Introduction
Laryngeal cancer is an aggressive malignant tumor of
the head and neck, the main pathological type of which is laryngeal
squamous cell carcinoma (LSCC) (1). It has been shown that there are
nearly 170,000 new cases of laryngeal cancer each year worldwide,
and >90,000 individuals die of laryngeal cancer (2). The incidence of hypopharyngeal
cancer is ~1/4 of that of laryngeal cancer, of which laryngeal and
hypopharyngeal squamous cell carcinoma (LHSCC) is one type with a
poor prognosis (2), making its
treatment difficult. Currently, surgical resection of the tumor is
the standard treatment, while radiotherapy and chemotherapy have
also shown efficacy in trials (3,4).
However, some patients who undergo surgery and postoperative
adjuvant chemoradiotherapy may have the possibility of recurrence
(5). Therefore, it is important
to improve the prognosis of LHSCC by searching for new therapeutic
agents and targets.
The occurrence and progression of cancer are
regulated by multiple genes, and long non-coding RNAs (lncRNAs) are
RNAs with a total length >200 nucleotides but without
protein-coding ability (6). An
increasing number of studies have shown that lncRNAs play key roles
in the treatment and diagnosis of various cancer types (7–9).
It has been reported that lncRNA HLA complex group 18 (HCG18)
expression was upregulated in gastric cancer and colorectal cancer
tissues, and knockdown of HCG18 expression notably decreased the
proliferation, migration and invasion of cancer cells (10,11). Moreover, HCG18 has been found to
act as an oncogene in lung adenocarcinoma and promote tumor growth
(12). In addition, HCG18, as an
oncogenic lncRNA involved in the progression of cancer, is
associated with poor prognosis in nasopharyngeal carcinoma
(13). Nevertheless, its detailed
biological functions and molecular mechanism in LHSCC are yet to be
fully elucidated.
Recent findings have suggested that lncRNAs could be
involved in the progression of cancer by regulating the expression
levels of microRNAs (miRNAs/miRs) (6). miRNAs are a class of evolutionarily
conserved non-coding small molecular RNAs that have the function of
regulating gene expression at the translation level (14). Furthermore, existing evidence has
indicated that HCG18 regulates the migration and proliferation of
bladder cancer cells by regulating the expression of miR-34c-5p
(15). It has also been revealed
that HCG18 promotes the development of nasopharyngeal carcinoma by
regulating the expression of miR-140 (13). In addition, miR-133b plays
important roles in inflammation and tumors, where it has been
revealed that miR-133b was aberrantly expressed in numerous tumor
types, including colorectal cancer, esophageal squamous cell
carcinoma and human tongue squamous cell carcinoma (16–19). However, to the best of our
knowledge, the specific effects of miR-133b in LHSCC remain
unknown.
Therefore, the present study aimed to examine the
expression levels of HCG18 and miR-133b in LHSCC, and determine the
mechanism underlying the action of HCG18 and miR-133b so as to
propose a potential approach for targeted therapy of LHSCC.
Materials and methods
Ethics statement
This study was discussed and approved by the
Zhongshan Hospital of Traditional Chinese Medicine Affiliated to
Guangzhou University of Chinese Medicine Ethics Committee (approval
no. JN2020020208), and all tissue samples were confirmed to be
LHSCC. Written informed consent was obtained from the participating
patients.
Patient tissue specimens
In this experiment, 50 patients (male, 27; female,
23; age, 40–65 years) with LHSCC who underwent surgical resection
at Zhongshan Hospital of Traditional Chinese Medicine Affiliated to
Guangzhou University of Chinese Medicine (Zhongshan, China) between
February 2020 and April 2020 were selected. All patients
participating in the present study met the following criteria: i)
Had not received any pre-surgical therapy, including chemotherapy
and radiotherapy; ii) had not been diagnosed with infectious
diseases, autoimmunity-related diseases, etc. A total of 50 LHSCC
tissues and 50 normal tissues adjacent to cancer tissues (>5 cm
from tumor tissue) were collected from the patients.
Bioinformatics analysis
In the present study, the targeting relationships of
HCG18 and miR-133b were predicted by starBase (http://starbase.sysu.edu.cn/index.php),
and the targeting relationships of miR-133b and FGFR1 were
predicted by TargetScanHuman (http://www.targetscan.org/vert_72/).
Cell culture
The laryngeal cancer cell line, TU212, was purchased
from Tongpai Biological Technology Co., Ltd., and TU177 (cat. no.
QCB1408) was obtained from Qincheng Biological Technology Co., Ltd.
Both cell lines were cultured in RPMI-1640 complete medium (cat.
no. PM150110B; Procell Life Science & Technology Co., Ltd.)
containing 10% FBS (cat. no. 164210; Procell Life Science &
Technology Co., Ltd.) and 1% penicillin-streptomycin (cat. no.
PB180120; Procell Life Science & Technology Co., Ltd.), and
cells were grown in a cell incubator (cat. no. BC-J160; Boxun) at
37°C with 5% CO2.
Cell line transfection
In this experiment, miR-133b mimic (M; cat. no.
miR10000770-1-5), miR-133b M control (MC; cat. no.
miR1N0000001-1-5), miR-133b inhibitor (I; cat. no. miR20000770-1-5)
and miR-133b I control (IC; cat. no. miR2N0000001-1-5) were
purchased from Guangzhou RiboBio Co., Ltd. Short hairpin RNAs
(shRNAs; sh) targeting HCG18 (shHCG18) or FGFR1 (shFGFR1), as well
as their negative control (shNC) were synthesized from Shanghai
GenePharma Co., Ltd., and used for the knockdown of HCG18 and
FGFR1, respectively.
TU212 and TU177 cells were collected, the
concentration of which was adjusted to 5×104/ml. Then,
cells were cultured in 6-well plates. When cell confluence reached
80%, the two cell lines were transfected with M, MC, I, IC,
shHCG18, shFGFR1 or shNC (2,500 ng/well) for 48 h at 37°C using the
Lipofectamine™ 3000 Transfection Reagent (cat. no. L3000150;
Invitrogen; Thermo Fisher Scientific, Inc.). Transfected cells were
collected after 12 h and used for subsequent experiments.
Dual-luciferase reporter assay
The wild-type (WT) sequence of HCG18 (HCG18-WT;
5′-AGGCUAGGACAUUUGGACCAAC-3′), mutant (MUT) sequence of HCG18
(HCG18-MUT; 5′-AGGAGAGGACAUUUGUCACAGC-3′), WT sequence of FGFR1
(FGFR1-WT; 5′-CCCCUCCCAGAUCUUGGACCAAC-3′) and MUT sequence of FGFR1
(FGFR1-MUT; 5′-CCCCUCCCAGAUCUUGCCCGAUC-3′) were constructed into
dual-luciferase reporter pmirGLO vectors (Promega Corporation).
TU212 and TU177 cells were collected, and miR-133b M or MC (100
ng/well) were co-transfected with HCG18 reporter vectors containing
the WT or the MUT (100 ng/well) into TU212 cells for 48 h at 37°C;
miR-133b M or MC (100 ng/well) were also co-transfected with FGFR1
reporter vectors containing the WT or the MUT (100 ng/well) into
TU177 cells for 48 h at 37°C. Cell line transfection was performed
using the Lipofectamine 3000 Transfection Reagent. The activity of
luciferase was detected on the dual luciferase reporter system
(cat. no. GM3000; Promega Corporation) with a Dual Luciferase
Reporter Gene Assay kit (cat. no. RG027; Beyotime Institute of
Biotechnology), and Renilla luciferase activity was used as
an internal reference.
Cell Counting Kit-8 (CCK-8) assay
TU212 and TU177 cell lines in the logarithmic phase
were collected and were cultured in a 96-well plate (2,000
cells/well) after adjusting the concentration. After the
transfection, the viability of the transfected cells was detected
using a CCK-8 assay (cat. no. C0038; Beyotime Institute of
Biotechnology). After the transfected cells were routinely cultured
for 24 h, 10 µl CCK-8 solution was added to the appropriate wells,
and then the cells were cultured for another 4 h in a 37°C
incubator, according to the manufacturer's instructions. Finally,
the absorbance of each well was measured using a microplate reader
(ELx808; BioTek Instruments, Inc.) with an excitation wavelength of
450 nm.
Wound healing assay
The transfected TU212 and TU177 cell lines were
collected and cell concentration was adjusted, following which
~1×106 cells were added to each well of a 6-well plate
and cultured in RPMI-1640 complete medium (containing 10% FBS and
1% penicillin-streptomycin). When the confluence of the cell
reached ~100%, straight scratches were made using a sterile pipette
(200 µl) on a 6-well plate. Then, the cells were washed with PBS
(cat. no. 10010023; Thermo Fisher Scientific, Inc.) to remove the
scratched cells and serum-free medium was added to 6-well plate.
The cells were incubated at 37°C in an incubator for 24 h. Next,
the wound was observed and imaged using an inverted microscope
(magnification, ×100; DMi8; Leica Microsystems GmbH) at 0 and 24 h
after scratching. Cell migration rate = (scratch width at 0
h-scratch width at 24 h)/scratch width at 0 h.
Transwell assay
Matrigel (cat. no. M8370; Beijing Solarbio Science
& Technology Co., Ltd.) was thawed at 4°C overnight, and was
then diluted with pre-cooled serum-free medium. Subsequently, 20 µl
diluted Matrigel was added to the upper chamber of Transwell (cat.
no. CLS3374; Merck KGaA), and incubated for 1 h in an incubator at
37°C. The transfected TU212 and TU177 cells were collected and
suspended in serum-free medium to adjust the concentration to
5×105 cell/ml. When Matrigel was gelled, 200 µl cell
suspension was added into each upper Transwell chamber, and then
700 µl medium containing 15% FBS was added to the corresponding
lower chamber. Next, the cells were incubated in an incubator at
37°C for 24 h. The chamber was removed and the residual cells were
wiped off with a cotton swab, after which the cells were fixed with
4% paraformaldehyde (cat. no. P6148; Sigma-Aldrich; Merck KGaA) and
stained with 0.1% crystal violet (cat. no. C0775; Sigma-Aldrich;
Merck KGaA) for 15 min at room temperature. Finally, the cells were
observed and counted under an inverted microscope (magnification,
×200).
