Introduction
Breast cancer is the most commonly diagnosed type of
cancer and the third leading cause of cancer-related mortality
among females worldwide (1). In
2020, 2.65 million new cases and 685,000 deaths occurred,
accounting for the second highest incidence of cancer and fifth
highest rate of cancer-related mortality (2). Moreover, the incidence and mortality
rates of breast cancer are rising annually (2). Triple-negative breast cancer (TNBC),
which is defined by a lack of expression of estrogen receptor,
progesterone receptor and HER-2, accounts for 11–20% of all breast
carcinomas (3,4). Notably, targeted therapy has a poor
effect on TNBC (5). Therefore, the
discovery of novel therapies for the treatment of TNBC is urgently
required.
MAPK4, also termed ERK4, p63 and Prkm4, is an
atypical MAPK as it is not phosphorylated by the dual
serine/threonine and tyrosine MAPK kinase due to a lack of the
threonine-X-tyrosine activation motif (6). Thus far, MAPK4 has been associated
with several diseases, such as numerous types of cancer and
methamphetamine addiction leading to psychosis (7). According to The Cancer Genome Atlas
(TCGA), lung adenocarcinoma, low-grade glioma, thyroid carcinoma
and urothelial carcinoma with upregulation of MAPK4 are associated
with poor prognosis (8). Moreover,
MAPK4 phosphorylates AKT at threonine 308 (AKTT308) and
serine 473 (AKTS473), thereby promoting cell
proliferation and inhibiting apoptosis (9). It has been verified that microRNA
(miR)-767-5p upregulates MAPK4, and that the circular (circ)RNA
circ_0000190 inhibits the progression of multiple myeloma by
regulating MAPK4 (10).
Furthermore, circRNA MAPK4 inhibits apoptosis by regulating
miR-125a-3p, which inhibits the MAPK signaling pathway (11). This evidence indicates the role of
MAPK4 as an oncogene. It has been shown that the AKT signaling
pathway is associated with the DNA damage repair mechanism
(12,13). Hence, we hypothesized that MAPK4
could regulate DNA damage repair by activating the AKT signaling
pathway.
In recent years, the DNA damage repair mechanism has
been utilized in the treatment of cancer (14). Poly ADP-ribose polymerase-1 (PARP1)
serves vital roles in DNA damage repair of single-strand and
double-strand breaks by catalyzing the formation of poly ADP-ribose
and maintaining genome integrity (15). PARP inhibitors, such as olaparib and
rucaparib, have been approved by the US Food and Drug
Administration, and it has been shown that these drugs are
effective for the treatment of ovarian and breast cancer,
particularly TNBC (16,17). However, the wide use of PARP
inhibitors can lead to the development of resistance (18). Thus, there is an urgent requirement
to discover novel targets or drugs that can be combined with PARP1
inhibitors.
As aforementioned, MAPK4 was identified to be
associated with several cancer types, and can promote DNA repair by
increasing AKT phosphorylation. Thus, small interfering (si)RNA
MAPK4 (siMAPK4) may be a novel molecule to combine with PARP1
inhibitors. Therefore, the present study aimed to investigate the
potential function and mechanism of siMAPK4 in enhancing the
efficacy of a PARP1 inhibitor, which could reduce the possibility
of PARP1 inhibitor resistance in TNBC and provide a new therapeutic
strategy for the treatment of TNBC.
Materials and methods
Materials and reagents
RPMI-1640 medium (cat. no. 11875093), FBS (cat. no.
10100139C) and 100 IU/ml penicillin-streptomycin (cat. no.
15140-122) were purchased from Gibco; Thermo Fisher Scientific,
Inc. MCF-10A-specific medium (cat. no. CM-0525) was from Procell
Life Science & Technology Co., Ltd. The PARP1 inhibitor
olaparib (cat. no. HY-10162) was purchased from MedChemExpress.
Antibodies against MAPK4 (cat. no. ab211501), p-AKTT308
(cat. no. ab38449), p-AKTS473 (cat. no. ab8932),
DNA-dependent protein kinase catalytic subunit (DNA-PK; cat. no.
ab44815), p-DNA-PK (cat. no. ab18192), RAD51 (cat. no. ab63801),
Bcl-2 (cat. no. ab59348), Bax (cat. no. ab53154), cleaved-casp-3
(cat. no. ab32042) and GAPDH (cat. no. ab245355) were from Abcam,
while HRP-labeled goat anti-mouse and anti-rabbit IgG (cat. nos.
ZB-2301 and ZB-2305, respectively) were from OriGene Technologies,
Inc. The anti-p-histone H2AX (γH2AX; phospho-Ser139; cat. no.
K001453M) antibody was purchased from Beijing Solarbio Science
& Technology Co., Ltd., while the anti-mouse IgG (H + L)
F(ab′)2 Fragment (Alexa Fluor® 488 Conjugate; cat. no.
4408S) for immunofluorescence was from Cell Signaling Technology,
Inc. The RNeasy Mini kit (cat. no. 74104), QuantiTect Rev
Transcription kit (cat. no. 205311) and miScript SYBR Green PCR kit
(cat. no. 218073) were obtained from Qiagen GmbH. siMAPK4-1
(5′-GGUGAGCUGUUCAAGUUCAGC-3′), siMAPK4-2
(5′-GGAAGGUCGCUGUGAAGAAGA−3′), siMAPK4-3
(5′-GGUUGGUAACAAAGUGGUACC−3′) and scramble siRNA
(5′-GGCGTGAATGGGTCTAACCTT−3′) were purchased from Addgene, Inc.
Overexpression-MAPK4 plasmid (cat. no. C05008), the pcDNA 3.1 empty
vector (cat. no. C06002), which was used as the
overexpression-negative control (NC), and constitutively active AKT
(AKT-CA) plasmids (cat. no. 53583) were designed and purchased from
Shanghai GenePharma Co., Ltd. The mRNA expression level of MAPK4
from 116 TNBC samples and 114 normal samples was obtained from
UALCAN (http://ualcan.path.uab.edu) (19), which could provide mRNA expression
data regarding cancer and normal samples in TCGA.
