Introduction
Osteosarcoma (OS) is the most common primary sarcoma
of the bone and mainly affects adolescents and children (1). Due to its high degree of malignancy,
early metastasis, low chance of surgery, easy recurrence and high
mortality, OS causes an unacceptable mortality rate (2). Although significant improvements have
been made in the treatment of OS over the past decade, the
prognosis of osteosarcoma remains poor (3). A previous study reported that the
5-year survival rate of OS patients without metastasis is ~60-70%
(4). However, the 5-year survival
rate of OS patients with distant metastasis is only 20–30%
(5). Therefore, it is of paramount
importance to investigate the molecular mechanisms underlying the
pathogenesis of OS development.
Long non-coding RNAs (lncRNAs) are a type of
non-coding nucleic acid with a length of >200 nucleotides and
diverse and largely uncharacterized biological functions (6). Recently, increasing evidence has
demonstrated that lncRNAs participate in fundamental cellular
processes, including proliferation, migration and apoptotic
processes, which are important in the development of cancer
(7,8). Previous studies have reported that
lncRNAs function as oncogenes or tumor suppressors and are
associated with cancer initiation and development, and lncRNAs may
be dysregulated in various types of human cancer, including OS
(9). For example, lncRNA SUMO1P3
promotes gastric cancer progression and invasion by regulating the
EMT signaling pathway (10), and
lncRNA AFAP1-AS1 accelerates nasopharyngeal carcinoma metastasis by
sponging miR-423-5p to regulate the Rho/Rac pathway (11). These findings indicated that lncRNAs
may be vital regulators during tumorigenesis and tumor
progression.
In recent years, pseudogene-derived lncRNA double
homeobox A pseudogene 8 (DUXAP8) has been shown to be upregulated
in various malignant tumor types. Previous studies have reported
that DUXAP8 works as an oncogene in renal cell carcinoma, gastric
cancer and other tumor types (12,13). A
recent study reported that in HCC, DUXAP8 repressed tumor
suppressor KLF2 transcription by interacting with EZH2 (14). However, the expression status and
prognostic value of DUXAP8 in OS remain unknown.
MicroRNAs (miRNAs) are ~ 22-nucleotide-long
non-coding RNA molecules that can regulate target gene expression
levels by binding to the 3′-untranslated regions (3′-UTRs) of
target genes at the posttranscriptional level and promoting
degradation or inhibiting translation (15). miR-635 is located in 17q and has
been recently identified in colorectal cancer (16). Weber et al (17) reported that miR-635 may
significantly accelerate the invasion of A375 melanoma cells.
However, the mechanism of miR-635 regulation in OS requires further
investigation. Topoisomerase alpha 2 (TOP2A) is a marker of
proliferation and chemotherapy resistance in different cancer
types, including adrenocortical carcinoma and breast carcinoma
(18,19). Furthermore, it has been reported
that several miRNAs serve a regulatory role by directly inhibiting
the target TOP2A in cancer (20).
In the present study, it was demonstrated for the
first time that DUXAP8 was enhanced in OS cell lines and tissues.
Downregulation of DUXAP8 markedly suppressed OS cell viability and
invasion. Additionally, it was confirmed that DUXAP8 may promote
the development of OS cells by modulating miR-635/TOP2A. The
results of the present study may offer a novel diagnostic and
therapeutic candidate for OS treatment.
Materials and methods
Patient samples
Patients with OS (n=35) who received surgery in the
Affiliated Hospital of Bei Hua University (Jilin, China) between
October 2018 and October 2019 were selected to obtain cancer tissue
samples and adjacent normal tissues. The patients were 31–73 years
old, including 19 males and 16 females and they had not received
chemotherapy or radiotherapy prior to surgery. The tissues were
subsequently stored in liquid nitrogen and then stored at −80°C
until extraction of RNA. All research protocols in the present
study were approved by the Ethics Committee of the Affiliated
Hospital of Bei Hua University. Written informed consent was
obtained from every patient.
Cell lines and cell culture
Human osteosarcoma cell lines, including KHOS-240S,
SaOS2, MG-63, SOSP-9607 and U2OS, and one normal osteoblastic cell
line (hFOB1.19) were obtained from the American Type Culture
Collection and the Cell Bank of the Chinese Academy of Sciences,
respectively. All cell lines were cultured according to the
manufacturer's protocols. Cells were cultured in Dulbecco's
modified Eagle's medium, supplemented with 10% fetal bovine serum,
and all incubations were performed at 37°C in a 5%
CO2-containing atmosphere.
Cell transfection
The short interfering RNAs that targeted lncRNA
DUXAP8 (si-lncRNA-DUXAP8), corresponding siRNA negative controls
(siNC), miR-635 mimic, negative control (NC) miRNA, miR-635
inhibitor and NC inhibitor were purchased from Shanghai GenePharma
Co., Ltd. Transfections were performed using the Lipofectamine 3000
kit (Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. RNAs (100 nM) or miR-635 mimics (50 nM) or
miR-635 inhibitor (150 nM) or plasmids (1.5 µg per well) were
transfected into cells. The sequences were as follows: si-DUXAP8,
5′-AAGAUAAAGGUGGUUUCCACAAGAATT-3′; si-NC,
5′-AGCUUGAUACGACAAAGCUTT-3′; miR-635 mimic,
5′-ACUUGGGCACUGAAACAAUGUCC-3′; miR-NC, 5′-CAGUACUUUUGUGUAGUACAA-3′;
miR-635 inhibitor, 5′-GGACAUUGUUUCAGUGCCCAAGU-3′; and inhibitor NC,
5′-CAGUACUUUUGUGUAGUACAA-3′.
Transfection was performed at room temperature for
30 min. The knockdown efficiency was assessed by reverse
transcription-quantitative (RT-q) PCR 48 h after transfection, when
the cells were collected for the subsequent experiments.
RNA extraction and RT-qPCR
Total RNA was extracted from tissues and cells using
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol. Total RNA was reverse transcribed into
cDNA using the Prime Script® RT Reagent kit. RT reaction
was conducted for 15 min at 42°C followed by 5 min at 98°C and the
reaction volume was 20 µl. qPCR was performed using SYBR Premix Ex
Taq (Takara Biotechnology Co., Ltd.) on an ABI 7500 RT-qPCR system.
Expression of DUXAP8 and TOP2A was detected using GAPDH as
endogenous control, respectively. High Pure miRNA Isolation kit
(Sigma-Aldrich; Merck KGaA) was used to extract miRNA. miRNA
reverse transcription was performed using MystiCq®
microRNA cDNA Synthesis mix (Sigma-Aldrich; Merck KGaA), and qPCR
was performed using MystiCq microRNA® SYBR®
Green qPCR ReadyMix® (Sigma-Aldrich; Merck KGaA) to
measure the level of miR-635 expression with U6 as an endogenous
control. The primer sequences were as follows: DUXAP8 forward,
5′-AGGATGGAGTCTCGCTGTATTGC-3′ and reverse,
5′-GGAGGTTTGTTTTCTTCTTTTTT-3′; TOP2A forward,
5′-GATTGATTATGACAAAGTATA-3′ and reverse,
5′-TACTTTGTCATAATCAATCAG-3′; GAPDH forward,
5′-CGCTCTCTGCTCCTCCTGTTC-3′ and reverse,
5′-ATCCGTTGACTCCGACCTTCAC-3′. miR-635 forward,
50-TATAGCATATGCAGGGTG-30; miR-635 reverse primer and U6 primers
were included in the kit. The thermocycling conditions were as
follows: Initial denaturation at 95°C for 10 min, followed by 40
cycles at 95°C for 15 sec and extension at 60°C for 1 min. The
relative expression of DUXAP8, miR-635 and TOP2A mRNA levels was
calculated using the 2−ΔΔCt method (21). The median value was the cut-off
between low and high DUXAP8 expression in patients with OS. The
median value was included in the low expression group.
