Introduction
Diabetes mellitus causes several complications,
including the formation and occurrence of cataracts (1). Despite the successful surgical
replacement of cataracts with intraocular lenses, cataracts remain
one of the leading causes of visual impairment and blindness
worldwide (2). Diabetic cataracts
(DC) characterized by high blood glucose levels usually occur
earlier and progress faster than cataract (3,4).
Human lens epithelial cells (HLECs) have been reported to play
essential roles in ocular health, as well as several diseases,
including DC (5). The
pathogenesis of DC is a multifactorial process, and gene
alterations that are associated with the proliferation,
differentiation and epithelial-to-mesenchymal transition (EMT) of
LECs may lead to the occurrence of cataract (6). It is therefore crucial to explore
the mechanism underlying LEC proliferation in DC.
Long non-coding RNAs (lncRNAs) are a group of
non-coding RNAs of >200 nts in length that lack protein-encoding
potential (7). In addition,
lncRNAs have been reported to participate in DC development. For
example, lncRNA metastasis-associated lung adenocarcinoma
transcript 1 has been shown to facilitate the apoptosis and
oxidative stress of HLECs through the p38 MAPK pathway in DC
(8). Sp1 transcription
factor-mediated lncRNA Pvt1 oncogene was shown to regulate LEC
viability and apoptosis in DC through the microRNA
(miRNA/miR)-214-3p/MMP2 axis (9).
Furthermore, lncRNA X-inactive specific transcript (XIST) has been
found to be associated with diabetic complications, such as
diabetic nephropathy and retinopathy (10,11). However, the biological role of
XIST during DC remains unclear.
miRNAs are small RNAs that can target the
3′-untranslated region (UTR) of mRNAs to modulate gene
transcription (12). miRNA
dysregulation has been observed in multiple diseases, including DC.
For example, miR-30a was shown to suppress autophagy by targeting
Beclin 1 in human DC (13).
miR-211 was found to promote apoptosis and repress proliferation of
LECs in DC mice by targeting SIRT1 (14). In addition, miR-34a inhibited LEC
viability and induced apoptosis by targeting E2F3 (15). Furthermore, SMAD family member 2
(SMAD2) has been reported to play a crucial role in posterior
capsular opacification (16).
Nevertheless, the regulatory mechanisms of miR-34a and SMAD2 during
DC remain largely unknown. In the present study, SRA01/04 cells
were stimulated by high glucose (HG) to establish a DC model, and
the biological effects of XIST on DC were then investigated.
Materials and methods
Samples
A total of 32 posterior capsular tissue samples from
patients with DC (18 men, 14 women; age range, 52–60 years) and
paired normal posterior capsular tissue samples without DC (17 men,
15 women; age range, 48–59 years) were obtained from the Shandong
Zaozhuang Municipal Hospital (Zaozhuang, China). The tissues were
rapidly frozen in liquid nitrogen at −80°C prior to use. This study
was approved by the Ethics Committee of Shandong Zaozhuang
Municipal Hospital and all participants signed informed consent
forms prior to surgery. Patients diagnosed with DC and provided
informed consent were included in this study. Patients with complex
cataracts with high myopia, ocular trauma and ocular inflammation
were excluded.
Cell culture and HG treatment
SRA01/04 HLECs were purchased from BeNa Culture
Collection (Beijing Beina Chuanglian Institute of Biotechnology).
The cells were incubated at 37°C with 5% CO2 in DMEM
(Thermo Fisher Scientific, Inc.) containing 10% FBS (Thermo Fisher
Scientific, Inc.) and 1% penicillin/streptomycin. To establish the
DC cell model, SRA01/04 cells were maintained in medium containing
HG (25 mM) for 24 h at 37°C, and cells in normal glucose (NG; 5.5
mM) were used as the control.
Cell transfection
Small hairpin (sh)RNA against XIST (shXIST,
5′-GUGCGUACAGUGCUGUACAGCAU-3′) and its negative control (NC; shNC,
5′-UACGCUCAGCAUGUGUCACUC-3′), miR-34a mimics
(5′-UCGUUCGUGAGCACUUGCGACG-3′), NC mimics
(5′-UCGUCGGAUCGACUGAGAUCU-3′), miR-34a inhibitors
(5′-AGCCUUGCUGCAGGUGCGCAU-3′) and NC inhibitors
(5′-UGCCUUACUGACGGUCGGAGA-3′) were obtained from Shanghai
GenePharma Co., Ltd. pcDNA3.1 vector (Thermo Fisher Scientific,
Inc.) was used to construct a XIST and SMAD2 overexpression vector.
SRA01/04 cells (1×105) were transfected with 50 nM
shXIST, 50 nM shNC, 50 nM miR-34a mimics, 50 nM miR-34a inhibitors,
50 nM NC mimics or 50 nM NC inhibitors using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) at
room temperature for ~30 min, according to the manufacturer's
protocol. After 48 h of transfection, the transfected cells were
used for subsequent experiments.
Reverse transcription-quantitative
(RT-q)PCR
TRIzol® reagent (Thermo Fisher
Scientific, Inc.) was used to isolate total RNA from the cultured
cells and tissue samples. PrimeScript™ RT reagent kit (Takara Bio,
Inc.) was used for cDNA generation of XIST and SMAD2, and TaqMan™
MicroRNA Reverse Transcription kit (Thermo Fisher Scientific, Inc.)
was used for cDNA generation of miR-34a, according to the
manufacturer's protocol. qPCR was performed on the ABI 7900
Detection System (Thermo Fisher Scientific, Inc.) using the
SYBR-Green PCR Master Mix kit (Thermo Fisher Scientific, Inc.). The
2−ΔΔCq method (17)
was used to calculate the relative expression. The relative miR-34a
expression was normalized to U6. GAPDH served as the control for
XIST and SMAD2 expression. The thermocycling conditions were as
follows: Pre-denaturation at 95°C for 1 min, followed by 40 cycles
of 95°C for 15 sec, 60°C for 30 sec and 72°C for 30 sec. The
following primers were used: XIST, forward
5′-TCAGCCCATCAGTCCAAGATC-3′ and reverse
5′-CCTAGTTCAGGCCTGCTTTTCAT-3′; miR-34a, forward
5′-ACCCAGTGCGATTTGTCA-3′ and reverse 5′-ACTGTACTGGAAGATGGACC-3′;
SMAD2, forward 5′-TCCTACTACCGCCTCACA-3′ and reverse
5′-ACCTCCTCCTCCTCCTCT-3′; GAPDH, forward
5′-TCGACAGTCAGCCGCATCTTCTTT-3′ and reverse
5′-ACCAAATCCGTTGACTCCGACCTT-3′; and U6, forward
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse
5′-CGCTTCACGAATTTGCGTGTCA-3′.