Cell culture
MCF-10A (normal human mammary epithelial cell line)
cells were purchased from The Cell Bank of Type Culture Collection
of Chinese Academy of Sciences and cultured in MCF-10A-specific
medium (Procell Life Science & Technology Co., Ltd.).
MDA-MB-468 and MDA-MB-231 (breast adenocarcinoma), HCC1937 (ductal
breast carcinoma) cells are TNBC cell lines (all from The Cell Bank
of Type Culture Collection of Chinese Academy of Sciences) and were
cultured in RPMI-1640 medium containing 10% FBS and 1% 100 IU/ml
penicillin-streptomycin at 37°C in an incubator with 5%
CO2.
Cell transfection
According to the instructions provided by the
manufacturer of Lipofectamine® 2000 (Thermo Fisher
Scientific, Inc.), HCC1937 cells were seeded in 6-well plates at
the density of 1.2×106 cells/well. The following day, 10
µl Lipofectamine 2000 and 4 µg plasmids were diluted with 500 µl
Opti-MEM (Thermo Fisher Scientific, Inc.) and incubated at room
temperature for 20 min. Subsequently, the Lipofectamine 2000 and
plasmids solution was added to the cells. Following 6 h of
incubation at 37°C, the medium was changed with fresh RPMI-1640
medium. After 24 h, the subsequent experiments were performed with
the transfected HCC1937 cells.
Western blotting
Western blotting was performed as previously
described (20,21). Briefly, total protein of HCC1937
cells was extracted using RIPA lysis buffer (Beyotime Institute of
Biotechnology), quantified using an Enhanced BCA Protein assay kit
(Beyotime Institute of Biotechnology), separated via 10% SDS-PAGE
(20 µg total protein/lane) and transferred to nitrocellulose
membranes (Whatman pcl). After blocking with 5% fat-free milk for 1
h at room temperature, the membranes were incubated with primary
antibodies at 4°C overnight. After washing three times with
TBS-Tween-20 (1%), the membranes were incubated with secondary
antibodies for 1 h at room temperature. Finally, the blots were
acquired using Supersignal WestFemto Maximum Sensitivity substrate
(Thermo Fisher Scientific, Inc.) and a ChemiDoc XRS+
system (Bio-Rad Laboratories). The antibodies used were as follows:
MAPK4 (1:1,000), p-AKTT308 (1:800), p-AKTS473
(1:800), DNA-PK (1:1,000), p-DNA-PK (1:200), RAD51 (1:3,000) Bcl-2
(1:800), Bax (1:800), cleaved-casp-3 (1:800), GAPDH (1:2,000), and
HRP-labeled goat anti-mouse and anti-rabbit IgG (1:2,000).
MTT assay
The MTT assay was performed as previously described
(21). Briefly, HCC1937 cells
transfected with siMAPK, overexpression-MAPK4 or scramble siRNA
were seeded into 96-well plates (5,000 cells/well), and 10 µM
olaparib was added the following day for 24 h. After incubating for
24, 48 and 72 h in a 37°C incubator, cells were incubated with MTT
(10 µg/ml) at 37°C for 4 h, then 150 µl DMSO was added to the wells
and the optical density values were measured.
Wound healing assay
The wound healing assay was performed as previously
described (20,21). Briefly, HCC1937 cells transfected
with siMAPK4 or scramble siRNA, and control HCC1937 cells were
seeded into 6-well plates at the density of 106 cells
per well, which had been marked with a straight line on the back of
the plates in advance. The following day, cells were treated with
10 µM olaparib for 24 h in a 37°C incubator. Following the
treatment, wounds were made in the cell monolayer by scratching
with 10-µl pipette tips. The cells were then incubated in DMEM
without FBS at 37°C in an incubator. Images were captured using a
light microscope (magnification, ×100) at 0 and 24 h after
treatment with olaparib and the migratory distance was measured
using ImageJ 1.49 (National Institutes of Health).
Immunofluorescence
Immunofluorescence was performed as previously
described (21). Briefly, cells
were fixed in 4% paraformaldehyde for 15 min at room temperature
and blocked with 2% BSA (Sigma-Aldrich; Merck KGaA) for 1 h at room
temperature. Primary antibody γH2AX (1:200) was incubated at 4°C
overnight and FITC-labelled secondary anti-mouse antibody (1:400;
cat. no. 4408S; Cell Signaling Technology, Inc.) was incubated for
1 h at room temperature. Nuclei were counterstained with DAPI at
room temperature. Finally, cells were sealed, and images were
acquired using a confocal microscope (magnification, ×100).
RT-quantitative (RT-q)PCR
mRNA was extracted using a RNeasy Mini kit and
reverse transcribed into cDNA according to the manufacturer's
instructions of the QuantiTect Rev Transcription kit. qPCR was
performed with the miScript SYBR Green PCR kit. The system was as
follows: 10 µl 2X SYPR Green Supermix, 1 µl forward primer, 1 µl
reverse primer, 2 µl cDNA and 6 µl RNase-free water. The
amplification was performed as follows: Initial denaturation at
95°C for 10 min; followed by 40 cycles of 95°C for 15 sec, 60°C for
30 sec; and a final extension at 72°C for 30 sec. The mRNA
expression levels were quantified using the 2−∆∆Cq
method (22). The primers used were
as follows: MAPK4 forward, 5′-ATCCTGGCTGAGATGCTTAC-3′ and reverse,
5′-CTTGTCTTCCTCCCGGATTAC-3′ (product length, 108 bp); and GAPDH
(NM_001289726.1) forward, 5′-CCTTCCGTGTTCCTACCC-3′ and reverse,
5′-AAGTCGCAGGAGACAACC-3′ (product length, 163 bp); and AKT
(NM_001382432) forward, 5′-TGGACTACCTGCACTCGGAGAA-3′ and reverse,
5′-GTGCCGCAAAAGGTCTTCATGG-3′ (product length, 154 bp).