Cell proliferation assay
Cell proliferation was quantified using Cell
Counting Kit-8 (CCK-8; Beyotime Institute of Biotechnology),
according to the manufacturer's protocols. In brief,
1×105/well cells were seeded and transfected into a
96-well plate (Corning Incorporated). At the indicated times, 10 µl
CCK-8 solution was added to each well, and the cells were incubated
for 4 h at 37°C. The absorbance was measured using a microplate
reader (BioTek Instruments, Inc.) at 450 nm.
Wound healing assay
To measure the migratory ability of OS cells, a
wound-healing assay was performed. Cells were seeded and cultured
to a confluent monolayer in a rectangular cell culture plate. The
medium was removed, and then the teeth of the cell comb were drawn
across the cell monolayer with sufficient force. Cells were washed,
replenished with fresh Dulbecco's modified Eagle's medium (DMEM;
Thermo Fisher Scientific, Inc.) and incubated for an additional 24
h. Wound closure was monitored with a light microscope
(magnification, ×100; BX51; Olympus Corporation). Gap distance was
quantified using NIH ImageJ software version 1.46 (National
Institutes of Health).
Transwell assay
Transwell membranes with 8-µm pore sizes (Corning
Incorporated) coated with Matrigel (BD Biosciences) were used for
the cancer cell invasion assay as previously described (22). Following the indicated transfection,
2×105 cells were resuspended in fresh serum-free DMEM
and replated into the upper chamber. Fresh DMEM containing 10%
fetal bovine serum (FBS) was directly added to the lower chamber.
After an additional 24 h of incubation at 37°C, the invasive cells
penetrated the lower surface, were fixed with 4% paraformaldehyde
in phosphate-buffered saline (PBS) at room temperature for 10 min
and stained with 0.1% crystal violet at room temperature for 10
min. (Sigma-Aldrich; Merck KGaA). The number of invasive cells was
counted under a light microscope (magnification, ×100; BX51;
Olympus Corporation).
Colony formation assay
A colony formation assay was also performed. Cells
were added to 6-well plates (2×103 cells/well) following
transfection for 2 weeks. Colonies were fixed with 100% methanol at
room temperature for 20 min and stained with 0.1% crystal violet
(Sigma-Aldrich; Merck KGaA) at 25°C for 30 min. The total number of
visible colonies was imaged and counted using a light microscope
(magnification, ×100). All experiments were repeated three
times.
Luciferase activity assay
The wild-type (WT) or mutant (MUT) DUXAP8 sequences
containing the miR-635 binding sites were cloned into the pmir-GLO
Dual-luciferase vector (Promega Corporation) and co-transfected
with the miR-635 mimics or corresponding control sequences into the
cells with Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.). The sequences were WT DUXAP8:
5′-UUUAAAACUCUUGAUGCUGGUU-3′; MUT DUXAP8:
5′-UUUAUUUGAGUUGAUGCUGGUU-3′ miR-635 mimic,
5′-ACUUGGGCACUGAAACAAUGUCC-3′; miR-NC, 5′-CAGUACUUUUGUGUAGUACAA-3′.
Following transfection, the cells were incubated for 48 h and the
luciferase activity was measured with the Dual Luciferase Reporter
Assay System (Promega Corporation). The Renilla luciferase
was used as an internal control to homogenize the detection of the
reporter gene.
Western blotting
Western blotting was performed following the
protocol as previously described (23). In brief, cells were lysed with RIPA
lysis buffer (Thermo Fisher Scientific, Inc.), incubated on ice for
30 min, and centrifuged at 11,000 × g for 30 min at 4°C. The
supernatant was then collected, and the protein concentration was
determined using a BCA protein quantitation kit (Beyotime Institute
of Biotechnology). Cell lysates (20 µg) were subjected to 10%
SDS-PAGE and transferred to polyvinylidene fluoride membranes (GE
Healthcare). Next, the membranes were blocked with 5% bovine serum
albumin in PBS for 1 h at room temperature and incubated with
primary antibodies TOP2A (cat. no. 12286, Cell Signaling
Technology, Inc.; dilution, 1:1,000) and GAPDH (cat. no. 5174, Cell
Signaling Technology, Inc.; dilution, 1:5,000) overnight at 4°C.
Next, the blots were washed and probed with a secondary antibody of
HRP Goat Anti-Rabbit (IgG; cat. no. ab6721, Abcam; dilution,
1:10,000) at room temperature for 1.5 h and visualized using an ECL
detection system (Thermo Fisher Scientific, Inc.). The relative
densities of the protein bands were analyzed with NIH ImageJ
software version 1.46 (National Institutes of Health).
Statistical analysis
Statistical analyses were performed using GraphPad
Prism software (version 6; GraphPad Software, Inc.). A normal
distribution and homogeneity of variance were tested. Measurement
data conforming to a normal distribution were expressed as the mean
± standard deviation. If the data did not conform to a normal
distribution or homogeneity of variance, quantile spacing was
applied. The sample size of each group for the cell experiments was
nine. The relationship between lncRNA DUXAP8 and miR-635 expression
levels in OS and normal adjacent tissues was analyzed by paired
Student's t-test. Survival curves were plotted using the
Kaplan-Meier method and log-rank tests were performed. An unpaired
t-test was applied for the other comparisons between two groups.
For the comparison of multiple groups, one-way ANOVA analysis was
performed followed by Tukey's post hoc test. Correlations were
analyzed using Pearson's correlation. Linear regression analysis
was performed to identify variables that significantly affected
these correlations. P<0.05 was considered to indicate a
statistically significant difference.
