Introduction
Lung cancer is the leading cause of
cancer-associated mortality worldwide (1). Non-small cell lung cancer (NSCLC)
accounts for ~80% of cases of lung cancer, contributing to the
majority of cases in terms of both incidence and mortality
(2). Tumor invasion and metastasis
are primary reasons for cancer progression and therapy failure
(3). Therefore, investigation of
the molecular mechanisms underlying NSCLC invasion may be of
significance for therapy in patients with metastatic NSCLC.
Long non-coding RNAs (lncRNAs) are a class of
non-coding transcripts that are >200 nucleotides in length
(4). lncRNAs serve essential roles
in cell physiological and pathological processes, including
proliferation, apoptosis, differentiation, migration and invasion
(5,6). A number of dysregulated lncRNAs have
been identified in cancer and characterized as oncogenes or tumor
suppressors (7,8). For example, maternally expressed gene
3 (MEG3) has been reported to act as an antitumor lncRNA in
multiple types of cancer, such as breast, liver, colorectal, lung
and gastric cancer (9). Previous
studies have suggested that MEG3 is significantly downregulated in
NSCLC tissues and cell lines, and that low MEG3 expression levels
are associated with poor prognosis (10,11).
In vitro experiments have demonstrated that MEG3
overexpression can inhibit cell proliferation, promote apoptosis
and enhance chemotherapy sensitivity in NSCLC cells (11,12).
However, little is known about the functions and underlying
mechanisms of MEG3 in lung cancer metastasis. Given that MEG3
participates in migration and invasion of numerous other types of
cancer, including glioma, as well as breast and ovarian cancer
(13–15), it was hypothesized that MEG3 may
mediate migration and invasion of NSCLC cells.
As another type of non-coding RNA, microRNAs (miRNAs
or miRs) are small (18–22 nucleotides in length) single-stranded
transcripts that are able to regulate gene expression levels at the
post-transcriptional level by specific binding to the
3′-untranslated region (UTR) of the target mRNA, leading to
translational repression or degradation (16,17).
Similarly to lncRNAs, miRNAs are also involved in numerous
biological behaviors of cancer cells, and aberrant expression
levels of miRNAs are an important indicator of cancer (18,19).
Previous studies have shown that miR-21-5p is significantly
increased in NSCLC cell lines and tissues (20–22);
this is positively associated with tumor size, metastasis and poor
prognosis of patients with NSCLC (23,24),
indicating the oncogenic properties of miR-21-5p. miR-21-5p has
been shown to promote NSCLC cell proliferation in vitro
(23,25). However, the involvement and
mechanisms of miR-21-5p in NSCLC metastasis have yet to be fully
elucidated.
Increasing evidence has shown that interactions
between lncRNAs and miRNAs have a critical role in potential
mechanisms of tumorigenic processes (26,27).
lncRNAs can serve as competing endogenous RNAs (ceRNAs) or natural
miRNA sponges to modulate miRNA expression levels or sequester
miRNAs away from target mRNAs via competitively combining with
miRNAs, which decreases mRNA expression levels (26,27).
Bioinformatics analysis here suggested putative binding sites for
miR-21-5p in MEG3; therefore, it was possible to hypothesize
whether MEG3 could function as a ceRNA for miR-21-5p to regulate
migration and invasion of NSCLC cells in vitro.
The present study assessed the ability of MEG3 to
function as an miR-21-5p sponge to inhibit miR-21-5p expression
levels, and, in turn, enhance the expression levels of phosphatase
and tensin homolog (PTEN) and suppress the PI3K/AKT signaling
pathway in NSCLC PC9 and H1299 cells. Functional analyses
demonstrated that MEG3 overexpression could inhibit cell migration,
invasion and epithelial-to-mesenchymal transition (EMT). These
effects, induced by MEG3, were attenuated by a miR-21-5p mimic. To
the best of our knowledge, the present study is the first to
identify the molecular mechanism underlying the involvement of the
MEG3/miR-21-5p/PTEN axis in NSCLC metastasis.
Materials and methods
Cell culture
NSCLC cell lines PC9 and H1299 (purchased from
Shanghai Cell Bank of Chinese Academy of Sciences) and human lung
bronchial epithelial BEAS-2B cells (American Type Culture
Collection) were cultured in RPMI-1640 medium supplemented with 10%
fetal bovine serum (both Gibco; Thermo Fisher Scientific, Inc.) and
1% (w/v) penicillin/streptomycin (Sigma-Aldrich; Merck KGaA) at
37°C in a 5% CO2 incubator. Cells in the exponential
growth phase were used for subsequent experiments.
Target prediction for miR-21-5p
Potential target genes of miR-21-5p were predicted
by TargetScan (targetscan.org/), which is a
web-based resource used for the prediction of biological targets of
miRNAs by searching for the presence of 8mer, 7mer and 6mer sites
that match the seed region of each miRNA.
Reverse transcription-quantitative
(RT-q)PCR
RNA was extracted from cells using a MagMAX™
MiRVana™ Total RNA Isolation kit (Thermo Fisher Scientific, Inc.),
and subsequently transcribed into cDNA using a PrimeScript™ RT
reagent kit (Takara Biotechnology Co., Ltd.) according to the
manufacturers' protocols. RT-qPCR was performed using SYBR Green
PCR Master Mix on an ABI 7300 Real-Time PCR System (both Applied
Biosystems; Thermo Fisher Scientific, Inc.) with customized primer
sets for MEG3, PTEN and miR-21-5p. RT-qPCR amplification included a
holding stage at 95°C for 10 min, followed by 40 cycles of 95°C for
15 sec and 60°C for 1 min. The subsequent melt curve stage
consisted of 95°C for 15 sec, 60°C °for 1 min and 95°C for 15 sec.
GAPDH was used as the internal control for MEG3 and PTEN, whereas
U6 was used as the internal control for miR-21-5p. The relative
fold change of gene expression levels was determined using the
2−ΔΔCq method (28).
Primers (synthesized by Sangon Biotech Co., Ltd.) are listed in
Table I.
 | Table I.Primers for reverse
transcription-quantitative PCR. |
Table I.