Western blot analysis
Total protein was extracted from cells using RIPA
lysis buffer (Sangon Biotech Co., Ltd.). Protein concentration was
detected using a BCA assay kit (Sangon Biotech Co., Ltd.). Next,
the proteins (20 µg) were separated via 10% SDS-PAGE and
transferred to PVDF membranes (MilliporeSigma). After blocking with
5% non-fat milk for 2 h, the membrane was incubated with primary
antibodies against SMAD2 (1:1,000; cat. no. ab40855; Abcam) and
GADPH (1:1,000; cat. no. ab9485; Abcam) at 4°C. Subsequently, the
membranes were washed with 0.1% Tween-20 and incubated with a
HRP-conjugated secondary antibody (1:1,000; cat. no. ab205718;
Abcam) for 2 h at room temperature. Finally, protein bands were
visualized using an ECL reagent (Beyotime Institute of
Biotechnology).
MTT assay
Cell proliferation was assessed using MTT assay.
Transfected SRA01/04 cells (1×105 cells/well) were
cultured in 96-well plates and incubated for 0, 24, 48 or 72 h. MTT
(5 mg/ml) was then added for 4 h at 37°C. Next, 150 µl DMSO (Thermo
Fisher Scientific, Inc.) was added to each well, and the optical
density 490 nm value was measured using a microplate reader (BioTek
Instruments, Inc.).
Transwell assay
SRA01/04 cell invasion was assessed by Transwell
assay (8.0-µm pore size; MilliporeSigma) with Matrigel (Corning
Inc.) precoating at room temperature for 1 h. SRA01/04 cells
(2×105 per well) were plated in the upper Transwell
chamber in serum-free DMEM. Next, 500 µl DMEM (10% FBS) was plated
in the lower chambers. Following incubation for 48 h at 37°C,
invading cells were stained with 0.1% crystal violet for 20 min at
room temperature and counted under a light microscope
(magnification, ×100; Olympus Corporation).
Wound healing assay
A total of 5×105 SRA01/04 cells were
cultured in DMEM supplemented without serum in 6-well plates and
allowed to reach 70% confluence for the wound healing assay. A
200-µl pipette tip was used to generate artificial scratches.
Images of migrated cells were captured at 0 and 24 h using a
microscope (magnification, ×100; Leica DMI4000B; Leica
Microsystems, Ltd.).
TUNEL assay
TUNEL Apoptosis Assay kit (Roche Diagnostics GmbH)
was used to assess cell apoptosis. Following fixation in 4%
paraformaldehyde for 1 h at 4°C, 1×105 SRA01/04 cells
were cultured with TUNEL reaction mixture (Roche Diagnostics GmbH)
for 1 h at room temperature. Nuclear staining with DAPI was then
performed for 15 min at room temperature. The TUNEL-positive cells
were counted in five randomly selected fields under a fluorescence
microscope (magnification, ×200; Nikon Corporation).
Dual-luciferase reporter assay
The potential binding sites of miR-34a and XIST or
SMAD2 were predicted using starBase 2.0 (http://starbase.sysu.edu.cn), as previously described