Results
The expression of lncRNA DUXAP8 is
upregulated in OS tissues and cell lines
To investigate the role of lncRNA DUXAP8 in OS, the
relative expression level of lncRNA DUXAP8 was investigated in 35
pairs of OS tissues and adjacent non-tumor tissues by RT-qPCR
analysis. Differences in expression level of DUXAP8 between OS and
non-tumor tissues were analyzed by paired t-test. As presented in
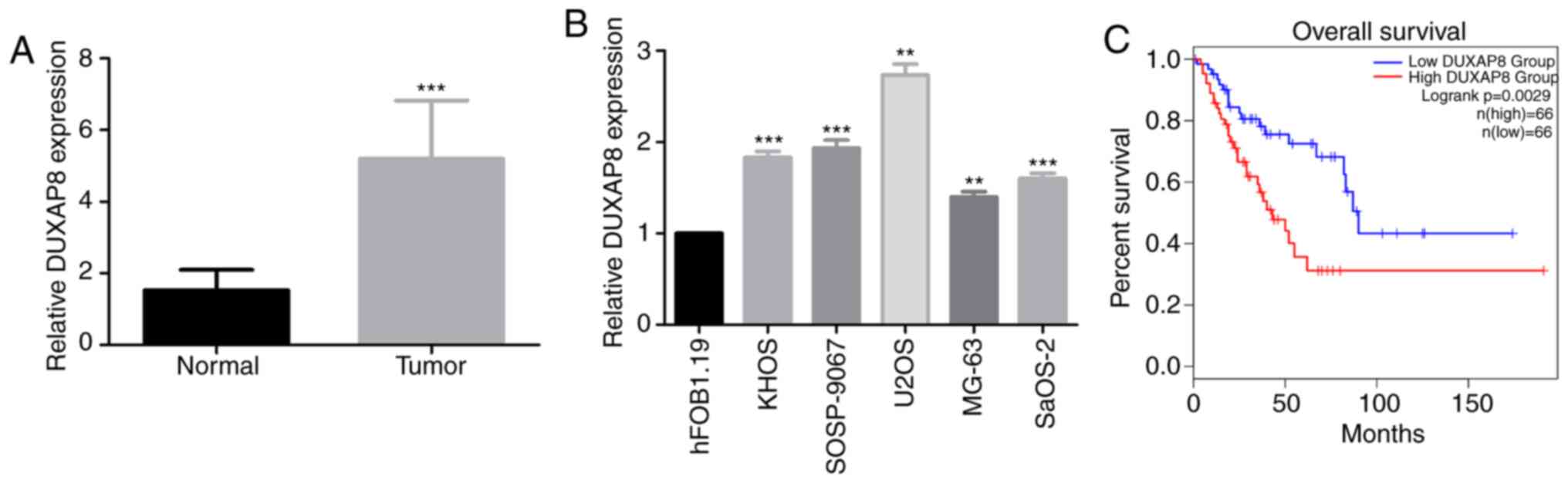
Fig. 1A (P<0.001), the level of
lncRNA DUXAP8 was higher in OS tissues than in non-cancerous
samples. Additionally, the expression of DUXAP8 was investigated in
5 human OS cell lines (KHOS, SOSP-9607, U2OS, MG-63 and SaOS-2) and
the normal osteoblastic hFOB1.19 cell line by RT-qPCR. The results
revealed that lncRNA DUXAP8 expression was markedly increased in
the five OS cell lines compared with the hFOB1.19 cell line
(Fig. 1B; P<0.001). To further
determine the association between DUXAP8 expression and the
long-term prognosis of patients, Kaplan-Meier analysis was
performed based on TCGA patients using GEPIA. Patients with higher
DUXAP8 expression levels had shorter overall survival times than
patients with lower DUXAP8 expression levels (Fig. 1C). These results suggested that
DUXAP8 was involved in the progression of OS.
DUXAP8 promotes the proliferation,
migration and invasion of OS cells
To determine the potential biological role of lncRNA
DUXAP8 in OS cells, two OS cell lines, U2OS and SaOS2 cells, with
higher expression of lncRNA DUXAP8 were selected to assess the
effects of siRNA-mediated knockdown of lncRNA DUXAP8 on cell
proliferation and colony formation. Following transfection with
lncRNA DUXAP8-specific siRNAs or a control siRNA, lncRNA DUXAP8
expression was revealed to be efficiently decreased by RT-qPCR
analysis (Fig. 2A; P<0.01). It
was observed that knockdown of lncRNA DUXAP8 caused a significant
decrease in cell proliferation, as measured using CCK-8 assays and
colony formation assays (Fig. 2B-D;
P<0.01). The migration ability of U2OS and SaOS2 cells were
further examined by scratch assay. The results demonstrated that
the migration ability of U2OS and SaOS2 cells was significantly
decreased following knockdown of DUXAP8 (Fig. 2E; P<0.001). The effects of DUXAP8
on the invasion of OS cells were investigated by Transwell assay.
Furthermore, the results demonstrated that knockdown of DUXAP8
inhibited the invasion of U2OS and SaOS2 cells (Fig. 2F; P<0.001). Taken together, these
results confirmed that DUXAP8 was an oncogenic lncRNA in OS.
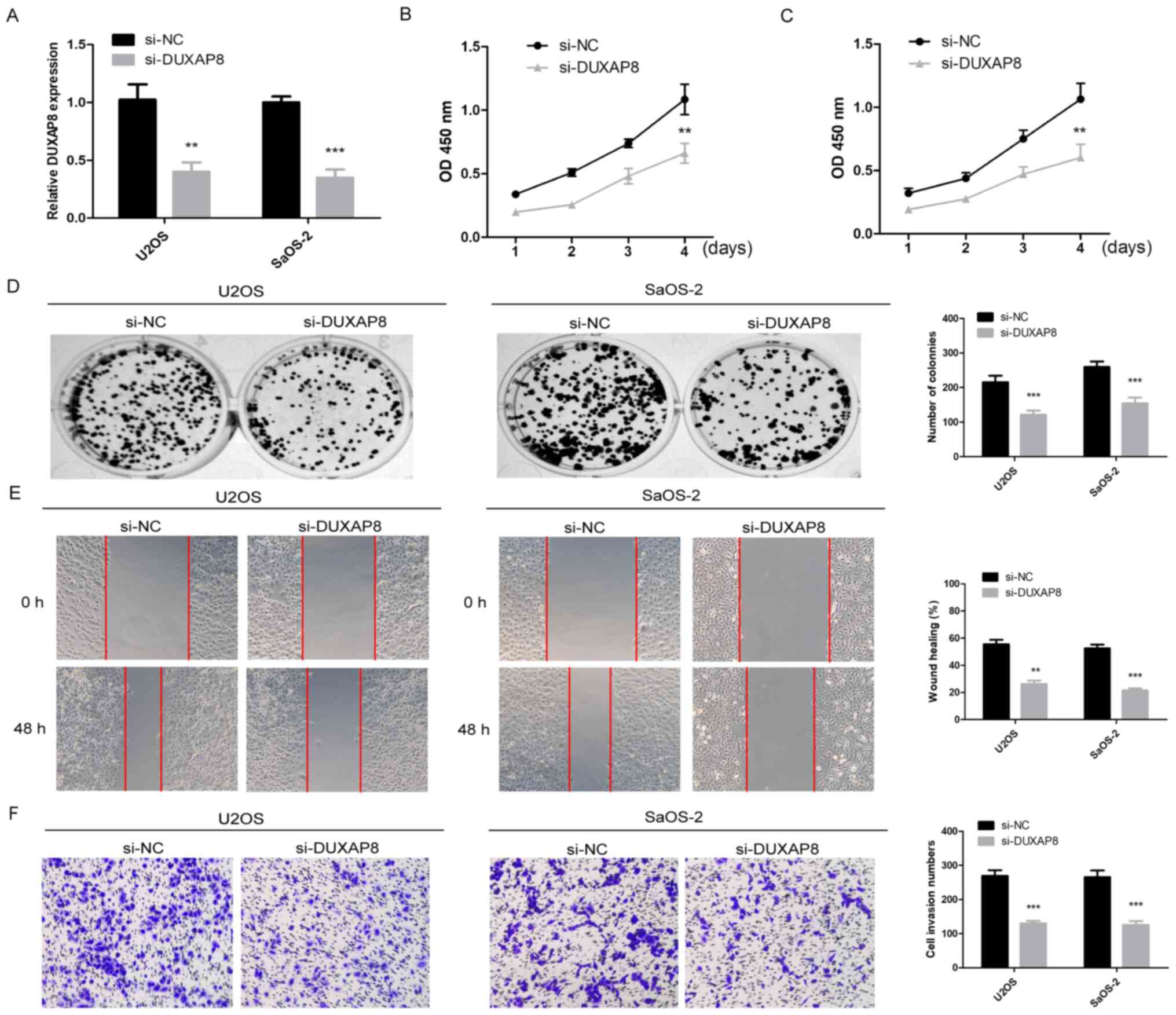 | Figure 2.DUXAP8 promotes OS proliferation,
migration and invasion. (A) lncRNA DUXAP8 expression was detected
in U2OS and SaOS-2 cells transduced with a lncRNA DUXAP8 siRNA
vector (si-lncRNA DUXAP8) or negative control siRNA vector (si-NC)
**P<0.01, ***P<0.001 vs. si-DUXAP8. (B-D) Cell viability was
measured using a Cell Counting kit-8 assay and colony formation
