Introduction
Tuberculosis (TB) remains a major health challenge
worldwide (1). Global estimates
report that, at any time, 25% of the world's population is infected
with Mycobacterium tuberculosis (Mtb). If undiagnosed
and untreated, 5–10% of patients with latent TB will progress to
the condition of active TB (2).
Therefore, accurate and reliable diagnostic tests are crucial for
the prevention of TB.
Exosomes are minuscule, phospholipid enclosed
membrane vesicles ranging from 30–150 nm in size that originate
from a variety of different cell types (3,4). These
cells are able to release exosomes into their surroundings
(5). Exosomes contain and release
DNA (3), RNA (5) and proteins (6,7), which
are commonly used as exosomal markers, serving major roles in
intercellular communication through the transfer of secreted
molecules and through direct cell-cell contact (8). Exosomes have been identified in the
supernatants of cultured cells and in biological fluids using
specific membrane proteins that serve as markers, notably CD9, CD63
and CD81 (9–13). Exosomes have also been shown to be
involved in the pathogenesis of various types of infection, and
have themselves been suggested to serve a role as valuable
biomarkers that may guide different anti infective therapies
(14). The identification of easily
measurable, accurate and reliable diagnostic markers would be
expected to have a significant impact on disease control. Exosomes
have biomarker potential, and yet, to fulfil this potential, their
careful characterization is required, since components within
exosomes vary according to the cell type that they originate from
(15). Therefore, the accurate
extraction of exosomes from cells and biological fluids is an
essential requirement for exosome research, and for ensuring their
eventual use in biomedical applications.
Mtb-infected human THP-1 macrophages have
been shown to release Mtb molecules within exosomes, as
determined from the fact that Mtb, which is an intracellular
pathogen, is able to release Mtb protein-containing exosomes
(6). Mtb-derived proteins
have been detected in cell-culture supernatants from cells infected
with the laboratory Mtb strain H37Rv (6) and in the biological fluids of patients
with TB (7,14). Exosomes could serve as diagnostic
markers for TB since they are carriers of the molecular
constituents of mycobacteria (16).
Therefore, the present study aimed to investigate the protein
composition of exosomes released by Mtb-infected human
macrophages.
To meet this end, the tetraspanin family membrane
proteins CD9, CD63, CD81 and lysosomal associated membrane
protein-1 (LAMP-1) were employed as markers for host-derived
molecules. Furthermore, mycobacterial antigen 85 (Ag85) and the
Mpt64 protein were selected to evaluate the presence of Mtb
molecules within exosomes. The majority of the published studies on
exosomes and protein surface markers to date have focused on the
use of cells cultured in media supplemented with 10% fetal bovine
serum (FBS) (17–19). Abramowicz et al (13) demonstrated that high abundance
exosome-contaminating serum proteins are potential sources of
contaminants that cause substantial drawbacks in terms of exosome
harvest, isolation and processing. Accordingly, a decision was made
in the present study to select serum-free ultra-centrifuged
dendritic cell medium for the purpose of culturing THP-1 cells for
subsequent exosome processing. Consequently, the human monocytic
cell line THP-1 was cultured in a serum-free ultracentrifuged
CellGenix® GMP DC medium to avoid interference with sera
proteins. Upon optimization of a sera free system for the
cultivation and infection of human THP-1 macrophages, the exosomes
were investigated, principally for their biophysical features,
surface markers and their content of mycobacterial proteins
following infection.
Materials and methods
Cell culture
The human monocytic cell line THP-1 (kindly provided
by the Institute of Clinical Immunology, University of Leipzig,
Leipzig, Germany) was maintained in RPMI-1640 medium (Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10% FBS (Invitrogen;
Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin
(Seromed; Biochrom KG), hereafter designated as complete RPMI
(cRPMI). To avoid contaminating serum-derived exosomes, four
different types of cell-culture media were investigated for their
suitability for THP-1 cell cultures. These media were cRPMI,
RPMI-1640 medium without 10% FBS (hereafter designated as RPMI SF),
CellGenix® GMP DC medium (CellGenix GmbH) that was not
ultra-centrifuged (here after designated as DCM) and DCM-UG medium,
which was DCM medium that was ultra-centrifuged at 120,000 × g for
20 h to remove particles or events that are found within the size
range of exosomes in DCM medium using NanoSight LM10 (Malvern
Panalytical Ltd.) (hereafter designated as DCM-UG).
Apoptotic assay
THP-1 cells were seeded at a density of
1×106 cells/ml in 6-well plates in cRPMI, RPMI SF, DCM
or DCM-UG media. The cells were incubated either for 16 or 48 h at
37°C in a humidified atmosphere with 5% CO2. Following
the incubation, cells were subjected to flow cytometric analysis
for assessment of apoptosis and necrosis using an Annexin V-FITC
and propidium iodide (PI) staining kit (cat. no. 640914; BioLegend,
Inc.). The cells were harvested and washed three times at 350 × g
for 5 min with cold phosphate-buffered saline (PBS); subsequently,
200 µl supernatant was resuspended in Annexin binding buffer at a
concentration of 1×106 cells/ml, and 100 µl cell
suspension was stained with 5 µl Annexin V and 10 µl PI for 15 min
in the dark at room temperature. A total of 200 µl Annexin binding
buffer was then added, and the extent of apoptosis or necrosis was
subsequently analyzed using a BD FACSCanto II flow cytometer and
FACSDiva software 8.0.1 (BD Biosciences). Live cells
(Annexin-V−/PI−), apoptotic cells
(Annexin-V+/PI−) and necrotic cells
(Annexin-V+/PI+) were evaluated for each
culture condition in triplicate, and the averaged values were then
calculated.
Mtb culture and infection of the THP-1
macrophages
The laboratory strain Mtb H37Rv (kindly
provided by Professor Stefan H.E. Kaufmann, Max Planck Institute
for Infection Biology, Berlin, Germany) was used for these
experiments. Bacteria were grown in Middlebrook 7H9 medium (Difco;
BD Biosciences) supplemented with 0.2% glycerol, 0.05% Tween-80
(MilliporeSigma) and 10% albumin-dextrose complex (ADC) (BD
Biosciences). Mtb cultures were subsequently incubated at
37°C in an orbital shaker (80 rev./min) to avoid clumping. For
infection of the THP-1 macrophages, bacteria in the early
logarithmic phase were used (when the OD600 of the cells
was in the range of 0.4–0.8). THP-1 cells (2–5×105/ml)
were seeded into a T75 flask containing 10 ml cRPMI, and
the cell population was expanded by sub culturing every 2–3 days.