(18,19). The wild-type (wt) or mutant (mut)
3′-UTR sequences of XIST or SMAD2 were cloned into the pmirGLO
vector (Promega Corporation). Site-directed mutagenesis was used to
create the mut 3′-UTR sequence. The pmirGLO-XIST-wt or XIST-mut and
pmirGLO-SMAD2-wt or SMAD2-mut reporter vector were co-transfected
with miR-34a mimics and NC mimics into SRA01/04 cells using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.).
Following a 48-h incubation, luciferase activity was measured by
Dual-Luciferase Reporter Assay System (Promega Corporation).
Firefly luciferase activity was normalized to Renilla
(Promega Corporation) luciferase gene activity.
RNA immunoprecipitation (RIP)
assay
RIP assay was performed using the Magna RIP
RNA-Binding Protein Immunoprecipitation kit (MilliporeSigma).
SRA01/04 cells were lysed in RIP lysis buffer (MilliporeSigma). The
obtained cell lysate (100 µl) was centrifuged at 40,000 × g at 4°C
for 10 min and incubated with 50 µl A/G magnetic beads conjugated
with 5 µg anti-AGO2 (5 µg; cat. no. ab32381; Abcam) or 5
µg anti-IgG antibodies (cat. no. ab172730; Abcam) for 1 h at 4°C.
Subsequently, the beads were washed three times using RIP Wash
Buffer. The beads were then incubated with proteinase K buffer at
55°C for 30 min to digest the protein. Finally, XIST and miR-34a
enrichment was measured via RT-qPCR.
Statistical analysis
Data are expressed as the mean ± SD and analysis was
performed using SPSS 17.0 (SPSS, Inc.). Each experiment was
performed in triplicate. The correlation between miR-34a and XIST
or SMAD2 was assessed by Pearson's correlation analysis.
Comparisons between two groups or among multiple groups were
assessed using an unpaired Student's t-test or one-way ANOVA
followed by Tukey's post hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
XIST is upregulated and miR-34a is
downregulated in DC tissues and cells
First, the levels of XIST and miR-34a were detected
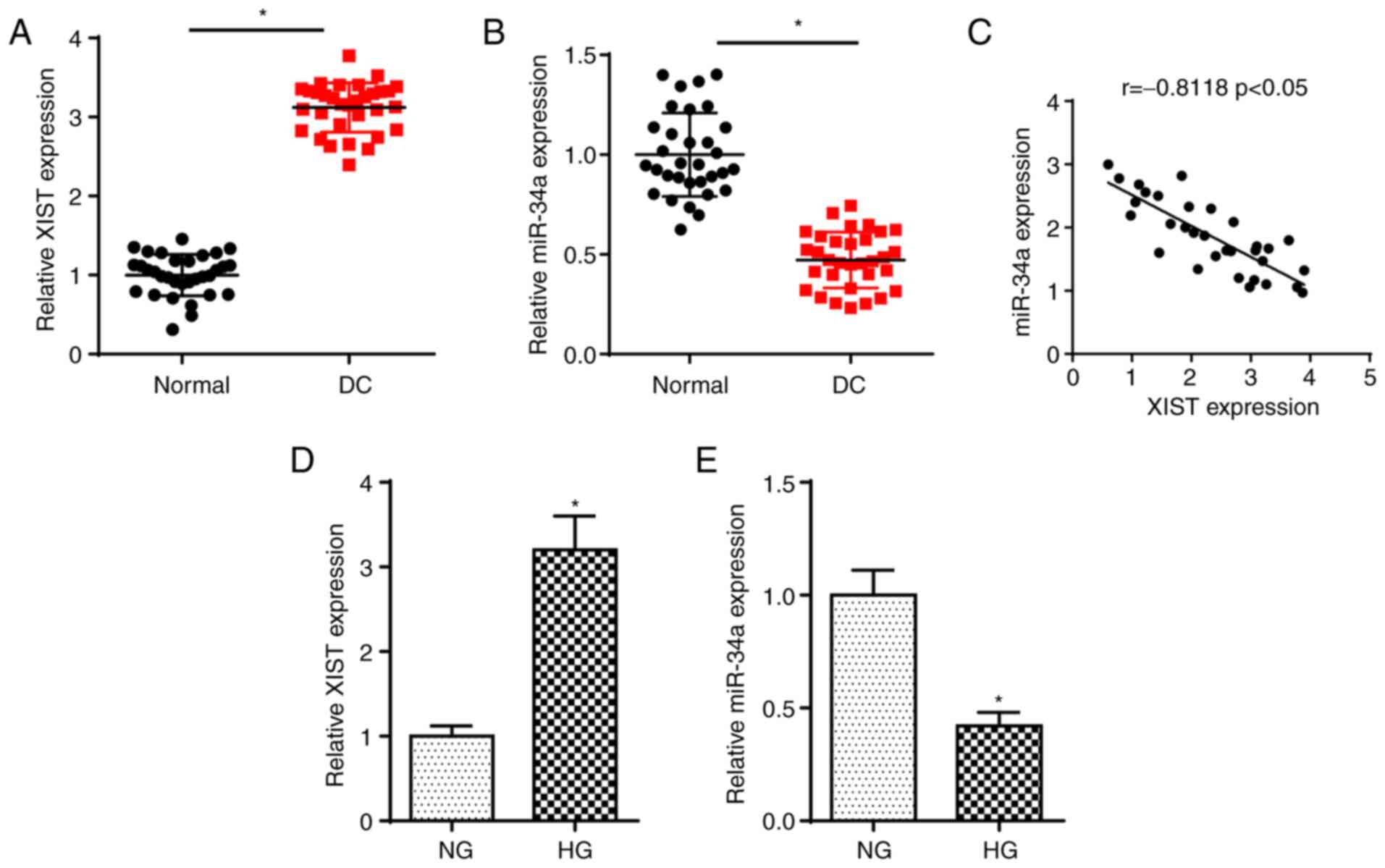
in DC tissues by RT-qPCR. As shown in Fig. 1A and B, the expression of XIST was
increased and that of miR-34a was reduced in DC samples. In
addition, an inverse correlation between the miR-34a and XIST
levels was observed in DC tissues (Fig. 1C). In addition, RT-qPCR analysis
determined that the XIST expression was significantly elevated and
miR-34a abundance was reduced in SRA01/04 cells treated with HG
compared with the NG group (Fig. 1D
and E). The data suggested that XIST was significantly
upregulated and miR-34a was significantly downregulated in DC.
XIST depletion inhibits DC development
in HG-treated LECs
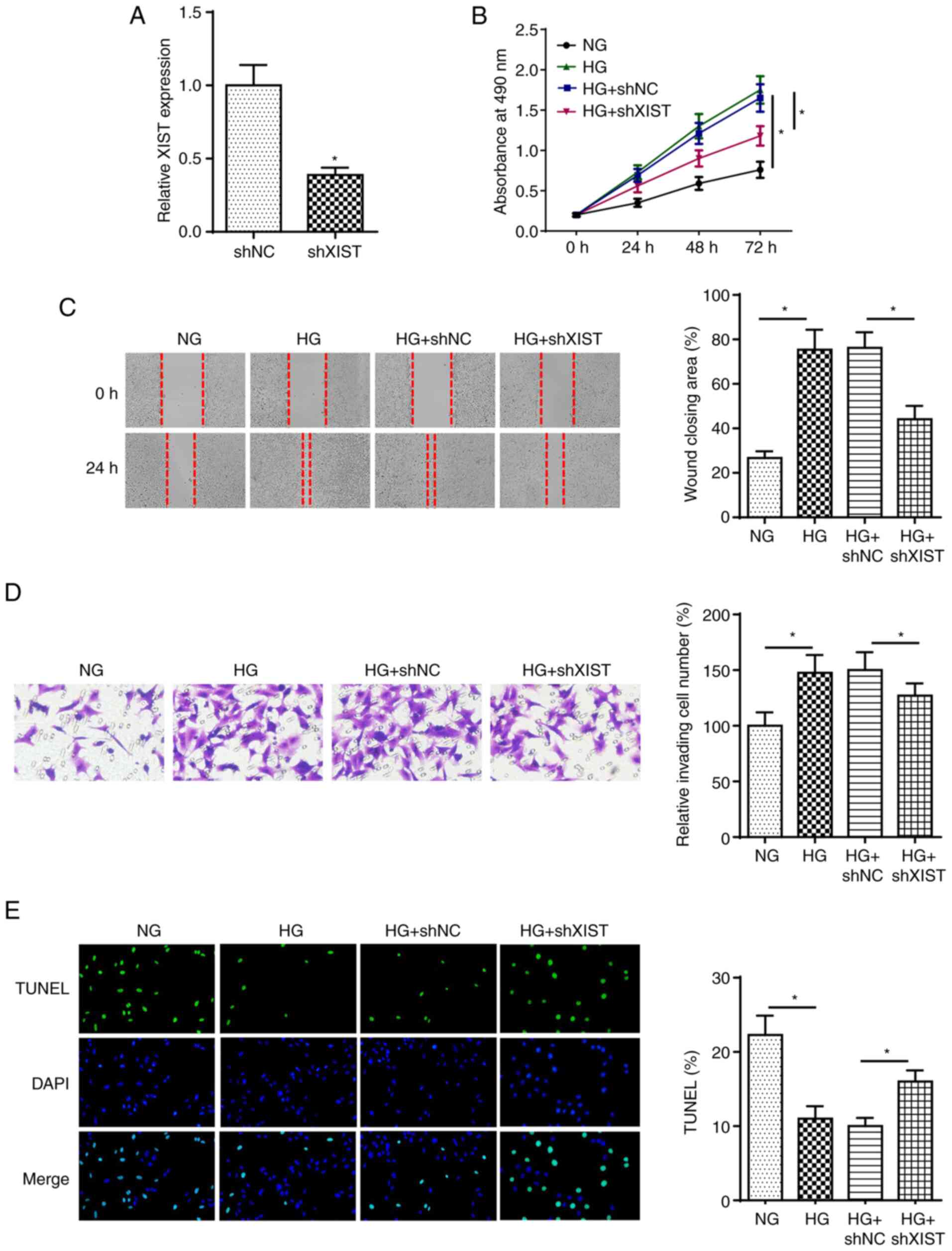
To explore the role of XIST in DC, SRA01/04 cells
were transfected with shXIST and shNC prior to HG treatment.