Immunohistochemistry
After collection, the specimens of LHSCC tissues and
normal tissues were fixed via immersion in 4% paraformaldehyde for
24 h at 4°C. Then, the tissues specimens were routinely embedded in
paraffin (cat. no. P3558; Sigma-Aldrich; Merck KGaA) and then
sectioned (thickness, 4 µm) using a paraffin slicer (cat. no.
E0972; Beyotime Institute of Biotechnology). The paraffin sections
were deparaffinized with xylene (cat. no. 9990501; Thermo Fisher
Scientific, Inc.) and hydrated with different concentrations of
alcohol (100, 95, 90, 80 and 70%; cat. no. E7023; Sigma-Aldrich;
Merck KGaA). The blocking solution was prepared using Triton X-100
(cat. no. P1080; Beyotime Institute of Biotechnology) and
H2O2 (cat. no. H112517; Shanghai Aladdin
Biochemical Technology Co., Ltd.). The paraffin sections were
immersed in blocking solution for 30 min at room temperature in the
dark, and then in 10% normal goat serum (cat. no. SL038; Beijing
Solarbio Science & Technology Co., Ltd.) for 20 min at 37°C.
Next, sections were incubated firstly with anti-FGFR1 antibody
(cat. no. ab10646; Abcam; 1:200) at 4°C for 12 h and then with
horseradish peroxidase-labeled secondary antibody (rabbit IgG; cat.
no. ab205718; Abcam; 1:2,000) at 37°C for 30 min. The sections were
washed with PBS, treated with DAB (cat. no. DA1015; Beijing
Solarbio Science & Technology Co., Ltd.) at room temperature
for 10 min, and counterstained with Mayer's hematoxylin solution
(cat. no. G1080; Beijing Solarbio Science & Technology Co.,
Ltd.) for 2 min at room temperature. Then, the sections were
treated with different concentrations of alcohol (50% for 2 min;
70% for 2 min; 95% for 1 min; 100% for 1 min) at room temperature,
rinsed using xylene and mounted with Neutral balsam (cat. no.
G8590; Beijing Solarbio Science & Technology Co., Ltd.).
Finally, the sections were imaged (magnification, ×200) under an
inverted microscope. Immunohistochemistry revealed that FGFR1 was
stained brownish-yellow. Briefly, the mean gray value (staining
intensity) of positive cells was analyzed using ImageJ software
(version 1.50i; National Institutes of Health), and the cells were
then classified as high-positive (+++), positive (++), low-positive
(+) and negative (0) according to the degree of staining.
Meanwhile, the positive area (staining area) of the sections was
determined using ImageJ software, and then the percentage of cells
with different grades of staining in the total staining area was
calculated.
Reverse transcription-quantitative PCR
(RT-qPCR)
The collected LHSCC tissues were minced and
transferred into centrifuge tubes, to which the transfected LHSCC
cells were transferred as well. Then, the extraction of total RNA
from cells and tissue samples was performed using
TRIzol® reagent (cat. no. 15596018; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. The
concentration and purity of the RNA were assessed by measuring the
absorbance values of the RNA solutions using a spectrophotometer
(ND-ONEC-W; Thermo Fisher Scientific, Inc.). The RT-PCR reaction
system was configured using the One Step RT-PCR SuperMix kit (cat.
no. T2240; Beijing Solarbio Science & Technology Co., Ltd.).
The RT-PCR reaction system was performed and the expressions of
genes were measured on the RT-PCR system (Bio-Rad iQ5; Bio-Rad
Laboratories, Inc.) under the conditions: 50°C, 20 min, 1 cycle;
95°C, 3 min, 1 cycle; 95°C, 10 sec, 60°C, 15 sec, 72°C, 30 sec, 40
cycles; and 72°C, 5 min, 1 cycle. GAPDH and U6 were used as
reference genes, and sequences of all primers used are shown in
Table I. The results of RT-qPCR
were analyzed using the 2−ΔΔCq method (12).
 | Table I.All primers used in reverse
transcription-quantitative PCR experiments. |
Table I.
All primers used in reverse
transcription-quantitative PCR experiments.
| Gene | Forward sequence
(5′-3′) | Reverse sequence
(5′-3′) |
|---|
| miR-133b |
AAACCTGGCGGCCACGCTAC |
GACCGTGGTCCACTGCAGGC |
| HCG18 |
TTGGCTTCAGTCCTGTTCATCAG |
ACCTTGCACACTGTCTCTTG |
| FGFR1 |
CCCGTAGCTCCATATTGGACA |
TTTGCCATTTTTCAACCAGCG |
| U6 |
CTCGCTTCGGCAGCACATATACT |
ACGCTTCACGAATTTGCGTGTC |
| GAPDH |
GCAAGTTCAACGGCACAG |
GCCAGTAGACTCCACGACAT |
Western blotting
The transfected TU212 and TU177 cell lines were
collected in centrifuge tubes, and the cells were washed twice with
precooled PBS (each time for 2 min), and then the appropriate
amount of RIPA lysis buffer (cat. no. P0013C; Beyotime Institute of
Biotechnology) was added to the centrifuge tubes to extract total
protein from the LHSCC cells, after which the concentration of the
total protein was measured using BCA protein kit (cat. no. P0010;
Beyotime Institute of Biotechnology). The SDS-PAGE gel (6, 8 or
10%) was prepared using an SDS-PAGE Gel Quick Preparation kit (cat.
no. P0012AC; Beyotime Institute of Biotechnology), and an
appropriate amount (20 µl, 2 µg/µl) of protein sample was added to
the sample well before electrophoresis. After the completion of
electrophoresis, the separated protein was transferred to a PVDF
membrane (cat. no. 88518; Thermo Fisher Scientific, Inc.), which
was then immersed in blocking solution containing 5% skimmed milk
at room temperature for 1 h. Subsequently, the PVDF membrane was
washed in 1X TBS-0.1% Tween-20 (TBST) solution (cat. no. ST673;
Beyotime Institute of Biotechnology) three times (each time for 10
min), and then immersed in diluted primary antibody solution at 4°C
overnight after the dilution of the primary antibody with TBST to
the appropriate concentration. The following day, the PVDF membrane
was soaked in diluted secondary antibody solution at 37°C for 1.5 h
after being washed with TBST solution. Then, the PVDF membrane was
washed with TBST solution again, and the Pierce™ ECL Western
Blotting substrate (cat. no. 32106; Thermo Fisher Scientific, Inc.)
was used for visualization following the collection of protein
bands. The signals were finally read using a ChemiDoc XRS+ system
(Bio-Rad Laboratories, Inc.). All the expressions of proteins were
normalized against GAPDH, and the information for the antibodies
used is listed in Table II.
 | Table II.All antibody information for those
used in western blotting assay. |
Table II.
All antibody information for those
used in western blotting assay.
| ID | Cat. no. | Company | Molecular weight
(kDa) | Dilution ratio |
|---|
| p-PI3K | ab182651 | Abcam | 84 | 1:1,000 |
| PI3K | 4249 | CST | 110 | 1:1,000 |
| p-AKT | 4060 | CST | 60 | 1:2,000 |
| AKT | 4685 | CST | 60 | 1:1,000 |
| p53 | 2527 | CST | 53 | 1:1,000 |
| Bax | 5023 | CST | 20 | 1:1,000 |
| Bcl-2 | 3498 | CST | 26 | 1:1,000 |
| FGFR1 | 9740 | CST | 120 | 1:1,000 |
| Rabbit IgG | ab205718 | Abcam | N/A | 1:5,000 |
| GAPDH | 5174 | CST | 36 | 1:1,000 |
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 8.0 (GraphPad Software, Inc.). Measurement data are presented
as the mean ± SD and were indicative of three experiments
independently. An independent sample t-test was used for comparison
between two groups (paired sample t-test was used for comparison
between cancer tissue and normal tissues), and one-way anova was
performed for comparison between multiple groups followed by
Tukey's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
HCG18 expression is increased and
miR-133b expression is decreased in LHSCC tissues
To confirm the roles of HCG18 and miR-133b in LHSCC,
the expression levels of miR-133b and HCG18 in LHSCC tissues and
normal tissues were examined. It was found that the expression
level of miR-133b was downregulated in LHSCC tissues in comparison
with adjacent normal tissue (P<0.001; Fig. 1A). However, the expression level
of HCG18 was upregulated in LHSCC tissues compared with adjacent
normal tissue (P<0.001; Fig.
1B).