The differentiation of THP-1 monocytes into macrophages was
achieved upon stimulation with 50 ng/ml
Phorbol-12-myristat-13-acetat (PMA) (Sigma-Aldrich; Merck KGaA),
with subsequent overnight incubation and further cultivation for 2
days in cRPMI. Differentiated THP-1 macrophages were infected with
Mtb strain H37Rv for 4 h in DCM-UG at a multiplicity of
infection (MOI) of 5, and then the cells were either processed for
their exosomes or subsequently washed twice with PBS at room
temperature and maintained in DCM-UG. Cell culture supernatants
were harvested at various time points after infection (4, 24 and 48
h). Naïve samples were processed in parallel with the
Mtb-infected samples. The cell culture supernatants were
filtered twice through a 0.22-µm filter membrane (Millex GP;
MilliporeSigma) and stored at −80°C until use.
Exosome isolation
DCM-UG culture supernatants (10 ml) from Mtb
H37Rv-infected (MOI 5) and naïve THP-1 cells were centrifuged at
2,000 × g for 30 min at room temperature to remove dead cells, cell
debris and large particles, and subsequently the supernatants were
transferred into sterile tubes. Cell culture supernatants (10 ml)
were mixed with 5 ml total exosome isolation reagent (TEIR kit;
Invitrogen; Thermo Fisher Scientific, Inc.) from cell culture media
and mixed thoroughly through vortexing until a homogenous solution
was formed. The samples were subsequently incubated at 4°C
overnight, and then centrifuged at 4°C at10,000 × g for 1 h. The
supernatant was discarded, and the exosome pellet was either
resuspended in NaCl buffer [for subsequent nanoparticle tracking
analysis (NTA)] or 1X RIPA buffer (Thermo Fisher Scientific, Inc.)
with protease inhibitor (Roche Diagnostics) for western blot
analysis.
NTA
The concentration and size of exosomes were analyzed
via NTA using a NanoSight LM10 (Malvern Panalytical, Ltd.) equipped
with a 450 nm laser to determine the size and concentration of
isolated particles, according to the manufacturer's protocol.
Samples were diluted 1:20 in sterile 0.9% NaCl. Each experimental
run was performed in triplicate (n=3), where each capture was made
over a 60 sec period with a frame rate of 25 frames/sec and a
camera level of 9. For each sample, the nanoparticle size
distribution curve, refractive index and the relative nanoparticle
concentration of a particular size were recorded, together with the
cumulative percentages of nanoparticles. Data were analyzed using
NTA 3.0 software (Malvern Panalytical, Ltd.) with a detection
threshold of four and a controlled temperature of 25°C.
Western blot analysis
After exosome isolation, the exosome pellets of
Mtb H37Rv-infected and naïve THP-1 macrophages were directly
lysed in RIPA buffer comprising 25 mM Tris/HCl (pH 7.6), 150 mM
NaCl, 1% NP-40, 1% sodium deoxycholate and 0.1% SDS (Invitrogen;
Thermo Fisher Scientific Inc.) with the protease inhibitor (Roche
Diagnostics) for 15 min at 4°C. Loading buffer (4X LDS; Invitrogen;
Thermo Fisher Scientific, Inc.) under reducing or non reducing
conditions was added, and the exosome preparation was boiled at
70°C for 10 min. Lysed exosome samples of 40 µl (typically
equivalent of 3.x107 vesicles) were then separated on
SDS-PAGE (12% gels) under non reducing (CD9, CD63, CD81 and LAMP-1
a positive loading control) and reducing (anti-Ag85 and anti-Mpt64)
conditions, and transferred onto PVDF membranes (Cytiva) with a wet
blot chamber (Bio Rad Laboratories, Inc.). After transfer, the
membrane was blocked with 5% dry milk and 0.1% Tween-20 in 1X TBS
for 1 h at room temperature, and then probed overnight at 4°C with
the following primary antibodies: CD9 (1:5,000, cat. no. 10626D)
CD63 (1:5,000, cat. no. 10628D), CD81 (1:5,000, cat. no. MA5-13548)
and LAMP-1 (1:5,000, cat. no. 14-1079-80) (all from Invitrogen;
Thermo Fisher Scientific, Inc.), and Ag85 (1:5,000, clone ab36731),
Mpt64 (1:1,000, cat. no. ab193435) and His tag Mpt64 protein
(1:1,000, cat. no. ab225589) (all from Abcam). Subsequently, HRP
conjugated secondary goat anti mouse (1:100,000, cat. no. 31430,
Invitrogen; Thermo Fisher Scientific, Inc.) and rabbit anti mouse
antibodies (1:5,000, cat. no. ab205718 Abcam) were used. Secondary
antibodies were incubated for 1 h in a shaker at room temperature.
After a consecutive washing step with 1X TBS/0.1% Tween-20, a
washing buffer was removed and a SuperSignal™ West Femto
Maximum Sensitivity substrate (Invitrogen; Thermo Fisher
Scientific, Inc.) was added. The membrane was exposed to Stella ray
test (Xstella software 2.1.8.415) and analyzed using Aida Image
Analyzer software 5.0 (all from Raytest Isotopenmessgeräte
GmbH).
Multiplex surface markers
Flow cytometric exosome analysis was performed using
the MACSPlex Exosome kit, human (Miltenyi Biotec GmbH) according to
the manufacturer's overnight incubation protocol. A combined sample
volume comprising 175 µl THP-1 exosomes and 15 µl exosome capture
beads, which were coated with 37 different antibodies, were added
to a well of the microplate and incubated overnight at room
temperature in the dark on an orbital shaker (450 rev./min). After
washing, 15 µl master mix of anti-CD9-, anti-CD63- and
anti-CD81-APC antibodies were added, and the mixture was incubated
for 1 h at room temperature on an orbital shaker. Flow cytometric
analysis was performed on a BD FACSCanto II flow cytometer and
FACSDiva software 8.0.1 (BD Biosciences). After subtracting the
values of the control (buffer only) from the measured samples, the
median APC-signal intensity of each specific population of single
beads was normalized to the average of the anti-CD9, anti-CD63 and
anti-CD81 beads. Surface marker values that were below the
corresponding control antibody included in the kit, considered as
the measurement threshold, were regarded as negative.
Statistical analysis
Experiments were independently repeated at least in
triplicates (n=3). Statistical analysis was performed using
Microsoft Excel 2016 software (Microsoft Corporation).
Additionally, statistically significant differences in measurements
between experiments with two groups were determined using a paired
Student's t-test, while one way analysis of variance was performed
for comparisons between multiple groups. The percentage coefficient
of variation (% CV) was calculated to indicate the precision of the
cell culture methods using GraphPad Prism version 5 (GraphPad
Software, Inc.). Descriptive data analyses were performed to
visualize differences within the data. P<0.05 was considered to
indicate a statistically significant difference. Error bars in all
figures represent the standard error of the mean (± SEM).
Results
Cell culture and assessment of
cellular viability by flow cytometry
To circumvent the possible presence of serum derived
exosomes, THP-1 cells were cultured in the four different types of
media outlined in the Materials and methods section, and cellular
viability was assessed at 16 h (Fig.
