Introduction
Nasopharyngeal carcinoma (NPC) is a malignant and
aggressive tumor originating in the nasopharyngeal epithelium,
which is characterized by a high incidence rate in Southern China
(1–3). Despite the great advances achieved in
the treatment of NPC, patients with NPC generally have a poor
prognosis (4). Previous studies
indicated that Epstein-Barr virus infection is a major pathogenic
factor for NPC (5). With the
development of second-generation sequencing, a large number of
genes or non-protein-coding transcripts, such as long non-coding
RNAs (lncRNAs) and microRNAs (miRNAs/miRs) have been identified in
NPC, and they may serve as biomarkers for the diagnosis, treatment
and prognosis of NPC (6–8).
lncRNAs are a family of non-coding RNAs that are
>200 nucleotides (nt) in length and lack protein-coding ability
(9). Accumulating evidence has
demonstrated the regulatory functions of lncRNAs in multiple types
of cancer, including NPC (10). For
example, the depletion of ZFAS1 suppressed the development of NPC
by regulating miR-135a (11). The
knockdown of SOX2 overlapping transcript suppressed the viability,
migration and invasion of NPC via the miR-146b-5p/heterogeneous
nuclear ribonucleoproteins A2/B1 axis (12). Small nucleolar RNA host gene 5
accelerated NPC progression by adsorbing miR-1179 (13). DLX6-AS1 upregulated the level of
hypoxia-inducible factor-1α and facilitated the progression of NPC
via sponging miR-199a-5p (14).
FBXL19-AS1 has been identified as an oncogene in various human
cancers, such as colorectal (15),
lung (16) and breast (17) cancers. However, the function of
FBXL19-AS1 in NPC remains unknown.
miRNAs are small non-coding RNAs that are ~22 nt in
length (18,19). Numerous studies have revealed that
the aberrant expression of miRNAs is implicated in the progression
of various tumors, including NPC (20,21).
For example, miR-449b-5p inhibited cell viability, migration and
invasion by regulating tumor protein D52 in NPC (22). miR-432 repressed the migration and
invasion of NPC cells by modulating transcription factor E2F3
expression (23). miR-100
suppressed cell viability and growth by modulating homeobox protein
Hox-A1 expression (24). miR-431
has been reported to inhibit the development and progression of
various cancers, such as melanoma (25), hepatocellular carcinoma (26) and papillary thyroid carcinoma
(27). However, the involvement of
miR-431 in the development of NPC remains unclear.
The present study was undertaken to investigate
whether FBXL19-AS1 contributes to the progression of NPC via the
miR-431/prostate and breast cancer overexpressed 1 (PBOV1) axis, in
the hope that the findings may uncover novel targets for NPC
treatment.
Materials and methods
Clinical specimens
A total of 30 NPC and 30 adjacent normal tissue
samples (distance from tumor margin, 2 cm) were obtained from
patients with NPC (age range, 33–67 years) at Zhangjiagang TCM
Hospital Affiliated to Nanjing University of Chinese Medicine
(Zhangjiagang, China) between March 2016 and September 2019. The
inclusion criteria were as follows: Patients were diagnosed as NPC
with no history of other tumor. The exclusion criteria were as
follows: i) diagnosed with other diseases; and ii) patients who had
received preoperative radiotherapy, chemotherapy or other adjuvant
treatments prior to admission. All fresh specimens were immediately
frozen in liquid nitrogen and stored at −80°C until RNA extraction.
Written informed consent was obtained from each participant. The
protocol of the present study was approved by the ethics committee
of Zhangjiagang Hospital of Traditional Chinese Medicine. The
clinical information of patients with NPC is presented in Table I.
 | Table I.Clinicopathological characteristics
of patients with nasopharyngeal carcinoma. |
Table I.
Clinicopathological characteristics
of patients with nasopharyngeal carcinoma.
| Clinicopathological
characteristics | Number of patients
(n=30) |
|---|
| Age, years |
|
|
<50 | 14 |
|
≥50 | 16 |
| Sex |
|
|
Male | 15 |
|
Female | 15 |
| Tumor size, cm |
|
|
<5 | 12 |
| ≥5 | 18 |
| Lymph node
status |
|
| N0 | 11 |
|
N1-3 | 19 |
| Distant
metastasis |
|
| No | 10 |
|
Yes | 20 |
| TNM stage |
|
|
I–II | 6 |
|
III–IV | 24 |
Cell culture
Human NPC cell lines (C666-1, SUNE1, 5–8F and 6–10B)
and nasopharyngeal epithelial cells (NP69) were obtained from
Shanghai Institutes for Biological Sciences, Chinese Academy of
Sciences. All cells were cultured in RPMI-1640 medium (Thermo
Fisher Scientific, Inc.) supplemented with 10% FBS (Invitrogen;
Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2.
Cell transfection
Short hairpin RNA (shRNA) targeting FBXL19-AS1
(sh-FBXL19-AS1; 5′-GCAGGCCCAUCUACGGUGUGU-3′) and control (sh-NC;
5′-AAUUAGCCGCAUGCGUCACAU-3′), miR-431 mimics
(5′-UGCAAUGUUAGAUGGUGUGAGG-3′), negative control (NC mimics;
5′-GACUCCUUACUCGCUCUACUG-3′), miR-431 inhibitor
(5′-UUCACCGGAUCUGUCACGUAU-3′) and control (NC inhibitor;
5′-CAGUAGAGAUUGAAAGUUGUC-3′) were obtained from Shanghai GenePharma
Co., Ltd. To establish FBXL19-AS1 or PBOV1-overexpression plasmids
(pcDNA3.1-FBXL19-AS1 or pcDNA3.1-PBOV1), the full-length FBXL19-AS1
or PBOV1 sequence was inserted into pcDNA3.1 vector (Thermo Fisher
Scientific, Inc.), with pcDNA3.1 as a control.
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used for transfection of C666-1 and SUNE1
cells (1×105) with sh-FBXL19-AS1 (10 nM), sh-NC (10 nM),
miR-431 mimics (10 nM), NC mimics (10 nM), miR-431 inhibitor (10
nM), NC inhibitor (10 nM) pcDNA3.1-FBXL19-AS1 (10 nM),
pcDNA3.1-PBOV1 (10 nM) or pcDNA3.1 (10 nM) at room temperature for
~30 min. At 48 h post-transfection, subsequent experiments were
performed.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Total RNA was extracted from NPC tissues and cells
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). Then, cDNA was established from total RNA using
the PrimeScript™ 1st strand cDNA Synthesis Kit (cat. no. 6110A;
Takara Biotechnology Co., Ltd.) according to the manufacturer's
protocol. qPCR was performed using SYBR Green Taq ReadyMix
(Sigma-Aldrich; Merck KGaA) on an Applied Biosystems 7500 Real-Time
PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.).
The following thermocycling conditions were used for qPCR: Initial
denaturation at 95°C for 15 sec; 40 cycles of denaturation at 94°C
for 30 sec, annealing at 60°C for 20 sec and extension at 72°C for
40 sec. The expression of genes was detected using the
2−ΔΔCq method (28).