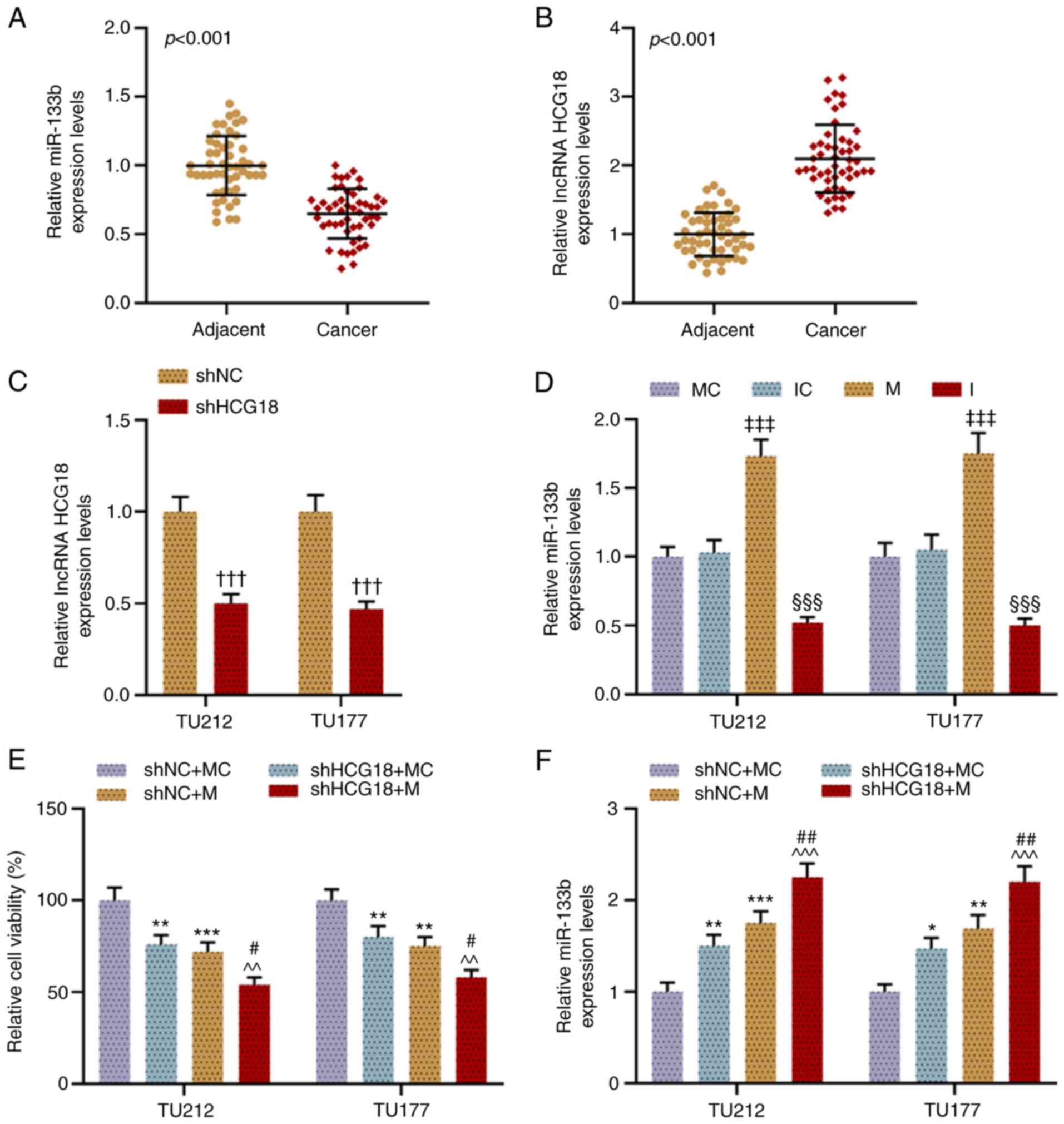 | Figure 1.HCG18 expression is increased and
that of miR-133b is decreased in LHSCC tissues, and shHCG18 and
miR-133b M inhibit LHSCC cell activity while enhancing miR-133b
expression. (A) Expression level of miR-133b in LHSCC tissues was
examined via RT-qPCR, and U6 was used as a reference gene.
Expression level of HCG18 in (B) LHSCC tissues and (C) transfected
LHSCC cells was examined via RT-qPCR, and GAPDH was used as a
reference gene. (D) Expression level of miR-133b in transfected
LHSCC cells was examined via RT-qPCR, and U6 was used as a
reference gene. (E) Viability of transfected TU212 and TU177 cell
lines was evaluated using a Cell Counting Kit-8 assay. (F)
Expression of miR-133b in transfected TU212 and TU177 cells was
examined via RT-qPCR, and U6 was used as a reference gene.
†††P<0.001 vs. shNC; ‡‡‡P<0.001 vs. MC;
§§§P<0.001 vs. IC; *P<0.05, **P<0.01,
***P<0.001 vs. shNC + MC; ^^P<0.01,
^^^P<0.001 vs. shHCG18 + MC; #P<0.05,
##P<0.01 vs. shHCG18 + M. LHSCC, laryngeal and
hypopharyngeal squamous cell carcinoma; RT-qPCR, reverse
transcription-quantitative PCR; M, mimic; MC, mimic control; IC,
inhibitor control; I, inhibitor; miR, microRNA; sh, short hairpin
RNA; shNC, short hairpin negative control; lncRNA, long non-coding
RNA; HCG18, HLA complex group 18. |
shHCG18 and miR-133b M inhibit LHSCC
cell viability, while enhancing miR-133b expression
As shown in Fig. 1C
and D, the transfection efficiency was examined via RT-qPCR,
which demonstrated that shHCG18 inhibited the expression of HCG18
in comparison with the shNC group (P<0.001). Moreover, the
miR-133b M promoted miR-133b expression (P<0.001), while the
miR-133b I knocked down its expression, compared with their
respective control groups (P<0.001). In addition, it was found
that HCG18 knockdown and miR-133b overexpression inhibited the
viability of LHSCC cell, while enhancing the expression level of
miR-133b, when compared with the shNC + MC group (P<0.05;
Fig. 1E and F). Moreover, when
miR-133b M and shHCG18 were co-transfected, the inhibitory effects
on cell viability and the promotive effects on the expression of
miR-133b were notably enhanced as compared with the shHCG18 + MC
group and shNC + M group (P<0.05; Fig. 1E and F).
HCG18 can competitively bind with
miR-133b in LHSCC cells, and miR-133b I promoted cell viability,
migration and invasion, which is reversed by shHCG18
The targeting relationship of miR-133b and HCG18 was
predicted using bioinformatics analysis (Fig. 2A). Next, a dual-luciferase
reporter assay was performed to confirm the relationship, from
which miR-133b was successfully determined as the candidate miRNA
that could competitively bind with HCG18. For instance, when TU212
cells were co-transfected with HCG18-WT and miR-133b M, the
luciferase activity of the cells was lower than that of cells
co-transfected with HCG18-WT and miR-133b MC, while there was no
notable difference in the luciferase activity of each group of
cells when TU212 cells were co-transfected with HCG18-MUT and
miR-133b M or MC (P<0.01; Fig.
2B). Moreover, it was found that shHCG18 increased miR-133b
expression, while the miR-133b I decreased this, as compared with
the shNC + IC group (P<0.05). After the co-transfection of
shHCG18 and miR-133b I into LHSCC cells, shHCG18 reversed the
inhibitory effect of miR-133b I on the expression level of miR-133b
(P<0.001; Fig. 2C).
Additionally, the viability (Fig.
2D), migration (Fig. 3A and
B) and invasion (Fig. 3C and
D) of LHSCC cells after transfection were evaluated, and
results suggested that shHCG18 decreased the viability, migration
and invasion of LHSCC cells, while miR-133b I increased cell
viability, migration and invasion as compared with the shNC + IC
group (P<0.05). It was found that shHCG18 counteracted the
promoting effect of the miR-133b I on the viability, migration and
invasion of transfected LHSCC cells in comparison with the shHCG18
+ IC group and I +shNC group (P<0.05).
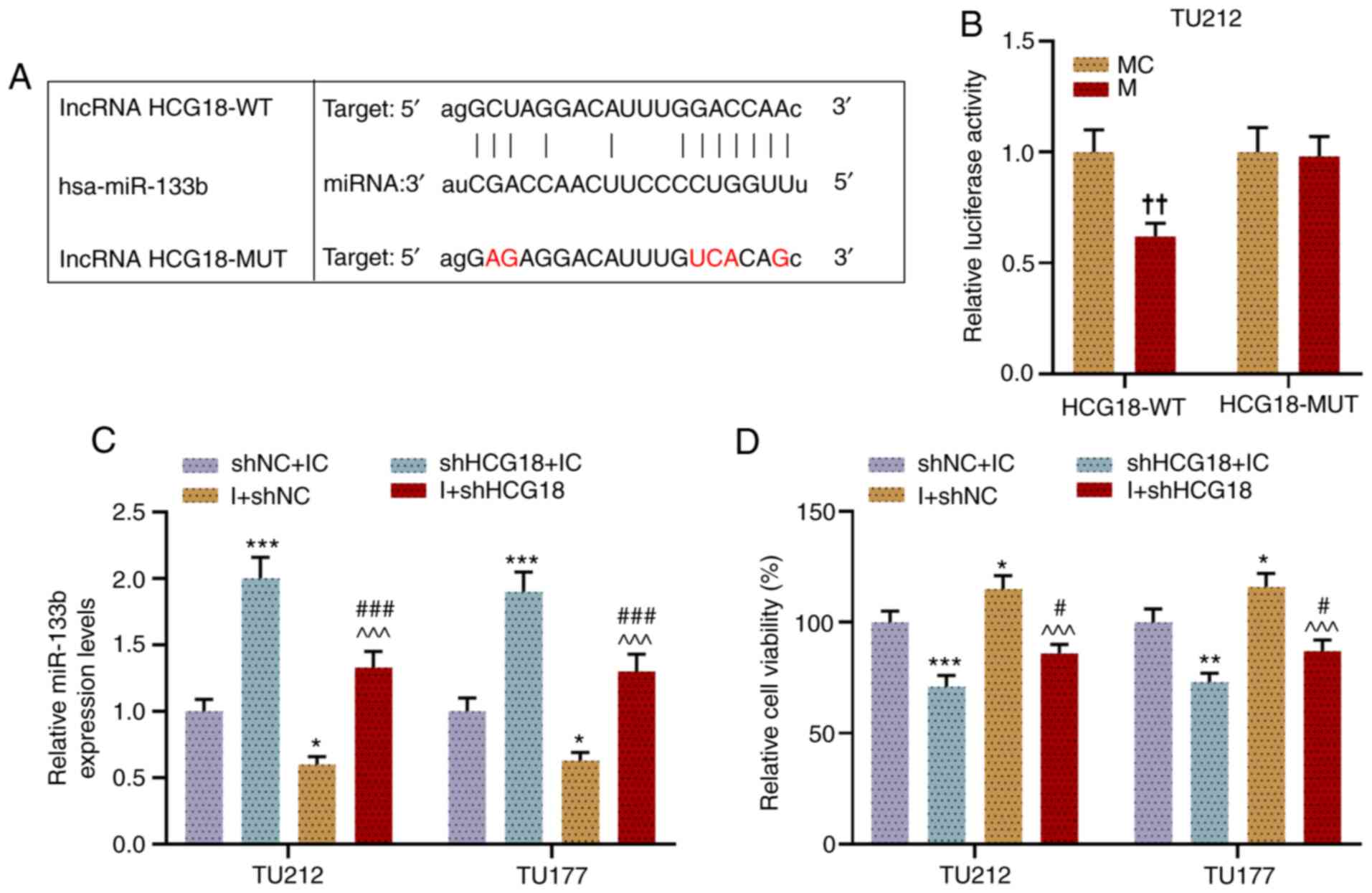 | Figure 2.HCG18 can competitively bind with
miR-133b in laryngeal and hypopharyngeal squamous cell cells, and
miR-133b I promotes cell viability, which is reversed by shHCG18.