TUNEL assay
The TUNEL assay was performed according to the
manufacturer's instructions (Roche Diagnostics). Cells were fixed
with 4% paraformaldehyde for 25 min at room temperature and washed
twice with PBS. Subsequently, 0.2% Triton X-100 was added for 5 min
at room temperature, the cells were washed with PBS and TUNEL
reaction reagent was added for 60 min at 37°C in a dark moisture
chamber. Of note, only dUTP solution and DNase 1 were added to the
negative and positive control groups, respectively. After the
slides were washed, converter-POD was added for 30 min at 37°C. The
cells were washed again thrice with PBS, and diaminobenzidine was
added at room temperature for 10 min. Next, the slides were washed
with PBS, captured and counterstained with hematoxylin for 1 min at
room temperature, washed with running water and dehydrated with
gradient alcohol (50, 70, 85, 95 and 100%) for 1 min at each
concentration. The process was completed by the addition of xylene
twice (5 min per time) and sealing of the slides with neutral gum
(Beijing Solarbio Science & Technology Co., Ltd.). The
apoptotic rate was calculated in 200 cells obtained from different
groups using a light microscope (magnification, ×100) (23).
Statistical analysis
All experiments were repeated in triplicate and data
are presented as the mean ± SEM, and statistical analyses were
performed using GraphPad Prism 5.01 software (GraphPad Software,
Inc.). An unpaired t-test and one-way ANOVA were used to determine
the significant differences, and multiple comparison between the
groups was performed using Bonferroni's method. P<0.05 was
considered to indicate a statistically significant difference.
Results
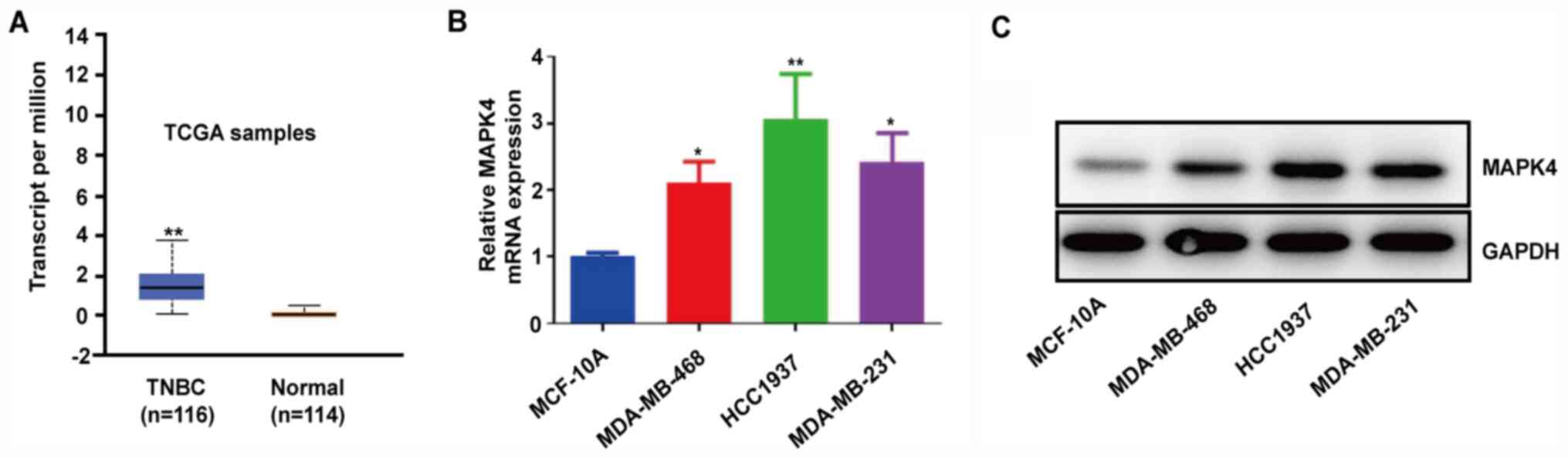
MAPK4 is highly expressed in TNBC
tissues and cell lines
MAPK4 is upregulated in numerous types of cancer,
including lung adenocarcinoma and bladder cancer (9). By analyzing 114 normal breast tissues
and 116 TNBC samples from TCGA database, it was found that TNBC
samples had higher MAPK4 mRNA expression compared with normal
samples (Fig. 1A). Moreover, the
TNBC cell lines MDA-MB-468, HCC1937 and MDA-MB-231 expressed higher
mRNA and protein levels of MAPK4 compared with the normal MCF-10A
cell line (Fig. 1B and C). These
findings indicated that the upregulation of MAPK4 may be associated
with TNBC.
MAPK4 promotes the proliferation of
HCC1937 cells
HCC1937 cells were transfected with siMAPK4 and an
overexpression-MAPK4 plasmid. As HCC1937 cells expressed higher
MAPK4 expression, these cells were chosen for knockdown
experiments. The siMAPK4 group had lower MAPK4 mRNA and protein
expression, while the overexpression-MAPK4 group had higher MAPK4
expression compared with the control groups (HCC1937 wild-type
cells) (Fig. 2A-D). Then their
proliferation was determined and the results revealed that
knockdown and overexpression of MAPK4 inhibited and promoted cell
proliferation, respectively, compared with the control group
(Fig. 2E).