GAPDH or U6 were used as the internal controls for FBXL19-AS1 and
PBOV1 or miR-431, respectively. The primer sequences were as
follows: FBXL19-AS1 forward, 5′-GGTACAACTACGGATATGA-3′ and reverse,
5′-TACGTCTCGACCATTACGCA-3′; miR-431 forward,
5′-TGTCTTGCAGGCCGTCATG-3′ and reverse,
5′-GCTGTCAACGATACGCTACCTA-3′; PBOV1 forward,
5′-TGAGTCCCCTCTCGGTAATG-3′ and reverse, 5′-GCCCCGAGTTAAGAACATCA-3′;
GAPDH forward, 5′-ACCACAGTCCATGCCATCAC-3′ and reverse,
5′-TCCACCACCCTGTTGCTGTA-3′; and U6 forward,
5′-ATTGGAACGATACAGAGAAGATT-3′ and reverse,
5′-GGAACGCTTCACGAATTTG-3′.
Cell Counting Kit-8 (CCK-8) assay
Cell viability was examined using a CCK-8 assay
(Dojindo Molecular Technologies, Inc.). The transfected C666-1 and
SUNE1 cells (1×105 cells/well) were plated into 96-well
plates for 48 h. Then, CCK-8 reagent was added to each well at 0,
24, 48 and 72 h and incubated at 37°C for 4 h. The absorbance at
450 nm was detected by a microplate reader (Olympus
Corporation).
Transwell assay
The Transwell assay was performed using Transwell
chambers (8.0-µm pore size; EMD Millipore). For the migration
assay, transfected C666-1 and SUNE1 cells (1×105) in
serum-free culture RPMI-1640 medium (Thermo Fisher Scientific,
Inc.) were added to the upper chamber. Subsequently, RPMI-1640
medium with 10% FBS was added to the lower chamber. After 24 h, 4%
paraformaldehyde and 0.1% crystal violet solution was used to fix
and stain the cells, respectively, that had migrated to the lower
surface of the membrane for 20 min at room temperature, and the
cells were counted under a light microscope (magnification, ×200).
For cell invasion, the upper chamber was pre-coated with Matrigel
for 1 h at room temperature. The remaining steps were consistent
with those for the cell migration assay.
Dual-luciferase reporter assay
Starbase (version 2.0; starbase.sysu.edu.cn/) was used to predict the binding
sites between FBXL19-AS1 and miR-431. TargetScan (www.targetscan.org/vert_72/) was used to predict
the binding sites between miR-431 and PBOV1. The mutated 3′-UTR was
generated using the QuikChange II Site Directed Mutagenesis kit
(Agilent Technologies, Inc.). The 3′-untranslated region (UTR) of
wild-type (WT) and mutant (Mut) FBXL19-AS1 (WT-FBXL19-AS1 and
Mut-FBXL19-AS1) or PBOV1 (WT-PBOV1 and Mut-PBOV1) were inserted
into the pmiR-GLO vector (Promega Corporation). Then, all plasmids
(0.6 µg) were transfected into C666-1 and SUNE1 cells
(1×105) with 10 nM miR-431 mimics or 10 nM NC mimics
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.). After 48 h, the luciferase activities were detected using
the Dual-Luciferase Reporter System (Promega Corporation). Firefly
luciferase activity was normalized to Renilla luciferase
gene activity.
RNA-binding protein
immunoprecipitation (RIP) assay
A RIP assay was utilized with Magna RIP RNA-Binding
Protein Immunoprecipitation Kit (EMD Millipore). The transfected
C666-1 and SUNE1 cells were dissolved in RIP lysis buffer (Beyotime
Institute of Biotechnology), and then cell lysate was centrifuged
at 40,000 × g at 4°C for 10 min and incubated with 50 µl magnetic
beads bound with the 5 µg Ago2 antibody (cat. no. 2897; Cell
Signaling Technology, Inc.). IgG (5 µg; cat. no. PP64B; EMD
Millipore) was used as a control group. Then, immunoprecipitated
RNA was detected via an RT-qPCR assay.
Statistical analysis
All experiments were repeated three times. All data
were analyzed using GraphPad Prism 7 (GraphPad Software, Inc.) and
presented as the mean ± SD. Paired/unpaired Student's t-test or
one-way ANOVA, followed by Tukey's post hoc test were used to
evaluate the differences. The correlation between FBXL19-AS1 and
PBOV1 expression was assessed by a Pearson's correlation analysis.
P<0.05 was considered to indicate a statistically significant
difference.
Results
FBXL19-AS1 is significantly
upregulated, whereas miR-431 is downregulated in NPC tissues and
cells
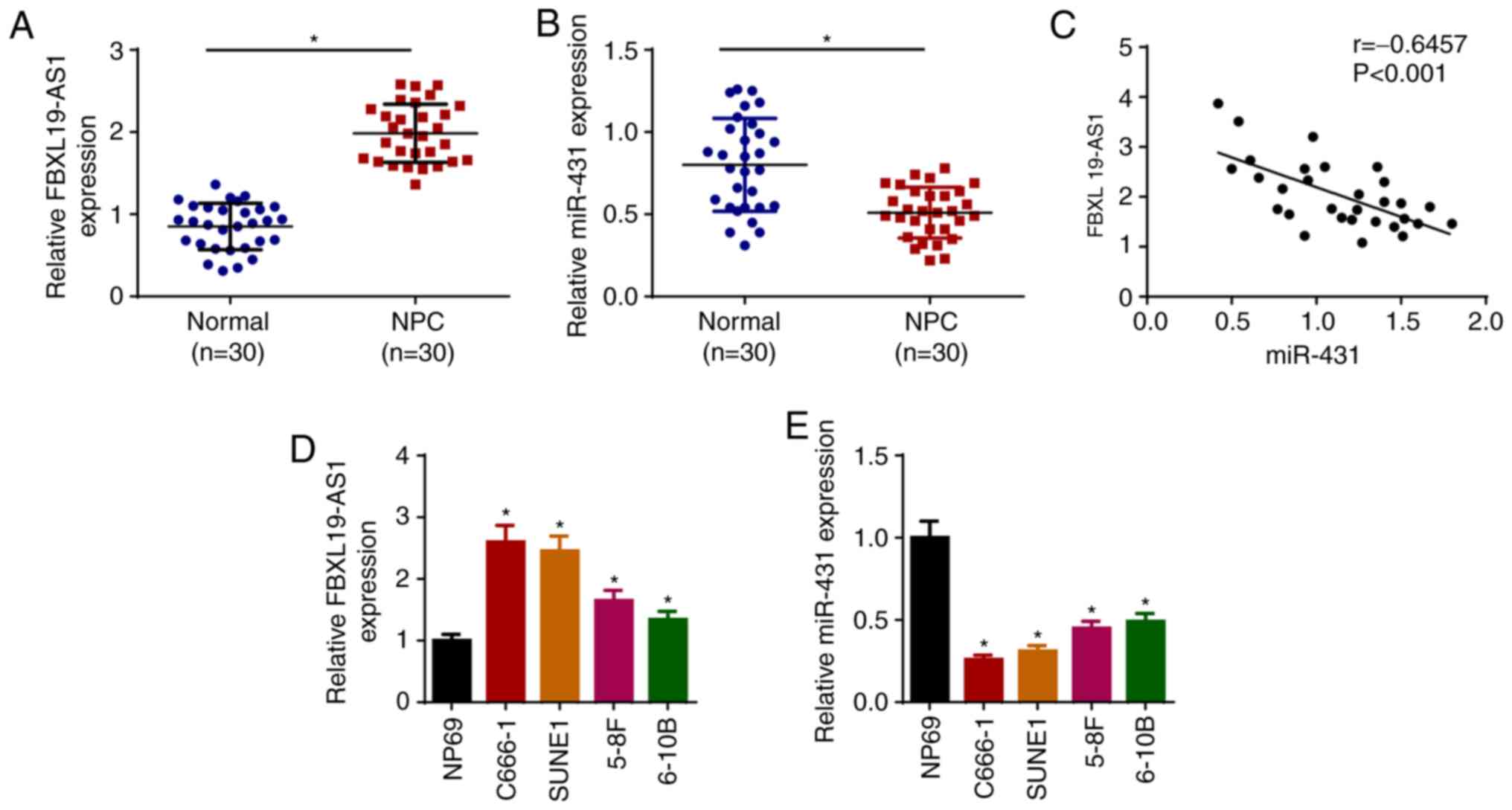
To explore the function of FBXL19-AS1 and miR-431
(derived from 5′ of pre-miR-431) in NPC, RT-qPCR was performed to
detect the levels of FBXL19-AS1 and miR-431 in NPC tissues. The
data indicated that FBXL19-AS1 was increased and miR-431 was
decreased in NPC tissues (n=30; Fig. 1A
and B), and that FBXL19-AS1 expression was inversely correlated
with miR-431 expression (Fig. 1C).