Primers for reverse
transcription-quantitative PCR.
| Name | Direction | Sequence
(5′-3′) |
|---|
| Maternally
expressed gene 3 | Forward |
TCCATGCTGAGCTGCTGCCAAG |
|
| Reverse |
AGTCGACAAAGACTGACACCC |
| PTEN | Forward |
CCCAGTCAGAGGCGCTATG |
|
| Reverse |
GGCAGACCACAAACTGAGGATT |
| GAPDH | Forward |
AATGGACAACTGGTCGTGGAC |
|
| Reverse |
CCCTCCAGGGGATCTGTTTG |
| MicroRNA-21-5p | Forward |
GCACCTAGCTTATCAGACTGA |
|
| Reverse |
GTGCAGGGTCCGAGGT |
| U6 | Forward |
GCTTCGGCAGCATATACTAAAAT |
|
| Reverse |
CGCTTCACGAATTTGCGTGTCAT |
Cell transfection
The pcDNA3.1-MEG3 plasmid (termed MEG3) was
constructed by sub-cloning MEG3 sequence into pcDNA 3.1 vector
(Shanghai GenePharma Co., Ltd.). The cells were transfected with
MEG3 and and pcDNA 3.1 empty vector (vector) to a final
concentration of 100 nM using Lipofectamine® 2000 (cat.
no. 11668019; Invitrogen; Thermo Fisher Scientific, Inc.) according
to the manufacturer's protocols. miR-21-5p inhibitor
(5′-UAGCUUAUCAGACUGAUGUUGA-3′), miR-21-5p mimic
(5′-UAGCUUAUCAGACUGAUGUUGA-3′) and their negative controls (NC,
5′-CCCAGAATGTTGACAGCTGCCTCTT-3′) were synthesized by Guangzhou
RiboBio Co., Ltd. The cells were transfected with miR-21-5p mimic
or inhibitor to a final concentration of 50 nM using Lipofectamine
2000 according to the manufacturer's protocols. Subsequent
experiments were performed 24 h after transfection.
In order to assess the effects of miR-21-5p on
MEG3-mediated cellular responses, cells were transfected with MEG3,
miR-21-5p mimic, miR-21-5p inhibitor, MEG3+miR-21-5p mimic and
their negative controls using Lipofectamine 2000, respectively.
Luciferase reporter assay
The 3′-UTR regions of MEG3 or PTEN containing the
predicted miR-21-5p specific binding sites were amplified by PCR
and cloned into the firefly luciferase reporter vector, pmirGLO
(Promega Corporation), to obtain the wild-type luciferase reporter
plasmids, wt-MEG3 and wt-PTEN, respectively. In order to generate
the mutant reporter plasmids mut-MEG3 and mut-PTEN, certain
nucleotides in MEG3 or PTEN 3′-UTR were mutated using PCR lacking
miR-21-5p-binding sites. The constructed luciferase reporter
plasmids (wt-MEG3, mut-MEG3, wt-PTEN and mut-PTEN) were separately
co-transfected with miR-21-5p mimic or mimic control into PC9 and
H1299 cells using Lipofectamine 2000. Cells were lysed and
luciferase activity was assayed at 24 h post-transfection using a
Dual-Luciferase Reporter Assay system (Promega Corporation),
according to the manufacturer's instructions.
Western blotting
Proteins were extracted from cells using ice-cold
RIPA lysis buffer (Beyotime Institute of Biotechnology). Following
quantification using a BCA kit (Thermo Fisher Scientific, Inc.), 40
µg total protein of each sample was denatured in a boiling water
bath and separated by SDS-PAGE (10% gels), then transferred to a
polyvinylidene fluoride membrane. Subsequently, the membranes were
probed with 5% non-fat milk in TBS at room temperature for 1 h, and
incubated with specific primary antibodies for PTEN (1:500; cat.
no. ab31392; Abcam), N-cadherin (N-cad; 1:500; cat. no. ab76011;
Abcam), E-cadherin (E-cad; 1:500; cat. no. ab1416; Abcam), Vimentin
(Vim; 1:500; cat. no. ab137321; Abcam), matrix metalloprotein
(MMP)9 (1:500; cat no. ab38898; Abcam), PI3K (1:500; cat. no.
GW21071; Sigma-Aldrich; Merck KGaA), phosphorylated (p)-PI3K
(1:1,000; cat. no. 4228; Cell Signaling Technology, Inc.), AKT
(1:1,000; cat. no. SAB4500797; Sigma-Aldrich; Merck KGaA), p-AKT
(1:1,000, cat. no. 9271; Cell Signaling Technology, Inc.) and
β-actin (1:1,000; cat. no. 3700; Cell Signaling Technology, Inc.)
at 4°C overnight. After being washed five times, the membranes were
incubated with horseradish peroxidase-conjugated goat anti-rabbit
IgG (1:2,000; cat. no. ab6721; Abcam) at room temperature for 2 h,
and detected using a Novex® ECL Chemiluminescent
Substrate Reagent kit (Thermo Fisher Scientific, Inc.) and
ChemiDoc™ XRS+imaging system (Bio-Rad Laboratories,
Inc.). The signal intensity was quantified using ImageJ software
(version 5.0; National Institutes of Health).
Migration and invasion assays
Cell migration was assessed using 24-well Transwell
filters (Corning Life Sciences). Briefly, a total of
5×104 cells was seeded into the upper chamber in
serum-free medium, while the lower chamber was filled with complete
medium containing 20% FBS. After 24 h incubation at 37°C, cells
that passed through the filter were fixed using 5% glutaraldehyde
at 4°C for 30 min and stained with 0.5% crystal violet solution at
37°C for 30 min. Images were captured using an inverted light
microscope (magnification, ×200; Olympus Corporation) and cells in
five randomly selected fields were counted. For cell invasion
assay, the filter membranes were pre-coated with Matrigel™ (Beijing
Solarbio Science & Technology Co., Ltd.) at 37°C for 30 min,
and other steps were performed as described in the migration
assay.