(A) Binding sites of miR-133b and HCG18 were predicted using
starBase. (B) Binding sites of miR-133b and HCG18 were validated
using a dual-luciferase reporter assay. (C) Expression level of
miR-133b in transfected TU212 and TU177 cells was examined via
reverse transcription-quantitative PCR, and U6 was used as a
reference gene. (D) Viability of transfected TU212 and TU177 cell
lines was evaluated using a Cell Counting Kit-8.
††P<0.01 vs. MC; *P<0.05, **P<0.01,
***P<0.001 vs. shNC + IC; ^^^P<0.001 vs. shHCG18 +
IC; #P<0.05, ###P<0.001 vs. shHCG18 +
I. M, mimic; MC, mimic control; IC, inhibitor control; I,
inhibitor; miR, microRNA; sh, short hairpin RNA; shNC, short
hairpin negative control; lncRNA, long non-coding RNA; HCG18, HLA
complex group 18; WT, wild-type; MUT, mutant. |
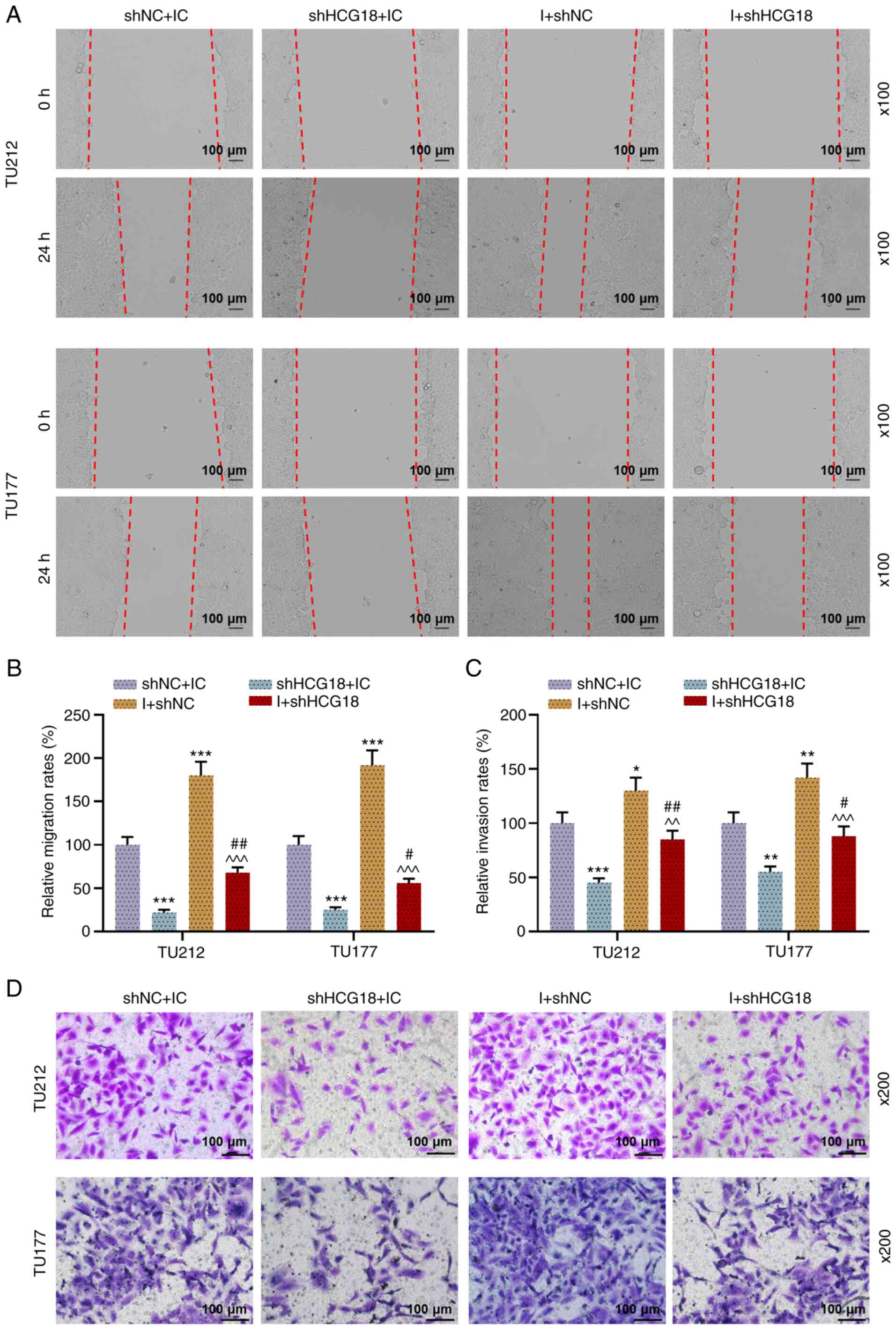 | Figure 3.miR-133b I promotes cell migration
and invasion, which is abrogated by shHCG18. (A and B) TU212 and
TU177 cell migration after transfection was examined using a wound
healing assay (magnification, ×100). (C and D) Invasion of TU212
and TU177 cell lines after transfection was examined using a
Transwell assay (magnification, ×200). *P<0.05, **P<0.01,
***P<0.001 vs. shNC + IC; ^^P<0.01,
^^^P<0.001 vs. shHCG18 + IC; #P<0.05,
##P<0.01 vs. shHCG18 + I. M, mimic; MC, mimic
control; IC, inhibitor control; I, inhibitor; miR, microRNA; sh,
short hairpin RNA; shNC, short hairpin negative control; lncRNA,
long non-coding RNA; HCG18, HLA complex group 18. |
miR-133b I promotes the expression
levels of Bcl-2, p-PI3K and p-AKT, yet inhibits the expression
levels of p53 and Bax in LHSCC cells, which are reversed by
shHCG18
Subsequently, the expression levels of p-PI3K, PI3K,
p-AKT, AKT, p53, Bax and Bcl-2, and the ratios of p-PI3K/PI3K and
p-AKT/AJT in transfected TU212 (Fig.
4A-E) and TU177 cells (Fig.
4F-J) were examined. It was found that the miR-133b I promoted
Bcl-2 expression and the ratios of p-PI3K/PI3K and p-AKT/AKT, yet
suppressed the expression levels of p53 and Bax compared with the
shNC + IC group, while knockdown of HCG18 had the opposite effects
in transfected LHSCC cells (P<0.05). Moreover, shHCG18 reversed
the effect of the miR-133b I on the expression levels of p-PI3K,
p-AKT, p53, Bax and Bcl-2 in transfected LHSCC cells, when compared
with the shHCG18 + IC group and I + shNC group (P<0.05).