Moreover, the levels of FBXL19-AS1 and miR-431 in NPC cell lines
(C666-1, SUNE1, 5–8F and 6–10B) and NP69 cells were detected.
Consistent with previous results, FBXL19-AS1 expression was
upregulated in NPC cells, whereas miR-431 expression was
downregulated, particularly in C666-1 and SUNE1 cell lines
(Fig. 1D and E). These data
suggested that FBXL19-AS1 may act as an oncogene and miR-431 as a
tumor suppressor in the progression of NPC. Since C666-1 and SUNE1
exhibited the highest and lowest expression of FBXL19-AS1 and
miR-431, respectively, these two cell lines were selected for
subsequent experiments.
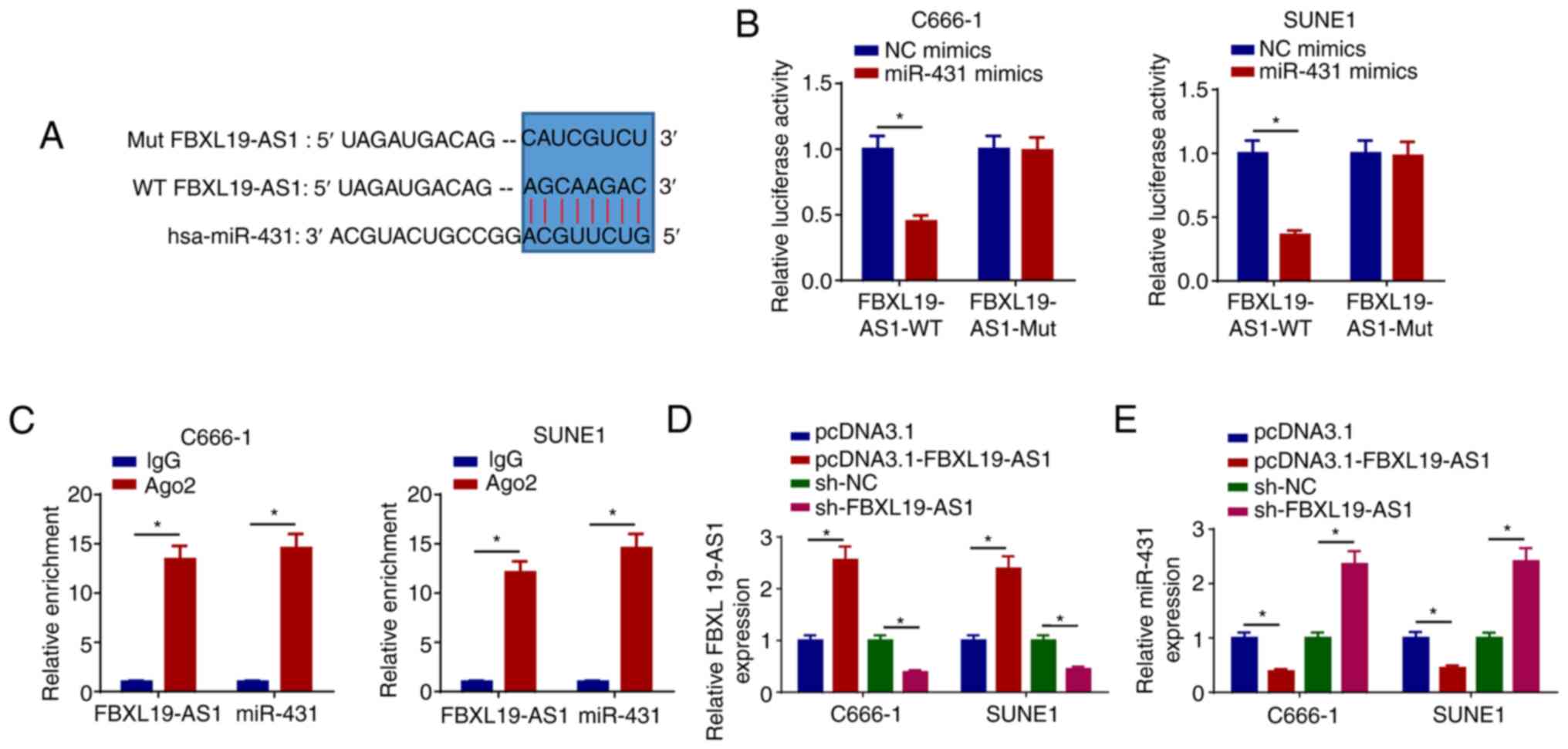
FBXL19-AS1 interacts with miR-431
A growing body of evidence has demonstrated that
lncRNAs can interact with miRNAs to modulate cancer development
(29,30). In the present study, it was observed
that FBXL19-AS1 acted as a molecular sponge for miR-431 (Fig. 2A). Moreover, the luciferase activity
of FBXL19-AS1-WT was reduced by miR-431 mimics in C666-1 and SUNE1
cells, whereas the activity of FBXL19-AS1-Mut was not changed
(Fig. 2B). Furthermore, the RIP
assay revealed that FBXL19-AS1 and miR-431 were markedly enriched
in the Ago2 group compared with the IgG group (Fig. 2C). To confirm whether FBXL19-AS1
could modulate the level of miR-431, pcDNA3.1-FBXL19-AS1 or
sh-FBXL19-AS1 was transfected into C666-1 and SUNE1 cells to
overexpress or suppress FBXL19-AS1, respectively (Fig. 2D), following which the expression of
miR-431 was determined. The results indicated that overexpression
of FBXL19-AS1 suppressed miR-431 expression and depletion of
FBXL19-AS1 enhanced miR-431 expression (Fig. 2E). These results suggested that
FBXL19-AS1 interacts with and negatively regulates the level of
miR-431.