Statistical analysis
All data were analyzed using GraphPad Prism software
(version 5; GraphPad Software, Inc.) and are expressed as the mean
± SD of three independent repeats. Significant differences were
analyzed by one-way ANOVA to compare >2 groups, followed by
Tukey's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
MEG3 suppresses miR-21-5p expression
levels and acts as a sponge of miR-21-5p
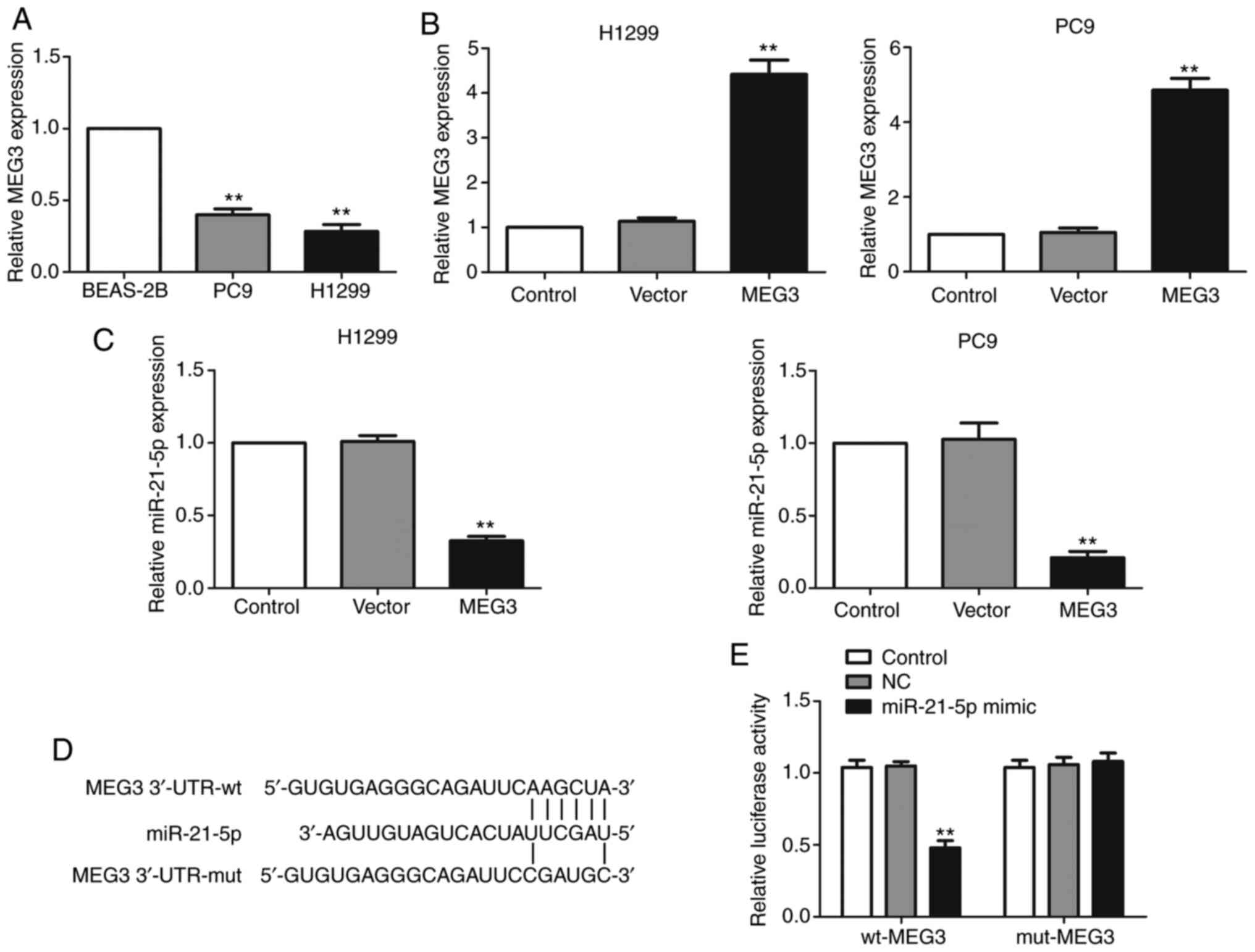
First, RT-qPCR was used to analyze the expression
levels of MEG3 in NSCLC cell lines. The results demonstrated that
MEG3 was downregulated in the NSCLC cell lines PC9 and H1299
compared with normal lung bronchial epithelial BEAS-2B cells
(Fig. 1A). Next, to investigate the
effect of MEG3 on miR-21-5p expression levels, MEG3 was
overexpressed by transfecting MEG3-overexpressing plasmid in PC9
and H1299 cells. The expression levels of MEG3 were shown to be
upregulated, assessed via RT-qPCR (Fig.
1B). MEG3 overexpression significantly suppressed the
expression levels of miR-21-5p (Fig.
1C).
In order to investigate whether MEG3 functions as a
sponge of miR-21-5p to regulate its expression levels, the online
database TargetScan (targetscan.org) was used to predict the association
between MEG3 and miR-21-5p. Bioinformatics analysis demonstrated
that miR-21-5p directly interacts with MEG3 via sequence
complementarity (Fig. 1D). In order
to verify the direct binding of miR-21-5p to MEG3, luciferase
reporter plasmids containing wt-MEG3 or mut-MEG3 were
co-transfected with either miR-21-5p mimic or NC-mimic into cells.
As expected, miR-21-5p mimic significantly repressed the luciferase
intensity of wt-MEG3, but had no effect on the luciferase activity
of mut-MEG3 (Fig. 1E). These
results demonstrated that MEG3 repressed miR-21-5p expression
levels via directly targeting miR-21-5p.
PTEN is a direct target of
miR-21-5p
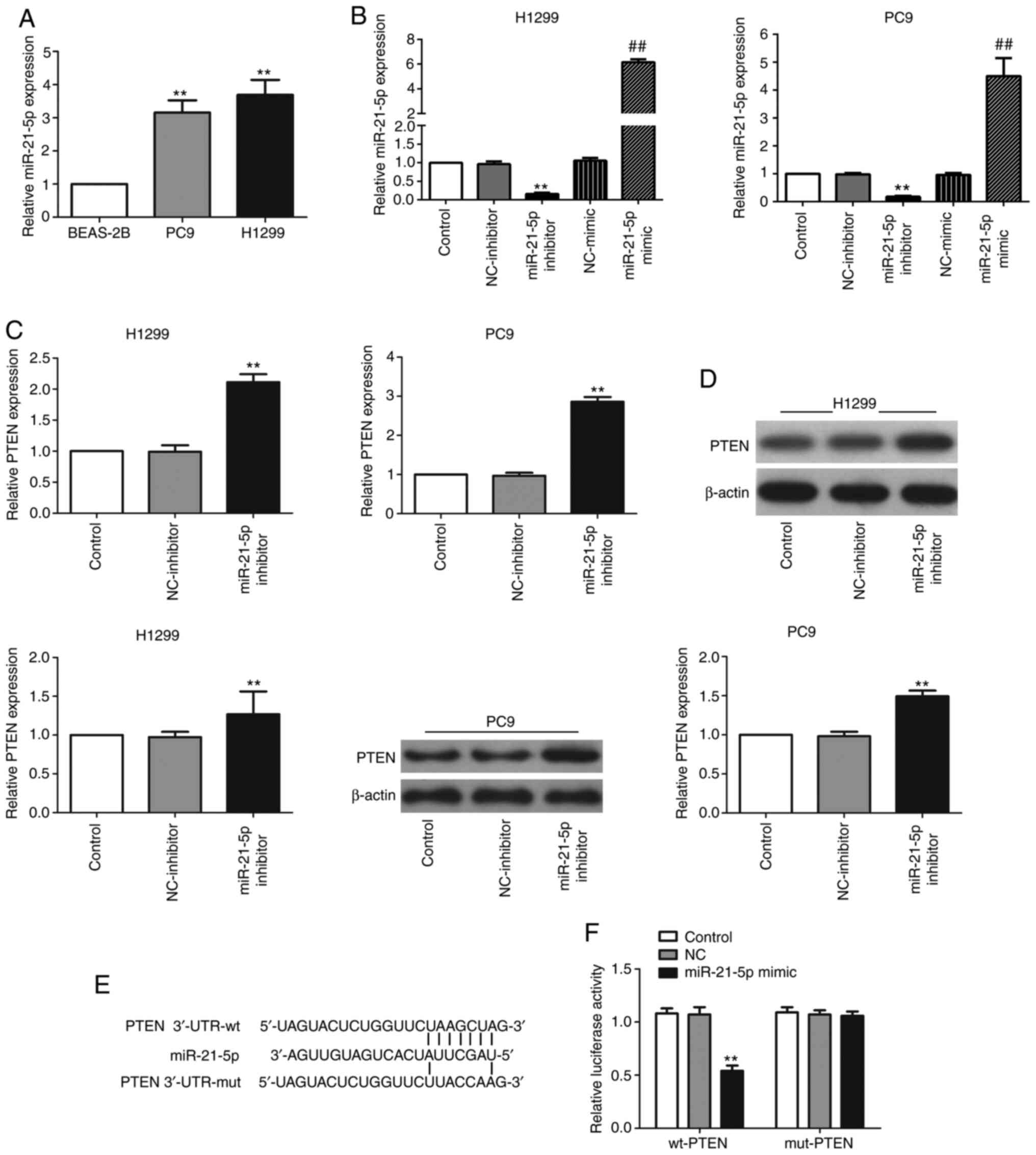
First, the expression levels of miR-21-5p in NSCLC
cell lines were evaluated via RT-qPCR; the results showed that
miR-21-5p was substantially increased in the two NSCLC cell lines
PC9 and H1299 in comparison with BEAS-2B cells (Fig. 2A). PTEN was predicted to be a
candidate target gene of miR-21-5p using TargetScan (targetscan.org) (Fig.