1A) and 48 h (Fig. 1B) by flow
cytometry. Although there were no statistically significant
differences between culture methods (P=0.87), flow cytometry for
cell growth assessment showed reduced number of viable cells
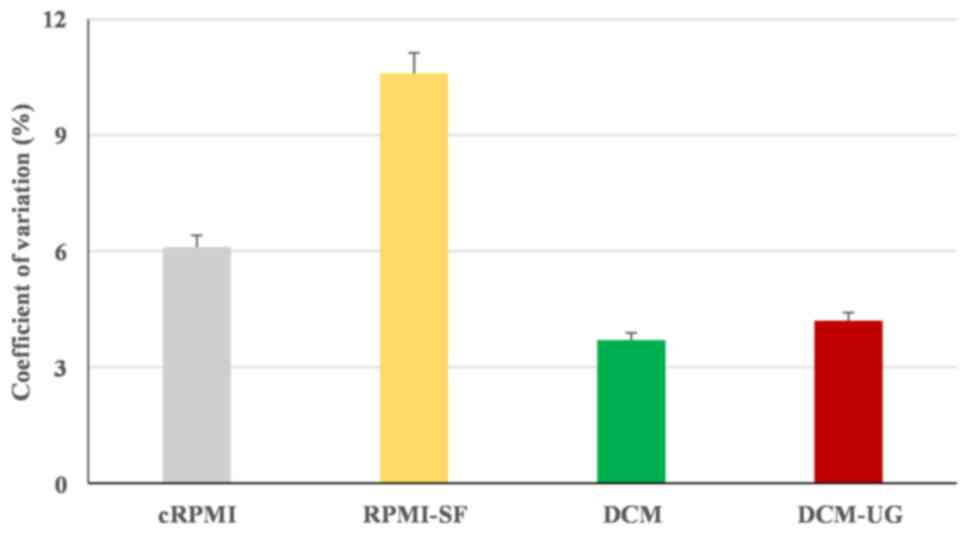
cultured in the RPMI-SF medium compared with the others, and this
was confirmed by the highest percentage coefficient value of
cRPMI-SF (% CV of 10.6%) (Fig. 2).
Overall, all the types of media (cRPMI, RPMI-SF, DCM and DCM-UG)
allowed the monocytic cells to replicate over a period of 16 h, but
serum-free media resulted in the highest variance amongst the
tested media. DCM (% CV of 3.7) and DCM-UG (% CV of 4) were found
to be suitable media based on their lower coefficient values. The
NTA analysis of DCM medium showed particles/events within the size
range of exosomes (data not shown). These events or particles in
DCM medium were poorly understood and their sources are also
unclear. In this regard, apoptosis and necrosis was not performed
at 48 h for DCM and RPMI-SF (due to its highest % CV; Fig. 2). Afterwards, the media cRPMI
(control) and DCM-UG were selected for further cell growth at 48 h
since the main objective of the present study was to use serum-free
medium for further exosome research and analysis. Taken together,
these data demonstrated that DCM-UG was consistently the best type
of medium in terms of cell viability and subsequent exosome purity,
and this was therefore selected for subsequent experiments.
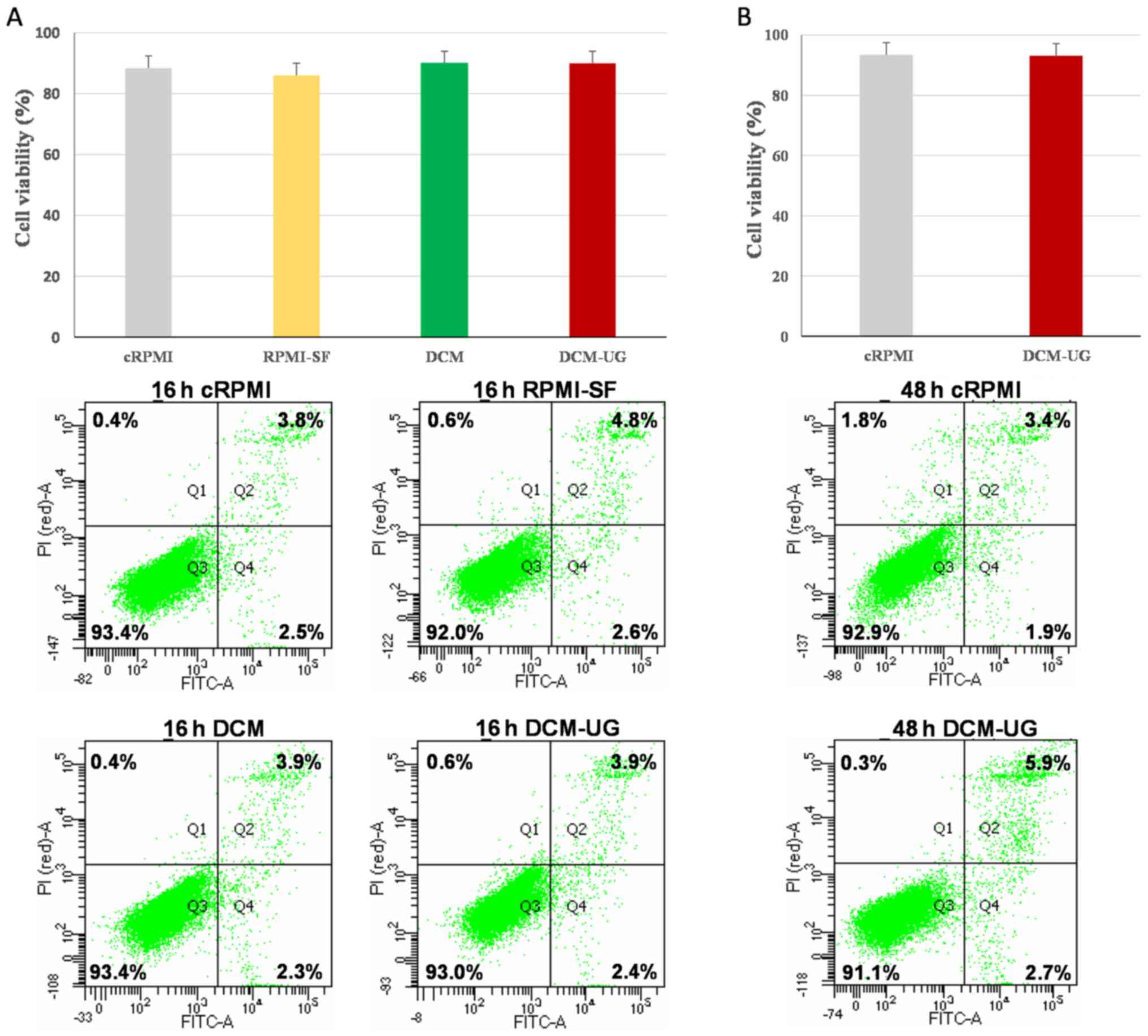 | Figure 1.Cell death patterns of THP-1 cells
maintained under distinct culture conditions, as determined using
flow cytometry. Cells were analyzed after (A) 16 h and (B) 48 h of
incubation at 37°C in a humidified atmosphere with 5%
CO2. The lower left quadrants (Q3) of the panels show
the viable cells, which were negative for Annexin V and PI staining
(Annex-V−/PI−). The lower right quadrants
(Q4) represent apoptotic cells, which were positive for Annexin V
and negative for PI (Annex-V+/PI−), whereas
the upper right quadrants (Q2) contain the non-viable, necrotic
cells, positive for both Annexin V and PI
(Annex-V+/PI+). Data included in the figures
[parts (A) and (B)] represent the mean readings of three
independent replicates, and error bars indicate standard error of
mean. cRPMI, RPMI 1640 complete media; RPMI SF, RPMI serum-free
media; DCM, CellGenix® GMP DC Medium; DCM-UG,
ultra-centrifuged CellGenix® GMP DC Medium. |
Exosome isolation, size and
concentration of isolated vesicles
Subsequently, exosomes were extracted from freshly
prepared THP-1 cell cultured supernatants using a precipitation
method, and an acceptable number of exosomes were recovered via
this process. The size and concentration of the exosomes were then
analyzed by NTA using a NanoSight LM10 apparatus. This instrument
is able to detect and analyze small particles <30 nm in size by
increasing the viscosity of the sample (19). This technology, however, is not able
to differentiate exosomes from other synthetic nanoparticles or
from large protein aggregates (20).
