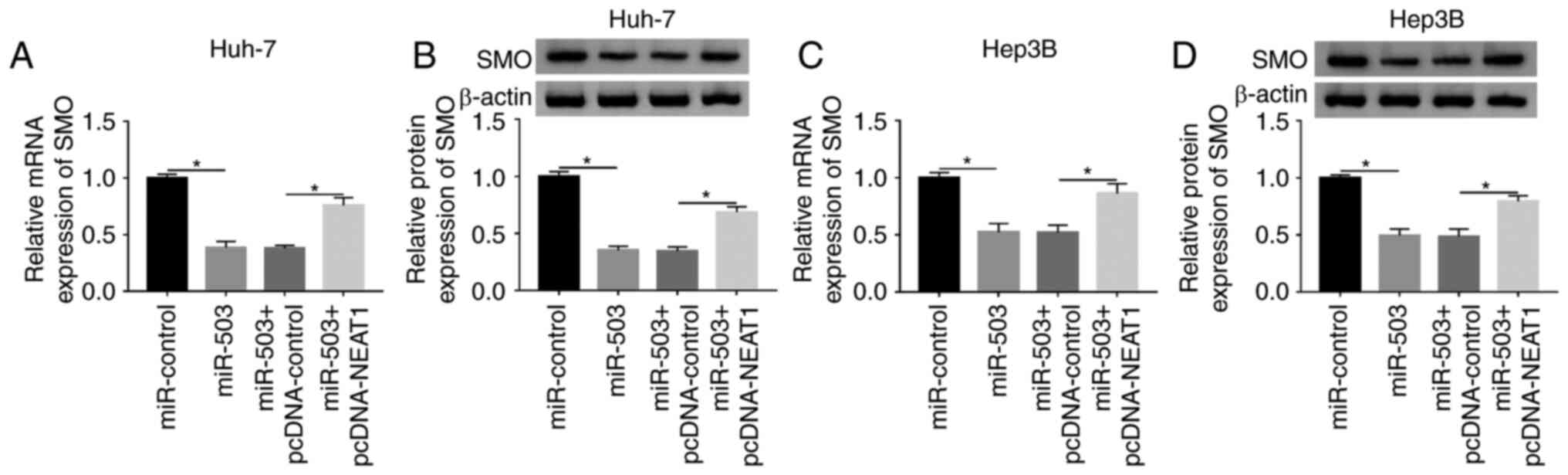
5B). In addition, SMO was found to be significantly upregulated
in HCC tissues compared with that in the normal adjacent tissues
(Fig. 5C). Further analysis
indicated that the expression level of SMO was negatively
correlated with miR-503 expression level (Fig. 5D), and the protein and mRNA
expression levels of SMO were elevated in both HCC tissues and
cells, respectively (Fig. 5E-G).
Subsequently, the expression level of miR-503 in transfected HCC
cells was investigated and the results showed that miR-503 was
significantly reduced in HCC cells transfected with the miRNA
inhibitor (anti-miR-503) compared with that in the control group
(anti-control) (Fig. 5H). To
investigate the association between SMO and miR-503 in the HCC cell
lines, the expression level of SMO in HCC cells transfected with
miR-503 or anti-miR-503, as well as with the corresponding
controls, was measured. The results revealed that downregulation of
miR-503 significantly increased the mRNA and protein expression
levels of SMO in the HCC cell lines (Fig. 5I-L), which are also consistent with
the finding that SMO mRNA expression level was negatively
correlated with miR-503 expression level. Collectively, these
results demonstrated that miR-503 could bind to the 3′-UTR of SMO
and negatively modulated the expression level of SMO in the HCC
cell lines.