2E). Subsequently, cells were transfected with miR-21-5p
inhibitor to determine whether the expression levels of PTEN were
regulated by miR-21-5p. Suppressed expression levels of miR-21-5p
were confirmed via RT-qPCR (Fig.
2B), and significant increases in PTEN expression, at both the
mRNA (Fig. 2C) and protein
(Fig. 2D) levels, were observed
following miR-21-5p inhibitor transfection.
Next, it was determined whether the enhanced PTEN
expression levels were caused by the direct binding of miR-21-5p to
the 3′-UTR of PTEN. To this end, PTEN 3′-UTR containing the
wild-type or mutated miR-21-5p target site was cloned in a firefly
luciferase reporter vector to obtain the reporter plasmids wt-PTEN
and mut-PTEN, respectively. Following miR-21-5p mimic and wt-PTEN
co-transfection, the luciferase activity was markedly attenuated,
whereas co-transfection with miR-21-5p and mut-PTEN had little
effect on luciferase activity (Fig.
2F). These data indicated that miR-21-5p inhibited PTEN
expression levels by directly binding to PTEN 3′-UTR.
MEG3 positively regulates PTEN
expression levels by sponging miR-21-5p
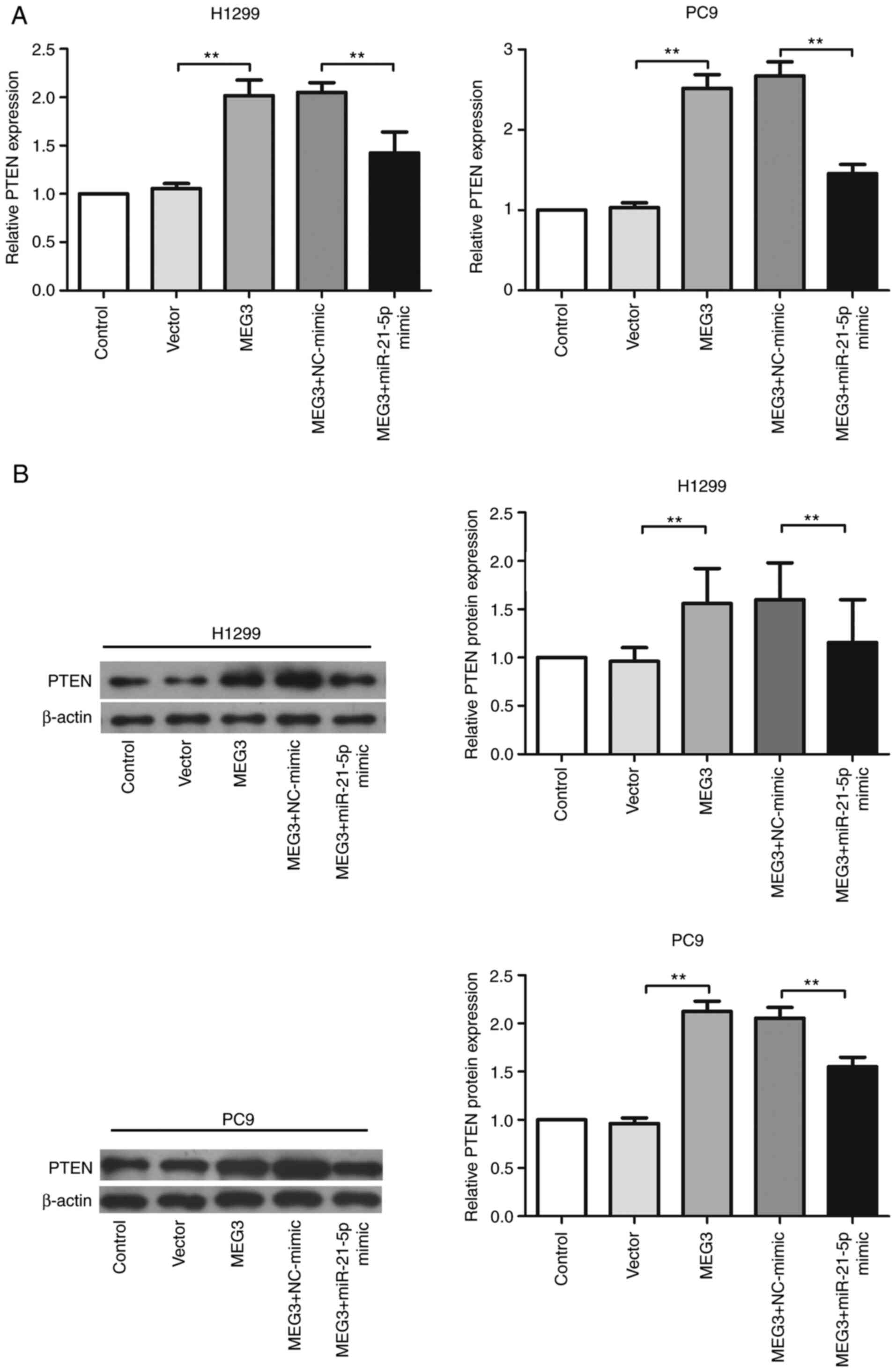
It was further investigated whether MEG3 targeted
PTEN expression levels by sponging miR-21-5p. miR-21-5p mimic and
MEG3-overexpressing plasmid were co-transfected into H1299 cells,
and PTEN expression at both the mRNA (Fig. 3A) and protein (Fig. 3B) levels was analyzed. The results
indicated that upregulation of miR-21-5p significantly repressed
MEG3-induced increased expression levels of PTEN.