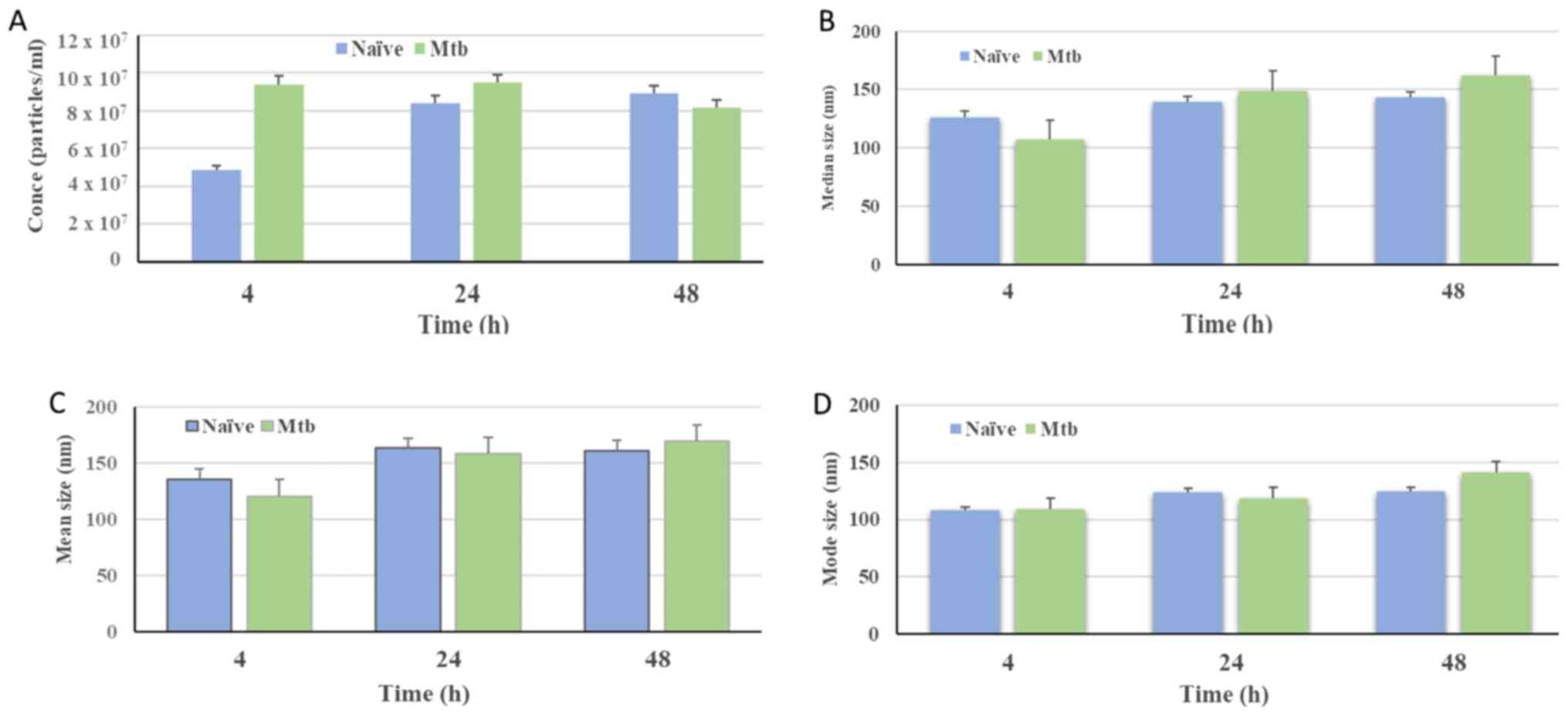
The concentration (particles/ml) of isolated
vesicles was found to be higher than the detection levels,
irrespective of the Mtb infection status (Fig. 3A). The particle distribution
observed for nanovesicles isolated from naïve THP-1 cells indicated
that 50% of the population were below 126.3±8.9 nm in size at 4 h,
with subsequent measurements of 138.8±20.8 and 142.5±7.2 nm at 24
and 48 h, respectively. By contrast, for the Mtb-infected
THP-1 cells, it was observed that 50% of the particles were below
106.8.2±5.6, 148.5±13.6 and 161.5±14.1 nm in size at the 4, 24 and
48 h times points, respectively (Fig.
3B). In addition, the mean size distribution of NTA
measurements of nanovesicles isolated from naïve THP-1 cells was at
136±11.0 nm, 161±12.9 nm and 162±30.8 nm in size at 4, 48 and 24 h
times points, respectively. However, for the Mtb-infected
THP-1 cells, it was 120±13.5 nm, for 4 h, 160±8.5 nm for 24 h and
171±18.6 nm for 48 h (Fig 3C).
Nanovesicles from naïve THP-1 cells at the 4 h incubation time
point comprised a population with a lower mode value compared with
vesicles obtained from naïve cells incubated for 24 and 48 h.
Similarly, nanovesicles isolated from infected macrophages at 48 h
presented a higher mode value compared with those detected at 4 and
24 h post-infection (Fig. 3D).