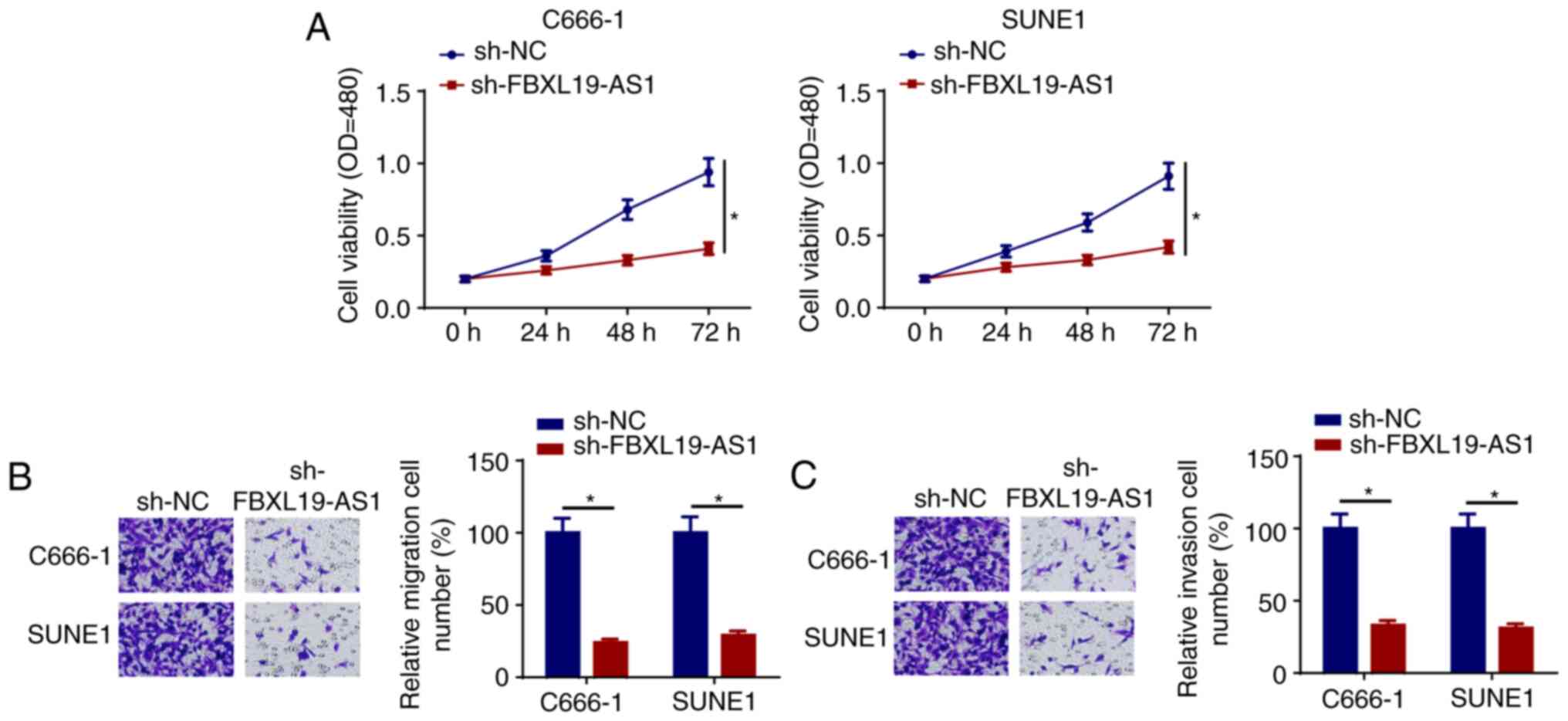
FBXL19-AS1 depletion suppresses the
progression of NPC
To further explore the function of FBXL19-AS1 in the
development of NPC, C666-1 and SUNE1 cells were transfected with
sh-FBXL19-AS1 or sh-NC. The CCK-8 assay indicated that the
interference of FBXL19-AS1 led to the inhibition of C666-1 and
SUNE1 cell viability (Fig. 3A).
Moreover, FBXL19-AS1 depletion markedly suppressed the migration
and invasion of NPC cells (Fig. 3B and
C). Thus, FBXL19-AS1 knockdown may inhibit the occurrence of
NPC.
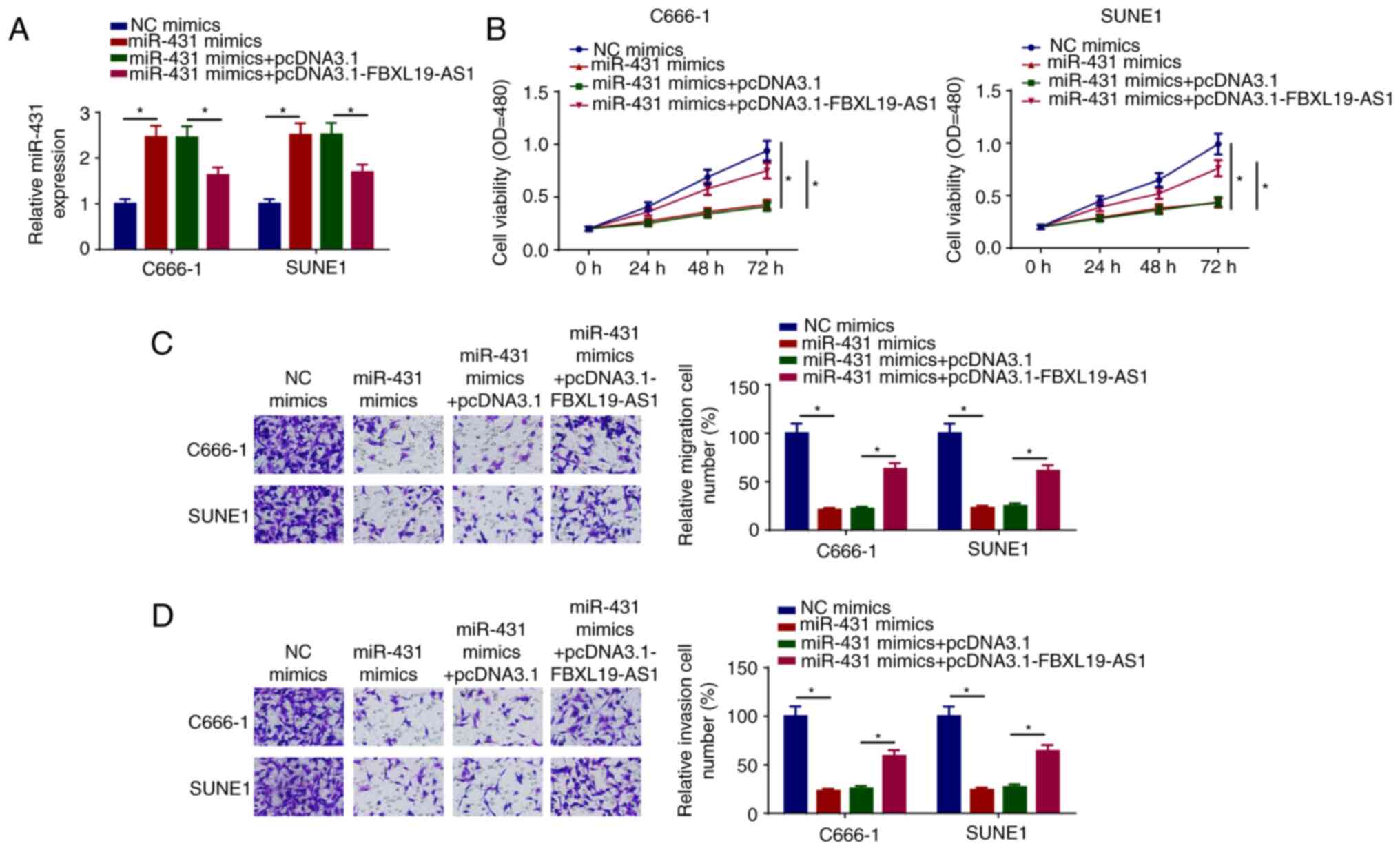
FBXL19-AS1 overexpression counteracts
the effects of miR-431 on NPC cells
To investigate whether pcDNA3.1-FBXL19-AS1 can
abolish the inhibition of NPC progression mediated by miR-431
mimics, the expression of miR-431 was assessed in C666-1 and SUNE1
cells transfected with NC mimics, miR-431 mimics, miR-431 mimics +
pcDNA3.1, and miR-431 mimics + pcDNA3.1-FBXL19-AS1. The results
indicated that the addition of FBXL19-AS1 diminished miR-431
expression (Fig. 4A). Further
studies revealed that the overexpression of miR-431 inhibited the
viability, migration and invasion of C666-1 and SUNE1 cells, which
was partially reversed by pcDNA3.1-FBXL19-AS1 transfection
(Fig. 4B-D). These results
suggested that the FBXL19-AS1/miR-431 axis may be implicated in NPC
progression.