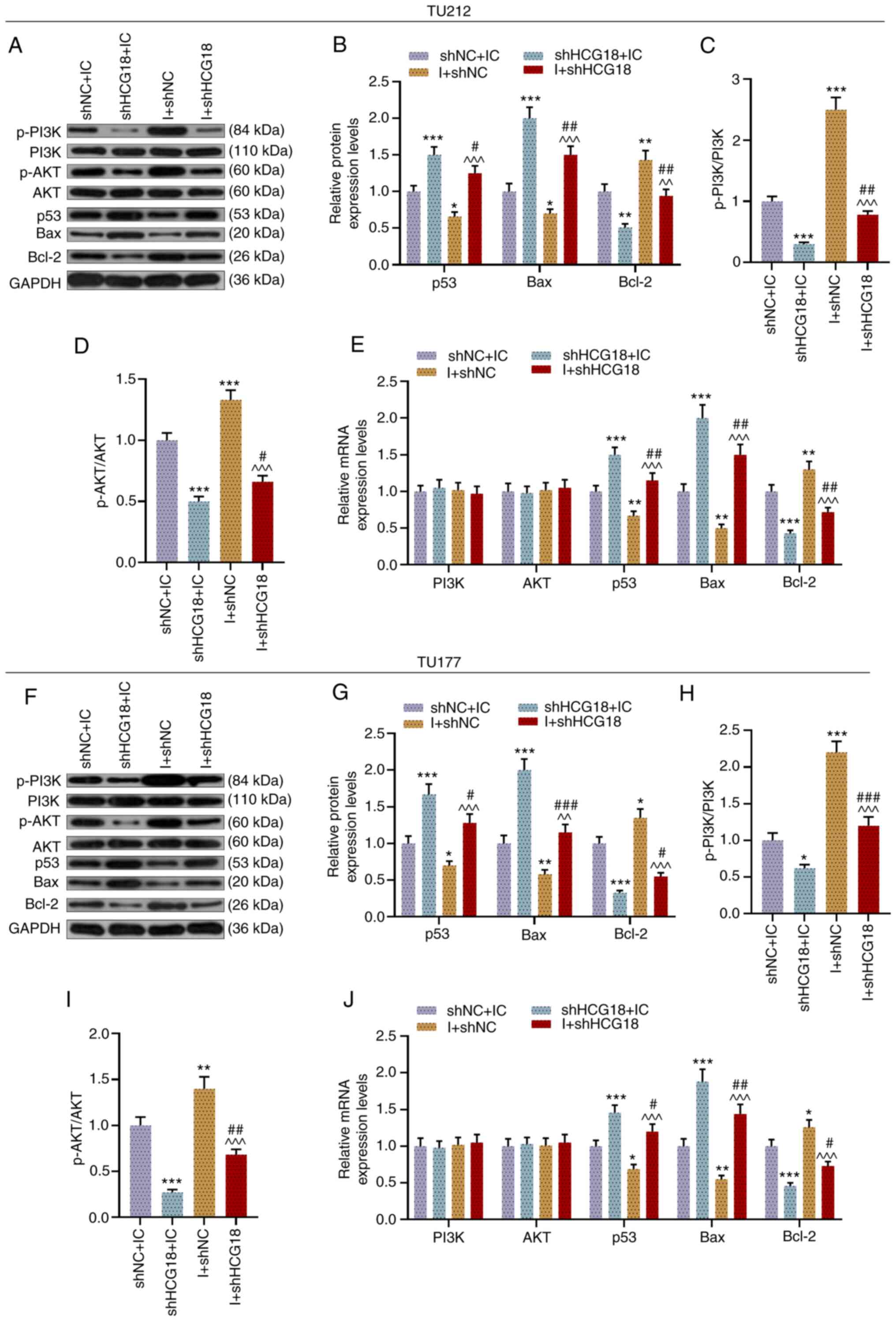 | Figure 4.miR-133b I promotes the expression
levels of Bcl-2, p-PI3K and p-AKT, but inhibits p53 and Bax
expression in laryngeal and hypopharyngeal squamous cell carcinoma
cells, while shHCG18 causes the opposite effects. (A) Protein
expression levels of p-PI3K, PI3K, p-AKT, AKT, p53, Bax and Bcl-2
in transfected TU212 cells were (B) semi-quantified via western
blotting, and GAPDH was used as an internal loading control. Ratios
of (C) p-PI3K to PI3K and (D) p-AKT to AKT in transfected TU212
cells were analyzed on the basis of the results of western blotting
assay. (E) Expression levels of PI3K, AKT, p53, Bax and Bcl-2 in
transfected TU212 cells were measured via RT-qPCR, and GAPDH was
used as a reference gene. (F and G) Expression levels of p-PI3K,
PI3K, p-AKT, AKT, p53, Bax and Bcl-2 in transfected TU177 cells
were calculated via western blotting, and GAPDH was used as an
internal loading control. Ratios of (H) p-PI3K to PI3K and (I)
p-AKT to AKT in transfected TU177 cells were analyzed using western
blotting. (J) Expression levels of PI3K, AKT, p53, Bax and Bcl-2 in
transfected TU177 cells were determined via RT-qPCR, and GAPDH was
used as a reference gene. *P<0.05, **P<0.01, ***P<0.001
vs. shNC + IC; ^^P<0.01, ^^^P<0.001 vs.
shHCG18 + IC; #P<0.05, ##P<0.01,
###P<0.001 vs. shHCG18 + I. p-, phosphorylated;
RT-qPCR, reverse transcription-quantitative PCR; M, mimic; MC,
mimic control; IC, inhibitor control; I, inhibitor; miR, microRNA;
sh, short hairpin RNA; shNC, short hairpin negative control;
lncRNA, long non-coding RNA; HCG18, HLA complex group 18. |
FGFR1 is increased and can
competitively bind with miR-133b in LHSCC cells
The expression level of FGFR1 in LHSCC tissues was
examined via RT-qPCR (Fig. 5A).
The results demonstrated that FGFR1 expression was significantly
higher in LHSCC tissues compared with in adjacent normal tissue
(P<0.001). Also, the level of FGFR1 was examined using
immunohistochemistry. Similarly, it was demonstrated that FGFR1
expression was significantly higher in LHSCC tissues compared with
that in adjacent normal tissue (Fig.
5B and C). Moreover, it was observed that shFGFR1 notably
decreased FGFR1 expression in comparison with the shNC group
(P<0.05; Fig. 5D-F).
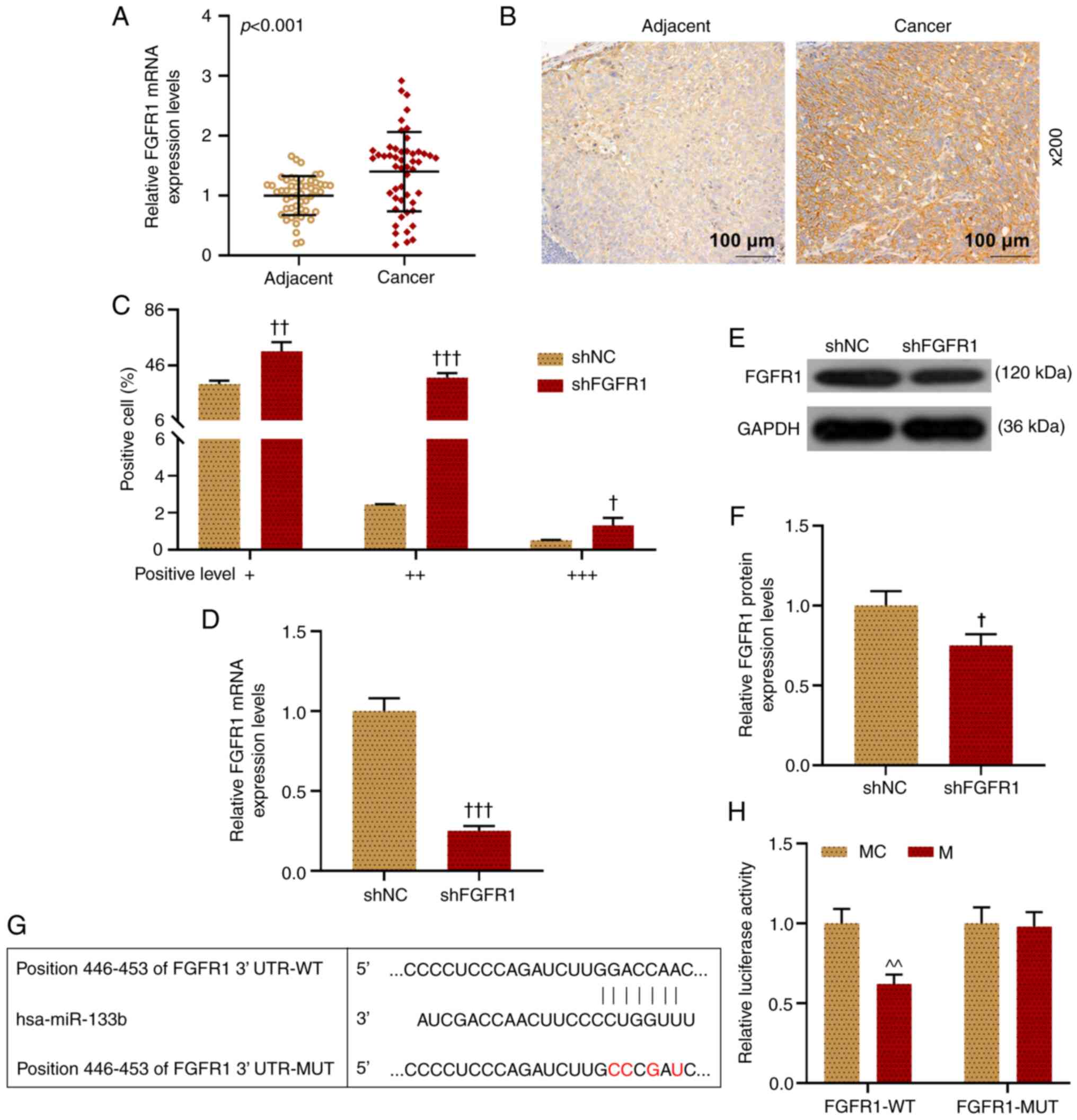 | Figure 5.FGFR1 expression is increased in
LHSCC, and miR-133b can bind with FGFR1 in TU212 cells. (A)
Expression level of FGFR1 in LHSCC tissues was examined via
RT-qPCR, and GAPDH was used as a reference gene. (B and C)
Expression level of FGFR1 in LHSCC tissues was examined using
immunohistochemistry. (D) Expression level of FGFR1 in transfected
TU212 cells was examined via RT-qPCR, and GAPDH was used as a
reference gene. (E and F) FGFR1 expression in transfected TU212
cells was examined via western blotting, and GAPDH was used as an
internal loading control. The binding sites of miR-133b and FGFR1
were predicted using (G) TargetScanHuman and (H) validated using a
dual-luciferase reporter assay. †P<0.05,
††P<0.01, †††P<0.001 vs. shNC;
^^P<0.01 vs. MC. LHSCC, laryngeal and hypopharyngeal
squamous cell carcinoma; RT-qPCR, reverse
transcription-quantitative PCR; M, mimic; MC, mimic control; IC,
inhibitor control; I, inhibitor; miR, microRNA; sh, short hairpin
RNA; shNC, short hairpin negative control; FGFR1, fibroblast growth
factor receptor 1; WT, wild-type; MUT, mutant; UTR, untranslated
region. |
Next, the current study successfully predicted
(Fig. 5G) and determined
(Fig. 5H) the targeting
relationship of miR-133b and FGFR1, as demonstrated in the results
that when TU212 cells were co-transfected with FGFR1-WT and
miR-133b M or MC, the luciferase activity after co-transfected with
FGFR1-WT and miR-133b M was lower than that of cells co-transfected
with FGFR1-WT and miR-133b MC, while there was no notable
difference in the luciferase activity of cells in each group when
TU212 cells were co-transfected with FGFR1-MUT and miR-133 M or MC
(P<0.01). In addition, it was identified that miR-133b M
significantly decreased the expression level of FGFR1 in
transfected TU212 cells in comparison with the MC group (P<0.05;
Fig. 6A-C).