MEG3 mediates the EMT process via
sponging miR-21-5p
Given the role of EMT in cancer migration and
invasion, it was investigated whether MEG3 or miR-21-5p mediated
EMT. Transfection with MEG3-overexpressing plasmid or miR-21-5p
inhibitor significantly increased levels of the epithelial marker
E-cad, but decreased levels of the mesenchymal markers N-cad, Vim
and MMP9 (Fig. 4A and C). Then, to
determine whether MEG3 mediated the EMT process via sponging
miR-21-5p, mir-21-5p mimic or NC-mimic was co-transfected with
MEG3-overexpressing plasmid in PC9 and H1299 cells. The alterations
in expression levels of epithelial and mesenchymal markers induced
by MEG3 were significantly abrogated by miR-21-5p mimic (Fig. 4A and C).
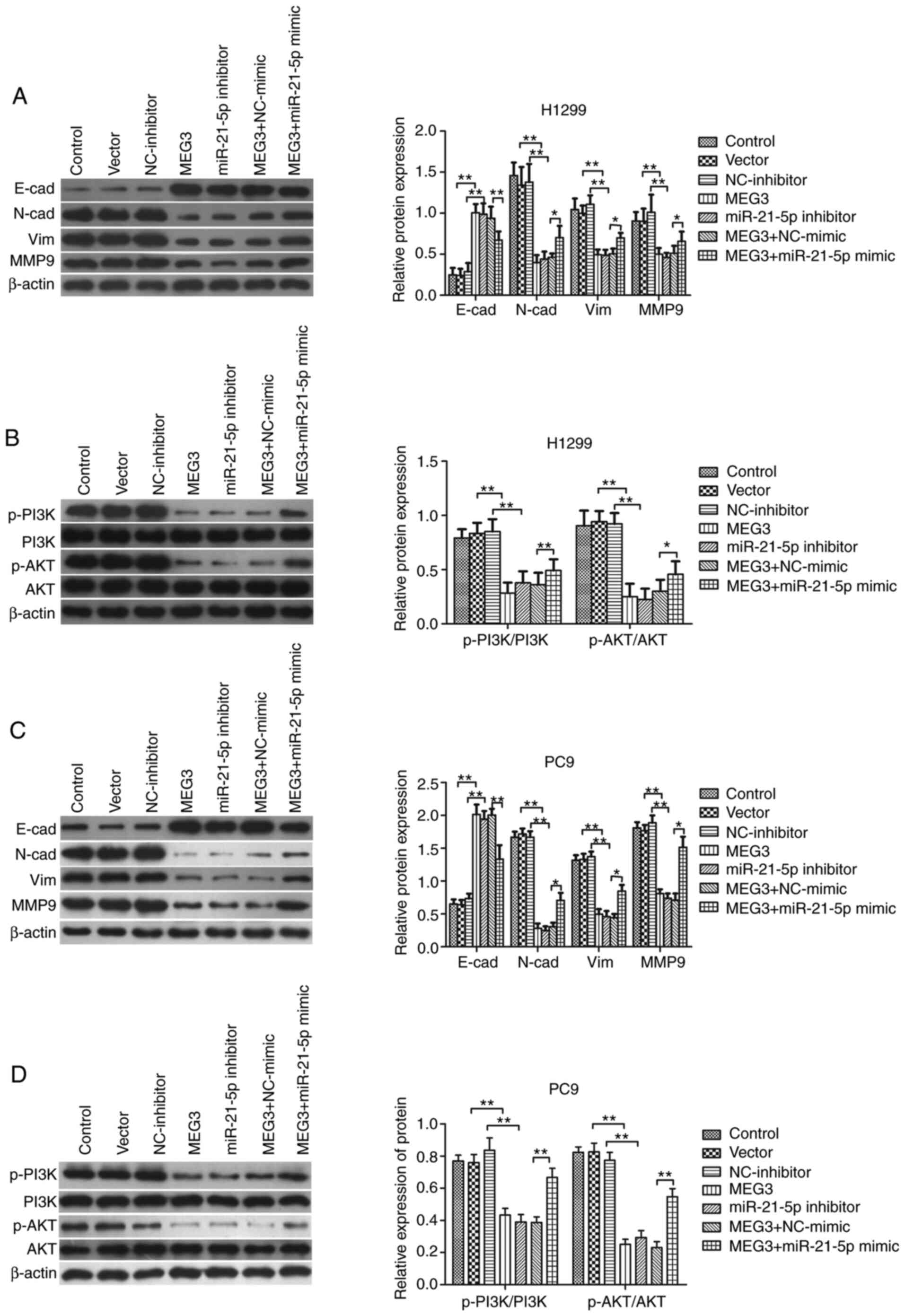 | Figure 4.MEG3 suppresses the PI3K/AKT pathway
and the epithelial-to-mesenchymal transition process via miR21-5p.
PC9 and H1299 cells were transfected with MEG3, empty vector,
miR-21-5p inhibitor, NC-inhibitor, MEG3+ miR-21-5p mimic and MEG3+
NC-mimic. Representative western blots, and semi-quantitative
analysis of relative changes in expression levels of E-cad, N-cad,
Vim and MMP9 in (A) H1299 and (C) PC9 cells. The protein expression
levels of p-PI3K, PI3K, p-AKT and AKT protein in (B) H1299 and (D)
PC9 cells are also shown. **P<0.01. *P<0.05. MEG3, maternally
expressed gene 3; miR, microRNA; NC, negative control; E-cad,
E-cadherin; N-cad, N-cadherin; Vim, vimentin; MMP, matrix
metalloprotein; p-, phosphorylated. |
MEG3/miR-21-5p/PTEN axis function may
be mediated via the PI3K/AKT signaling pathway
PTEN has been reported to be an antagonist regulator
of the PI3K/AKT pathway, which is involved in the regulation of
cancer migration and invasion (29). It was therefore speculated that the
MEG3/miR-21-5p/PTEN axis may function via the PI3K/AKT signaling
pathway. In order to investigate this, phosphorylation of PI3K and
AKT in cells transfected with MEG3-overexpressing plasmid was
analyzed. MEG3 overexpression significantly decreased
phosphorylation of both PI3K and AKT (Fig. 4B and D). Similar patterns of p-PI3K
and p-AKT were observed in cells transfected with miR-21-5p
inhibitor (Fig. 4B and D). The
participation of miR-21-5p in MEG3-mediated regulation of the
PI3K/AKT pathway was further investigated by co-transfecting
miR-21-5p mimic or NC-mimic with MEG3-overexpressing plasmid into
PC9 and H1299 cells. The results showed that the decreases in
phosphorylation levels of PI3K/AKT induced by MEG3 were
significantly abrogated by miR-21-5p (Fig. 4B and D).