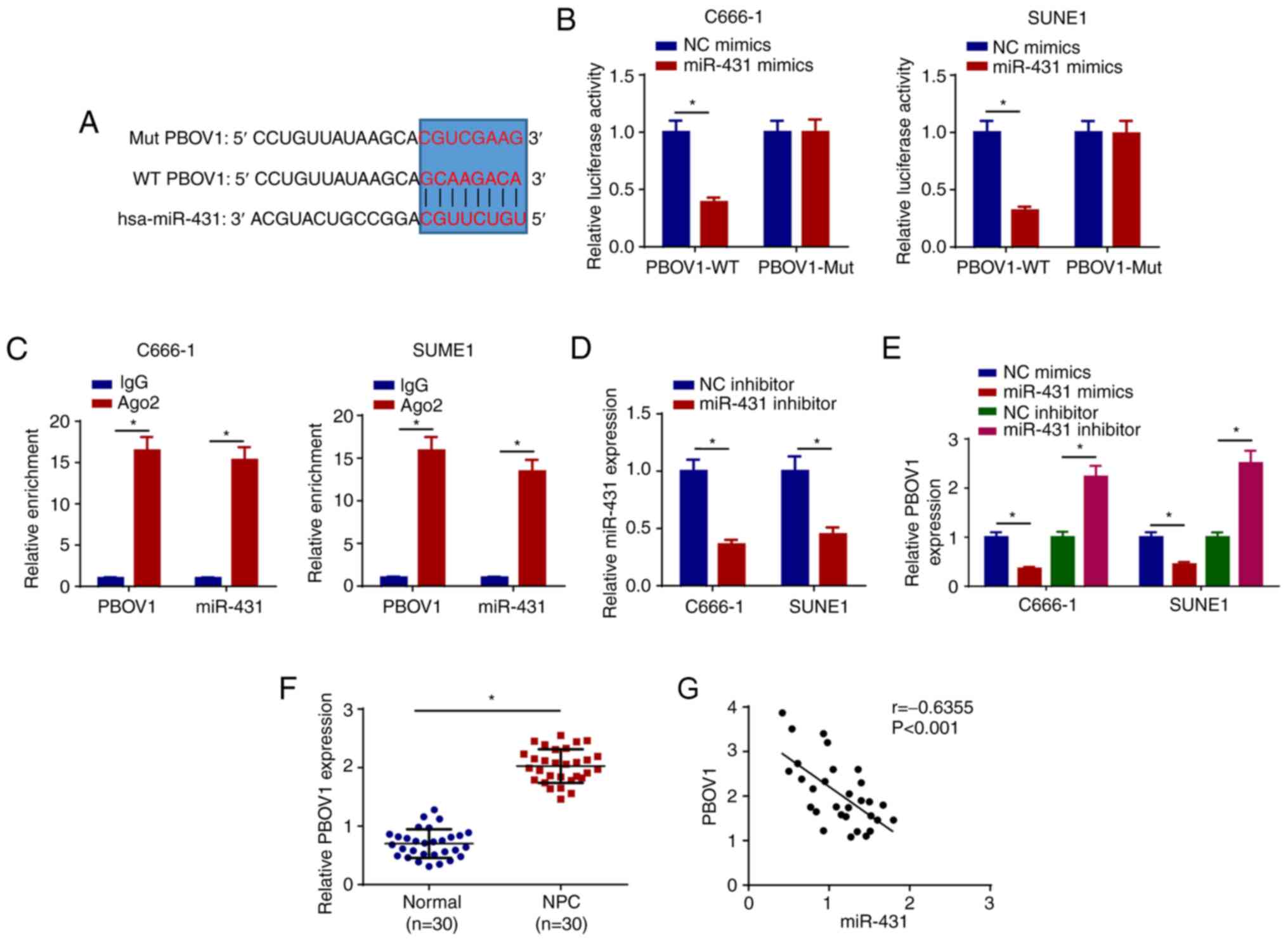
PBOV1 is directly targeted by
miR-431
A putative miR-431 binding site located in the
3′-UTR of PBOV1 mRNA was predicted by TargetScan (Fig. 5A). As shown in Fig. 5B, miR-431 mimics led to a
significant reduction in relative luciferase activity of PBOV1-WT,
but not in PBOV1-Mut. miR-431 and PBOV1 were markedly enriched by
Ago2, while IgG enrichment was not obvious (Fig. 5C). Next, RT-qPCR results showed that
miR-431 expression was reduced by transfecting miR-431 inhibitor in
NPC cells (Fig. 5D). Moreover, the
overexpression of miR-431 inhibited PBOV1 expression and the
knockdown of miR-431 enhanced PBOV1 expression (Fig. 5E). Furthermore, PBOV1 was markedly
enhanced in NPC tissues compared with normal tissues (Fig. 5F). In addition, PBOV1 expression was
negatively correlated with miR-431 expression (Fig. 5G). These results suggested that
PBOV1 is a direct target of miR-431.
FBXL19-AS1 regulates PBOV1 by sponging
miR-431 in NPC cells
To further study the association between PBOV1 and
miR-431 in NPC, the expression of PBOV1 was detected in C666-1
cells transfected with NC mimics, miR-431 mimics, miR-431 mimics +
pcDNA3.1 and miR-431 mimics + pcDNA3.1-PBOV1. The transfection
efficiency of PBOV1-overexpression plasmid was confirmed by
observing the upregulation of PBOV1 in C666-1 cells (Fig. 6A). pcDNA3.1-PBOV1 partially reversed
the suppressive effects of miR-431 mimics on PBOV1 expression
(Fig. 6B). Moreover, the
overexpression of miR-431 suppressed the viability, migration and
invasion of C666-1 cells, which was counteracted following
pcDNA3.1-PBOV1 transfection (Fig.
6C-E). FBXL19-AS1 expression was positively correlated with
PBOV1 expression (Fig. 6F).
Furthermore, C666-1 cells were treated with sh-NC, sh-FBXL19-AS1,
sh-FBXL19-AS1 + NC inhibitor and sh-FBXL19-AS1 + miR-431 inhibitor,
and the expression of PBOV1 was detected in transfected cells. The
results demonstrated that FBXL19-AS1 knockdown decreased the
expression of PBOV1, whereas these effects could be abolished by
miR-431 inhibitor transfection in NPC cells (Fig. 6G). Taken together, the
aforementioned results indicated that FBXL19-AS1 may drive the
progression of NPC via modulation of the miR-431/PBOV1 axis.