assay at the indicated times following transfection, **P<0.01,
***P<0.001 vs. si-DUXAP8. (E) The migration of OS cells
following DUXAP8-knockdown was detected by a scratch healing assay,
**P<0.01, ***P<0.001 vs. si-DUXAP8. Magnification, ×100. (F)
The invasion of OS cells following DUXAP8-knockdown was detected by
a Transwell assay, ***P<0.001 vs. si-DUXAP8. Magnification,
×100. Data are expressed as the mean ± standard deviation,
Student's t-test. DUXAP8, double homeobox A pseudogene 8; OS,
osteosarcoma; lncRNA, long non-coding RNA; si, small interfering
RNA; NC, negative control; OD, optical density. |
DUXAP8 targets miR-635 in OS
cells
lncRNAs may act as molecular sponges to regulate the
expression of downstream genes by absorbing miRNAs. By searching
for the potential binding miRNA of DUXAP8 in the StarBase online
database (www.starbase.sysu.edu.cn), miR-635 was selected as a
predictive target for DUXAP8 because of its high binding potential
(Fig. 3A). The expression of
miR-635 was significantly downregulated in OS tissues (Fig. 3B; P<0.001). Notably, the
expression correlation between DUXAP8 and miR-635 was analyzed in
OS tissue samples, and their expression was revealed to be
negatively correlated (Fig. 3C;
P<0.001). The expression of miR-635 was significantly
downregulated in OS cells (Fig. 3D;
P<0.01). Additionally, an increase in miR-635 expression was
observed in U2OS and SaOS2 cells with DUXAP8-knockdown, suggesting
that DUXAP8 may negatively regulate miR-635 expression in OS
(Fig. 3E; P<0.01). U2OS and
SaOS2 cells were transfected with miR-635 or negative control and
the efficiency of transfection was determined by RT-qPCR. Results
demonstrated that the ectopic transfection significantly
upregulated miR-635 expression levels (Fig. 3F; P<0.001). Additionally, U2OS
and SaOS2 cells were transfected with NC inhibitor/miR-635
inhibitor and the efficiency of transfection was determined by
RT-qPCR. Results demonstrated that the ectopic transfection
significantly downregulated miR-635 expression levels (Fig. 3G; P<0.01). To further validate
the binding relationship between miR-635 and DUXAP8, a
dual-luciferase reporter assay was performed and demonstrated that
luciferase activity decreased significantly following U2OS or SaOS2
cells being co-transfected with miR-635 and DUXAP8-WT reporter
plasmids; however, the luciferase activity did not decrease
significantly when the cells were co-transfected with miR-635 and
DUXAP8-MUT reporter plasmids (Fig. 3H
and I; P<0.01). Taken together, these results indicated that
DUXAP8 negatively regulated miR-635 in OS.
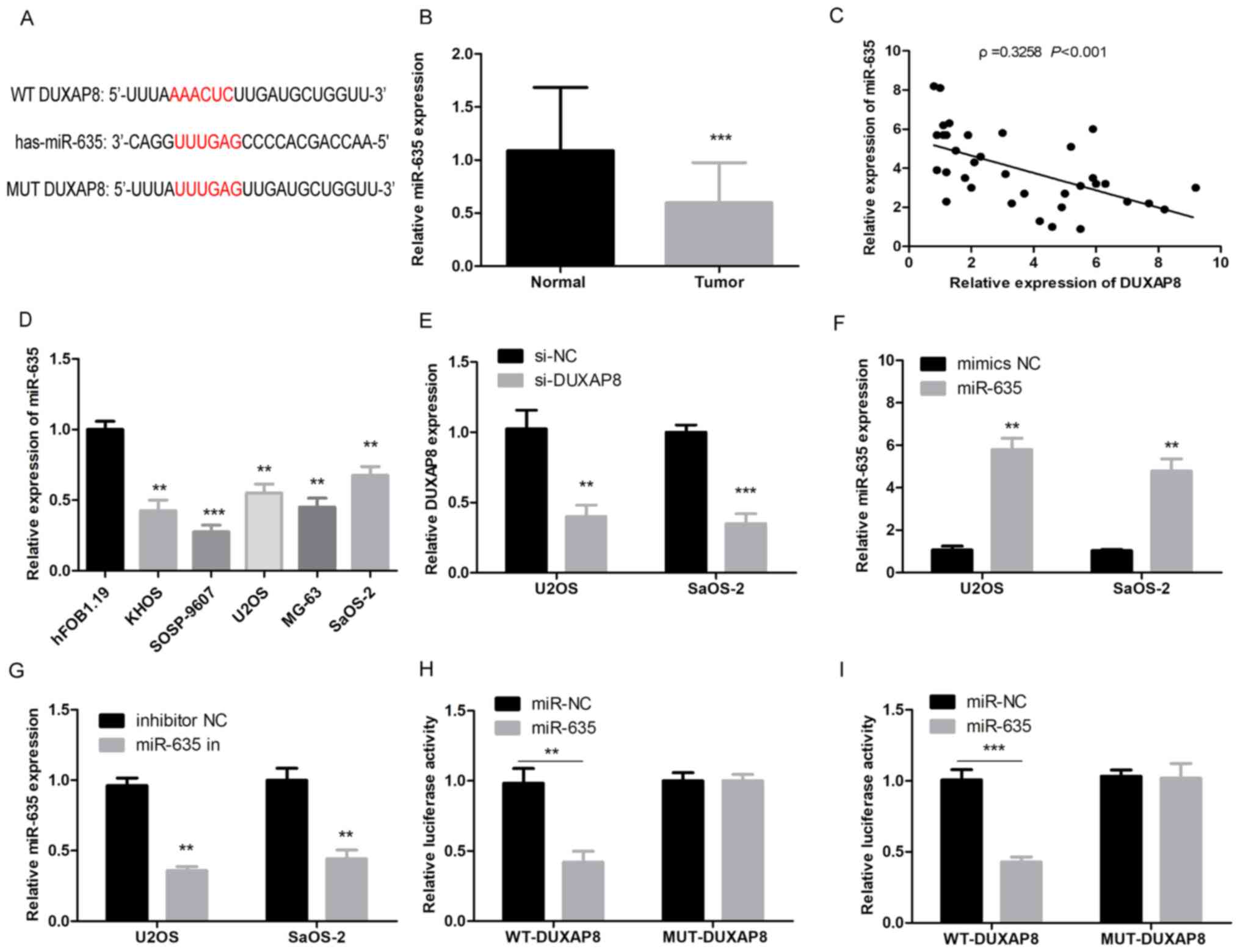 | Figure 3.DUXAP8 targets miR-635 in OS. (A)
Prediction of binding sites between miR-635 and DUXAP8. (B) The
expression levels of miR-635 in 35 matched OS and adjacent
non-tumor controls were detected by RT-qPCR, ***P<0.001 vs.