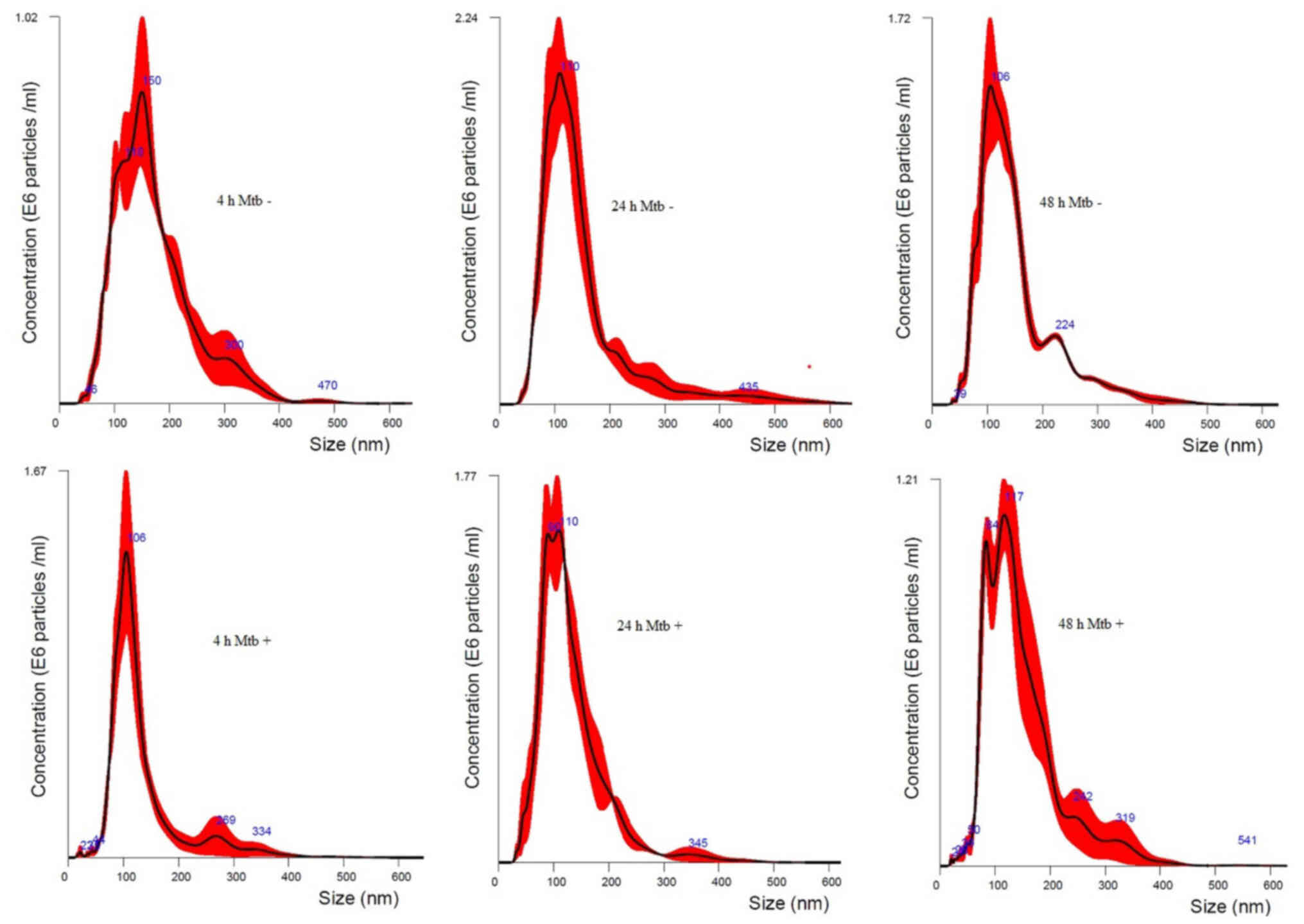
The size of isolated particles from all time points
in naïve and infected THP-1 macrophages was found to be <400 nm.
Besides, the size distribution of vesicles showed a size range
peaked at 106, 110 and 150 nm in size for naïve THP-1 cells at 48,
24 and 4 h and 106, 110 and 117 nm for Mtb-infected THP-1
cells at 4, 24 and 48 h times points, respectively. The majority of
the isolated vesicles were within the expected size range of 30–150
nm. Nano tracked particle size distribution curves measured at 4,
24 and 48 h for the Mtb-infected and naïve THP-1 macrophages
from one representative experiment out of at least three
independently performed experiments is shown in Fig. 4.
Western blot analysis
Isolated exosomes were then subjected to western
blot analysis to ascertain the presence of selected exosome markers
CD9, CD63, CD81 and LAMP-1 in the conditioned THP-1 cell culture
medium supernatant, as per the manufacturer's instructions. The
results revealed that CD9, CD63, CD81 and LAMP-1 were expressed in
exosomes, both from Mtb H37Rv-infected and naïve macrophages
(Fig. 5A and B). His-tag Mpt64
protein was also detected (data has not shown). However, blots run
with anti-Ag85 and anti-Mpt64 antibodies did not yield positive
signals in exosomes collected from infected THP-1 cells at the
exosome input recommended, specifically, 10 ml conditioned cell
culture medium supernatant.