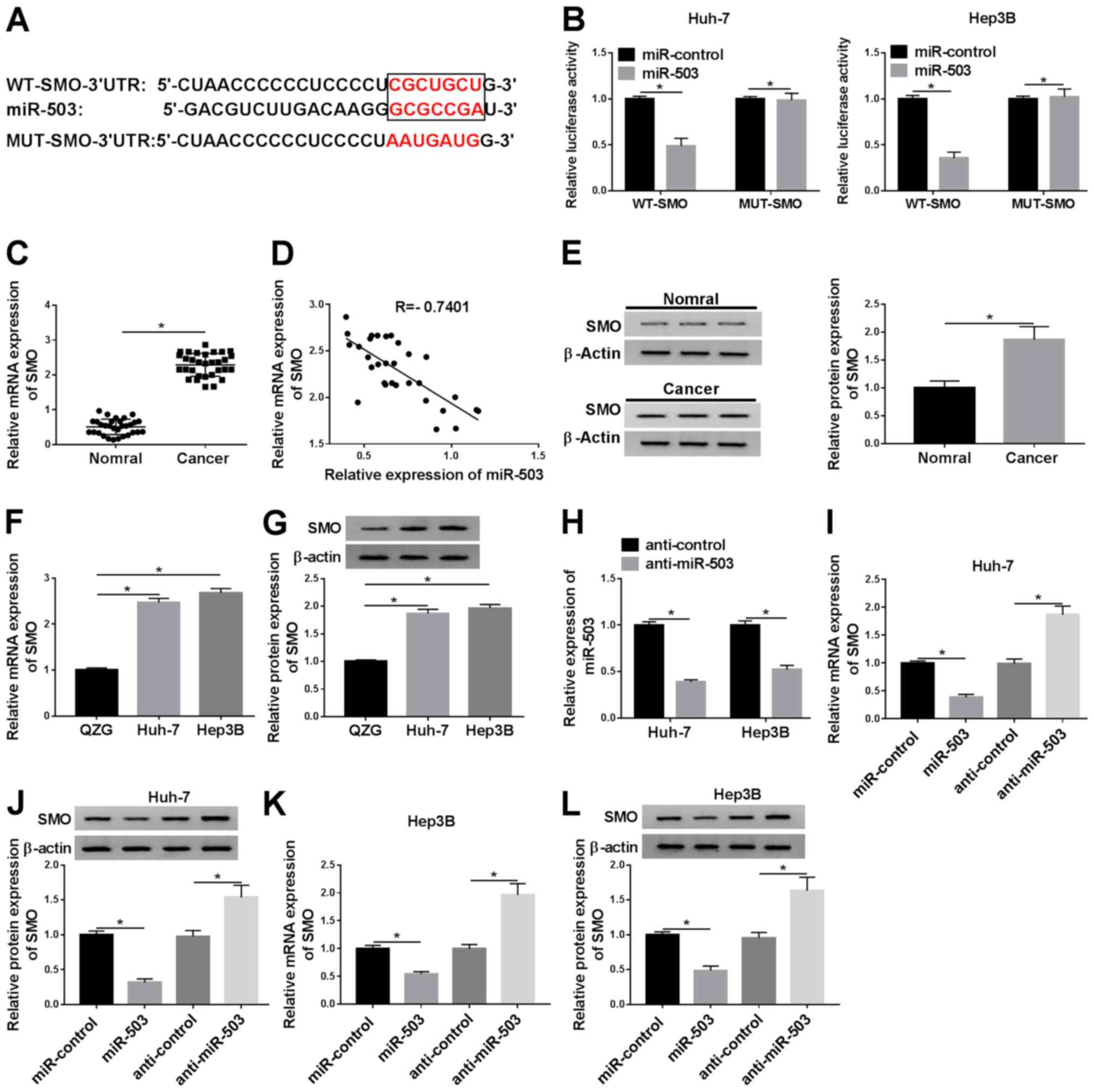 | Figure 5.miR-503 binds to SMO and negatively
regulates the expression level of SMO. (A) The binding sites
between miR-503 and SMO were predicted using the TargetScan online
tool. (B) The luciferase activity of cells cotransfected with the
miR-503 and WT-SMO or MUT-SMO was investigated using a
dual-luciferase assay. (C) The expression level of SMO in HCC
tissues (n=31) and adjacent normal tissues (n=31) was measured
using RT-qPCR. (D) The correlation between the expression of SMO
and miR-503 in HCC tissues was determined. (E) The protein
expression level of SMO was detected using western blot analysis.
The (F) mRNA and (G) protein expression levels of SMO were
evaluated using RT-qPCR and western blot analysis, respectively.