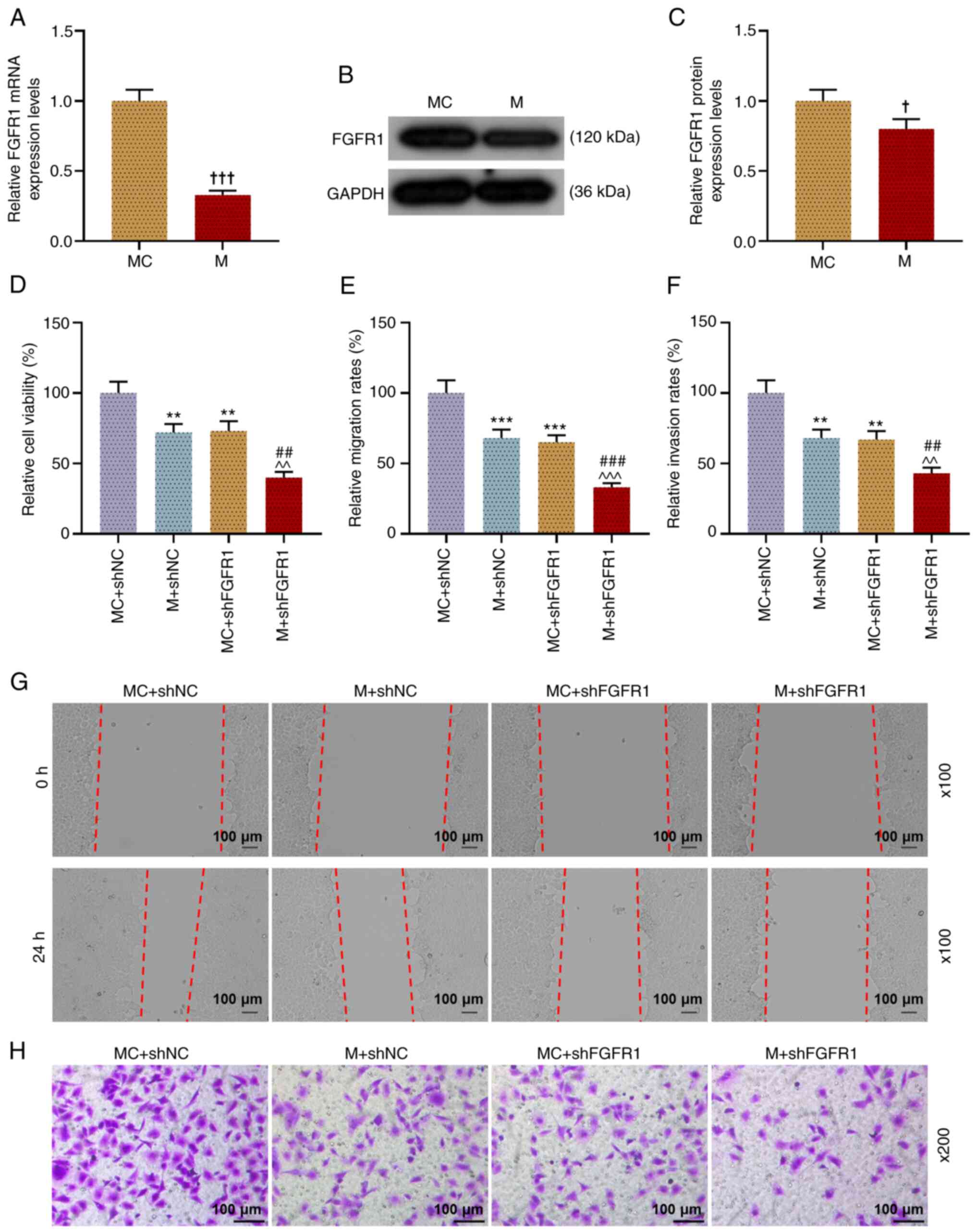 | Figure 6.miR-133b can competitively bind with
FGFR1 in TU212 cells, and miR-133b M and shFGFR1 inhibit cell
viability, migration and invasion. (A) FGFR1 expression in
transfected TU212 cells was examined via RT-qPCR, and GAPDH was
used as a reference gene. (B and C) FGFR1 expression in transfected
TU212 cells was examined via western blotting, and GAPDH was used
as an internal loading control. (D) Viability of transfected TU212
cells was evaluated using a Cell Counting Kit-8 assay. (E and G)
Migration of TU212 cell line after transfection was examined using
a wound healing assay (magnification, ×100). (F and H) Invasion of
TU212 cell line after transfection was examined using a Transwell
assay (magnification, ×200). †P<0.05,
†††P<0.001 vs. MC; **P<0.01, ***P<0.001 vs.
shNC + MC; ^^P<0.01, ^^^P<0.001 vs.
shNC + M; ##P<0.01, ###P<0.001 vs.
shHCG18 + MC. RT-qPCR, reverse transcription-quantitative PCR; M,
mimic; MC, mimic control; IC, inhibitor control; I, inhibitor; miR,
microRNA; sh, short hairpin RNA; shNC, short hairpin negative
control; FGFR1, fibroblast growth factor receptor 1. |
shFGFR1 enhanced the inhibitory effect
of miR-133b M on the viability, migration and invasion of LHSCC
cells
Next, the present study determined the viability
(Fig. 6D), migration (Fig. 6E and G), and invasion (Fig. 6F and H) of TU212 cells after
transfection. These experimental results indicated that miR-133b M
and shFGFR1 significantly inhibited the viability, migration and
invasion of cells (P<0.01); however, when TU212 cells were
co-transfected with miR-133b M and shFGFR1, the viability,
migration and invasion of transfected TU212 cells were lower
compared with those of cells in the M + shNC and MC + shFGFR1
groups (P<0.01). Thus, it was suggested that miR-133b regulated
the viability, migration and invasion of cell by regulating the
expression of FGFR1 in LHSCC cells.
shFGFR1 and miR-133b M promote the
expression levels of p53 and Bax, while inhibiting the expression
levels of Bcl-2, p-PI3K and p-AKT in LHSCC cells
As shown in Fig.
7A-D, shFGFR1 and miR-133b M promoted the expression levels of
p53 and Bax, while inhibiting those of Bcl-2 and the ratios of
p-PI3K/PI3K and p-AKT/AKT in transfected TU212 cells (P<0.01).
Moreover, when cells were co-transfected with shFGFR1 and miR-133b
M, the effects of shFGFR1 and miR-133b M on inhibiting Bcl-2
expression and the ratios of p-PI3K/PI3K and p-AKT/AKT and on
promoting the expression levels of p53 and Bax were notably
enhanced as compared with the M + shNC and MC + shFGFR1 groups
(P<0.01). Similarly, the results of RT-qPCR showed that shFGFR1
and miR-133b M promoted the expression levels of p53 and Bax yet
inhibited that of Bcl-2, whereas they had no effect on the
expression levels of PI3K and AKT (P<0.05). When the cells were
co-transfected with miR-133b M and shFGFR1, the effects of shFGFR1
and miR-133b M on promoting the expression levels of p53 and Bax,
as well as inhibiting Bcl-2 expression, were significantly enhanced
as compared with the M + shNC group and MC + shFGFR1 group
(P<0.01; Fig. 7E). These data
suggested that HCG18 may promote the viability, migration and
invasion of LHSCC cells by regulating the FGFR1/miR-133b
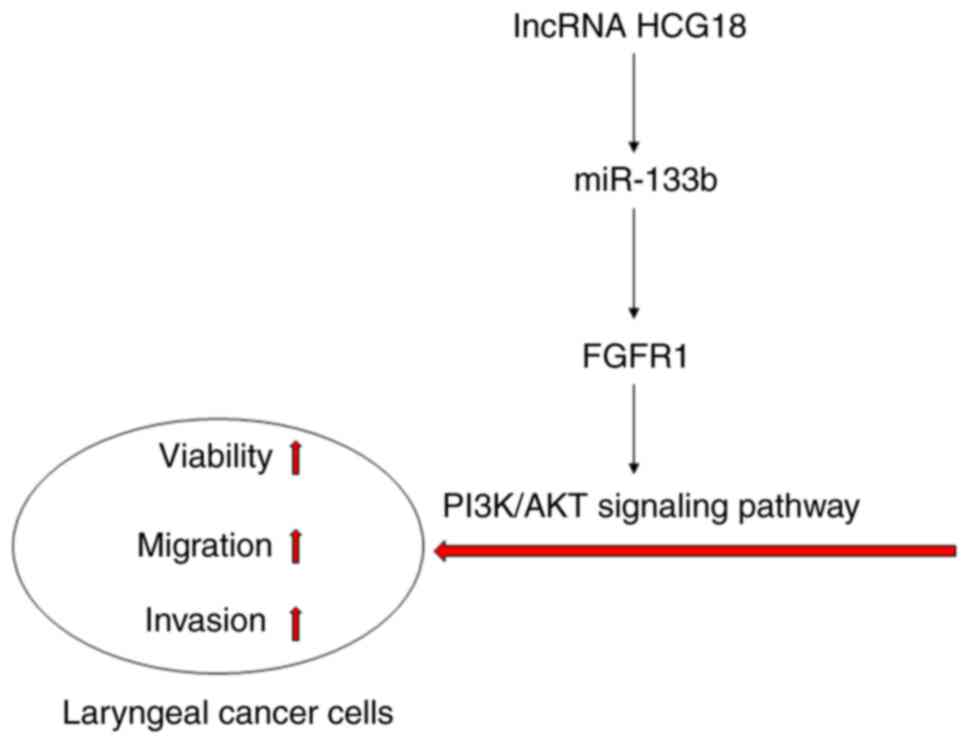
axis-mediated PI3K/AKT signaling pathway (Fig. 8).