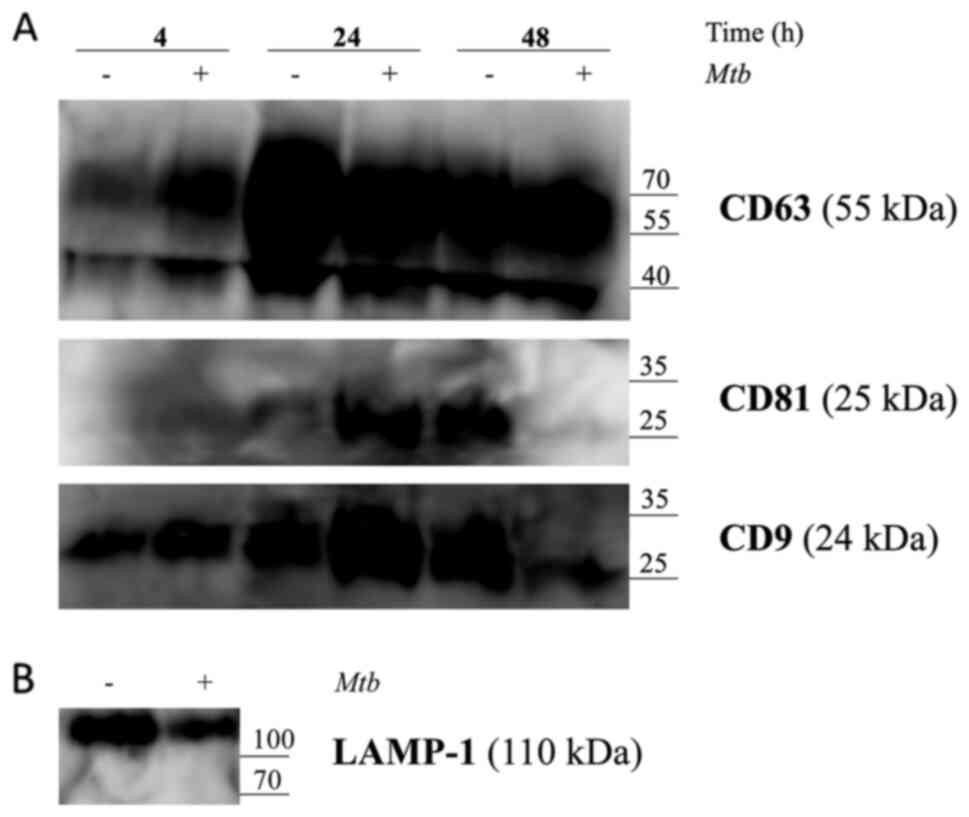 | Figure 5.Release of exosomes from Mtb
H37Rv-infected or naïve THP-1 cells. THP-1 macrophages were
infected with Mtb H37Rv (Mtb+) for 4 h (MOI 5) in
DCM-UG medium for 4, 24 and 48 h at 37°C and in the presence of 5%
CO2. Exosomes were extracted from cell culture
supernatants using total exosome isolation reagent and were further
analyzed by western blot analysis for the exosomal protein markers,
(A) CD63, CD81, CD9 and (B) LAMP-1, which served as a positive
control for exosomes signal and Mtb proteins [Ag85 and Mpt64
and recombinant Mtb Mpt64 protein (His-tag)]. Data from one
representative experiment out of at least three independent
experiments are shown. MOI, multiplicity of infection; Mtb,
Mycobacterium tuberculosis; DCM-UG, ultra-centrifuged
CellGenix® GMP DC Medium; LAMP-1, lysosomal associated
membrane protein-1; Ag85, mycobacterial antigen 85. |
Flow cytometry analysis of
exosomes
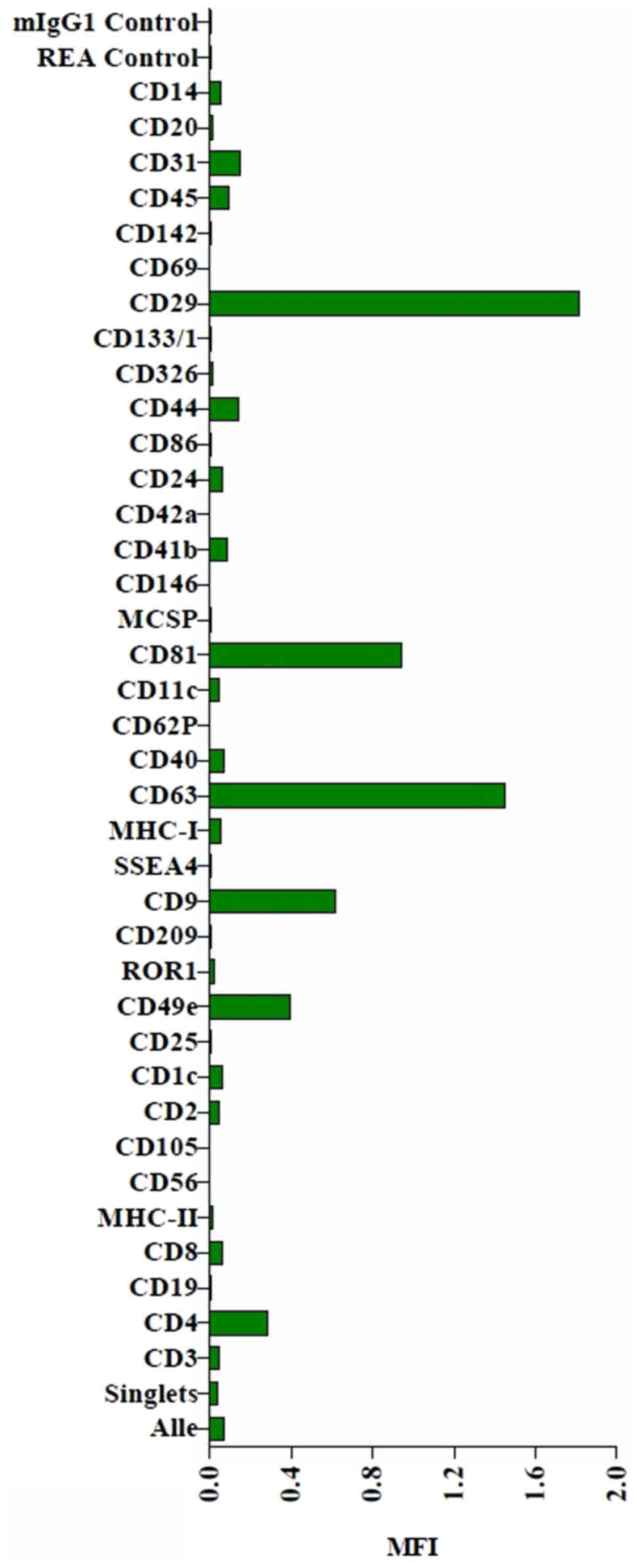
The MACSPlex exosome protein marker analysis of
naïve THP-1 cell culture supernatants revealed surface marker
profiles characterized by strong signals for the exosome associated
markers, CD9, CD63 and CD81. The MACSPlex exosome capture bead data
additionally indicated the presence of other markers at
intermediate-to-low positive APC fluorescence intensity levels,
with the highest intensity shown for CD29, indicating the presence
of other positive signals in the particular surface epitope within
the exosome population (Figs. 6 and
S1).
Discussion
The present study primarily aimed to identify an
appropriate particle free culture media amenable for the
cultivation of THP-1 cells, thereby enabling isolation of high
quality exosomes for further investigations. Apoptotic and necrotic
cell death are known to alter the quality of exosomes. Cells
maintained under inappropriate experimental conditions, i.e.,
subjected to nutritional stress subsequent to FBS withdrawal, may
undergo cell death (20). Whilst
the cell membrane of apoptotic cells remain undamaged, necrotic
cells lose their membrane integrity and release endogenous material
(13). Such events are critical for
establishing exosomes' features, and require careful assessment of
the most appropriate conditions, in particular, sera free media
that would have a minimal impact on cell viability. In the present
study, it was observed that a small proportion of THP-1 monocytes
underwent apoptosis and necrosis after 48 h of cultivation in the
serum-free, ultracentrifuged dendritic cell medium, DCM-UG. This is
consistent with other reports (9,10,13),
underlining that isolation of serum-free exosomes from THP-1 cells
is possible and that the use of DCM-UG additionally circumvents
erroneous results due to decreased cell viability. The present
study has reported, to the best of our knowledge for the first
time, the suitability of DCM-UG for exosome targeted studies in
THP-1 monocytes or macrophages, and has generated a schema for
downstream characterization of the exosomes.