RT-qPCR analysis revealed that XIST knockdown decreased the
expression of XIST in SRA01/04 cells (Fig. 2A). Next, MTT, wound healing and
Transwell assays revealed that XIST knockdown suppressed the
proliferation, migration and invasion of SRA01/04 cells treated by
HG (Fig. 2B-D). XIST depletion
induced the apoptosis of HG-stimulated SRA01/04 cells (Fig. 2E). These results indicated that
XIST regulated HG-treated LEC proliferation, metastasis and
apoptosis in DC.
miR-34a overexpression reduces
HG-induced decrease in proliferation, migration and invasion in
LECs
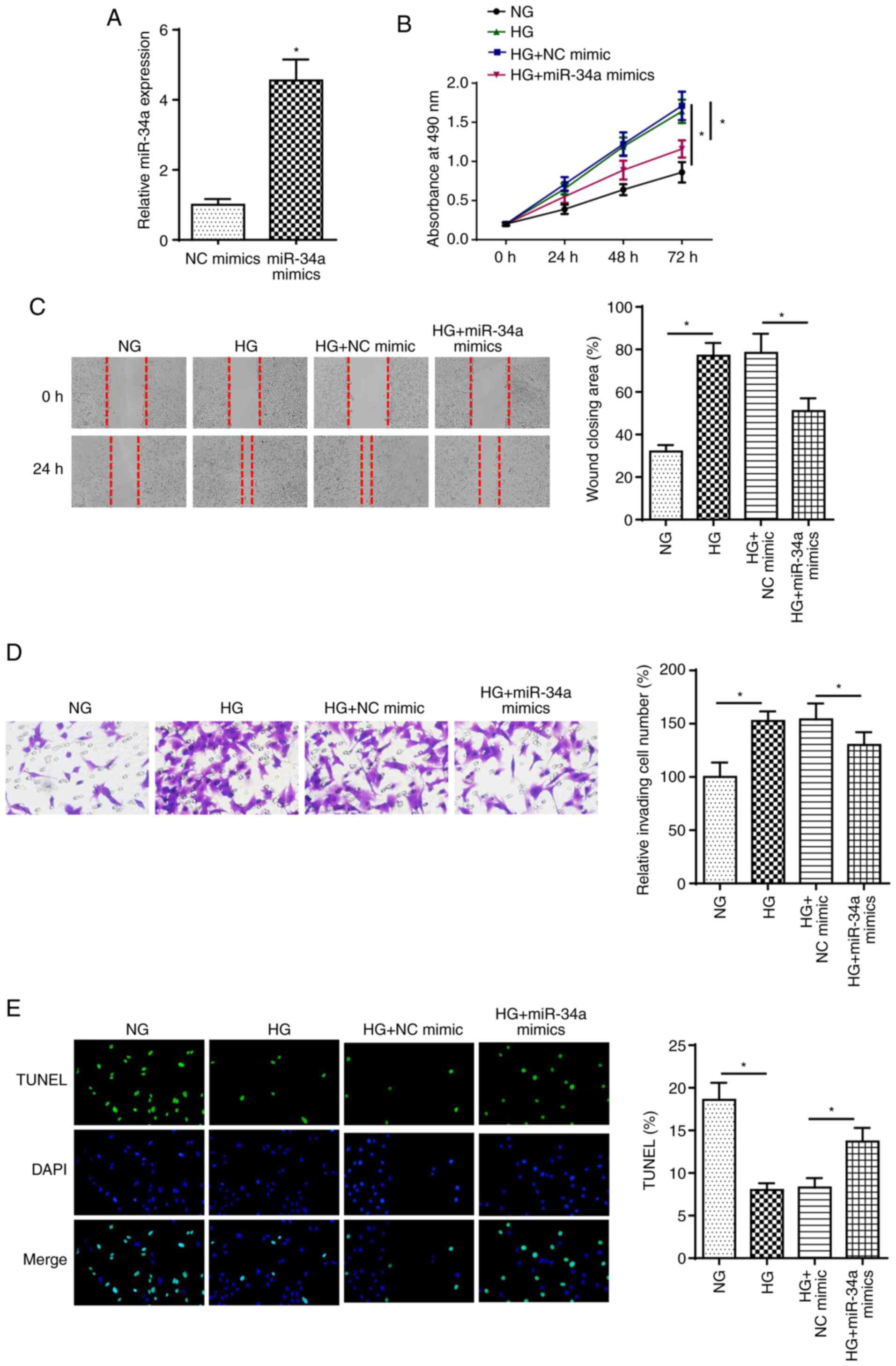
To investigate the effect of miR-34a on DC, SRA01/04
cells were transfected with miR-34a mimics. As shown in Fig. 3A, miR-34a was highly expressed
following miR-34a overexpression. Furthermore, HG stimulation
increased SRA01/04 cell proliferation, migration and invasion,
while miR-34a overexpression reversed these effects (Fig. 3B-D). In addition, the HG
treatment-induced decrease in cell apoptosis was rescued by miR-34a
overexpression (Fig. 3E). These
findings indicated that miR-34a played an essential role in
HG-treated LECs.
XIST deletion modulates DC progression
by targeting miR-34a in HG-treated LECs
Subsequently, it was explored whether XIST modulates
DC progression through miR-34a. As shown in Fig. 4A, the binding sequences of XIST
and miR-34a were predicted using starBase. Next, dual-luciferase
reporter assay revealed that miR-34a overexpression repressed the
luciferase activity of the XIST-wt group, while that of the
XIST-mut group was unchanged (Fig.
4B). In addition, RIP assay discovered that XIST and miR-34a
enrichment was increased in LECs cells, which confirmed that XIST
could combine with miR-34a (Fig.
4C). Furthermore, RT-qPCR demonstrated that the expression of
miR-34a was reduced by XIST overexpression in SRA01/04 cells and
enhanced by XIST knockdown (Fig.
4D). Functional assay suggested that miR-34a deficiency rescued
the repressive effects of XIST knockdown on the proliferation,
migration and invasion of HG-stimulated cells (Fig. 4E-G). Furthermore, XIST silencing
induced cell apoptosis, which was abrogated by miR-34a deletion in
HG-treated SRA01/04 cells (Fig.
4H). Therefore, the findings suggested that miR-34a is sponged
by XIST and participates in the regulation of XIST during DC
progression.