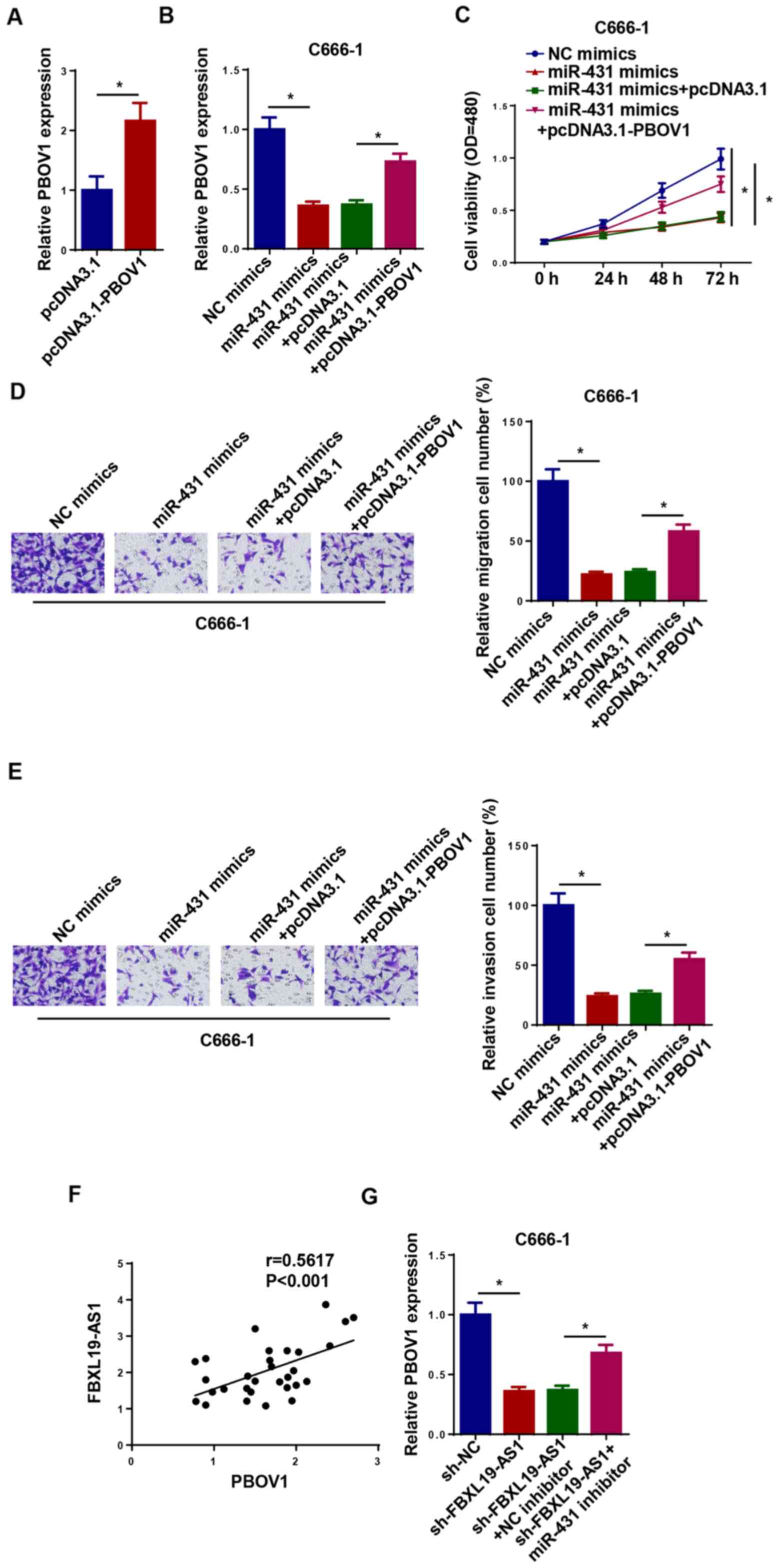 | Figure 6.FBXL19-AS1 regulates PBOV1 by
sponging miR-431 in nasopharyngeal carcinoma cells. (A) The
expression of PBOV1 in C666-1 cells transfected with pcDNA3.1 and
pcDNA3.1-PBOV1 was detected via RT-qPCR. (B) The expression of
PBOV1 in C666-1 cells transfected with NC mimics, miR-431 mimics,
miR-431 mimics + pcDNA3.1 and miR-431 mimics + pcDNA3.1-PBOV1 was
detected via RT-qPCR. (C) Cell Counting Kit-8 assay was used to
detect the viability of C666-1 cells transfected with NC mimics,
miR-431 mimics, miR-431 mimics + pcDNA3.1 and miR-431 mimics +
pcDNA3.1-PBOV1. Transwell assay was performed to measure the (D)
migration and (E) invasion of C666-1 cells transfected with NC
mimics, miR-431 mimics, miR-431 mimics + pcDNA3.1 and miR-431
mimics + pcDNA3.1-PBOV1. (F) The correlation between PBOV1 and
FBXL19-AS1 expression was analyzed using Pearson's correlation
analysis. (G) The expression of PBOV1 in C666-1 cells transfected
with sh-NC, sh-FBXL19-AS1, sh-FBXL19-AS1 + NC inhibitor and
sh-FBXL19-AS1 + miR-431 inhibitor was detected via RT-qPCR.
*P<0.05. PBOV1, prostate and breast cancer overexpressed 1; miR,
microRNA; NC, negative control; RT-qPCR, reverse
transcription-quantitative PCR; sh, short hairpin RNA. |
Discussion
Previous studies have revealed that aberrant
expression levels of lncRNAs are involved in the progression of
various malignancies, including NPC (31,32).
The present study demonstrated that FBXL19-AS1 interference may
suppress the progression of NPC via sponging miR-431 and inhibiting
PBOV1. To the best of our knowledge, the present study was the
first to demonstrate that the FBXL19-AS1/miR-431/PBOV1 axis may be
implicated in the regulation of NPC.
FBXL19-AS1 has been identified as an oncogene in
various cancers. For example, FBXL19-AS1 accelerated cell viability
and suppressed cell apoptosis by regulating miR-876-5p in breast
cancer (33). FBXL19-AS1
accelerated the progression of lung adenocarcinoma by inhibiting
miR-203a (34). In the present
study, FBXL19-AS1 was found to be upregulated in NPC tissues and
cells. Functionally, the depletion of FBXL19-AS1 inhibited the
viability, migration and invasion of NPC cells. These results
indicated that FBXL19-AS1 may function as an oncogene in NPC.
lncRNAs may act as competing endogenous RNAs
(ceRNAs) by specifically adsorbing miRNAs, and then regulating the
levels of target genes (35,36).
Multiple lncRNAs, such as PVT1 (37), ZNF667-AS1 (38) and MSC-AS1 (39), have been reported to serve as ceRNAs
in the development of NPC. The present study confirmed that miR-431
contained predicted binding sites for FBXL19-AS1. It was previously
demonstrated that miR-431 plays a key role in human malignancies.
For example, miR-431 was found to suppress breast cancer
progression via regulating fibroblast growth factor 9 (40). In addition, the upregulation of
miR-431 markedly suppressed colon cancer development via targeting
autophagy related 3 (41).