normal tissues. (C) The correlation between the expression levels
of DUXAP8 and miR-635 in OS samples was analyzed (***P<0.001).
(D) The expression of miR-635 in OS cell lines and the normal
osteoblastic hFOB1.19 cell line was detected by RT-qPCR,
**P<0.01 or ***P<0.001 vs. hFOB1.19. (E) The expression
levels of miR-635 in OS cell lines were detected by RT-qPCR
following DUXAP8-knockdown, **P<0.01, ***P<0.001 vs.
si-DUXAP8. (F) The expression levels of U2OS and SaOS-2 cells
transfected with miR-635 or mimic negative control, **P<0.01 vs.
miR-635. (G) The expression levels of U2OS and SaOS-2 cells
transfected with NC inhibitor/miR-635 inhibitor, **P<0.01 vs.
miR-635 in. (H and I) Dual-luciferase reporter assay indicated that
miR-635 mimics could decrease the luciferase activity of the
wild-type DUXAP8 reporter in U2OS and SaOS-2 cells, **P<0.01,
***P<0.001 vs. miR-635. DUXAP8, double homeobox A pseudogene 8;
miR, microRNA; OS, osteosarcoma; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; NC, negative
control; si, small interfering RNA; WT, wild-type. |
miR-635 may reverse the function of
DUXAP8 in OS cells
Next, miR-635 mimics were transfected into U2OS or
SaOS2 OS cells with DUXAP8-knockdown. It was revealed that
overexpression of miR-635 attenuated the effect of overexpressing
DUXAP8 on proliferation by CCK-8 assays and colony formation assays
(Fig. 4A-C, P<0.05). Next,
scratch and Transwell assays were conducted. Overexpression of
miR-635 attenuated the effect of overexpressing DUXAP8 on the
migration and invasion of OS cells (Fig. 4D and E, P<0.01). These findings
indicated that miR-635 reversed the function of DUXAP8 in OS
cells.
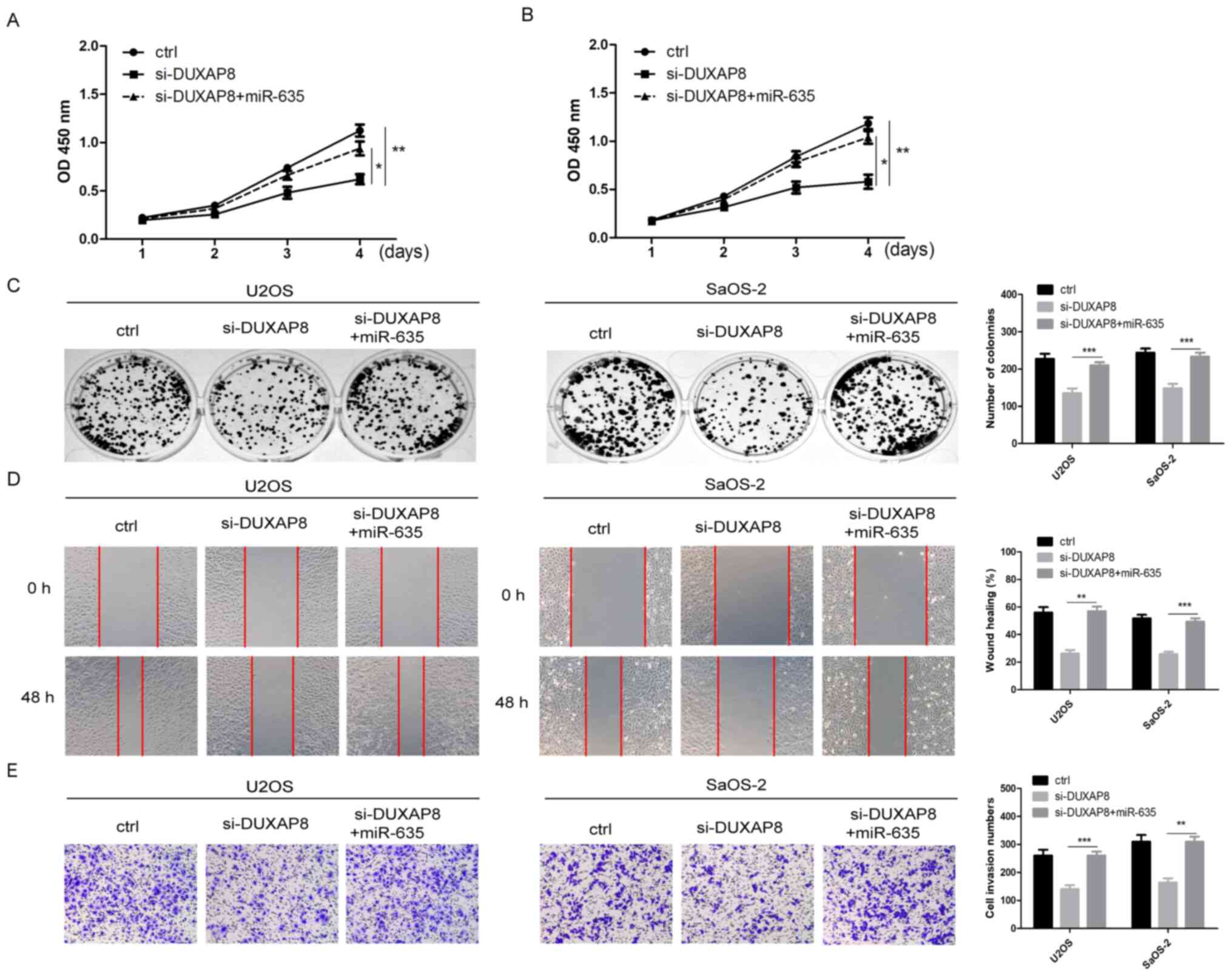 | Figure 4.miR-635 reverses the function of
DUXAP8 in OS. (A and B) miR-635 mimics and DUXAP8 siRNA were
co-transfected into OS cells, and the proliferation of OS cells in
each group was detected by Cell Counting kit-8 assay, *P<0.05
vs. si DUXAP8+miR-635, **P<0.01 vs. control. (C) The cell
viability in each group was measured using a colony formation
assay, **P<0.01, ***P<0.001 vs. si-DUXAP8. (D) The migration
in each group was evaluated by scratch healing assay, **P<0.01,
***P<0.001 vs. si-DUXAP8. Magnification, ×100. (E) The invasion
of OS cells in each group was evaluated by Transwell assay,
**P<0.01, ***P<0.001 vs. si-DUXAP8. Magnification, ×100. Data
are expressed as the mean ± standard deviation, Student's t-test.