(H) The expression level of miR-503 in the Huh-7 and Hep3B cells
transfected with anti-control or anti-miR-503 were detected using
RT-qPCR. The mRNA and protein expression levels of SMO in (I and J)
Huh-7 and (K and L) Hep3B HCC cells transfected with miR-503 or
anti-miR-503 and their matched controls were determined using
RT-qPCR and western blot analysis, respectively. *P<0.05. WT,
wild-type; MUT, mutant; RT-qPCR, reverse transcription-quantitative
PCR; SMO, smoothened, frizzled class receptor; miR, microRNA;
anti-miR-503, microRNA inhibitor; anti-control, microRNA inhibitor
control; HCC, hepatocellular carcinoma. |
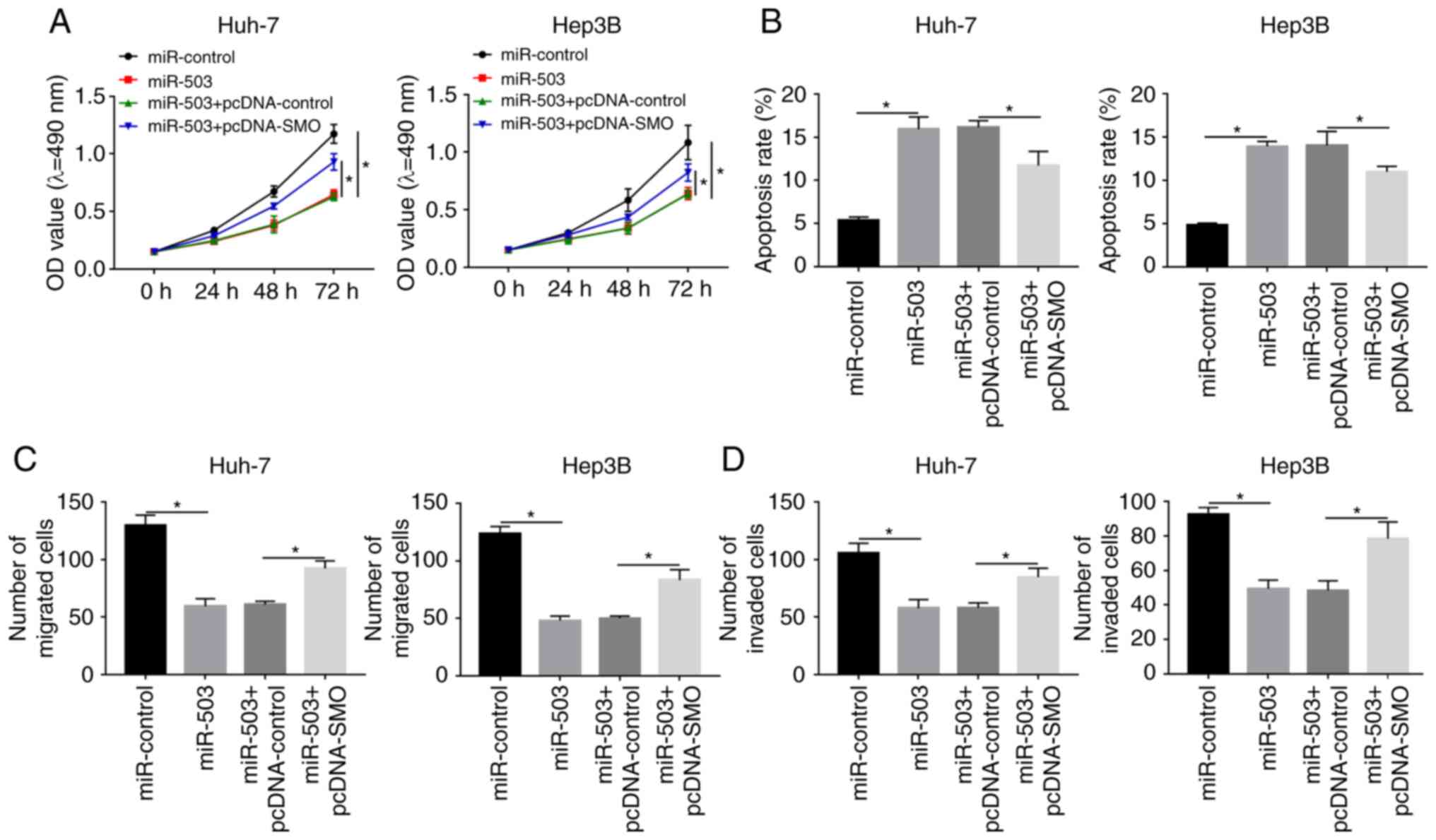
SMO overexpression reverses the
miR-503-mediated effects on viability, apoptosis, migration and
invasion of the HCC cell lines
The association between SMO and miR-503 was further
investigated with respect to HCC progression. MTT assays indicated
that SMO overexpression reduced the effects of miR-503-mediated
suppression on the viability of the HCC cell lines (Fig. 6A). Further analysis indicated that
the miR-503-mediated promotion on apoptosis in the HCC cell lines
was alleviated by the overexpression of SMO (Figs. 6B; S2A
and B). Similarly, the miR-508-mediated inhibition of migration
and invasion of the HCC cells could also be reduced by SMO
overexpression (Figs. 6C and D;
S2C-F). Collectively, these
results indicated that miR-503 may serve as a tumor suppressor and
SMO could function as an oncogene in the progression of HCC.