MEG3 attenuates cell migration and
invasion via sponging miR-21-5p
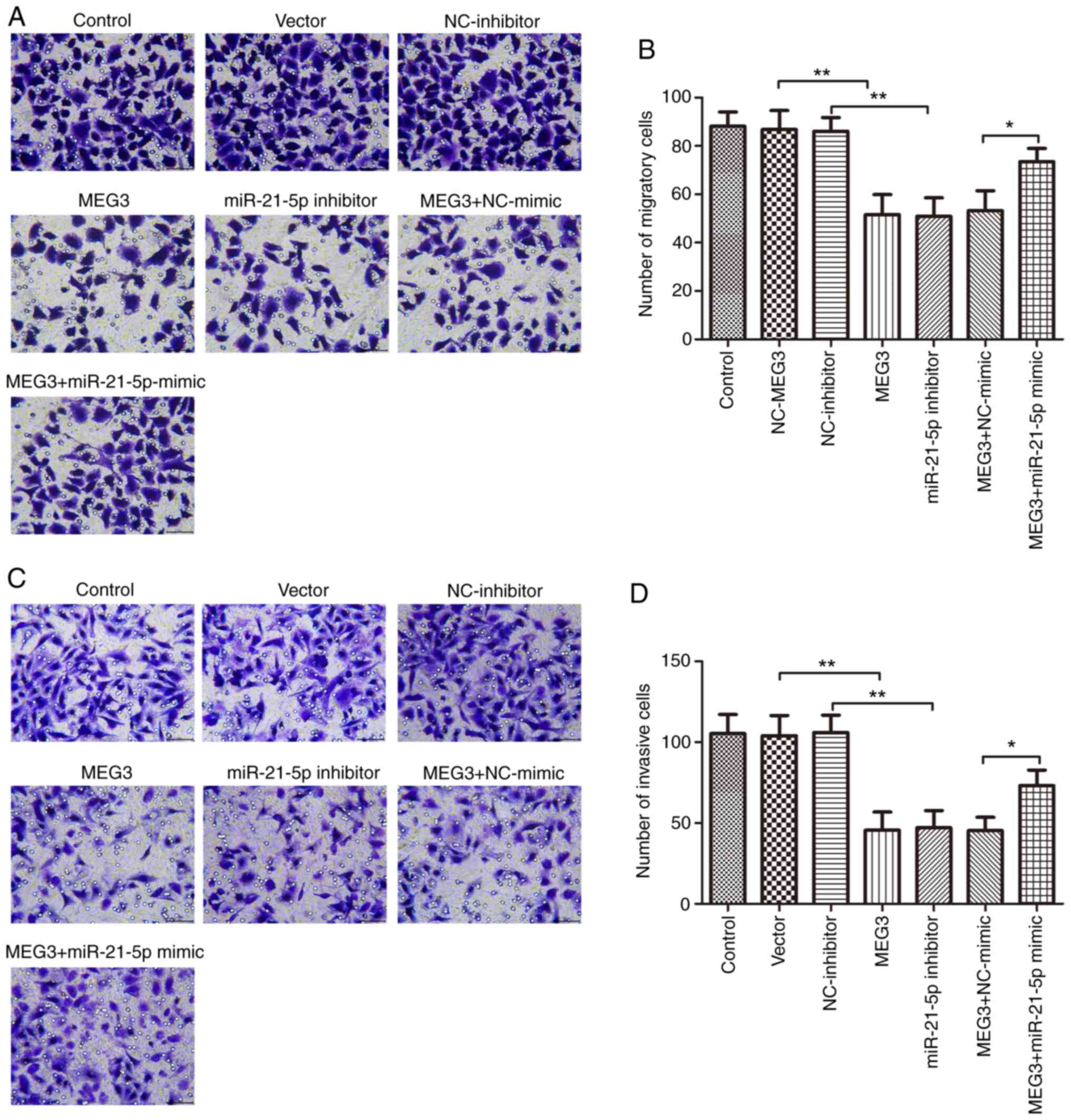
The effect of MEG3 on cell migration and invasion
was evaluated by transfecting MEG3-overexpressing or empty vector
plasmid into PC9 and H1299 cells. Transwell migration and invasion
assays showed that MEG3 overexpression induced a significant
decrease in the number of the migratory and invasive cells compared
with empty vector control (Fig.
5A-D). Similar results were observed in cells transfected with
miR-21-5p inhibitor; miR-21-5p attenuation significantly decreased
migration and invasion of PC9 and H1299 cells compared with
NC-inhibitor (Fig. 5A-D).
The contribution of miR-21-5p to the effect of MEG3
on biological behaviors of PC9 and H1299 cells was next
investigated. miR-21-5p mimic or NC-mimic was co-transfected with
MEG3-overexpressing plasmid into PC9 and H1299 cells, and the
migration and invasion abilities were assessed. The results
demonstrated that miR-21-5p mimic significantly mitigated the
inhibitory effects of MEG3 on cell migration and invasion (Fig. 5A-D). These results indicated that
MEG3 involvement in NSCLC cell migration and invasion was mediated,
at least partially, by sponging and suppressing miR-21-5p.
Discussion
lncRNAs have been implicated in cancer development
(7,8). MEG3 has been reported to function as a
tumor suppressor in a number of types of cancer (9), such as lung (10), breast (13) and ovarian cancer (15). Numerous studies have identified low
expression levels of MEG3 in NSCLC tissues and cell lines (10,11);
similarly, the present study demonstrated that MEG3 was
downregulated in NSCLC PC9 and H1299 cells. Previous studies have
also suggested that MEG3 overexpression may inhibit proliferation
and induce apoptosis in NSCLC cells (10,11).
In the present study, MEG3 overexpression suppressed the migration
and invasion abilities of NSCLC PC9 and H1299 cells in
vitro. EMT is a key process associated with cancer cell
migration and invasion. The present results showed that MEG3
overexpression inhibited the EMT process by upregulating the
expression levels of the epithelial marker, E-cad, and
downregulating the expression levels of the mesenchymal markers,
N-cad, Vim and MMP9, in PC9 and H1299 cells. MEG3 has been
previously reported to show similar effects on the migration and
invasion abilities of pancreatic cancer cells in vitro
(30).
lncRNAs regulate cancer development via sponging
miRNAs (26,27). MEG3 functions via sponging multiple
miRNAs in numerous types of cancer (31,32).