miR, microRNA; DUXAP8, double homeobox A pseudogene 8; OS,
osteosarcoma; siRNA, small interfering RNA; Ctrl, control; OD,
optical density. |
Regulation of DUXAP8/miR-635 on TOP2A
expression in OS cells
After confirming that DUXAP8 can regulate miR-635
expression, the downstream targets of miR-635 in OS were
investigated. A recent study (24)
demonstrated that TOP2A, an oncogene, is a target of miR-635;
therefore, the present study investigated whether DUXAP8 can
regulate TOP2A expression in OS. DUXAP8-knockdown significantly
decreased the mRNA and protein expression of TOP2A (Fig. 5A and B; P<0.001). Furthermore,
the results of the present study revealed that overexpression of
miR-635 decreased TOP2A expression and abolished the DUXAP8-induced
upregulation of TOP2A (Fig. 5C and
D; P<0.05). Furthermore, it was found that the expression of
TOP2A mRNA and DUXAP8 was positively correlated in OS tissue
samples, while the expression of TOP2A mRNA was negatively
correlated with the expression of miR-635 (Fig. 5E and F; P<0.001). These results
indicated that DUXAP8 may upregulate TOP2A expression in OS and
promote OS progression, possibly by modulating miR-635.
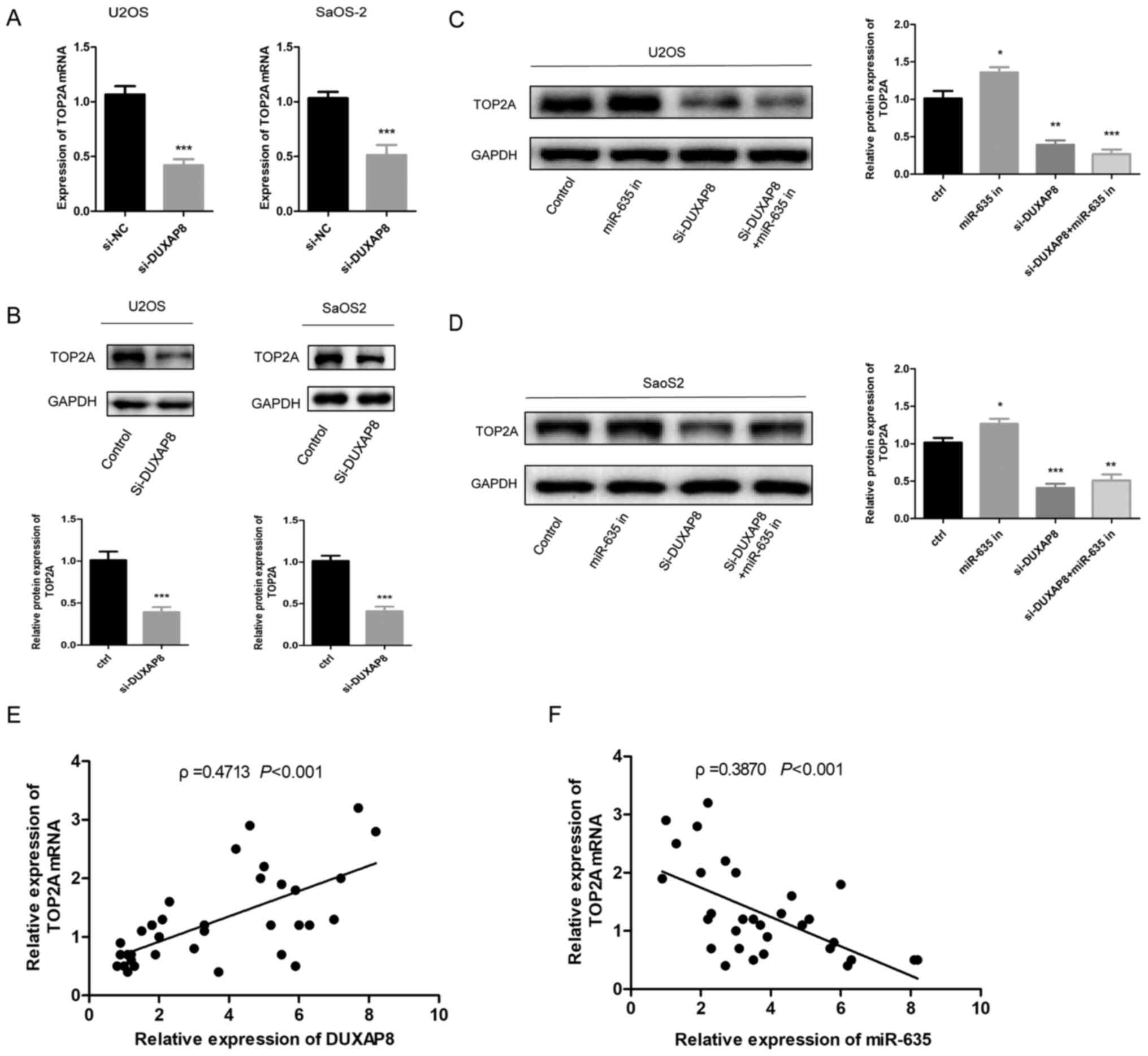 | Figure 5.Regulation of DUXAP8/miR-635 on
TOP2A. (A) Reverse transcription-quantitative polymerase chain
reaction was used to detect the effect of DUXAP8-knockdown on TOP2A
mRNA expression in OS cell lines, ***P<0.001 vs. control. (B)
Western blotting was used to detect the effect of knockdown or
overexpression of DUXAP8 on TOP2A protein expression in OS cell
lines, ***P<0.001 vs. control. (C and D) Western blotting was
used to detect the effect of DUXAP8 and miR-635 on TOP2A protein
expression in U2OS and SaoS2 cell lines, *P<0.05, **P<0.01,
***P<0.001 vs. control. (E) The correlation between the
expression levels of DUXAP8 and TOP2A mRNA in OS samples was
analyzed. (F) The correlation between the expression levels of
miR-635 and TOP2A mRNA in OS samples was analyzed. miR, microRNA;
OS, osteosarcoma; siRNA, small interfering RNA; DUXAP8, double
homeobox A pseudogene 8; TOP2A, topoisomerase alpha 2; NC, negative
control. |
Discussion
OS is thought to be one of most common causes of
cancer-related mortality worldwide, with an 8% 5-year survival rate
(25). Increasing evidence has
suggested that lncRNAs are involved in the development and
progression of a diverse range of cancer types, including gastric
cancer, hepatocellular carcinoma, clear cell renal cell carcinoma,
colorectal cancer, breast cancer, non-small cell lung cancer and OS
(26–29). For example, the lncRNA OSA3, which
is specifically upregulated in prostate cancer, has been approved
by the Food and Drug Administration for the diagnosis of prostate
cancer (30). Downregulation of
lncRNA HOST2 represses cell proliferation and promotes cell
apoptosis in OS, which may offer a potential therapeutic target for
OS (31). The regulation of lncRNA
on OS proliferation and metastasis has been studied, but not for
lncRNA DUXAP8. lncRNA DUXAP8 has been previously reported to be
upregulated and may serve as a potential therapeutic target in
several types of cancer (32). In a
recent study, it was reported that lncRNA DUXAP8 enhances renal
cell carcinoma progression by downregulating miR-126 (33). In the present study, it was found
that lncRNA DUXAP8 was expressed at significantly higher levels in
OS cell lines and tissues, suggesting that lncRNA DUXAP8 may
contribute toward the progression of OS. Furthermore, the present
study demonstrated that lncRNA DUXAP8-silencing significantly
inhibited OS cell growth, cell migration and invasion ability,
implying that lncRNA DUXAP8 serves an important role in OS
progression.