NEAT1 regulates SMO by sponging
miR-503 in the HCC cells
As NEAT1 could directly interact with miR-503, and
miR-503 could target the 3′-UTR of SMO, the underlying interactions
between these nucleic acids were further investigated. The mRNA and
protein expression levels of SMO in the Huh-7 and Hep3B cell lines
transfected with miR-503 or miR-503 + pcDNA-NEAT1 were measured
using RT-qPCR and western blot analysis, respectively (Fig. 7A-D). The results suggested that
miR-503 overexpression significantly reduced SMO expression level.
However, the suppression could be rescued following the
transfection of pcDNA-NEAT1. These results indicated that NEAT1
might act as a competitive endogenous sponge, interacting with and
downregulating miR-503, which affected the translational inhibition
of SMO, which was targeted by miR-503.
Discussion
HCC is one of the leading causes of
cancer-associated deaths worldwide (2,30).
Numerous studies have investigated the molecular mechanisms of HCC;
however, further investigation is required to develop more
effective treatments. The present study aimed to investigate the
underlying mechanism of NEAT1 in the regulation of HCC
progression.
Unusual expression of lncRNAs has been found in
different types of cancer and has been associated with cancer
progression (4,6,13).
Previous studies have indicated that the expression level of the
lncRNA NEAT1 was significantly elevated in numerous types of human
cancer, including non-small cell lung cancer, hepatocellular
carcinoma and cervical cancer (15,16,31),
while Zhu et al (16) found
that upregulated NEAT1 expression level promoted the malignant
biological properties of HCC. The results of the present study are
consistent with the study by Zhu et al (16), as it was observed that NEAT1 was
markedly upregulated in HCC tissues compared with that in adjacent
normal tissues, which indicated the potential importance of NEAT1
in the progression of HCC. In addition, further results from the
present study demonstrated that NEAT1 silencing could reduce cell
viability, migration and invasion and promoted apoptosis in the HCC
cell lines. These results suggested that NEAT1 played an oncogenic
role in the progression of HCC.
Previous studies have indicated that lncRNAs may
serve as sponges to interact with miRNAs, thereby regulating cancer
development (28,29). Results from the present study
identified miR-503 as a target of NEAT1. miR-503 has been
previously reported to be reduced in HCC tissues, and inhibited
cell proliferation and promoted apoptosis (22), the present study found the same
results and also revealed that the expression level of miR-503 was
also notably downregulated in 31 HCC tissues. Furthermore, the
expression level of miR-503 was negatively correlated with NEAT1 in
HCC tissues, and miR-503 was found to be downregulated by NEAT1.
miR-503 overexpression suppressed cell viability, migration,
invasion and induced apoptosis of the HCC cells, whereas its
effects could be restricted by the overexpression of NEAT1. From
these results, it was hypothesized that NEAT1 modulated HCC
progression by targeting miR-503 in vitro.
SMO was been found to be abnormally, highly
expressed in numerous types of cancer, including sporadic
basal-cell carcinoma and breast cancer, and might function as an
oncogene (23,24,32). A
previous report indicated that SMO was upregulated in pancreatic
cancer (33) and the results in the
present study revealed that SMO was also markedly increased in HCC
tissues and cells. Notably, SMO was predicted to be targeted by
miR-503, and the interaction was subsequently validated using a
dual-luciferase assay. Further analysis showed that miR-503
silencing increased the expression level of SMO, which clarified
the negative correlation between SMO and miR-503. In addition, SMO
overexpression could rescue the effects of miR-503-mediated
suppression on cell viability, migration and invasion, and promoted
the apoptosis of the HCC cells, which indicated that there was a
tumor-promoting effect of SMO on HCC. The results from the present
study suggested that miR-503 targeted the 3′-UTR of SMO in
vitro and negatively regulated SMO expression in the
progression of HCC.