In NSCLC, for instance, MEG3 has been reported to act as a
molecular sponge of miR-7-5p and miR-3163 to inhibit cell growth
(10,33). miR-21-5p has been reported to be an
oncogene in NSCLC (20–24). The present study showed a strong
association between miR-21-5p and MEG3 in PC9 and H1299 cells. MEG3
overexpression significantly inhibited miR-21-5p expression levels
in PC9 and H1299 cells, and dual luciferase assays demonstrated
that MEG3 directly interacted with miR-21-5p. These results are in
agreement with a previous report by Wang et al (34). Furthermore, an miR-21-5p mimic
significantly restored the changes induced by MEG3 on cell
migration, invasion and the EMT process. Additionally, miR-21-5p
attenuation also suppressed cell migration, invasion and the EMT
process in PC9 and H1299 cells. These results suggested that MEG3
inhibited migration and invasion of H1299 cells via acting as a
miR-21-5p sponge. Previous studies have also indicated the
involvement of the MEG3/miR-21-5p axis in cell proliferation and
apoptosis in NSCLC and cervical cancer (34,35).
PTEN has proved to be a powerful tumor suppressor,
and low levels of PTEN are one of the most frequent events observed
in a variety of types of cancer (36,37).
Numerous studies have observed decreased expression levels of PTEN
in NSCLC tissues and cell lines (29,38),
and PTEN overexpression has been reported to inhibit the migration
and invasion of NSCLC cells (39).
Notably, evidence suggests that PTEN expression levels may be
downregulated by miRNAs involved in cancer development, including
those involved in NSCLC (29,40).
In the present study, dual luciferase assay demonstrated that PTEN
was a direct target of miR-21-5p in H1299 lung cancer cells, and
attenuation of miR-21-5p enhanced PTEN expression levels. The
miR-21-5p/PTEN axis has also been reported in other types of
cancer, such as breast and epithelial ovarian cancer (41,42).
Furthermore, in the present study, MEG3 overexpression resulted in
increased PTEN expression levels, which was mitigated by an
miR-21-5p mimic in PC9 and H1299 cells. In addition, PTEN has been
reported to function by negatively regulating the PI3K/AKT
signaling pathway, which is activated in NSCLC development
(38). As expected, MEG3
overexpression also caused suppression of the PI3K/AKT signaling
pathway, whereas these effects were reversed by miR-21-5p in PC9
and H1299 cells. These data suggested that MEG3 upregulated PTEN
expression levels and subsequently attenuated the PI3K/AKT
signaling pathway, partially by sponging miR-21-5p, and thus
inhibited the migration and invasion of NSCLC PC9 and H1299
cells.
Taken together, the results of the present study
have demonstrated that MEG3 inhibited migration and invasion of
NSCLC PC9 and H1299 cells, partially by regulating the
miR-21-5p/PTEN axis and the PI3K/AKT signaling pathway. Therefore,
the MEG3/miR-21-5p/PTEN axis may represent a novel target pathway
for NSCLC therapy.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DL, YL and MZ performed the experiments, and
collected and interpreted data. YL, QB and JD participated in the
design and coordination of experimental work, and acquisition of
data. DL and NW participated in the study design, data collection,
analysis of data and preparation of the manuscript. SH designed the
study, analyzed and interpreted data and drafted the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
MEG3
|
maternally expressed gene 3
|
|
PTEN
|
phosphatase and tensin homolog
|
|
E-cad
|
E-cadherin
|
|
N-cad
|
N-cadherin
|
|
Vim
|
vimentin
|
|
MMP
|
matrix metalloprotein
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
References
|
1
|
Torre LA, Siegel RL and Jemal A: Lung
cancer statistics. Adv Exp Med Biol. 893:1–19. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zarogoulidis K, Zarogoulidis P, Darwiche
K, Boutsikou E, Machairiotis N, Tsakiridis K, Katsikogiannis N,
Kougioumtzi I, Karapantzos I, Huang H, et al: Treatment of
non-small cell lung cancer (NSCLC). J Thorac Dis. 5 (Suppl
4):S389–396. 2013.PubMed/NCBI
|
|
3
|
Domoto T, Pyko IV, Furuta T, Miyashita K,
Uehara M, Shimasaki T, Nakada M and Minamoto T: Glycogen synthase
kinase-3β is a pivotal mediator of cancer invasion and resistance
to therapy. Cancer Sci. 107:1363–1372. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jeong S, Lee J, Kim D, Seol MY, Lee WK,
Jeong JJ, Nam KH, Jung SG, Shin DY, Lee EJ, et al: Relationship of
focally amplified long noncoding on chromosome 1 (FAL1) lncRNA with
E2F transcription factors in thyroid cancer. Medicine (Baltimore).
95:e25922016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lian Y, Xiao C, Yan C, Chen D, Huang Q,
Fan Y, Li Z and Xu H: Knockdown of pseudogene derived from lncRNA
DUXAP10 inhibits cell proliferation, migration, invasion, and
promotes apoptosis in pancreatic cancer. J Cell Biochem.
119:3671–3682. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dong X, Chen R, Lin H, Lin T and Pan S:
lncRNA BG981369 inhibits cell proliferation, migration, and
invasion, and promotes cell apoptosis by SRY-related high-mobility
group box 4 (SOX4) signaling pathway in human gastric Cancer. Med
Sci Monit. 24:718–726. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ponzio G, Rezzonico R, Bourget I, Allan R,
Nottet N, Popa A, Magnone V, Rios G, Mari B and Barbry P: A new
long noncoding RNA (lncRNA) is induced in cutaneous squamous cell
carcinoma and down-regulates several anticancer and cell
differentiation genes in mouse. J Biol Chem. 292:12483–12495. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao JH, Sun JX, Song YX, Chen XW, Yang
YC, Ma B, Wang J, Gao P and Wang ZN: A novel long noncoding
RNA-LOWEG is low expressed in gastric cancer and acts as a tumor
suppressor by inhibiting cell invasion. J Cancer Res Clin Oncol.
142:601–609. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Al-Rugeebah A, Alanazi M and Parine NR:
MEG3: An oncogenic long non-coding RNA in different cancers. Pathol
Oncol Res. 25:859–874. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu JL, Meng FM and Li HJ: High expression
of lncRNA MEG3 participates in non-small cell lung cancer by
regulating microRNA-7-5p. Eur Rev Med Pharmacol Sci. 22:5938–5945.
2018.PubMed/NCBI
|
|
11
|
Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu
WQ, Xie WP and Hou YY: Long non-coding RNA MEG3 inhibits NSCLC
cells proliferation and induces apoptosis by affecting p53
expression. BMC Cancer. 13:4612013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xia Y, He Z, Liu B, Wang P and Chen Y:
Downregulation of Meg3 enhances cisplatin resistance of lung cancer
cells through activation of the WNT/β-catenin signaling pathway.