MicroRNAs (miRNAs/miRs) are non-coding RNAs that
serve an important regulatory role by acting as tumor promoters or
suppressors in various cancer types. For example, miRNA-214
suppression contributes toward cell migration, invasion and EMT in
gastric cancer by targeting FGFR (34). Previous studies have focused on the
role of miRs in OS cells. However, few reports have demonstrated
the effect of miR-635 in OS. A recent report demonstrated that
miR-635 may accelerate the invasion of A375 melanoma cells
(35). The present study
demonstrated that miR-635 may function as a tumor suppressor in OS,
as determined by experiments with human specimens and OS cell lines
in an in vitro study. The present study demonstrated that
miR-635 may be sponged by lncRNA DUXAP8 in OS. The binding
relationship between DUXAP8 and miR-635 was validated using a
dual-luciferase reporter gene assay. Additionally, knockdown of
DUXAP8 significantly induced the expression of miR-635 in OS.
Notably, with functional experiments, it was demonstrated that
miR-635 could reverse the function of DUXAP8 in OS cells. These
results not only explained the mechanism of miR-635 dysregulation
in OS but also proved that miR-635 was a crucial effector during
DUXAP8 regulation of the malignant phenotypes of OS cells. TOP2A
has been previously reported to be upregulated in hepatocellular
carcinoma (36,37). The majority of studies have focused
on TOP2A and reported the involvement of tumor chemoresistance to
DNA-damaging agents in acute myeloid leukemia (AML) as well as
other tumor types (38–40). The present study demonstrated that
TOP2A may be involved in the development of OS cells as a
carcinogenic factor and may be one of the candidate targets for
miR-635. Knockdown of DUXAP8 significantly decreased TOP2A mRNA and
protein expression, and DUXAP8 was positively correlated with TOP2A
expression in OS tissues. Notably, the expression of miR-635
partially reversed the promotion of TOP2A expression caused by
DUXAP8. These results demonstrated that DUXAP8 may target miR-635
to indirectly regulate TOP2A and thus affect the development of OS
cells.
In conclusion, the results of the present study
demonstrated that DUXAP8 is upregulated in OS tissues and cells.
lncRNA DUXAP8-knockdown may suppress cell proliferation, cell
migration and cell invasion in U2OS or SaOS2 cells. The present
study also elucidated the mechanism of the DUXAP8/miR-635/TOP2A
axis in the development of OS. With more in-depth research, DUXAP8
is likely to become a marker for clinical diagnosis and prognosis
and potentially a therapeutic target for the treatment of OS.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Science and
Technology Research Project of the Jilin Provincial Department of
Education (2020; grant no JJKH20200070KJ) and the Health Technology
Innovation Project of Jilin Province (grant no. 2019J045).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
TY was responsible for conceiving the study. TY, JPG
and FL curated the data. TY performed the formal analysis. TY and
XLD undertook the investigations. TY and XLD were responsible for
the methodology. HW, CX, JPG and FL performed the experiments. TY
wrote the original draft. XLD reviewed and edited the manuscript.
TY and XLD confirm the authenticity of all the raw data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
All research protocols in the present study were
approved by the Ethics Committee of the Affiliated Hospital of Bei
Hua University. Written informed consent was obtained from every
patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Heare T, Hensley MA and Dell'Orfano S:
Bone tumors: Osteosarcoma and Ewing's sarcoma. Curr Opin Pediatr.
21:365–372. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Harrison DJ, Geller DS, Gill JD, Lewis VO
and Gorlick R: Current and future therapeutic approaches for
osteosarcoma. Expert Rev Anticancer Ther. 18:39–50. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Strobel O, Neoptolemos J, Jäger D and
Büchler MW: Optimizing the outcomes of osteosarcoma surgery. Nat
Rev Clin Oncol. 16:11–26. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Isakoff MS, Bielack SS, Meltzer P and
Gorlick R: Osteosarcoma: Current treatment and a collaborative
pathway to success. J Clin Oncol. 33:3029–3035. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen LL and Carmichael GG: Long noncoding
RNAs in mammalian cells: What, where, and why? Wiley Interdiscip
Rev RNA. 1:2–21. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rafiee A, Riazi-Rad F, Havaskary M and
Nuri F: Long noncoding RNAs: Regulation, function and cancer.
Biotechnol Genet Eng Rev. 34:153–180. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mei D, Song H, Wang K, Lou Y, Sun W, Liu
Z, Ding X and Guo J: Up-regulation of SUMO1 pseudogene 3 (SUMO1P3)
in gastric cancer and its clinical association. Med Oncol.
30:7092013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lian Y, Xiong F, Yang L, Bo H, Gong Z,
Wang Y, Wei F, Tang Y, Li X, Liao Q, et al: Long noncoding RNA
AFAP1-AS1 acts AS a competing endogenous RNA of miR-423-5p to
facilitate nasopharyngeal carcinoma metastasis through regulating
the Rho/Rac pathway. J Exp Clin Cancer Res. 37:2532018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu LJ, Yu XJ, Wei B, Hui HX, Sun Y, Dai J
and Chen XF: Long non-coding RNA DUXAP8 regulates proliferation and
invasion of esophageal squamous cell cancer. Eur Rev Med Pharmacol
Sci. 22:2646–2652. 2018.PubMed/NCBI
|
|
13
|
Ma HW, Xie M, Sun M, Chen TY, Jin RR, Ma
TS, Chen QN, Zhang EB, He XZ, De W and Zhang ZH: The pseudogene
derived long noncoding RNA DUXAP8 promotes gastric cancer cell
proliferation and migration via epigenetically silencing PLEKHO1
expression. Oncotarget. 8:52211–52224. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jiang H, Shi X, Ye G, Xu Y, Xu J, Lu J and
Lu W: Up-regulated long non-coding RNA DUXAP8 promotes cell growth
through repressing Krüppel-like factor 2 expression in human
hepatocellular carcinoma. Onco Targets Ther. 11:7429–7436. 2019.