Furthermore, miR-431 repressed lung cancer progression via
regulating RAF proto-oncogene serine/threonine-protein kinase
(16). In the present study, the
expression of miR-431 was decreased in NPC tissues and cells, and
it was a downstream target of FBXL19-AS1. The overexpression of
FBXL19-AS1 inhibited miR-431 expression and interference of
FBXL19-AS1 enhanced miR-431 expression. The transfection of miR-431
mimics suppressed the progression of NPC, which was counteracted
following pcDNA3.1-FBXL19-AS1 transfection. The aforementioned data
demonstrated that FBXL19-AS1 promoted the progression of NPC by
regulating the expression of miR-431.
PBOV1 is a human protein-coding gene with a 2,501-bp
single-exon mRNA (42). PBOV1 has
been reported to be upregulated in several cancers. For example,
the overexpression of PBOV1 accelerated the tumorigenicity of
prostate cancer cells (43). PBOV1
facilitated the viability and metastasis of hepatocellular
carcinoma cells in vitro (44). Moreover, PBOV1 may be valuable as a
biomarker for the treatment of prostate cancer (45). In the present study, PBOV1 was
directly targeted by miR-431 and PBOV1 expression was found to be
negatively associated with miR-431 expression. Overexpression of
miR-431 inhibited the viability, migration and invasion of NPC
cells, whereas these effects could be abolished by upregulating
PBOV1 in NPC. Furthermore, the suppressive effects of FBXL19-AS1
silencing on PBOV1 expression were partly neutralized by miR-431
inhibitor transfection in NPC cells.
However, there were several limitations to the
present study. Firstly, there may be other downstream targets of
miR-143, which may also act as crucial regulators in the
pathogenesis of NPC. Secondly, increasing the patient sample size
may further verify the conclusion that FBXL19-AS1 is highly
expressed and acts as an oncogene in NPC.
To the best of our knowledge, the present study was
the first to demonstrate the molecular mechanism underlying the
role of FBXL19-AS1 in NPC. The findings demonstrated that
FBXL19-AS1 was upregulated in NPC tissues and cells, and FBXL19-AS1
silencing suppressed viability, migration and invasion of NPC
cells, which suggested the oncogenic role of FBXL19-AS1 in NPC. In
addition, accumulating studies have indicated that lncRNAs can
serve as tools in the diagnosis, treatment and prognosis of NPC
(46,47). Therefore, the
FBXL19-AS1/miR-431/PBOV1 axis may be a novel promising therapeutic
strategy for patients with NPC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
JH performed the experiments. HD and CH analyzed the
data and wrote the manuscript. All authors confirm the authenticity
of all the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from each
participant. The protocol of the present study was approved by the
Ethics Committee of Zhangjiagang TCM Hospital Affiliated to Nanjing
University of Chinese Medicine (Zhangjiagang, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wei KR, Zheng RS, Zhang SW, Liang ZH, Li
ZM and Chen WQ: Nasopharyngeal carcinoma incidence and mortality in
China, 2013. Chin J Cancer. 36:902017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang W, Guo Q, Liu G, Zheng F, Chen J,
Huang D, Ding L, Yang X, Song E, Xiang Y and Yao H: NKILA represses
nasopharyngeal carcinoma carcinogenesis and metastasis by NF-κB
pathway inhibition. PLoS Genet. 15:e10083252019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Razak AR, Siu LL, Liu FF, Ito E,
O'Sullivan B and Chan K: Nasopharyngeal carcinoma: The next
challenges. Eur J Cancer. 46:1967–1978. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cao C, Zhou S and Hu J: Long noncoding RNA
MAGI2-AS3/miR-218-5p/GDPD5/SEC61A1 axis drives cellular
proliferation and migration and confers cisplatin resistance in
nasopharyngeal carcinoma. Int Forum Allergy Rhinol. 10:1012–1023.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Young LS and Dawson CW: Epstein-Barr virus
and nasopharyngeal carcinoma. Chin J Cancer. 33:581–590.
2014.PubMed/NCBI
|
|
6
|
Zhao CX, Zhu W, Ba ZQ, Xu HJ, Liu WD, Zhu
B, Wang L, Song YJ, Yuan S and Ren CP: The regulatory network of
nasopharyngeal carcinoma metastasis with a focus on EBV, lncRNAs
and miRNAs. Am J Cancer Res. 8:2185–2209. 2018.PubMed/NCBI
|
|
7
|
Wang Y, Chen W, Lian J, Zhang H, Yu B,
Zhang M, Wei F, Wu J, Jiang J, Jia Y, et al: The lncRNA PVT1
regulates nasopharyngeal carcinoma cell proliferation via
activating the KAT2A acetyltransferase and stabilizing HIF-1α. Cell
Death Differ. 27:695–710. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zheng ZQ, Li ZX, Zhou GQ, Lin L, Zhang LL,
Lv JW, Huang XD, Liu RQ, Chen F, He XJ, et al: Long noncoding RNA
FAM225A promotes nasopharyngeal carcinoma tumorigenesis and
metastasis by acting as ceRNA to sponge miR-590-3p/miR-1275 and
upregulate ITGB3. Cancer Res. 79:4612–4626. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lou T, Ke K, Zhang L, Miao C and Liu Y:
LncRNA PART1 facilitates the malignant progression of colorectal
cancer via miR-150-5p/LRG1 axis. J Cell Biochem. 121:4271–4281.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen
X, Zhang Q, Yan G and Cui Q: LncRNADisease: A database for
long-non-coding RNA-associated diseases. Nucleic Acids Res.
41((Database Issue)): D983–D986. 2013.PubMed/NCBI
|
|
11
|
Wang M, Ji YQ, Song ZB, Ma XX, Zou YY and
Li XS: Knockdown of lncRNA ZFAS1 inhibits progression of
nasopharyngeal carcinoma by sponging miR-135a. Neoplasma.
66:939–945. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang E and Li X: LncRNA SOX2-OT regulates
proliferation and metastasis of nasopharyngeal carcinoma cells
through miR-146b-5p/HNRNPA2B1 pathway. J Cell Biochem.
120:16575–16588. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu D, Wang Y, Zhao Y and Gu X: LncRNA
SNHG5 promotes nasopharyngeal carcinoma progression by regulating
miR-1179/HMGB3 axis. BMC Cancer. 20:1782020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang B, Jia L, Ren H, Jin C, Ren Q, Zhang
H, Hu D, Zhang H, Hu L and Xie T: LncRNA DLX6-AS1 increases the
expression of HIF-1α and promotes the malignant phenotypes of
nasopharyngeal carcinoma cells via targeting MiR-199a-5p. Mol Genet
Genomic Med. 8:e10172020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shen B, Yuan Y, Zhang Y, Yu S, Peng W,
Huang X and Feng J: Long non-coding RNA FBXL19-AS1 plays oncogenic
role in colorectal cancer by sponging miR-203. Biochem Biophys Res
Commun. 488:67–73. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jiang Q, Cheng L, Ma D and Zhao Y:
FBXL19-AS1 exerts oncogenic function by sponging miR-431-5p to
regulate RAF1 expression in lung cancer. Biosci Rep.
39:BSR201818042019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang Y, Xiao X, Zhou W, Hu J and Zhou D:
LIN28A-stabilized FBXL19-AS1 promotes breast cancer migration,
invasion and EMT by regulating WDR66. In Vitro Cell Dev Biol Anim.