As NEAT1 directly interacted with miR-503, and
miR-503 could target the 3′-UTR of SMO, the potential mechanisms
were further investigated. The results demonstrated that enhancing
the expression level of miR-503 led to a significant reduction in
SMO expression, however, the suppression was reduced by the
subsequent overexpression of NEAT1. The results indicated that
NEAT1 could act as a competitive endogenous sponge, which impaired
the translational inhibition of SMO by interacting with and
downregulating miR-503. Taken together, to the best of our
knowledge, the present study has elucidated a new mechanism that
NEAT1 regulated SMO by sponging miR-503 in HCC cells.
In summary, the present study revealed that NEAT1
and SMO were significantly elevated in HCC tissues and cells. In
addition, NEAT1 could directly interact miR-503, which could target
the 3′-UTR of SMO. Furthermore, downregulation of NEAT1 expression
impaired cell viability, migration and invasion, and induced the
apoptosis of HCC cells through the NEAT1/miR-503/SMO axis. The
identification of the new NEAT1/miR-503/SMO axis might facilitate
the development of novel therapeutic approaches for HCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CS conceived and designed the experiments. TX and YL
performed the experiments. YX analyzed the data and wrote the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
All patients provided written informed consent and
this research was authorized by the Ethics Committee of Weifang
People's Hospital (Shandong, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
GBD 2016 Causes of Death Collaborators, .
Global, regional, and national age-sex specific mortality for 264
causes of death, 1980–2016: A systematic analysis for the Global
Burden of Disease Study 2016. Lancet. 390:1151–1210. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
GBD 2015 Mortality and Causes of Death
Collaborators: Global, regional, and national life expectancy,
all-cause mortality, and cause-specific mortality for 249 causes of
death, 1980–2015: A systematic analysis for the Global Burden of
Disease Study 2015. Lancet. 388:1459–1544. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu
XY, Yu ZW, Jia YH, Bai XF, Li L, et al: The lncRNA CRNDE promotes
colorectal cancer cell proliferation and chemoresistance via
miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol
Cancer. 16:92017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hao NB, He YF, Li XQ, Wang K and Wang RL:
The role of miRNA and lncRNA in gastric cancer. Oncotarget.
8:81572–81582. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Niknafs YS, Han S, Ma T, Speers C, Zhang
C, Wilder-Romans K, Iyer MK, Pitchiaya S, Malik R, Hosono Y, et al:
The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1
in breast cancer progression. Nat Commun. 7:127912016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li X, Wang S, Li Z, Long X, Guo Z, Zhang
G, Zu J, Chen Y and Wen L: The lncRNA NEAT1 facilitates cell growth
and invasion via the miR-211/HMGA2 axis in breast cancer. Int J
Biol Macromol. 105:346–353. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu X, Li Z, Zheng H, Chan MT and Wu WK:
NEAT1: A novel cancer-related long non-coding RNA. Cell Prolif.
50:e123292017. View Article : Google Scholar
|
|
9
|
Raveh E, Matouk IJ, Gilon M and Hochberg
A: The H19 Long non-coding RNA in cancer initiation, progression
and metastasis-a proposed unifying theory. Mol Cancer. 14:1842015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xie H, Liao X, Chen Z, Fang Y, He A, Zhong
Y, Gao Q, Xiao H, Li J, Huang W and Liu Y: LncRNA MALAT1 inhibits
apoptosis and promotes invasion by antagonizing miR-125b in bladder
cancer cells. J Cancer. 8:3803–3811. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang G, Lu X and Yuan L: LncRNA: A link
between RNA and cancer. Biochim Biophys Acta. 1839:1097–1109. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Clemson CM, Hutchinson JN, Sara SA,
Ensminger AW, Fox AH, Chess A and Lawrence JB: An architectural
role for a nuclear noncoding RNA: NEAT1 RNA is essential for the
structure of paraspeckles. Mol Cell. 33:717–726. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
He C, Jiang B, Ma J and Li Q: Aberrant
NEAT1 expression is associated with clinical outcome in high grade
glioma patients. APMIS. 124:169–174. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zeng C, Xu Y, Xu L, Yu X, Cheng J, Yang L,
Chen S and Li Y: Inhibition of long non-coding RNA NEAT1 impairs
myeloid differentiation in acute promyelocytic leukemia cells. BMC
Cancer. 14:6932014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun C, Li S, Feng Z, Xi Y and Li D: Long
non-coding RNA NEAT1 promotes non-small cell lung cancer
progression through regulation of miR-377-3p-E2F3 pathway.