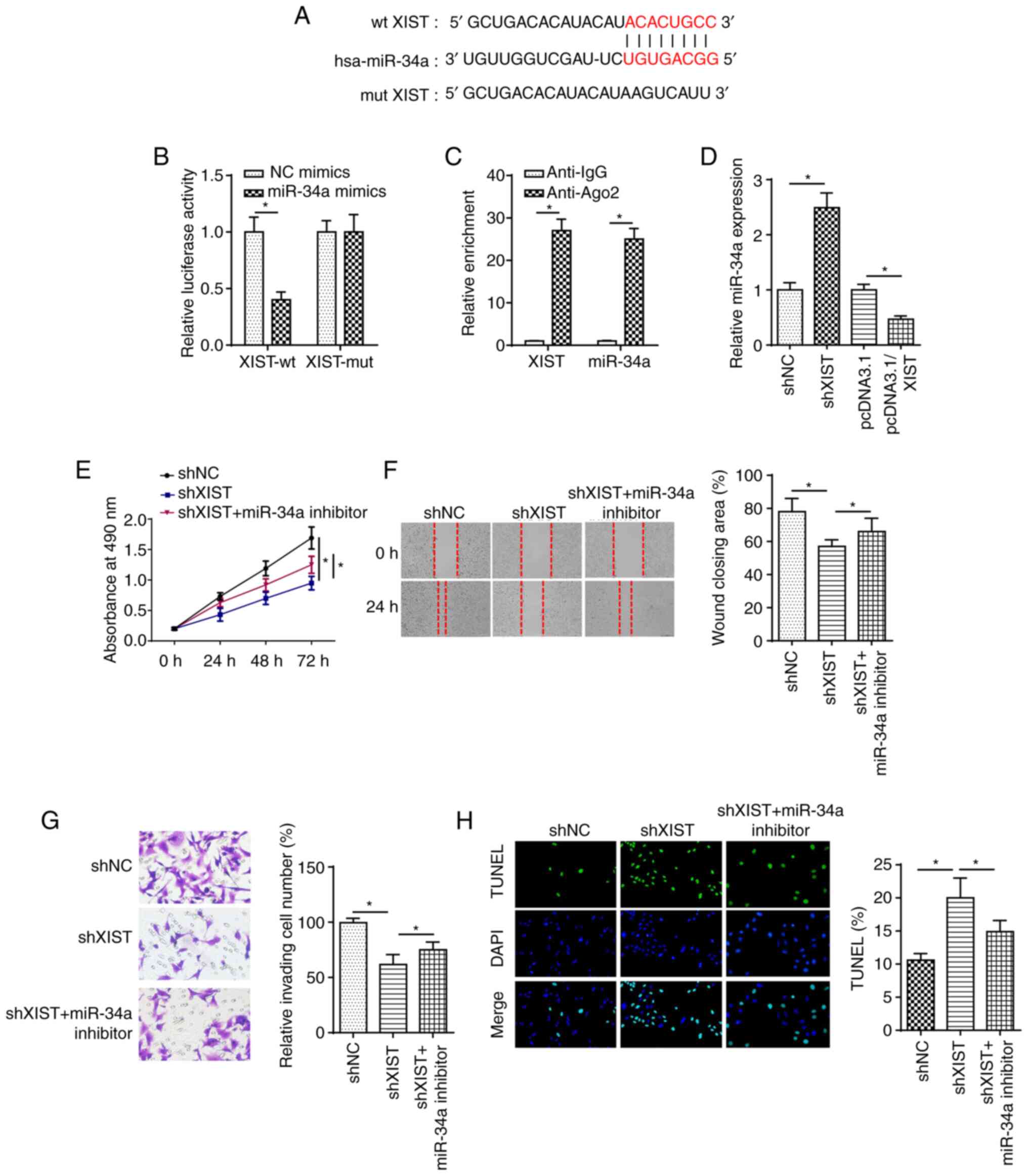 | Figure 4.XIST knockdown modulates diabetic
cataracts progression by targeting miR-34a in HG-induced lens
epithelial cells. (A) StarBase website was used to predict the
binding site between XIST and miR-34a. (B) Luciferase reporter
assay showed luciferase activity of XIST-wt or XIST-mut in SRA01/04
cells transfected with NC mimics or miR-34a mimics. (C) RIP assay
was performed to determine the enrichment of XIST and miR-34a in
Anti-IgG and Anti-Ago2. (D) Reverse transcription-quantitative PCR
showed the relative miR-34a expression in SRA01/04 cells
transfected with pcDNA3.1, pcDNA3.1/XIST, shNC or shXIST. (E) MTT,
(F) wound healing and (G) Transwell assays were used to analyze the
proliferation, migration and invasion of SRA01/04 cells transfected
with shNC, shXIST or shXIST + miR-34a inhibitor after treatment of
HG. (H) TUNEL assay determined the apoptosis in SRA01/04 cells
transfected with shNC, shXIST or shXIST + miR-34a inhibitor after
treatment of HG. n=3. *P<0.05. XIST, X-inactive specific
transcript; miR, microRNA; wt, wild-type; mut, mutant; sh, short
hairpin RNA; NG, normal glucose; HG, high glucose; NC, negative
control. |
SMAD2 is a target of miR-34a
To explore the downstream mechanism of miR-34a in DC
progression, the putative binding sites of miR-34a and SMAD2 were
predicted using the starBase website (Fig. 5A). Next, the luciferase reporter
assay revealed that miR-34a overexpression inhibited the luciferase
activity of SMAD2-wt, but did not change that of SMAD2-mut
(Fig. 5B). SMAD2 expression in DC
tissues was elevated and was likely correlated with that of XIST
(Fig. 5C and D). Furthermore, a
higher expression of SMAD2 was observed in HG-treated SRA01/04
cells compared with NG group cells (Fig. 5E). RT-qPCR indicated that miR-34a
expression was decreased in SRA01/04 cells transfected with miR-34a
inhibitor (Fig. 5F). In addition,
the mRNA and protein expression of SMAD2 in SRA01/04 cells was
decreased by miR-34a overexpression and increased by miR-34a
knockdown (Fig. 5G and H). In
combination, these findings indicated that miR-34a may target SMAD2
in SRA01/04 cells.