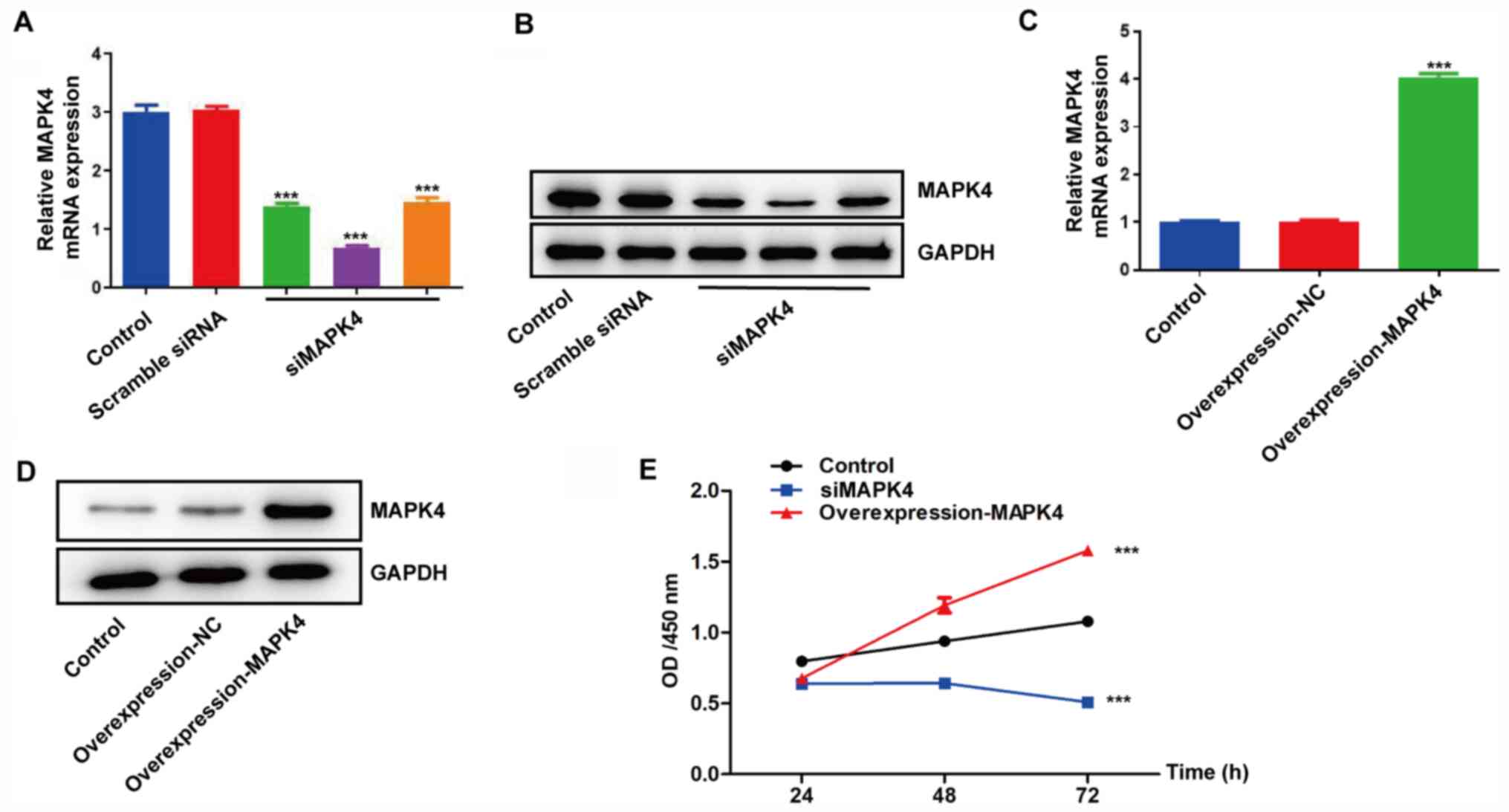 | Figure 2.MAPK4 promotes the proliferation of
HCC1937 cells. (A) Extraction of total mRNA from siMAPK4 cells,
scramble siRNA cells and control cells. RNA was reverse transcribed
to cDNA, and the relative mRNA expression levels of MAPK4 were
determined via RT-qPCR. (B) The total protein of siMAPK4 cells,
scramble siRNA cells and control cells was extracted, and the
expression level of MAPK4 was measured via western blotting. (C)
Extraction of total mRNA from control cells, overexpression-NC
cells and overexpression-MAPK4 cells. RNA was reverse transcribed
to cDNA, and the relative mRNA expression levels of MAPK4 were
determined using RT-qPCR. (D) The total protein of control cells,
overexpression-NC cells and overexpression-MAPK4 cells was
extracted, and the expression level of MAPK4 was measured via
western blotting. (E) Control cells, overexpression-MAPK4 cells and
siMAPK4 cells were seeded in 96-well plates (5,000 cells/well). MTT
assays were performed at 24, 48 and 72 h. ***P<0.001 vs.
control. NC, negative control; OD, optical density; RT-qPCR,
reverse transcription-quantitative PCR; siRNA, small interfering
RNA; siMAPK4, siRNA-MAPK4. |
siMAPK4 enhances the sensitivity of
TNBC cells to olaparib, a PARP1 inhibitor
Next, it was investigated how silencing of MAPK4
could affect the efficacy of the PARP1 inhibitor, olaparib. For
this purpose, HCC1937 cells were treated with 10 µM olaparib and
siMAPK4 separately or in combination at 37°C in an incubator. Both
siMAPK and olaparib inhibited cell proliferation and migration, and
promoted apoptosis. Moreover, as expected, the combination groups
showed increased toxicity (Fig.
3A-E).