Ultracentrifugation has been traditionally employed
for the isolation of exosomes; however, this method is time
consuming (8–30 h), laborious, requires costly equipment, increases
the risk of exosomal damage through the generation of large protein
aggregates, and offers variable recovery rates (11,12,15).
This method also grossly affects the quality of the isolated
exosomes. In the present study, the TEIR kit provided by Thermo
Fisher Scientific, Inc. was employed (17), which is a precipitation technique
for the isolation of exosomes that provides a higher particle yield
within a shorter processing time, which thereby decreases the use
of a higher volume of starting material that has been used in
previous studies (10,18). NTA, flow cytometric MACSPlex exosome
analysis of the THP-1 human monocytic cell line and western blot
analysis were also used to characterize and confirm that the
isolated vesicles were exosomes. In agreement with previous studies
(5,19,21),
the aforementioned methodologies allowed for the recovery of intact
exosomes at high yields from THP-1 macrophages. Similar results
have been reported for the human lung cancer cell line A549
(18), also with the same 10 ml
sample volume input (13). The
present study also confirmed that the isolated vesicles were
exosomes that fell mostly within the size range of 30–150 nm at all
time points. The exosomes isolated from naïve and infected
macrophages using the TEIR kit, according to the NTA assay,
presented comparable concentrations (particles/ml), mean and mode
sizes and values that were within the range of those reported by
other groups for THP-1 and other cells (10,18).
It was observed in the current study that ~50% of the vesicle
populations fell below 200 nm in diameter in both the Mtb
H37Rv-infected and uninfected THP-1 macrophages. However, vesicles
were also observed with wider size ranges that were >200 nm.
This probably resulted from the precipitation based techniques that
were used for exosome isolation in this study. The TEIR kit also
yielded extracellular vesicles (EVs) other than exosomes and
aggregates of soluble proteins from cell culture media, and similar
findings have been reported by other groups who also employed the
precipitation method (18,20,22).
The NTA technique does have certain limitations, however, regarding
adequate differentiation among vesicles released by precipitation
reagents, synthetic nanoparticles, large protein aggregates and
biological vesicles (18,23), indicating that the method may
overestimate the numbers of exosomes through detecting
contaminating proteins that are not associated with exosomes
(24).
The present study also characterized the expression
of proteins enriched in exosomes, including CD63, CD9 and CD81. All
samples expressed CD63 and CD9, similarly to what has been reported
previously (25). However, CD81 was
detected only in selected samples, at distinct time points in
infected and naïve macrophages. The identification of LAMP-1 in the
present study was inconsistent with another study (26), and a number of previously published
studies have documented a variety of exosome populations based on
their surface protein markers (21,27,28).
Furthermore, the marker profiles have been shown to vary with the
type of origin cell. In the present study, the Mtb antigen
Ag85 and Mpt64 protein were not detected in infected THP-1
macrophages. The inability to detect Mtb proteins could have
been due to the small sample volume, as only a 10 ml sample was
investigated, which therefore may have meant that the amounts of
mycobacterial proteins present were below the detection limits of
the western blotting technique. Giri et al (29) isolated Ag85 from Mtb-infected
J774 cells using western blotting. The absence of Ag85 and Mpt64
protein in Mtb-infected THP-1 cells in the present study may
have also been due to an absence of Mtb proteins in the
exosomes released under the culture conditions employed in the
present study, i.e., a lack of serum factors. On the other hand,
the choice of the sampling time points may have had an impact on
the abundance of exosomes observed during Mtb infection,
with more exosomes containing mycobacterial proteins possibly being
released during the late infection. Further investigations are
required to elucidate the thresholds for mycobacterial proteins in
macrophage derived exosomes, and also the kinetics of detection
during in vitro infection.
Flow cytometry analysis is a well-established method
that is used to characterize EV subpopulations in cells. In the
present study, the presence of CD9, CD63 and CD81 in exosomes
released by a human monocytic cell line was investigated by using a
multiplex bead-based flow cytometry assay. Predominantly four bead
populations, i.e., CD9, CD63, CD81 and CD29, were detected as being
strongly positive in the exosomes of THP-1 cell culture
supernatants, a finding that is in agreement with a study by
Wiklander et al (21), who
reported the presence of very strong signals of CD63, CD81, CD29
and CD49e from the cell lines of culture supernatants using flow
cytometry. The data obtained in the present study have confirmed
that the assay enabled us to detect EV surface markers from a low
volume of sample using a fast method.
The strength of this study was the ability to
optimize a sera-free system for the cultivation and infection of
human THP-1 macrophages for exosome quality and purity. Thus,
indicating that DCM-UG could be an alternative sera-free system to
analyze exosomes in further study. Despite this strength, the
current study has some drawbacks, which are subjects for future
studies. As a result of limited resources and time, a higher volume
of starting material to further investigate the Mtb proteins
over the manufacturers' recommendations for the use of the
precipitation reagent was not performed in this study. Besides,
this study did not evaluate the shape of the isolated exosomes due
to the lack of transmission electron microscope. Another limitation
of this study was the lack of LAMP-1 as a positive control at all
time points.
In conclusion, the present study provided evidence
in support of the fact that DCM-UG may be an alternative approach
for generating high purity exosome preparations. These culture
conditions allowed for the detection of surface protein markers
unique to exosomes in naïve and Mtb-infected THP-1
macrophages. However, more sensitive methods are necessary to
further investigate the Mtb proteins in exosome samples
obtained from cells maintained in DCM-UG.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank staff of Institute
of Clinical Immunology, Medical Faculty, University of Leipzig,
D-04103 Leipzig; Dr Roland Weiss for his help in editing figures
and his unreserved support for the present study and Ms Katrin
Bauer and Ms Kerstin Wenk for their assistance during the present
study.