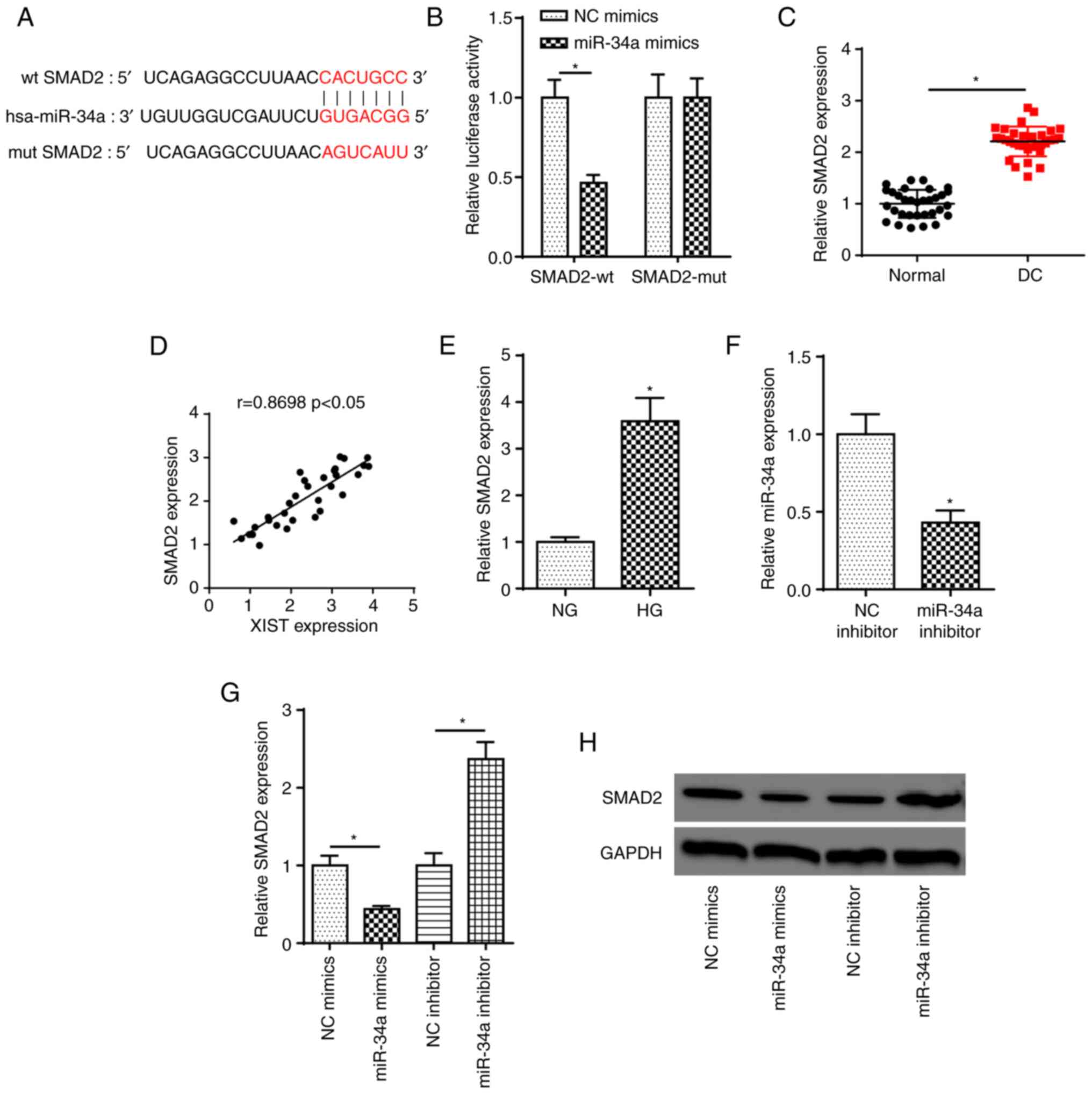 | Figure 5.SMAD2 is a target of miR-34a. (A)
StarBase website was used to predict the binding site between
miR-34a and SMAD2. (B) Luciferase reporter assay showed luciferase
activity of SMAD2-wt or SMAD2-mut in SRA01/04 cells transfected
with NC mimics or miR-34a mimics. *P<0.05. (C) RT-qPCR showed
the relative SMAD2 expression in DC tissues and normal samples.
*P<0.05. (D) Pearson's correlation analysis showed the
correlation between XIST and SMAD2 expression in DC tissues. (E)
RT-qPCR showed the relative SMAD2 expression in SRA01/04 cells
treated with HG. *P<0.05 vs. NG group. (F) RT-qPCR showed the
expression of miR-34a in SRA01/04 cells transfected with NC
inhibitor and miR-34a inhibitor. *P<0.05 vs. NC inhibitor. (G
and H) RT-qPCR and western blot assays showed the relative SMAD2
expression in SRA01/04 cells transfected with NC mimics or miR-34a
mimics and NC inhibitor or miR-34a inhibitor. n=3. *P<0.05.
SMAD2, SMAD family member 2; miR, microRNA; wt, wild-type; mut,
mutant; NC, negative control; RT-qPCR, reverse
transcription-quantitative PCR; DC, diabetic cataracts; XIST,
X-inactive specific transcript; NG, normal glucose; HG, high
glucose. |
XIST accelerates DC progression
through SMAD2 by sponging miR-34a in HG-treated LECs
Firstly, RT-qPCR showed that XIST and SMAD2
expression levels were significantly increased in SRA01/04 cells
transfected with XIST and SMAD2 overexpression plasmids,
respectively (Fig. 6A and B). The
mRNA and protein expression levels of SMAD2 were decreased by
miR-34a overexpression, which was rescued by XIST overexpression in
HG-treated cells (Fig. 6C and D).
To determine whether XIST mediated DC progression by modulating
SMAD2, SRA01/04 cells were transfected with shNC, shXIST and shXIST
+ pcDNA3.1/SMAD2. Functional assays revealed that XIST silencing
inhibited cell proliferation, migration and invasion in
HG-stimulated SRA01/04 cells, but these effects were abrogated by
SMAD2 overexpression (Fig. 6E-G).
Furthermore, XIST silencing promoted HG-stimulated SRA01/04 cell
apoptosis, which was reversed by SMAD2 overexpression (Fig. 6H). In combination, these findings
indicated that XIST may contribute to DC development by modulating
SMAD2 in HG-treated LECs.
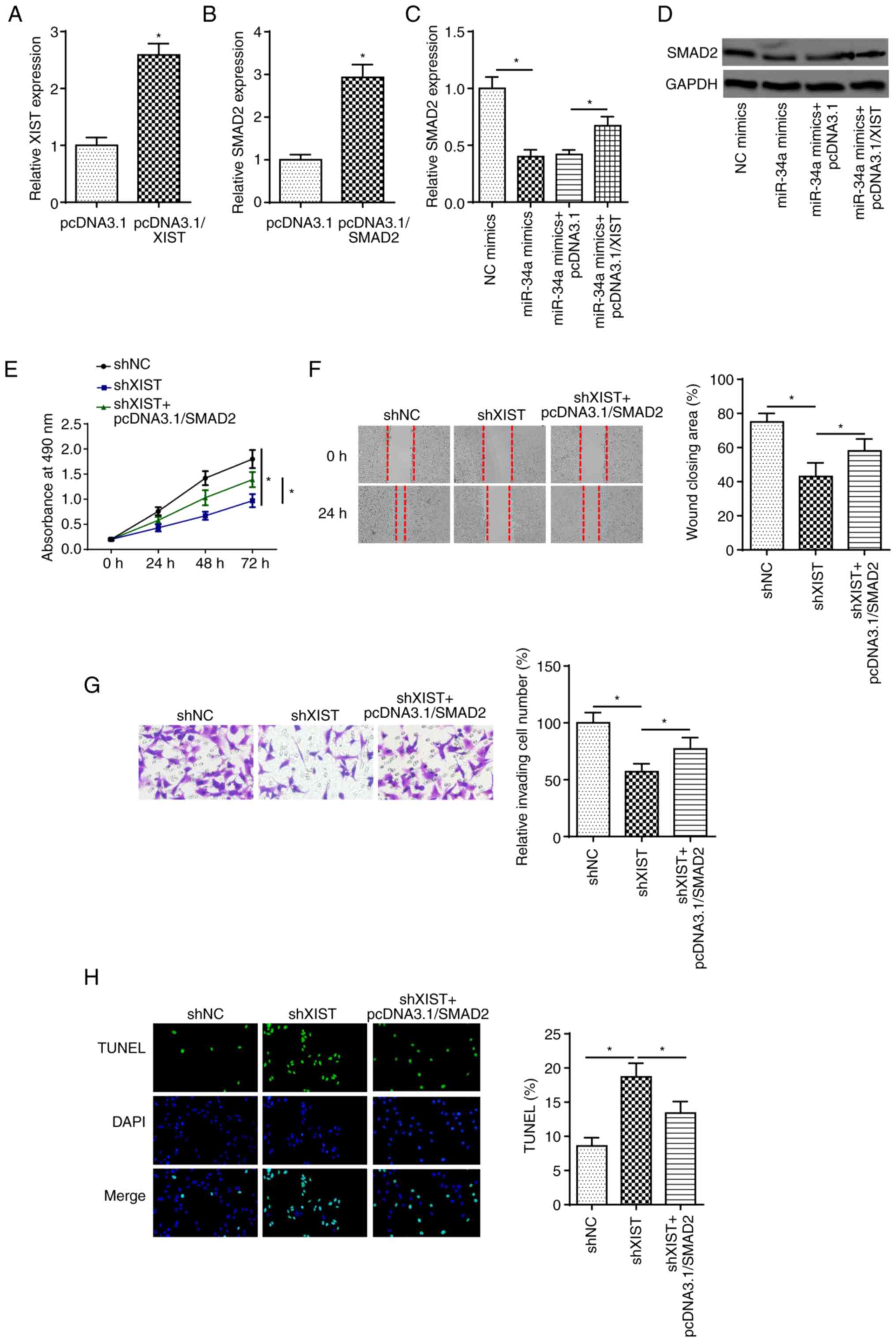 | Figure 6.XIST accelerates diabetic cataracts
progression through SMAD2 by sponging miR-34a in HG-treated lens
epithelial cells. (A) RT-qPCR showed the expression of XIST in
SRA01/04 cells transfected with pcDNA3.1 and pcDNA3.1/XIST. (B)
RT-qPCR showed the expression of SMAD2 in SRA01/04 cells
transfected pcDNA3.1 and pcDNA3.1/SMAD2. (C and D) RT-qPCR and
western blot assays determined SMAD2 expression in SRA01/04 cells
transfected with NC mimics, miR-34a mimics, miR-34a mimics +
pcDNA3.1 and miR-34a mimics + pcDNA3.1/XIST. (E) MTT, (F) wound
healing and (G) Transwell assays were used to analyze the
proliferation, migration and invasion of SRA01/04 cells transfected
with shNC, shXIST and shXIST + pcDNA3.1/SMAD2 after treatment of
HG. (H) TUNEL assay determined the apoptosis in SRA01/04 cells
transfected with shNC, shXIST and shXIST + pcDNA3.1/SMAD2 after
treatment of HG. n=3. *P<0.05. XIST, X-inactive specific
transcript; SMAD2, SMAD family member 2; miR, microRNA; HG, high
glucose; RT-qPCR, reverse transcription-quantitative PCR; NC,
negative control; sh, short hairpin RNA. |
Discussion
DC is an early ocular complication in diabetic
patients and one of the main causes of blindness (20). Certain studies have indicated that
lncRNAs affect multiple biological processes in DC (21,22). Therefore, understanding the
function of lncRNAs in LECs under HG conditions may enable the
discovery of novel targets for DC treatment. Previous studies
indicated that enhanced proliferation, migratory capacity,
invasiveness and resistance to apoptosis contribute to the
occurrence of cataracts (23).
Therefore, the present study investigated the role of XIST in
regulating cell proliferation, migration and invasion during DC