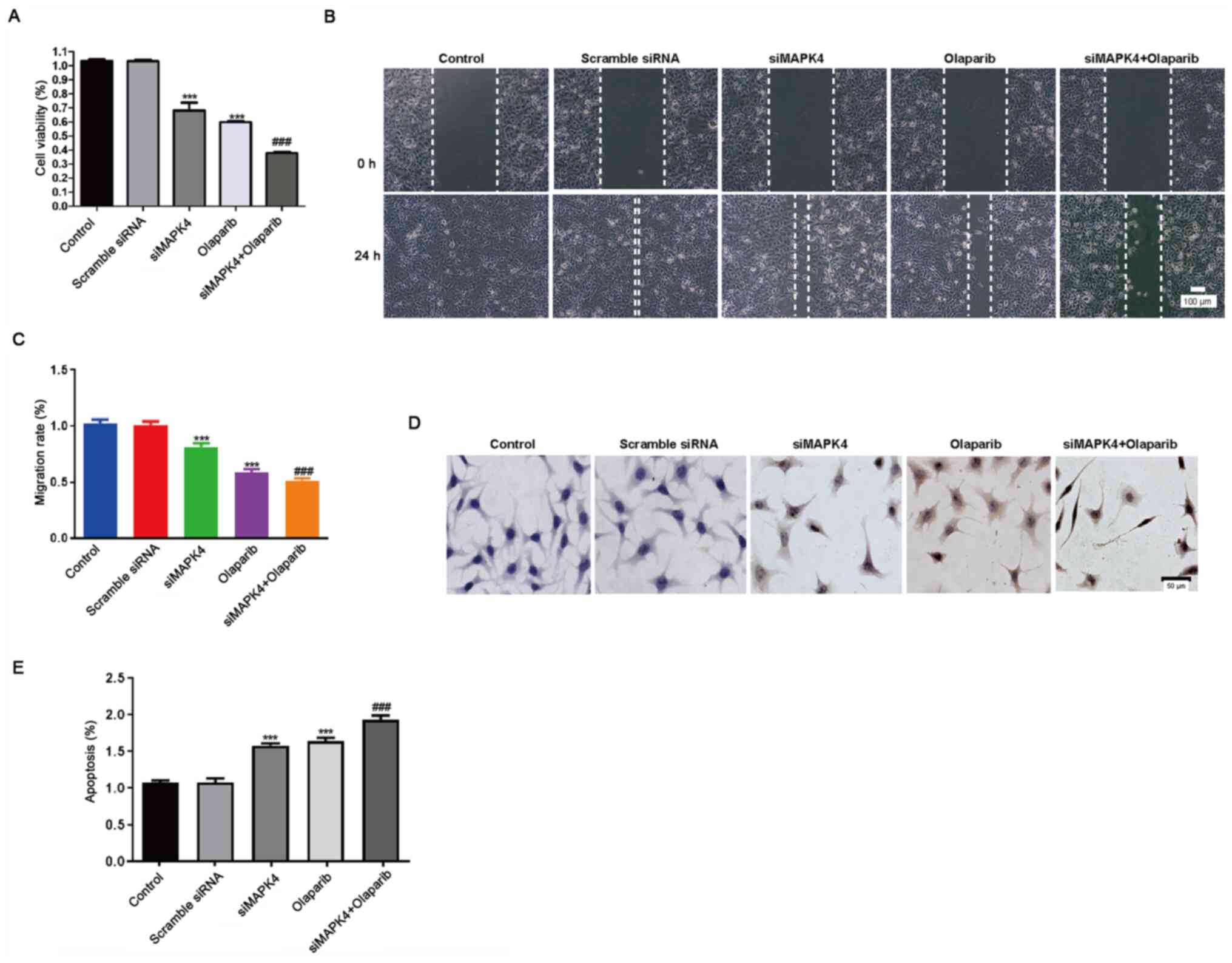 | Figure 3.siMAPK4 enhances the sensitivity of
TNBC cells to the poly ADP-ribose polymerase-1 inhibitor olaparib.
Cells were seeded in 6-well plates overnight, and transfected with
scramble siRNA and siMAPK4 plasmids for 48 h. Subsequently, cells
were treated with olaparib for 24 h. (A) A Cell Counting Kit-8
assay was performed to test the cell viability. (B) The transfected
cells were cultured, and the medium was changed with RPMI-1640
medium the following day. Next, the wound healing assay was
performed. Scale bar, 100 µm. (C) The migration rate was calculated
from the wound healing assay. (D) The apoptotic rate of cells
treated with control, siRNA-scramble, siMAPK4, olaparib and a
combination of siMAPK4 + olaparib was tested using the TUNEL assay.
Scale bar, 50 µm. (E) Statistical analysis of the results of the
TUNEL assay. ***P<0.001 vs. control; ###P<0.001
vs. olaparib. siMAPK4, siRNA-MAPK4; siRNA, small interfering
RNA. |
MAPK4 exerts its DNA damage repair
function by regulating the AKT signaling pathway
A previous study reported that MAPK4 could promote
tumor progression by phosphorylating AKTS473 and
AKTT308 to exert its DNA damage and repair function
(9). To verify this, the
phosphorylation of AKTS473 and AKTT308 was
detected in HCC1937 cells. p-AKTT308 expression was
markedly decreased in the siMAPK4 group, while no significant
differences were observed in p-AKTS473 expression
between the siMAPK4 and control groups. The results demonstrated
that MAPK4 could only phosphorylate AKTT308 (Fig. 4A). Thus, it was concluded that MAPK4
could exert its DNA damage and repair function by phosphorylating
AKTT308.
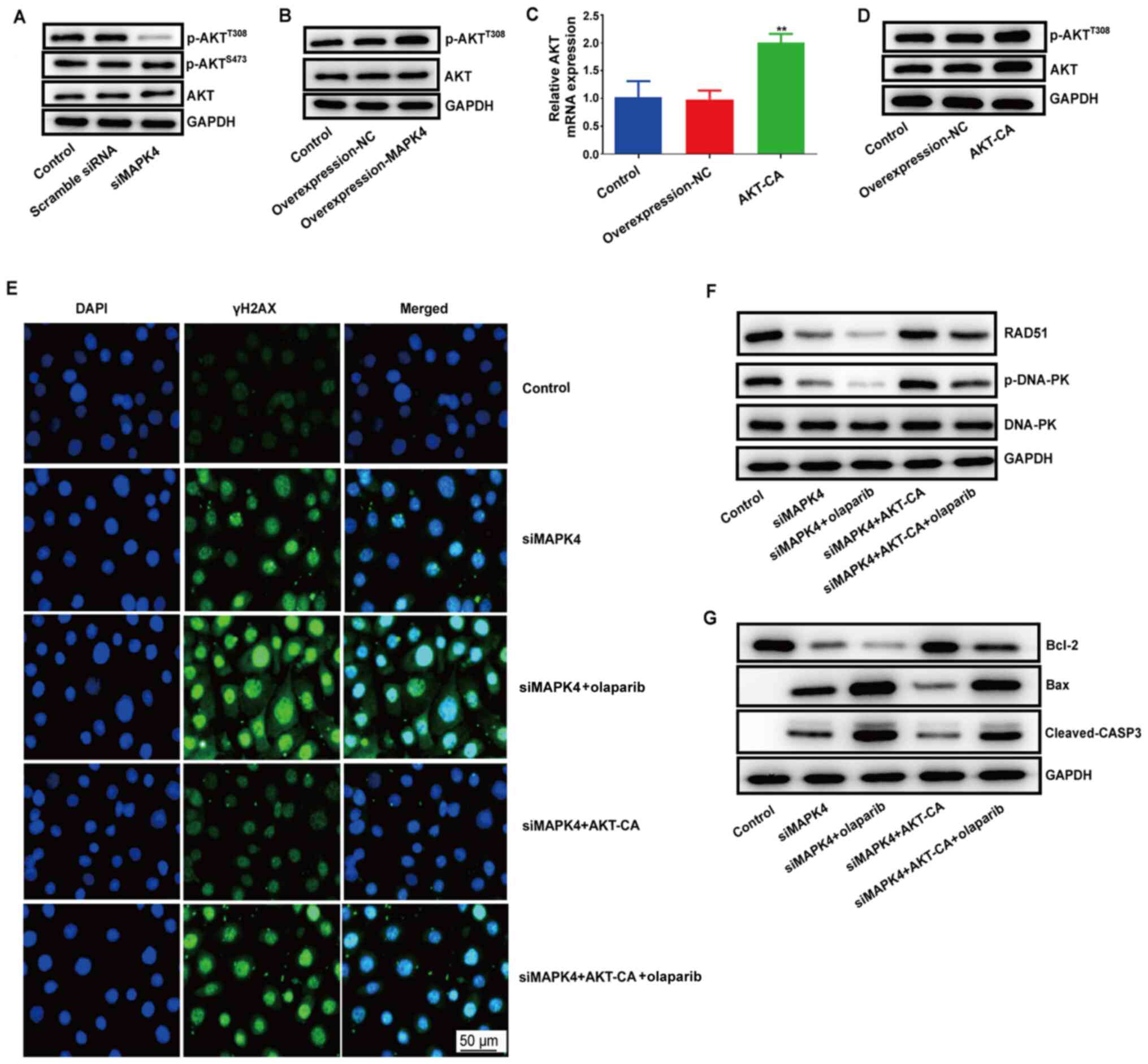 | Figure 4.MAPK4 exerts its DNA damage repair
function by regulating the AKT signaling pathway. (A) Western
blotting was performed to test the protein expression levels of
p-AKTT308 and p-AKTS473 in control cells,
cells transfected with scramble siRNA and cells transfected with
siMAPK4. (B) Protein expression level of p-AKTT308 in
MAPK4-overexpressing cells vs. control cells was measured via
western blotting. (C) mRNA expression level of AKT in AKT-CA cells
was tested via reverse transcription-quantitative PCR. **P<0.01.
(D) Western blotting was performed to examine the protein
expression levels of p-AKTT308 and total AKT. (E)
Immunofluorescence was used to test the protein expression level
and location of γH2AX in the control, siMAPK, siMAPK with olaparib
siMAPK, siMAPK + olaparib, siMAPK + AKT-CA and siMAPK + olaparib +
AKT-CA groups. (F) Protein expression levels of RAD51 and p-DNA-PK
were tested using western blotting in the control, siMAPK, siMAPK +
olaparib, siMAPK + AKT-CA and siMAPK + olaparib + AKT-CA groups.
(G) Protein expression levels of Bcl2 and Bax were tested via
western blotting. AKT-CA, constitutively active AKT; NC, negative
control; siMAPK4, siRNA-MAPK4; siRNA, small interfering RNA; γH2AX,
phosphorylated histone H2AX; casp, caspase; p-, phosphorylated;
DNA-PK, DNA-dependent protein kinase catalytic subunit. |
Subsequently, the phosphorylation of
AKTT308 was measured in MAPK4-overexpressing HCC1937
cells. The results indicated that MAPK4 overexpression increased