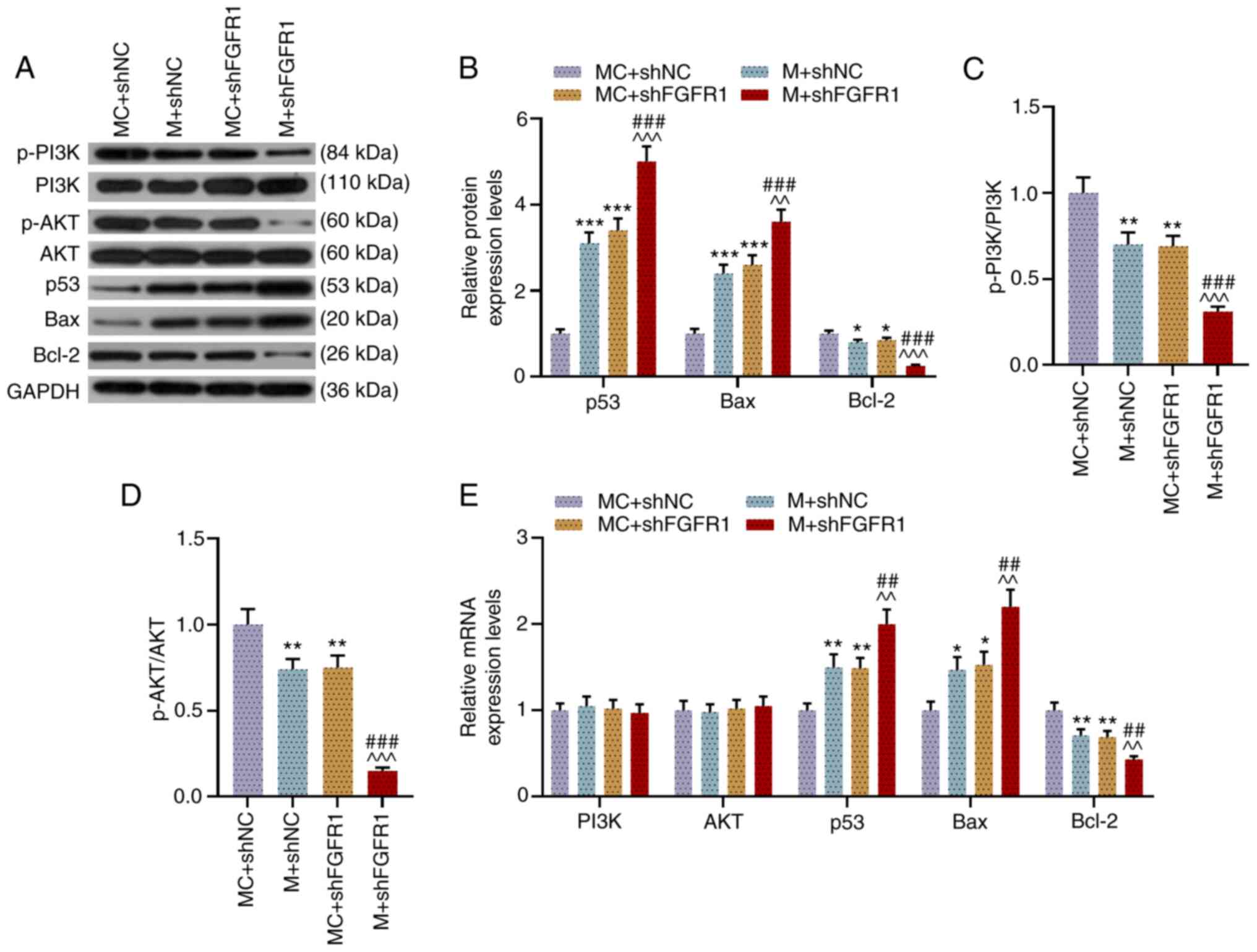 | Figure 7.shFGFR1 and miR-133b M promote the
expression levels of p53 and Bax, while inhibiting those of Bcl-2,
p-PI3K and p-AKT in TU212 cells. (A and B) Expression levels of
p-PI3K, PI3K, p-AKT, AKT, p53, Bax and Bcl-2 in transfected TU212
cells were determined via western blotting, and GAPDH was used as
an internal loading control. Ratios of (C) p-PI3K to PI3K and (D)
p-AKT to AKT in transfected TU212 cells were analyzed using western
blotting assays. (E) Expression levels of PI3K, AKT, p53, Bax and
Bcl-2 in transfected TU212 cells were measured via reverse
transcription-quantitative PCR, and GAPDH was used as a reference
gene. *P<0.05, **P<0.01, ***P<0.001 vs. shNC + MC;
^^P<0.01, ^^^P<0.001 vs. shNC + M;
##P<0.01, ###P<0.001 vs. shHCG18 + MC.
p-, phosphorylated; M, mimic; MC, mimic control; IC, inhibitor
control; I, inhibitor; miR, microRNA; sh, short hairpin RNA; shNC,
short hairpin negative control; FGFR1, fibroblast growth factor
receptor 1. |
Discussion
Although the exact etiology and mechanism of LHSCC
remain to be further clarified, tumor metastasis and recurrence are
the main reasons for the poor prognosis of LHSCC (3–5).
Inhibition of cancer progression by repressing the migration and
invasion of cancer cells is therefore an effective therapeutic
option (20). Moreover, the
migration and invasion of cancer cells are regulated by multiple
genes (21).
lncRNAs play multifaceted roles in regulating gene
expression in pathological processes, and dysregulation of lncRNAs
is associated with a variety of pathological conditions, including
preeclampsia, cancer and cerebrovascular pathologies (22–24). It has been reported that the
expression of HCG18 was promoted in gastric cancer tissues and
colorectal cancer tissues, and knockdown the expression of HCG18
evidently decreased the proliferation, migration and invasion of
cancer cells (10,11). To the best of our knowledge, the
present study demonstrated for the first time that HCG18 expression
was increased in LHSCC tissues and promoted the progression of
LHSCC, but its specific action mechanisms require further
investigation.
Apart from lncRNAs, recent studies have shown that
numerous miRNAs are differentially expressed during tumorigenesis
(25). Previous studies have
revealed that miR-133b was aberrantly expressed in several tumors,
including colorectal cancer, esophageal squamous cell carcinoma and
human tongue squamous cell carcinoma, and that lncRNAs are involved
in cancer progression by targeting and regulating miRNAs
expressions (16–19). The present study identified that
miR-133b expression was decreased in LHSCC tissues, and that
shHCG18 and miR-133b M inhibited the activity of LHSCC cells. In
addition, several studies have reported that HCG18 acts as an
oncogenic factor by regulating the expressions of miRNAs (13). To the best of our knowledge, the
present study demonstrated for the first time that HCG18 could
competitively bind with miR-133b in LHSCC cells, and that a
miR-133b I promoted cell viability, migration, and invasion, while
shHCG18 reversed the promotive effects of the miR-133b I. Thus, it
was suggested that HCG18 promoted LHSCC progression by regulating
miR-133b expression.
The interactions among lncRNA, miRNAs and mRNA, as
well as complex molecular regulatory networks, have been evidenced
in cancer (6). The FGFR1 gene is
located on chromosome 8p12, and upregulation of FGFR1 expression is
one of the most common genetic changes in tumors (26,27). In recent years, the upregulation
of FGFR1 has been found in breast cancer, ovarian cancer and tongue
squamous cell carcinoma (28–30). Similarly, the amplification of
FGFR1 may serve as an independent prognostic factor for
disease-free survival in LHSCC (31). Related studies have shown that
HCG18 promoted the progression of lung adenocarcinoma by regulating
miR-34a-5p-mediated hyaluronan mediated motility receptor
expression (12). The present
study found that FGFR1 expression was increased in LHSCC, miR-133b
could competitively bind with FGFR1 in LHSCC cells and shHCG18
enhanced the inhibitory effect of the miR-133b M. Collectively,
these data suggested that HCG18 may facilitate LHSCC progression by
upregulating FGFR1 via miR-133b. However, it remains necessary to
identify the roles of HCG18, miR-133b and FGFR1 in LHSCC at the
molecular level.
An increasing number of studies have shown that the
PI3K/AKT and p53 signaling pathways have important roles in
targeted therapy of human cancer types (32–34). In non-small cell lung cancer,
miR-133b has the potential to be involved in the progression by
regulating its target-mediated PI3K/AKT and p53 signaling pathways
(35). In laryngeal cancer,
lncRNA growth arrest-specific transcript 5 inhibits cancer cell
invasion and proliferation via the PI3K/AKT/mTOR signaling pathway
(36). Downregulation of sterol
regulatory element binding transcription factor 1 inhibits the
invasion by regulation of Bcl-2 and Bax and inhibition of the
PI3K/AKT pathway in non-small-cell lung cancer (37). Both Bcl-2 and Bax belong to the
Bcl-2 family, and Bcl-2 has the effect of inhibiting apoptosis,
while Bax can promote apoptosis (38). p53 is one of the most important
tumor-suppressor genes in human cancer (39). The present study identified that
shHCG18 inhibited the expression levels of Bcl-2, p-PI3K and p-AKT,
yet promoted those of p53 and Bax in LHSCC cells, while the
miR-133b I reversed the effect of shHCG18; however, shFGFR1 further
reversed the effect of the miR-133b I. The experimental results
demonstrated that HCG18/miR-133b axis contributes LHSCC progression
by regulating the FGFR1-mediated PI3K/AKT signaling pathway.
In conclusion, the present study identified that
HCG18 expression was upregulated and that there was a relationship
between HCG18 and miR-133b in LHSCC. The current results indicated
that HCG18 facilitated LHSCC progression by upregulating FGFR1 via
miR-133b, which may provide a new research direction and
theoretical basis for targeted therapy of LHSCC.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HBP made substantial contributions to study
conception and design. PJG performed data acquisition, data
analysis and interpretation. HBP drafted the article and critically
revised it for important intellectual content. Both authors read
and approved the final manuscript, as well as agreed to be
accountable for all aspects of the work in ensuring that questions
related to the accuracy or integrity of the work are appropriately
investigated and resolved. HBP and PJG confirmed the authenticity
of all the raw data.