development. Collectively, these findings demonstrated that XIST
regulated SMAD2 expression to participate in the occurrence and
development of DC by sponging miR-34a.
XIST dysregulation has been observed in multiple
biological processes, such as cell proliferation, invasion and
apoptosis (24,25). XIST has been reported to
facilitate retinoblastoma progression by sponging miR-101 to
regulate the levels of zinc finger E-box binding homeobox 1 and 2
(26). In addition, XIST
knockdown repressed retinoblastoma development through the
miR-124/STAT3 axis (27). In the
present study, XIST expression was found to be increased in
posterior capsular tissues and HG-stimulated SRA01/04 cells. XIST
knockdown attenuated cell proliferation, migration and invasion,
and induced apoptosis in HG-treated LECs. These findings suggested
that XIST and miR-34a may represent promising targets for DC
treatment.
Extensive evidence has demonstrated that XIST may
serve as a ceRNA for miRNAs to regulate various diseases. For
example, lncRNA XIST facilitated cell proliferation and invasion by
targeting miR-137 to upregulate paxillin in non-small cell lung
cancer (28). lncRNA XIST
accelerated extracellular matrix degradation by acting as a ceRNA
of miR-1277-5p in osteoarthritis (29). A previous study showed that
miR-34a could restrain LEC proliferation and migration by reducing
c-Met (30). In addition, Han
et al (31) reported that
miR-34a suppressed the EMT of LECs by targeting notch receptor 1.
In the present study, miR-34a was found to be downregulated in DC
tissues and HG-stimulated SRA01/04 cells, and miR-34a
overexpression in HG-treated LECs attenuated DC development.
Furthermore, miR-34a deficiency reversed the effects of XIST
deletion on cell viability, migration, invasion and apoptosis in
HG-treated cells.
The nuclear translocation of SMAD2 has been
confirmed as a crucial regulator of cell viability, migration and
EMT (32,33). The present study confirmed that
SMAD2 was the downstream target of miR-34a. In addition, SMAD2 was
also found to be upregulated in DC tissues and HG-stimulated
SRA01/04 cells. XIST knockdown suppressed cell proliferation,
migration and invasion and accelerated apoptosis in HG-treated
SRA01/04 cells. However, these effects were weakened by SMAD2
overexpression. In combination, the results of the present study
demonstrated that XIST modulated DC development by regulating SMAD2
in HG-treated SRA01/04 cells.
In conclusion, this study revealed a novel mechanism
of XIST in DC progression. XIST facilitated cell proliferation,
migration and invasion, and decreased apoptosis in HG-treated LECs
through the miR-34a/SMAD2 axis, providing a novel biomarker for DC
treatment. However, this study was not without its limitations.
XIST expression was only examined in HG-treated LECs and DC
tissues. Further studies detecting XIST levels in LECs from
diabetic patients without cataracts are required.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CW and SZ designed the present study. CW, RZ and SZ
performed the experiments. CW and RZ analyzed the data and prepared
the figures. CW and SZ drafted the initial manuscript. All have
authors read and approved the final manuscript. CW and SZ confirm
the authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Shandong Zaozhuang Municipal Hospital (Zaozhuang, China) and all
participants signed informed consent forms prior to surgery.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
McCarty CA and Taylor HR: Recent
developments in vision research: Light damage in cataract. Invest
Ophthalmol Vis Sci. 37:1720–1723. 1996.PubMed/NCBI
|
|
2
|
Pollreisz A and Schmidt-Erfurth U:
Diabetic cataract-pathogenesis, epidemiology and treatment. J
Ophthalmol. 2010:6087512010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Harding JJ, Egerton M, van Heyningen R and
Harding RS: Diabetes, glaucoma, sex, and cataract: Analysis of
combined data from two case control studies. Br J Ophthalmol.