the expression level of p-AKTT308 (Fig. 4B). HCC1937 cells were also
transfected with the AKT-CA plasmid at the AKTT308 site
to upregulate AKT phosphorylation (Fig.
4C and D). It was also identified that siMAPK4 increased the
expression level of γH2AX (marker of a double-stranded break)
(Fig. 4E). Moreover, siMAPK4
decreased the expression level of RAD51 and the phosphorylation of
DNA-PK (Fig. 4F), which reflected
DNA damage repair. These effects were blocked by AKT-CA.
Collectively, these results indicated that MAPK4 exerted its DNA
damage repair function by regulating the AKT signaling pathway.
Moreover, olaparib aggravated the function of siMAPK4, but weakened
the function of AKT by lowering RAD51 and p-DNA-PK expression in
the siMAPK4 group and the combination of siMAPK4 + AKT-CA group
(Fig. 4F). These effects resulted
in failure of DNA damage repair. Furthermore, the cells underwent
apoptosis, as evident by the decrease in the expression level of
Bcl-2 and the increase in that of Bax and cleaved-casp-3 when
treated with olaparib and siMAPK4 (Fig.
4G).
Discussion
TNBC is intractable due to the absence of HER-2,
estrogen receptor and progesterone receptor expression, which
renders the currently available drugs ineffective (24). Previous studies have reported that
PARP1 inhibitors are highly effective against breast cancer type 1
and 2 susceptibility proteins (BRCA1/2) and MCF-7 and MDA-MB-231
cells (25–27). It has also been revealed that TNBC
has a lower expression or mutation rate of BRCA1/2 (28). Hence, the use of PARP1 inhibitors
for the treatment of TNBC is reasonable. In fact, PARP1 inhibitors,
such as olaparib, have demonstrated high effectiveness against
TNBC. However, the extensive use of PARP1 inhibitors can lead to
the occurrence of resistance (29),
partly due to the restoration of homologous recombination and
upregulation of the ataxia-telangiectasia-mutated-and-Rad3-related
kinase/checkpoint kinase 1 signaling pathway (30). Thus, it is important to identify
other drugs or genes that can overcome this resistance.
MAPK4 is an oncogene that is upregulated in numerous
types of cancer and contributes to poor prognosis. For instance, in
cervical cancer, high MAPK4 mRNA expression is associated with a
lower survival rate (31). The
present data indicated that MAPK4 was highly expressed in patients
with TNBC, suggesting that downregulating MAPK4 expression may
enhance the effect of PARP1 inhibitors on TNBC. To verify this, the
current study first determined the expression level of MAPK4 in
TNBC tissues and cell lines. The results identified a higher MAPK4
expression in TNBC tissues and cell lines (MDA-MB-468, HCC1937 and
MDA-MB-231) compared with that in normal tissues and cells
(MCF-10A). Furthermore, MAPK4 overexpression promoted the
proliferation of HCC1937 cells, while knockdown of MAPK4 inhibited
cell viability. Next, the effect of combination of siMAPK4 and
olaparib was determined using cell viability, wound healing and
TUNEL assays. It was observed that MAPK4 knockdown in combination
with olaparib treatment elicited a markedly greater inhibitory
effect on cell proliferation, migration and apoptosis compared with
either treatment alone.