Oncotarget. 7:51784–51814. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu L, Yang N, Li C, Liu G, Pan W and Li
X: Long noncoding RNA NEAT1 promotes cell proliferation, migration,
and invasion in hepatocellular carcinoma through interacting with
miR-384. J Cell Biochem. 120:1997–2006. 2018. View Article : Google Scholar
|
|
17
|
Jovanovic M and Hengartner MO: miRNAs and
apoptosis: RNAs to die for. Oncogene. 25:6176–6187. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shamsizadeh S, Goliaei S and Moghadam ZR:
CAMIRADA: Cancer microRNA association discovery algorithm, a case
study on breast cancer. J Biomed Inform. 94:1031802019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kogure A, Kosaka N and Ochiya T:
Cross-talk between cancer cells and their neighbors via miRNA in
extracellular vesicles: An emerging player in cancer metastasis. J
Biomed Sci. 26:72019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Božinović K, Sabol I, Dediol E, Milutin
Gašperov N, Manojlović S, Vojtechova Z, Tachezy R and Grce M:
Genome-wide miRNA profiling reinforces the importance of miR-9 in
human papillomavirus associated oral and oropharyngeal head and
neck cancer. Sci Rep. 9:23062019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xiao F, Zhang W, Chen L, Chen F, Xie H,
Xing C, Yu X, Ding S, Chen K, Guo H, et al: MicroRNA-503 inhibits
the G1/S transition by downregulating cyclin D3 and E2F3 in
hepatocellular carcinoma. J Transl Med. 11:1952013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xiao Y, Tian Q, He J, Huang M, Yang C and
Gong L: miR-503 inhibits hepatocellular carcinoma cell growth via
inhibition of insulin-like growth factor 1 receptor. Onco Targets
Ther. 9:3535–3544. 2016.PubMed/NCBI
|
|
23
|
Xie J, Murone M, Luoh SM, Ryan A, Gu Q,
Zhang C, Bonifas JM, Lam CW, Hynes M, Goddard A, et al: Activating
Smoothened mutations in sporadic basal-cell carcinoma. Nature.
391:90–92. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Y, Zhou Z, Walsh CT and Mcmahon AP:
Selective translocation of intracellular Smoothened to the primary
cilium in response to Hedgehog pathway modulation. Proc Natl Acad
Sci USA. 106:2623–2628. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T))method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
Starbase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale clip-seq data. Nucleic Acids
Res. 42((Database Issue)): D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar
|
|
28
|
Du Z, Sun T, Hacisuleyman E, Fei T, Wang
X, Brown M, Rinn JL, Lee MG, Chen Y, Kantoff PW and Liu XS:
Integrative analyses reveal a long noncoding RNA-mediated sponge
regulatory network in prostate cancer. Nat Commun. 7:109822016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shao Y, Ye M, Li Q, Sun W, Ye G, Zhang X,
Yang Y, Xiao B and Guo J: LncRNA-RMRP promotes carcinogenesis by
acting as a miR-206 sponge and is used as a novel biomarker for
gastric cancer. Oncotarget. 7:37812–37824. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bruix J, Qin S, Merle P, Granito A, Huang
YH, Bodoky G, Pracht M, Yokosuka O, Rosmorduc O, Breder V, et al:
Regorafenib for patients with hepatocellular carcinoma who
progressed on sorafenib treatment (RESORCE): A randomised,
double-blind, placebo-controlled, phase 3 trial. Lancet. 389:56–66.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xie Q, Lin S, Zheng M, Cai Q and Tu Y:
Long noncoding RNA NEAT1 promotes the growth of cervical cancer
cells via sponging miR-9-5p. Biochem Cell Biol. 97:100–108. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang L, Duan W, Kang L, Mao J, Yu X, Fan
S, Li L and Tao Y: Smoothened activates breast cancer stem-like
cell and promotes tumorigenesis and metastasis of breast cancer.
Biomed Pharmacother. 68:1099–1104. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Walter K, Omura N, Hong SM, Griffith M,
Vincent A, Borges M and Goggins M: Overexpression of smoothened
activates the sonic hedgehog signaling pathway in pancreatic
cancer-associated fibroblasts. Clin Cancer Res. 16:1781–1789. 2010.
View Article : Google Scholar : PubMed/NCBI
|