Funding
The present study was funded by the Alexander von
Humboldt Foundation.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FB was the primary researcher, who conceived and
designed the study with guidance from YGJDS, BK, ACR, US and AD. FB
performed the laboratory experiments, interpreted the results,
conducted data analysis and wrote the manuscript. PR participated
in laboratory experiments and contributed reagents. UZ performed
the Mtb infection studies. FB and PR confirm the
authenticity of all the raw data. FB, BK, ACR AD and US searched
for relevant literature. PR, YGJDS, BK, ACR, AD and US reviewed the
initial and final drafts of the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
World Health Organization (WHO), . Global
Tuberculosis Report 2018. WHO; Geneva: pp. 2312018
|
|
2
|
World Health Organization (WHO), . WHO
Guidelines on Tuberculosis Infection Prevention and Control: 2019
Update. WHO; Geneva: pp. 622019
|
|
3
|
van der Pol E, Böing AN, Harrison P, Sturk
A and Nieuwland R: Classification, functions, and clinical
relevance of extracellular vesicles. Pharmacol Rev. 64:676–705.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee S-S, Won J-H, Lim GJ, Han J, Lee JY,
Cho K-O and Bae Y-K: A novel population of extracellular vesicles
smaller than exosomes promotes cell proliferation. Cell Commun
Signal. 17:952019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Patel GK, Khan MA, Zubair H, Srivastava
SK, Khushman M, Singh S and Singh AP: Comparative analysis of
exosome isolation methods using culture supernatant for optimum
yield, purity and downstream applications. Sci Rep. 9:53352019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Diaz G, Wolfe LM, Kruh-Garcia NA and Dobos
KM: Changes in the Membrane-Associated Proteins of Exosomes
Released from Human Macrophages after Mycobacterium
tuberculosis Infection. Sci Rep. 6:379752016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kruh-Garcia NA, Wolfe LM, Chaisson LH,
Worodria WO, Nahid P, Schorey JS, Davis JL and Dobos KM: Detection
of Mycobacterium tuberculosis peptides in the exosomes of
patients with active and latent M. tuberculosis infection using
MRM-MS. PLoS One. 9:e1038112014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang Y, Liu Y, Liu H and Tang WH:
Exosomes: Biogenesis, biologic function and clinical potential.
Cell Biosci. 9:192019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lobb RJ, Becker M, Wen SW, Wong CSF,
Wiegmans AP, Leimgruber A and Möller A: Optimized exosome isolation
protocol for cell culture supernatant and human plasma. J Extracell
Vesicles. 4:270312015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zeringer E, Li M, Barta T, Schageman J,
Pedersen KW, Neurauter A, Magdaleno S, Setterquist R and Vlassov
AV: Methods for the extraction and RNA profiling of exosomes. World
J Methodol. 3:11–18. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rosas LE, Elgamal OA, Mo X, Phelps MA,
Schmittgen TD and Papenfuss TL: In vitro immunotoxicity assessment
of culture-derived extracellular vesicles in human monocytes. J
Immunotoxicol. 13:652–665. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tauro BJ, Greening DW, Mathias RA, Ji H,
Mathivanan S, Scott AM and Simpson RJ: Comparison of
ultracentrifugation, density gradient separation, and
immunoaffinity capture methods for isolating human colon cancer
cell line LIM1863-derived exosomes. Methods. 56:293–304. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Abramowicz A, Marczak L, Wojakowska A,
Zapotoczny S, Whiteside TL, Widlak P and Pietrowska M:
Harmonization of exosome isolation from culture supernatants for
optimized proteomics analysis. PLoS One. 13:e02054962018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Singhal N, Sharma P, Kumar M, Joshi B and
Bisht D: Analysis of intracellular expressed proteins of
Mycobacterium tuberculosis clinical isolates. Proteome Sci.
10:142012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Théry C, Amigorena S, Raposo G and Clayton
A: Isolation and characterization of exosomes from cell culture
supernatants and biological fluids. Curr Protoc Cell Biol Chapter.
3:Unit 3.22. 2006.doi.org/10.1002/0471143030.cb0322s30. PubMed/NCBI
|
|
16
|
Hu X, Liao S, Bai H, Wu L, Wang M, Wu Q,
Zhou J, Jiao L, Chen X, Zhou Y, et al: Integrating exosomal
microRNAs and electronic health data improved tuberculosis
diagnosis. EBioMedicine. 40:564–573. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Biadglegne F, König B, Rodloff AC, Dorhoi
A and Sack U: Composition and Clinical Significance of Exosomes in
Tuberculosis: A Systematic Literature Review. J Clin Med.
10:E1452021. View Article : Google Scholar
|
|
18
|
Tang Y-T, Huang Y-Y, Zheng L, Qin S-H, Xu
X-P, An T-X, Xu Y, Wu Y-S, Hu X-M, Ping B-H, et al: Comparison of
isolation methods of exosomes and exosomal RNA from cell culture
medium and serum. Int J Mol Med. 40:834–844. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Arteaga-Blanco LA, Mojoli A, Monteiro RQ,
Sandim V, Menna-Barreto RFS, Pereira-Dutra FS, Bozza PT, Resende RO
and Bou-Habib DC: Characterization and internalization of small
extracellular vesicles released by human primary macrophages
derived from circulating monocytes. PLoS One. 15:e02377952020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Atha DH and Ingham KC: Mechanism of
precipitation of proteins by polyethylene glycols. Analysis in
terms of excluded volume. J Biol Chem. 256:12108–12117. 1981.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wiklander OPB, Bostancioglu RB, Welsh JA,
Zickler AM, Murke F, Corso G, Felldin U, Hagey DW, Evertsson B,
Liang X-M, et al: Systematic Methodological Evaluation of a
Multiplex Bead-Based Flow Cytometry Assay for Detection of
Extracellular Vesicle Surface Signatures. Front Immunol.
9:13262018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Helwa I, Cai J, Drewry MD, Zimmerman A,
Dinkins MB, Khaled ML, Seremwe M, Dismuke WM, Bieberich E, Stamer
WD, et al: A Comparative Study of Serum Exosome Isolation Using
Differential Ultracentrifugation and Three Commercial Reagents.
PLoS One. 12:e01706282017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gercel-Taylor C, Atay S, Tullis RH,
Kesimer M and Taylor DD: Nanoparticle analysis of circulating
cell-derived vesicles in ovarian cancer patients. Anal Biochem.
428:44–53. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Caradec J, Kharmate G, Hosseini-Beheshti
E, Adomat H, Gleave M and Guns E: Reproducibility and efficiency of
serum-derived exosome extraction methods. Clin Biochem.
47:1286–1292. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Keller BO, Sui J, Young AB and Whittal RM:
Interferences and contaminants encountered in modern mass
spectrometry. Anal Chim Acta. 627:71–81. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Smith VL, Cheng Y, Bryant BR and Schorey
JS: Exosomes function in antigen presentation during an in vivo
Mycobacterium tuberculosis infection. Sci Rep. 7:435782017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Koliha N, Wiencek Y, Heider U, Jüngst C,
Kladt N, Krauthäuser S, Johnston ICD, Bosio A, Schauss A and Wild
S: A novel multiplex bead-based platform highlights the diversity
of extracellular vesicles. J Extracell Vesicles. 5:299752016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Brahmer A, Neuberger E, Esch-Heisser L,
Haller N, Jorgensen MM, Baek R, Möbius W, Simon P and Krämer-Albers
E-M: Platelets, endothelial cells and leukocytes contribute to the
exercise-triggered release of extracellular vesicles into the
circulation. J Extracell Vesicles. 8:16158202019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Giri PK, Kruh NA, Dobos KM and Schorey JS:
Proteomic analysis identifies highly antigenic proteins in exosomes
from M. tuberculosis-infected and culture filtrate
protein-treated macrophages. Proteomics. 10:3190–3202. 2010.
View Article : Google Scholar : PubMed/NCBI
|