Subsequently, the present study investigated the
mechanism underlying how siMAPK4 enhances the sensitivity of TNBC
cells to olaparib. Previous studies have reported that MAPK4 can
phosphorylate AKT at T308 and S473 without influencing total AKT
levels, thereby enhancing the efficacy of PARP1 inhibitors by
suppressing the repair of radiation-induced DNA damage in cervical
cancer cell lines (9,31). Nevertheless, the current data
indicated that MAPK4 could only activate AKTT308 in
HCC1937 cells. It was also found that the expression levels of
RAD51 and p-DNA-PK, which reflected the DNA damage repair, were
inhibited by the PARP1 inhibitor and siMAPK. Moreover, the high
γH2AX expression indicated that the PARP1 inhibitor and siMAPK
could increase the double-stranded breaks. The present results also
indicated that cells became apoptotic, due to the decreased
expression level of Bcl-2 and increased Bax and cleaved-casp-3
expression when cells were treated with olaparib and siMAPK in the
siMAPK4 + olaparib group.
Currently, there are several clinical trials
investigating the role of olaparib and BRCA1/2 mutations in breast
and ovarian cancer (32,33). The present study demonstrated that
siMAPK4 can enhance the sensitivity of TNBC cells to the PARP1
inhibitor by inhibiting AKTT308 phosphorylation and DNA
repair, thereby promoting cell apoptosis in TNBC with BRCA1/2
mutations.
While the present study revealed that siMAPK4 can
enhance the sensitivity of HCC1937 cells to a PARP1 inhibitor,
olaparib, and identified the underlying mechanism in vitro,
these results require further verification in animal models and
human trials. Additionally, although it is known that MAPK4 can
phosphorylate AKT at both T308 and S473 in bladder urothelial
carcinoma, low-grade glioma, lung adenocarcinoma, cervical cancer,
thyroid carcinoma and prostate cancer (9,30,34),
the present study found that only T308 was phosphorylated in TNBC
cells and this was not further investigated. siRNAs can have poor
stability and specificity, which may limit their usage in
vivo (35). To solve these
problems, in vivo experiments should be performed, and the
use of nanoparticles should be considered to increase siRNA
stability and ensure the effective delivery of these molecules to
the appropriate target cells.
In conclusion, the present study demonstrated that
MAPK4 was highly expressed in TNBC tissues and cell lines.
Furthermore, knockdown of MAPK4 inhibited the proliferation of
HCC1937 cells. By contrast, overexpression of MAPK4 increased the
viability of HCC1937 cells. It was identified that knockdown of
MAPK4 and treatment with the PARP1 inhibitor inhibited the
proliferation and migration of HCC1937 cells by inhibiting DNA
damage repair, increasing double-stranded breaks and promoting cell
apoptosis. Therefore, MAPK4 may be another target for the treatment
of TNBC in combination with PARP1 inhibitors.
Acknowledgements
Not applicable.
Funding
This work was funded by the President Foundation of
Nanfang Hospital, Southern Medical University, Guangzhou, China
(grant nos. 2016L007 and 2020C032).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XZ, SJ, CY and JD contributed to the study
conception and design. XZ, SR and ZG performed the experiments. JG
and ML contributed to the analysis and interpretation of data. XZ,
SR and SJ contributed to the drafting of the manuscript. JD and XZ
contributed to the project administration and writing, reviewing
and editing the manuscript. JD and XZ confirm the authenticity of
all the raw data. All authors contributed to the critical revision
of the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
TNBC
|
triple-negative breast cancer
|
|
PARP1
|
poly ADP-ribose polymerase-1
|
|
BRCA1/2
|
breast cancer type 1 and 2
susceptibility protein
|
|
TCGA
|
The Cancer Genome Atlas
|
|
AKT-CA
|
constitutively active AKT
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
casp-3
|
caspase-3
|
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
World Health Organization (WHO), . Cancer.
https://www.who.int/news-room/fact-sheets/detail/cancerMarch
3–2021
|
|
3
|
Gluz O, Liedtke C, Gottschalk N, Pusztai
L, Nitz U and Harbeck N: Triple-negative breast cancer-current
status and future directions. Ann Oncol. 20:1913–1927. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vagia E, Mahalingam D and Cristofanilli M:
The landscape of targeted therapies in TNBC. Cancers (Basel).
12:9162020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Coulombe P and Meloche S: Atypical
mitogen-activated protein kinases: Structure, regulation and
functions. Biochim Biophys Acta. 1773:1376–1387. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee BD, Park JM, Lee YM, Moon ES, Jeong
HJ, Chung YI and Rim HD: A pilot study for discovering candidate
genes of chromosome 18q21 in methamphetamine abusers: Case-control
association study. Clin Psychopharmacol Neurosci. 12:54–64. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cancer Genome Atlas Research Network, ;
Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA,
Ellrott K, Shmulevich I, Sander C and Stuart JM: The cancer genome
atlas pan-cancer analysis project. Nat Genet. 45:1113–1120. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang W, Shen T, Dong B, Creighton CJ, Meng
Y, Zhou W, Shi Q, Zhou H, Zhang Y, Moore DD and Yang F: MAPK4
overexpression promotes tumor progression via noncanonical
activation of AKT/mTOR signaling. J Clin Invest. 129:1015–1029.