Ethics approval and consent to
participate
This study was discussed and approved by the
Zhongshan Hospital of Traditional Chinese Medicine Affiliated to
Guangzhou University of Chinese Medicine Ethics Committee (approval
no. JN2020020208), and all tissue samples were confirmed to be
LHSCC. Written informed consent was obtained from the participating
patients. All procedures performed in studies involving human
participants were in accordance with the ethical standards of the
institutional and/or national research committee and with the 1964
Helsinki declaration and its later amendments or comparable ethical
standards.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Song L, Zhang S, Yu S, Ma F, Wang B, Zhang
C, Sun J, Mao X and Wei L: Cellular heterogeneity landscape in
laryngeal squamous cell carcinoma. Int J Cancer. 147:2879–2890.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Abdeyrim A, He S, Zhang Y, Mamtali G, Asla
A, Yusup M and Liu J: Prognostic value of lymph node ratio in
laryngeal and hypopharyngeal squamous cell carcinoma: A systematic
review and meta-analysis. J Otolaryngol Head Neck Surg. 49:312020.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Colevas AD, Yom SS, Pfister DG, Spencer S,
Adelstein D, Adkins D, Brizel DM, Burtness B, Busse PM, Caudell JJ,
et al: NCCN guidelines insights: Head and neck cancers, Version
1.2018. J Natl Compr Canc Netw. 16:479–490. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dietz A, Wiegand S, Kuhnt T and Wichmann
G: Laryngeal preservation approaches: Considerations for new
selection criteria based on the DeLOS-II trial. Front Oncol.
9:6252019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang L, Luo D, Yi J, Li L, Zhao Y, Lin M,
Guo W, Hu L and Zhou C: Therapy effects of advanced hypopharyngeal
and laryngeal squamous cell carcinoma: Evaluated using dual-energy
CT quantitative parameters. Sci Rep. 8:90642018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tang XJ, Wang W and Hann SS: Interactions
among lncRNAs, miRNAs and mRNA in colorectal cancer. Biochimie.
163:58–72. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang Y and Tang L: The application of
lncRNAs in cancer treatment and diagnosis. Recent Pat Anticancer
Drug Discov. 13:292–301. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nandwani A, Rathore S and Datta M: lncRNAs
in cancer: Regulatory and therapeutic implications. Cancer Lett.
501:162–171. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Taniue K and Akimitsu N: The functions and
unique features of lncRNAs in cancer development and tumorigenesis.
Int J Mol Sci. 22:6322021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ma P, Li L, Liu F and Zhao Q:
HNF1A-induced lncRNA HCG18 facilitates gastric cancer progression
by upregulating DNAJB12 via miR-152-3p. Onco Targets Ther.
13:7641–7652. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li S, Wu T, Zhang D, Sun X and Zhang X:
The long non-coding RNA HCG18 promotes the growth and invasion of
colorectal cancer cells through sponging miR-1271 and upregulating
MTDH/Wnt/β-catenin. Clin Exp Pharmacol Physiol. 47:703–712. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li W, Pan T, Jiang W and Zhao H:
HCG18/miR-34a-5p/HMMR axis accelerates the progression of lung
adenocarcinoma. Biomed Pharmacother. 129:1102172020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li L, Ma TT, Ma YH and Jiang YF: lncRNA
HCG18 contributes to nasopharyngeal carcinoma development by
modulating miR-140/CCND1 and Hedgehog signaling pathway. Eur Rev
Med Pharmacol Sci. 23:10387–10399. 2019.PubMed/NCBI
|
|
14
|
Testa U, Pelosi E, Castelli G and Labbaye
C: miR-146 and miR-155: Two key modulators of immune response and
tumor development. Noncoding RNA. 3:222017.PubMed/NCBI
|
|
15
|
Xu Z, Huang B, Zhang Q, He X, Wei H and
Zhang D: NOTCH1 regulates the proliferation and migration of
bladder cancer cells by cooperating with long non-coding RNA HCG18
and microRNA-34c-5p. J Cell Biochem. 120:6596–6604. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Georgantas RW, Streicher K, Greenberg SA,
Greenlees LM, Zhu W, Brohawn PZ, Higgs BW, Czapiga M, Morehouse CA,
Amato A, et al: Inhibition of myogenic microRNAs 1, 133, and 206 by
inflammatory cytokines links inflammation and muscle degeneration
in adult inflammatory myopathies. Arthritis Rheumatol.
66:1022–1033. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lv L, Li Q, Chen S, Zhang X, Tao X, Tang
X, Wang S, Che G, Yu Y and He L: miR-133b suppresses colorectal
cancer cell stemness and chemoresistance by targeting
methyltransferase DOT1L. Exp Cell Res. 385:1115972019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zeng W, Zhu JF, Liu JY, Li YL, Dong X,
Huang H and Shan L: miR-133b inhibits cell proliferation, migration
and invasion of esophageal squamous cell carcinoma by targeting
EGFR. Biomed Pharmacother. 111:476–484. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang K, Zhou H, Yan B and Cao X:
TUG1/miR-133b/CXCR4 axis regulates cisplatin resistance in human
tongue squamous cell carcinoma. Cancer Cell Int. 20:1482020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xia L, Li S, Liu Y, Huang Y, Ni B, Wan L,
Mei H, Li X, Cai Z and Li Z: NDNF inhibits the migration and
invasion of human renal cancer cells through epithelial-mesenchymal
transition. Oncol Lett. 17:2969–2975. 2019.PubMed/NCBI
|
|
21
|
Bian Q: Circular RNA PVT1 promotes the
invasion and epithelial-mesenchymal transition of breast cancer
cells through serving as a competing endogenous RNA for miR-204-5p.
Onco Targets Ther. 12:11817–11826. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Song X, Luo X, Gao Q, Wang Y, Gao Q and
Long W: Dysregulation of lncRNAs in placenta and pathogenesis of
preeclampsia. Curr Drug Targets. 18:1165–1170. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lou W, Ding B, Zhong G, Du C, Fan W and Fu
P: Dysregulation of pseudogene/lncRNA-hsa-miR-363-3p-SPOCK2 pathway
fuels stage progression of ovarian cancer. Aging (Albany NY).
11:11416–11439. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang X, Tang X, Liu K, Hamblin MH and Yin
KJ: Long noncoding RNA malat1 regulates cerebrovascular pathologies
in ischemic stroke. J Neurosci. 37:1797–1806. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mamoori A, Gopalan V and Lam AK: Role of
miR-193a in cancer: Complexity and factors control the pattern of
its expression. Curr Cancer Drug Targets. 18:618–628. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Malchers F, Ercanoglu M, Schütte D,
Castiglione R, Tischler V, Michels S, Dahmen I, Brägelmann J, Menon
R, Heuckmann JM, et al: Mechanisms of primary drug resistance in
FGFR1-amplified lung cancer. Clin Cancer Res. 23:5527–5536. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Haq F, Sung YN, Park I, Kayani MA, Yousuf
F, Hong SM and Ahn SM: FGFR1 expression defines clinically distinct
subtypes in pancreatic cancer. J Transl Med. 16:3742018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen S, Qiu Y, Guo P, Pu T, Feng Y and Bu
H: FGFR1 and HER1 or HER2 co-amplification in breast cancer
indicate poor prognosis. Oncol Lett. 15:8206–8214. 2018.PubMed/NCBI
|
|
29
|
Uusi-Kerttula H, Legut M, Davies J, Jones
R, Hudson E, Hanna L, Stanton RJ, Chester JD and Parker AL:
Incorporation of peptides targeting EGFR and FGFR1 into the
adenoviral fiber knob domain and their evaluation as targeted
cancer therapies. Hum Gene Ther. 26:320–329. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jiao J, Zhao X, Liang Y, Tang D and Pan C:
FGF1-FGFR1 axis promotes tongue squamous cell carcinoma (TSCC)
metastasis through epithelial-mesenchymal transition (EMT). Biochem
Biophys Res Commun. 466:327–332. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim EK, Cho YA, Koh YW, Shin HA, Cho BC
and Yoon SO: Prognostic implications of Fibroblast growth factor
receptor 1 (FGFR1) gene amplification and protein overexpression in
hypopharyngeal and laryngeal squamous cell carcinoma. BMC Cancer.
20:3482020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Murugan AK: Special issue: PI3K/Akt
signaling in human cancer. Semin Cancer Biol. 59:1–2. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Narayanankutty A: PI3K/Akt/mTOR pathway as
a therapeutic target for colorectal cancer: A review of preclinical
and clinical evidence. Curr Drug Targets. 20:1217–1226. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Khan H, Reale M, Ullah H, Sureda A, Tejada
S, Wang Y, Zhang ZJ and Xiao J: Anti-cancer effects of polyphenols
via targeting p53 signaling pathway: Updates and future directions.
Biotechnol Adv. 38:1073852020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen GY and Ruan L: Downregulation Of
microRNA-133b and its clinical value in non-small cell lung cancer.
Onco Targets Ther. 12:9421–9434. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu W, Zhan J, Zhong R, Li R, Sheng X, Xu
M, Lu Z and Zhang S: Upregulation of long noncoding RNA_GAS5
suppresses cell proliferation and metastasis in laryngeal cancer
via regulating PI3K/AKT/mTOR signaling pathway. Technol Cancer Res
Treat. 20:15330338219900742021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang B, Wu J, Guo P, Wang Y, Fang Z, Tian
J, Yu Y, Teng W, Luo Y and Li Y: Down-regulation of SREBP via
PI3K/AKT/mTOR pathway inhibits the proliferation and invasion of
non-small-cell lung cancer cells. Onco Targets Ther. 13:8951–8961.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bumbat M, Wang M, Liang W, Ye P, Sun W and
Liu B: Effects of Me2SO and trehalose on the cell viability,
proliferation, and Bcl-2 family gene (BCL-2, BAX, and BAD)
expression in cryopreserved human breast cancer cells. Biopreserv
Biobank. 18:33–40. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hong B, van den Heuvel AP, Prabhu VV,
Zhang S and El-Deiry WS: Targeting tumor suppressor p53 for cancer
therapy: Strategies, challenges and opportunities. Curr Drug
Targets. 15:80–89. 2014. View Article : Google Scholar : PubMed/NCBI
|