55:426–435. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fromm B, Billipp T, Peck LE, Johansen M,
Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E and
Peterson KJ: A uniform system for the annotation of vertebrate
microRNA Genes and the evolution of the human microRNAome. Annu Rev
Genet. 49:213–242. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rong X, Gao W, Yang X and Guo J:
Downregulation of hsa_circ_0007534 restricts the proliferation and
invasion of cervical cancer through regulating miR-498/BMI-1
signaling. Life Sci. 235:1167852019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fridrichova I and Zmetakova I: MicroRNAs
contribute to breast cancer invasiveness. Cells. 8:13612019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Aghajani M, Mansoori B, Mohammadi A,
Asadzadeh Z and Baradaran B: New emerging roles of CD133 in cancer
stem cell: Signaling pathway and miRNA regulation. J Cell Physiol.
234:21642–21661. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yin W, Shi L and Mao Y: MicroRNA-449b-5p
suppresses cell proliferation, migration and invasion by targeting
TPD52 in nasopharyngeal carcinoma. J Biochem. 166:433–440. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang T, Du M, Zhang W, Bai H, Yin L, Chen
W, He X and Chen Q: MicroRNA-432 suppresses invasion and migration
via E2F3 in nasopharyngeal carcinoma. Onco Targets Ther.
12:11271–11280. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He W, Huang Y, Jiang CC, Zhu Y, Wang L,
Zhang W, Huang W, Zhou T and Tang S: miR-100 inhibits cell growth
and proliferation by targeting HOXA1 in nasopharyngeal carcinoma.
Onco Targets Ther. 13:593–602. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yin D, Wei G, Yang F and Sun X: Circular
RNA has circ 0001591 promoted cell proliferation and metastasis of
human melanoma via ROCK1/PI3K/AKT by targeting miR-431-5p. Hum Exp
Toxicol. 40:310–324. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li MF, Li YH, He YH, Wang Q, Zhang Y, Li
XF, Meng XM, Huang C and Li J: Emerging roles of hsa_circ_0005075
targeting miR-431 in the progress of HCC. Biomed Pharmacother.
99:848–858. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu Y, Li L, Liu Z, Yuan Q and Lu X:
Downregulation of miR-431 expression associated with lymph node
metastasis and promotes cell invasion in papillary thyroid
carcinoma. Cancer Biomark. 22:727–732. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang Y, Qian W, Feng F, Cao Q, Li Y, Hou
Y, Zhang L and Fan J: Upregulated lncRNA CASC2 may inhibit
malignant melanoma development through regulating miR-18a-5p/RUNX1.
Oncol Res. 27:371–377. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gao W, Lin S, Cheng C, Zhu A, Hu Y, Shi Z,
Zhang X and Hong Z: Long non-coding RNA CASC2 regulates Sprouty2
via functioning as a competing endogenous RNA for miR-183 to
modulate the sensitivity of prostate cancer cells to docetaxel.
Arch Biochem Biophys. 665:69–78. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu D, Gong H, Tao Z, Chen S, Kong Y and
Xiao B: LncRNA IUR downregulates miR-144 to regulate PTEN in
nasopharyngeal carcinoma. Arch Physiol Biochem. Aug 14–2020.(Epub
ahead of print). View Article : Google Scholar
|
|
32
|
Tang T, Yang L, Cao Y, Wang M, Zhang S,
Gong Z, Xiong F, He Y, Zhou Y, Liao Q, et al: LncRNA AATBC
regulates Pinin to promote metastasis in nasopharyngeal carcinoma.
Mol Oncol. 14:2251–2270. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dong G, Pan T, Zhou D, Li C, Liu J and
Zhang J: FBXL19-AS1 promotes cell proliferation and inhibits cell
apoptosis via miR-876-5p/FOXM1 axis in breast cancer. Acta Biochim
Biophys Sin (Shanghai). 51:1106–1113. 2019.PubMed/NCBI
|
|
34
|
Wang L, Zhang X, Liu Y and Xu S: Long
noncoding RNA FBXL19-AS1 induces tumor growth and metastasis by
sponging miR-203a-3p in lung adenocarcinoma. J Cell Physiol.
235:3612–3625. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang J, Qiu Q, Qian X, Yi J, Jiao Y, Yu M,
Li X, Li J, Mi C, Zhang J, et al: Long noncoding RNA LCAT1
functions as a ceRNA to regulate RAC1 function by sponging
miR-4715-5p in lung cancer. Mol Cancer. 18:1712019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Thomson DW and Dinger ME: Endogenous
microRNA sponges: Evidence and controversy. Nat Rev Genet.
17:272–283. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cui M, Chang Y, Fang QG, Du W, Wu JF, Wang
JH, Liu ST and Luo SX: Non-Coding RNA Pvt1 promotes cancer stem
cell-like traits in nasopharyngeal cancer via inhibiting miR-1207.
Pathol Oncol Res. 25:1411–1422. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen X, Huang Y, Shi D, Nie C, Luo Y, Guo
L, Zou Y and Xie C: LncRNA ZNF667-AS1 promotes ABLIM1 expression by
adsorbing micro RNA-1290 to suppress nasopharyngeal carcinoma cell
progression. Onco Targets Ther. 13:4397–4409. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yao H, Yang L, Tian L, Guo Y and Li Y:
LncRNA MSC-AS1 aggravates nasopharyngeal carcinoma progression by
targeting miR-524-5p/nuclear receptor subfamily 4 group A member 2
(NR4A2). Cancer Cell Int. 20:1382020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang W, Dong Y, Li X, Pan Y, Du J and Liu
D: MicroRNA-431 serves as a tumor inhibitor in breast cancer
through targeting FGF9. Oncol Lett. 19:1001–1007. 2020.PubMed/NCBI
|
|
41
|
Huang W, Zeng C, Hu S, Wang L and Liu J:
ATG3, a Target of miR-431-5p, promotes proliferation and invasion
of colon cancer via promoting autophagy. Cancer Manag Res.
11:10275–10285. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
An G, Ng AY, Meka CS, Luo G, Bright SP,
Cazares L, Wright GL Jr and Veltri RW: Cloning and characterization
of UROC28, a novel gene overexpressed in prostate, breast, and
bladder cancers. Cancer Res. 60:7014–7020. 2000.PubMed/NCBI
|
|
43
|
Pan T, Wu R, Liu B, Wen H, Tu Z, Guo J,
Yang J and Shen G: PBOV1 promotes prostate cancer proliferation by
promoting G1/S transition. Onco Targets Ther. 9:787–795. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Guo Y, Wu Z, Shen S, Guo R, Wang J, Wang
W, Zhao K, Kuang M and Shuai X: Nanomedicines reveal how PBOV1
promotes hepatocellular carcinoma for effective gene therapy. Nat
Commun. 9:34302018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Carleton NM, Zhu G, Gorbounov M, Miller
MC, Pienta KJ, Resar LMS and Veltri RW: PBOV1 as a potential
biomarker for more advanced prostate cancer based on protein and
digital histomorphometric analysis. Prostate. 78:547–559. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu Z, Wu K, Wu J, Tian D, Chen Y, Yang Z
and Wu A: NEAT1 is a potential prognostic biomarker for patients
with nasopharyngeal carcinoma. J Cell Biochem. 120:9831–9838. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yao L, Wang T and Wang X: LncRNA FOXP4-AS1
serves as a biomarker for nasopharyngeal carcinoma diagnosis and
prognosis. 3 Biotech. 11:252021. View Article : Google Scholar : PubMed/NCBI
|