2019. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Feng Y, Zhang L, Wu J, Khadka B, Fang Z,
Gu J, Tang B, Xiao R, Pan G and Liu J: CircRNA circ_0000190
inhibits the progression of multiple myeloma through modulating
miR-767-5p/MAPK4 pathway. J Exp Clin Cancer Res. 38:542019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He J, Huang Z, He M, Liao J, Zhang Q, Wang
S, Xie L, Ouyang L, Koeffler HP, Yin D and Liu A: Circular RNA
MAPK4 (circ-MAPK4) inhibits cell apoptosis via MAPK signaling
pathway by sponging miR-125a-3p in gliomas. Mol Cancer. 19:172020.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Piscitello D, Varshney D, Lilla S, Vizioli
MG, Reid C, Gorbunova V, Seluanov A, Gillespie DA and Adams PD: AKT
overactivation can suppress DNA repair via p70S6 kinase-dependent
downregulation of MRE11. Oncogene. 37:427–438. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Y, Xie C, Li A, Vizioli MG, Reid C,
Gorbunova V, Seluanov A, Gillespie DA and Adams PD: PKI-587
enhances chemosensitivity of oxaliplatin in hepatocellular
carcinoma through suppressing DNA damage repair pathway (NHEJ and
HR) and PI3K/AKT/mTOR pathway. Am J Transl Res. 11:5134–5149.
2019.PubMed/NCBI
|
|
14
|
Ali R, Rakha EA, Madhusudan S and Bryant
HE: DNA damage repair in breast cancer and its therapeutic
implications. Pathology. 49:156–165. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gallmeier E and Kern SE: Absence of
specific cell killing of the BRCA2-deficient human cancer cell line
CAPAN1 by poly(ADP-ribose) polymerase inhibition. Cancer Biol Ther.
4:703–706. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim G, Ison G, McKe AE, Zhang H, Tang S,
Gwise T, Sridhara R, Lee E, Tzou A, Philip R, et al: FDA approval
summary: Olaparib monotherapy in patients with deleterious germline
BRCA-mutated advanced ovarian cancer treated with three or more
lines of chemotherapy. Clin Cancer Res. 21:4257–4261. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Balasubramaniam S, Beaver JA, Horton S,
Fernandes LL, Tang S, Horne HN, Liu J, Liu C, Schrieber SJ, Yu J,
et al: FDA approval summary: Rucaparib for the treatment of
patients with deleterious BRCA mutation-associated advanced ovarian
cancer. Clin Cancer Res. 23:7165–7170. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Noordermeer SM and van Attikum H: PARP
inhibitor resistance: A tug-of-war in BRCA-mutated cells. Trends
Cell Biol. 29:820–834. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin H, Li N, He H, Ying Y, Sunkara S, Luo
L, Lv N, Huang D and Luo Z: AMPK inhibits the stimulatory effects
of TGF-β on Smad2/3 activity, cell migration, and
epithelial-to-mesenchymal transition. Mol Pharmacol. 88:1062–1071.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang S, Wang Y, Luo L, Shi F, Zou J, Lin
H, Ying Y, Luo Y, Zhan Z, Liu P, et al: AMP-activated protein
kinase regulates cancer cell growth and metabolism via nuclear and
mitochondria events. J Cell Mol Med. 23:3951–3961. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kwon KY, Jan JH, Kwon SY, Cho CH, Oh HK
and Kim SP: Cadmium induced acute lung injury and TUNEL expression
of apoptosis in respiratory cells. J Korean Med Sci. 18:655–662.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lyons TG: Targeted therapies for
triple-negative breast cancer. Curr Treat Options Oncol. 20:822019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bryant HE, Schultz N, Thomas HD, Parker
KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ and Helleday T:
Specific killing of BRCA2-deficient tumours with inhibitors of
poly(ADP-ribose) polymerase. Nature. 434:913–917. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dziadkowiec KN, Gąsiorowska E,
Nowak-Markwitz E and Jankowska A: PARP inhibitors: Review of
mechanisms of action and BRCA1/2 mutation targeting. Prz
Menopauzalny. 15:215–219. 2016.PubMed/NCBI
|
|
27
|
Lee JM, Ledermann JA and Kohn EC: PARP
inhibitors for BRCA1/2 mutation-associated and BRCA-like
malignancies. Ann Oncol. 25:32–40. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ryu JM, Choi HJ, Kim I, Nam SJ, Kim SW, Yu
J, Lee SK, Choi DH, Park YH, Kim JW, et al: Prevalence and
oncologic outcomes of BRCA 1/2 mutations in unselected
triple-negative breast cancer patients in Korea. Breast Cancer Res
Treat. 173:385–395. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
D'Andrea AD: Mechanisms of PARP inhibitor
sensitivity and resistance. DNA Repair (Amst). 71:172–176. 2018.
View Article : Google Scholar
|
|
30
|
Mak JPY, Ma HT and Poon RYC: Synergism
between ATM and PARP1 inhibition involves DNA damage and abrogating
the G2 DNA damage checkpoint. Mol Cancer Ther. 19:123–134. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tian S, Lou L, Tian M, Lu G, Tian J and
Chen X: MAPK4 deletion enhances radiation effects and triggers
synergistic lethality with simultaneous PARP1 inhibition in
cervical cancer. J Exp Clin Cancer Res. 39:1432020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tutt A, Robson M, Garber JE, Domchek SM,
Audeh MW, Weitzel JN, Friedlander M, Arun B, Loman N, Schmutzler
RK, et al: Oral poly(ADP-ribose) polymerase inhibitor olaparib in
patients with BRCA1 or BRCA2 mutations and advanced breast cancer:
A proof-of-concept trial. Lancet. 376:235–244. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Audeh MW, Carmichael J, Penson RT,
Friedlander M, Powell B, Bell-McGuinn KM, Scott C, Weitzel JN,
Oaknin A, Loman N, et al: Oral poly(ADP-ribose) polymerase
inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and
recurrent ovarian cancer: A proof-of-concept trial. Lancet.
376:245–251. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shen T, Wang W, Zhou W, Coleman I, Cai Q,
Dong B, Ittmann MM, Creighton CJ, Bian Y, Meng Y, et al: MAPK4
promotes prostate cancer by concerted activation of androgen
receptor and AKT. J Clin Invest. 131:e1354652021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cho YS, Lee GY, Sajja HK, Qian W, Cao Z,
He W, Karna P, Chen X, Mao H, Wang YA and Yang L: Targeted delivery
of siRNA-generating DNA nanocassettes using multifunctional
nanoparticles. Small. 9:1964–1973. 2013. View Article : Google Scholar : PubMed/NCBI
|