Mol Med Rep. 12:4530–4537. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun L, Li Y and Yang B: Downregulated long
non-coding RNA MEG3 in breast cancer regulates proliferation,
migration and invasion by depending on p53's transcriptional
activity. Biochem Biophys Res Commun. 478:323–329. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qin N, Tong GF, Sun LW and Xu XL: Long
Noncoding RNA MEG3 Suppresses Glioma Cell Proliferation, Migration,
and Invasion by Acting as a Competing Endogenous RNA of miR-19a.
Oncol Res. 25:1471–1478. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang J, Xu W, He Y, Xia Q and Liu S:
lncRNA MEG3 impacts proliferation, invasion, and migration of
ovarian cancer cells through regulating PTEN. Inflamm Res.
67:927–936. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hujie G, Zhou SH, Zhang H, Qu J, Xiong XW,
Hujie O, Liao CG and Yang SE: MicroRNA-10b regulates
epithelial-mesenchymal transition by modulating KLF4/KLF11/Smads in
hepatocellular carcinoma. Cancer Cell Int. 18:102018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li C, Jiang Y, Miao R, Qu K, Zhang J and
Liu C: MicroRNA-1271 functions as a metastasis and
epithelial-mesenchymal transition inhibitor in human HCC by
targeting the PTP4A1/c-Src axis. Int J Oncol. 52:536–546.
2018.PubMed/NCBI
|
|
18
|
Lages E, Ipas H, Guttin A, Nesr H, Berger
F and Issartel JP: MicroRNAs: Molecular features and role in
cancer. Front Biosci (Landmark Ed). 17:2508–2540. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang C, Sun C, Liang X, Xie S, Huang J and
Li D: Integrative analysis of microRNA and mRNA expression profiles
in non-small-cell lung cancer. Cancer Gene Ther. 23:90–97. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tian F, Li R, Chen Z, Shen Y, Lu J, Xie X
and Ge Q: Differentially Expressed miRNAs in tumor, adjacent, and
normal tissues of lung adenocarcinoma. Biomed Res Int.
2016:14282712016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang K, Chen M and Wu W: Analysis of
microRNA (miRNA) expression profiles reveals 11 key biomarkers
associated with non-small cell lung cancer. World J Surg Oncol.
15:1752017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li X and Wu X: MiR-21-5p promotes the
progression of non-small-cell lung cancer by regulating the
expression of SMAD7. Onco Targets Ther. 11:8445–8454. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li C, Yin Y, Liu X, Xi X, Xue W and Qu Y:
Non-small cell lung cancer associated microRNA expression
signature: Integrated bioinformatics analysis, validation and
clinical significance. Oncotarget. 8:24564–24578. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yan L, Ma J, Wang Y, Zan J, Wang Z, Zhu Y,
Zhu Y, Ling L, Cao L, Liu X, et al: miR-21-5p induces cell
proliferation by targeting TGFBI in non-small cell lung cancer
cells. Exp Ther Med. 16:4655–4663. 2018.PubMed/NCBI
|
|
26
|
Liz J and Esteller M: lncRNAs and
microRNAs with a role in cancer development. Biochim Biophys Acta.
1859:169–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ling C, Wang X, Zhu J, Tang H, Du W, Zeng
Y, Sun L, Huang JA and Liu Z: MicroRNA-4286 promotes cell
proliferation, migration, and invasion via PTEN regulation of the
PI3K/Akt pathway in non-small cell lung cancer. Cancer Med.
8:3520–3531. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sun Y, Zhu Q, Zhou M, Yang W, Shi H, Shan
Y, Zhang Q and Yu F: Restoration of miRNA-148a in pancreatic cancer
reduces invasion and metastasis by inhibiting the Wnt/beta-catenin
signaling pathway via downregulating maternally expressed gene-3.
Exp Ther Med. 17:639–648. 2019.PubMed/NCBI
|
|
31
|
Xu G, Meng L, Yuan D, Li K, Zhang Y, Dang
C and Zhu K: MEG3/miR21 axis affects cell mobility by suppressing
epithelialmesenchymal transition in gastric cancer. Oncol Rep.
40:39–48. 2018.PubMed/NCBI
|
|
32
|
Long J and Pi X: lncRNA-MEG3 suppresses
the proliferation and invasion of melanoma by regulating CYLD
expression mediated by sponging miR-499-5p. Biomed Res Int.
2018:20865642018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Su L, Han D, Wu J and Huo X: Skp2
regulates non-small cell lung cancer cell growth by Meg3 and
miR-3163. Tumour Biol. 37:3925–3931. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang P, Chen D, Ma H and Li Y: lncRNA MEG3
enhances cisplatin sensitivity in non-small cell lung cancer by
regulating miR-21-5p/SOX7 axis. Onco Targets Ther. 10:5137–5149.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang J, Yao T, Wang Y, Yu J, Liu Y and
Lin Z: Long noncoding RNA MEG3 is downregulated in cervical cancer
and affects cell proliferation and apoptosis by regulating miR-21.
Cancer Biol Ther. 17:104–113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Alvarez-Garcia V, Tawil Y, Wise HM and
Leslie NR: Mechanisms of PTEN loss in cancer: It's all about
diversity. Semin Cancer Biol. 59:66–79. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Papa A and Pandolfi PP: The PTEN-PI3K Axis
in Cancer. Biomolecules. 9:1532019. View Article : Google Scholar
|
|
38
|
Perez-Ramirez C, Canadas-Garre M, Molina
MA, Faus-Dader MJ and Calleja-Hernandez MA: PTEN and PI3K/AKT in
non-small-cell lung cancer. Pharmacogenomics. 16:1843–1862. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dong L, Li G, Li Y and Zhu Z: Upregulation
of Long Noncoding RNA GAS5 Inhibits Lung Cancer Cell Proliferation
and Metastasis via miR-205/PTEN Axis. Med Sci Monit. 25:2311–2319.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang H, Ma Z, Liu X, Zhang C, Hu Y, Ding
L, Qi P, Wang J, Lu S and Li Y: MiR-183-5p is required for
non-small cell lung cancer progression by repressing PTEN. Biomed
Pharmacother. 111:1103–1111. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yu X, Chen Y, Tian R, Li J, Li H, Lv T and
Yao Q: miRNA-21 enhances chemoresistance to cisplatin in epithelial
ovarian cancer by negatively regulating PTEN. Oncol Lett.
14:1807–1810. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fang H, Xie J, Zhang M, Zhao Z, Wan Y and
Yao Y: miRNA-21 promotes proliferation and invasion of
triple-negative breast cancer cells through targeting PTEN. Am J
Transl Res. 9:953–961. 2017.PubMed/NCBI
|