77:2–6. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kahn HA, Leibowitz HM, Ganley JP, Kini MM,
Colton T, Nickerson RS and Dawber TR: The Framingham Eye Study. II.
Association of ophthalmic pathology with single variables
previously measured in the framingham heart study. Am J Epidemiol.
106:33–41. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Martinez G and de Iongh RU: The lens
epithelium in ocular health and disease. Int J Biochem Cell Biol.
42:1945–1963. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang Y, Zhang G, Kang L and Guan H:
Expression profiling of DNA methylation and transcriptional
repression associated genes in lens epithelium cells of age-related
cataract. Cell Mol Neurobiol. 37:537–543. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mathieu EL, Belhocine M, Dao LT, Puthier D
and Spicuglia S: Functions of lncRNA in development and diseases.
Med Sci (Paris). 30:790–796. 2014.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gong W, Zhu G, Li J and Yang X: lncRNA
MALAT1 promotes the apoptosis and oxidative stress of human lens
epithelial cells via p38MAPK pathway in diabetic cataract. Diabetes
Res Clin Pract. 144:314–321. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang J, Zhao S and Tian F: SP1-mediated
lncRNA PVT1 modulates the proliferation and apoptosis of lens
epithelial cells in diabetic cataract via miR-214-3p/MMP2 axis. J
Cell Mol Med. 24:554–561. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dong Y, Wan G, Peng G, Yan P, Qian C and
Li F: Long non-coding RNA XIST regulates hyperglycemia-associated
apoptosis and migration in human retinal pigment epithelial cells.
Biomed Pharmacother. 125:1099592020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Q: XIST silencing alleviated
inflammation and mesangial cells proliferation in diabetic
nephropathy by sponging miR-485. Arch Physiol Biochem. Jul
15–2020.(Epub ahead of print). doi: 10.1080/13813455.2020.1789880.
View Article : Google Scholar
|
|
12
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang L, Cheng R and Huang Y: miR-30a
inhibits BECN1-mediated autophagy in diabetic cataract. Oncotarget.
8:77360–77368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zeng K, Feng QG, Lin BT, Ma DH and Liu CM:
Effects of microRNA-211 on proliferation and apoptosis of lens
epithelial cells by targeting SIRT1 gene in diabetic cataract mice.
Biosci Rep. 37:BSR201706952017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xiang W, Lin H, Wang Q and Chen W, Liu Z,
Chen H, Zhang H and Chen W: miR34a suppresses proliferation and
induces apoptosis of human lens epithelial cells by targeting E2F3.
Mol Med Rep. 14:5049–5056. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li H, Song H, Yuan X, Li J and Tang H:
miR-30a reverses TGF-β2-induced migration and EMT in posterior
capsular opacification by targeting Smad2. Mol Biol Rep.
46:3899–3907. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42((Database Issue)): D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang JH, Li JH, Shao P, Zhou H, Chen YQ
and Qu LH: starBase: A database for exploring microRNA-mRNA
interaction maps from Argonaute CLIP-Seq and Degradome-Seq data.
Nucleic Acids Res. 39((Database Issue)): D202–D209. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou Y, Li L, Li S, Li S, Zhao M, Zhou Q,
Gong X, Yang J and Chang J: Autoregenerative redox nanoparticles as
an antioxidant and glycation inhibitor for palliation of diabetic
cataracts. Nanoscale. 11:13126–13138. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ye W, Ma J, Wang F, Wu T, He M, Li J, Pei
R, Zhang L, Wang Y and Zhou J: lncRNA MALAT1 regulates miR-144-3p
to facilitate epithelial-mesenchymal transition of lens epithelial
cells via the ROS/NRF2/Notch1/Snail pathway. Oxid Med Cell Longev.
2020:81843142020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Y, Jiang SH, Liu S and Wang Q: Role of
lncRNA NEAT1 mediated by YY1 in the development of diabetic
cataract via targeting the microRNA-205-3p/MMP16 axis. Eur Rev Med
Pharmacol Sci. 24:5863–5870. 2020.PubMed/NCBI
|
|
23
|
Wang H and Zheng G: lncRNA NEAT1 promotes
proliferation, migration, invasion and epithelial-mesenchymal
transition process in TGF-β2-stimulated lens epithelial cells
through regulating the miR-486-5p/SMAD4 axis. Cancer Cell Int.
20:5292020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang H, Li H, Yu Y, Jiang Q, Zhang R, Sun
H, Xing W and Li Y: Long non-coding RNA XIST promotes the
progression of esophageal squamous cell carcinoma through sponging
miR-129-5p and upregulating CCND1 expression. Cell Cycle. 20:39–53.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang N, He JX, Jia GZ, Wang K, Zhou S, Wu
T and He XL: The lncRNA XIST promotes colorectal cancer cell growth
through regulating the miR-497-5p/FOXK1 axis. Cancer Cell Int.
20:5532020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cheng Y, Chang Q, Zheng B, Xu J, Li H and
Wang R: lncRNA XIST promotes the epithelial to mesenchymal
transition of retinoblastoma via sponging miR-101. Eur J Pharmacol.
843:210–216. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hu C, Liu S, Han M, Wang Y and Xu C:
Knockdown of lncRNA XIST inhibits retinoblastoma progression by
modulating the miR-124/STAT3 axis. Biomed Pharmacother.
107:547–554. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jiang H, Zhang H, Hu X and Li W: Knockdown
of long non-coding RNA XIST inhibits cell viability and invasion by
regulating miR-137/PXN axis in non-small cell lung cancer. Int J
Biol Macromol. 111:623–631. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang T, Liu Y, Wang Y, Huang X, Zhao W and
Zhao Z: Long non-coding RNA XIST promotes extracellular matrix
degradation by functioning as a competing endogenous RNA of
miR-1277-5p in osteoarthritis. Int J Mol Med. 44:630–642.
2019.PubMed/NCBI
|
|
30
|
Feng D, Zhu N, Yu C and Lou D:
MicroRNA-34a suppresses human lens epithelial cell proliferation
and migration via downregulation of c-Met. Clin Chim Acta.
495:326–330. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Han R, Hao P, Wang L, Li J, Shui S, Wang
Y, Ying M, Liu J, Tang X and Li X: MicroRNA-34a inhibits
epithelial-mesenchymal transition of lens epithelial cells by
targeting Notch1. Exp Eye Res. 185:1076842019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li H, Yuan X, Li J and Tang X: Implication
of Smad2 and Smad3 in transforming growth factor-β-induced
posterior capsular opacification of human lens epithelial cells.
Curr Eye Res. 40:386–397. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li J, Tang X and Chen X: Comparative
effects of TGF-β2/Smad2 and TGF-β2/Smad3 signaling pathways on
proliferation, migration, and extracellular matrix production in a
human lens cell line. Exp Eye Res. 92:173–179. 2011. View Article : Google Scholar : PubMed/NCBI
|