View Article : Google Scholar
|
|
15
|
AmbRoS V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cummins JM, He Y, Leary RJ, Pagliarini R,
Diaz LA Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE,
Labourier E, et al: The colorectal microRNAome. Proc Natl Acad Sci
USA. 103:3687–3692. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Weber CE, Luo C, Hotz-Wagenblatt A,
Gardyan A, Kordass T, Holland-Letz T, Osen W and Eichmüller SB:
miR-339-3p is a tumor suppressor in melanoma. Cancer Res.
76:3562–3571. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jain M, Zhang L, He M, Zhang YQ, Shen M
and Kebebew E: TOP2A is overexpressed and is a therapeutic target
for adrenocortical carcinoma. Endocr Relat Cancer. 20:361–370.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ejlertsen B, Jensen MB, Nielsen KV,
Balslev E, Rasmussen BB, Willemoe GL, Hertel PB, Knoop AS,
Mouridsen HT and Brünner N: HER2, TOP2A, and TIMP-1 and
responsiveness to adjuvant anthracycline-containing chemotherapy in
high-risk breast cancer patients. J Clin Oncol. 28:984–990. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang S, Jiang H, Xu Z, Jiang Y, She Y,
Huang X, Feng S, Chen W, Chen S, Chen Y, et al: The resistance of
esophageal cancer cells to paclitaxel can be reduced by the
knockdown of long noncoding RNA DDX11-AS1 through TAF1/TOP2A
inhibition. Am J Cancer Res. 9:2233–2248. 2019.PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu H, Xu Y, Zhang D and Liu G: Long
noncoding RNA LUCAT1 promotes malignancy of ovarian cancer through
regulation of miR-612/HOXA13 pathway. Biochem Biophys Res Commun.
503:2095–2100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rojo AI, Medina-Campos ON, Rada P,
Zúñiga-Toala A, López-Gazcón A, Espada S, Pedraza-Chaverri J and
Cuadrado A: Signaling pathways activated by the phytochemical
nordihydroguaiaretic acid contribute to a Keap1-independent
regulation of Nrf2 stability: Role of glycogen synthase kinase-3.
Free Radic Biol Med. 52:473–487. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Golabchi K, Soleimani-Jelodar R, Aghadoost
N, Momeni F, Moridikia A, Nahand JS, Masoudifar A, Razmjoo H and
Mirzaei H: MicroRNAs in retinoblastoma: Potential diagnostic and
therapeutic biomarkers. J Cell Physiol. 233:3016–3023. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lulla RR, Costa FF, Bischof JM, Chou PM,
de F Bonaldo M, Vanin EF and Soares MB: Identification of
differentially expressed microRNAs in osteosarcoma. Sarcoma.
2011:7326902011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang C, Wu K, Wang S and Wei G: Long
non-coding RNA XIST promotes osteosarcoma progression by targeting
YAP via miR-195-5p. J Cell Biochem. 119:5646–5656. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu X, Wang Y, Sun L, Min J, Liu J, Chen
D, Zhang H, Zhang H, Zhang H, Zhou Y and Liu L: Long noncoding RNA
BC005927 upregulates EPHB4 and promotes gastric cancer metastasis
under hypoxia. Cancer Sci. 109:988–1000. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liang Y, Ma Y, Li L, Shen X, Xin T, Zhao Y
and Ma R: Effect of long non-coding RNA LINC01116 on biological
behaviors of non-small cell lung cancer cells via the hippo
signaling pathway. J Cell Biochem. 119:63102018. View Article : Google Scholar
|
|
29
|
Zhang Y, Tao Y, Li Y, Zhao J, Zhang L,
Zhang X, Dong C, Xie Y, Dai X, Zhang X and Liao Q: The regulatory
network analysis of long noncoding RNAs in human colorectal cancer.
Funct Integr Genomics. 18:261–275. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
An N and Cheng D: The Long Noncoding RNA
HOST2 promotes gemcitabine resistance in human pancreatic cancer
cells. Pathol Oncol Res. 26:425–431. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lu W, Zhang H, Niu Y, Wu Y, Sun W, Li H,
Kong J, Ding K, Shen HM, Wu H, et al: Long non-coding RNA
lincDUXAP8 regulated non-small cell lung cancer proliferation,
migration, invasion and epithelial mesenchymal transition by
sponging miR-150-5p. Mol Cancer. 16:1182017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lin MG, Hong YK, Zhang Y, Lin BB and He
XJ: Mechanism of lncRNA DUXAP8 in promoting proliferation of
bladder cancer cells by regulating PTEN. Eur Rev Med Pharmacol Sci.
22:3370–3377. 2018.PubMed/NCBI
|
|
33
|
Huang T, Wang X, Yang X, Ji J, Wang Q, Yue
X and Dong Z: Long non-coding RNA DUXAP8 enhances renal cell
carcinoma progression via downregulating miR-126. Med Sci Monit.
14:7340–7347. 2018. View Article : Google Scholar
|
|
34
|
Wang R, Sun Y, Yu W, Yan Y, Qiao M, Jiang
R, Guan W and Wang L: Downregulation of miRNA-214 in
cancer-associated fibroblasts contributes to migration and invasion
of gastric cancer cells through targeting FGF9 and inducing EMT. J
Exp Clin Cancer Res. 38:202019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Weber CE, Luo C, Hotz-Wagenblatt A,
Gardyan A, Kordass T, Holland-Letz T, Osen W and Eichmüller SB:
miR-339-3p is a tumor suppressor in melanoma. Cancer Res.
76:3562–3571. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wong N, Yeo W, Wong WL, Wong NL, Chan KY,
Mo FK, Koh J, Chan SL, Chan AT, Lai PB, et al: TOP2A overexpression
in hepatocellular carcinoma correlates with early age onset,
shorter patients' survival and chemoresistance. Int J Cancer.
124:644–652. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Panvichian R, Tantiwetrueangdet A,
Angkathunyakul N and Leelaudomlipi S: TOP2A amplification and
overexpression in hepatocellular carcinoma tissues. Biomed Res Int.
2015:3816022015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chekerov R, Klaman I, Zafrakas M, Könsgen
D, Mustea A, Petschke B, Lichtenegger W, Sehouli J and Dahl E:
Altered expression pattern of topoisomerase IIalpha in ovarian
tumor epithelial and stromal cells after platinum-based
chemotherapy. Neoplasia. 8:38–45. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen CC, Gau JP, You JY, Lee KD, Yu YB, Lu
CH, Lin JT, Lan C, Lo WH, Liu JM and Yang CF: Prognostic
significance of beta-catenin and topoisomerase IIalpha in de novo
acute myeloid leukemia. Am J Hematol. 84:87–92. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen CF, He X, Arslan AD, Mo YY, Reinhold
WC, Pommier Y and Beck WT: Novel regulation of nuclear factor-YB by
miR-485-3p affects the expression of DNA topoisomerase IIα and drug
responsiveness. Mol Pharmacol. 79:735–741. 2011. View Article : Google Scholar : PubMed/NCBI
|