Introduction
Gallbladder cancer (GBC) is the most common
malignant tumor of the bile duct system, accounting for 80–95% of
all malignant biliary tract tumor cases worldwide (1). Due to the insidious onset of GBC and
the lack of specific symptoms at the early stages, the majority of
GBC cases are diagnosed in the middle and late stages of the
disease (2). GBC is often
misdiagnosed as biliary colic clinically (3). The current main treatment option for
GBC is radical cholecystectomy, but the resectability of the tumor
must be evaluated prior to surgery to determine eligibility. If the
GBC can be resected, the liver function should be evaluated and
adjuvant treatment should be administered after surgery. If the
cancer is unresectable, chemotherapy or radiotherapy can be
prescribed for the treatment of the disease (3). The diagnosis of cholangiocarcinoma,
which is also a type of biliary tract cancer, is obtained through
imaging techniques and invasive examinations, followed by
pathological confirmation. These diagnostic approaches are roughly
the same as those used for GBC, and so are the treatments regimens
(4). For patients with advanced
GBC, it is difficult to achieve the desired therapeutic effect
using surgery, radiotherapy and chemotherapy (5,6).
Although, several molecular prognostic markers for GBC have been
discovered, including KRAS, HER2, tumor protein 53 (p53) and p16,
the reduced specificity and instability of these markers limit
their application (7,8). Therefore, the current treatment
strategy for GBC is still based on surgery combined with adjuvant
radiotherapy and chemotherapy (9).
Until recently, the basic molecular mechanisms underlying the
occurrence, development and metastasis of GBC have not been fully
clarified. Therefore, further studies on these mechanisms are
urgently required.
The high mobility group AT-hook 2 (HMGA2) protein is
a non-histone, architectural transcription factor that modulates
the transcription of numerous genes (10), including snail family
transcriptional repressor 2 (Slug) (11), SRY-related HMG-box (12) and twist family bHLH transcription
factor (13). It has been reported
that HMGA2 regulates several biological processes, including cell
cycle progression, DNA damage repair, apoptosis,
epithelial-mesenchymal transition (EMT) and cell senescence
(14). Furthermore, the
upregulation of HMGA2 is a common feature of malignancy, and its
increased expression has been associated with a poor prognosis and
reduced chemotherapeutic efficacy in several types of cancer, such
as colorectal, gastric and ovarian cancer (10,15).
Accumulating evidence has indicated that determining HMGA2
expression could be used as a routine procedure in clinical tumor
analysis (10,16,17). A
previous study demonstrated that HMGA2 was upregulated in
gallbladder adenocarcinoma tissues compared with adjacent normal
tissues, and its expression was closely associated with a poor
clinical prognosis and the metastasis of gallbladder adenocarcinoma
(18). Another study demonstrated
that microRNA-26a acted as a tumor suppressor by inhibiting GBC
cell proliferation via directly targeting and negatively regulating
HMGA2 expression (19). The
aforementioned findings suggested that HMGA2 may play a role in
promoting the carcinogenesis and progression of GBC. However, to
the best of our knowledge, the effect of HMGA2 on promoting GBC
cell invasion, migration and angiogenesis has not been previously
investigated.
Metastasis is the leading cause of cancer-related
mortality (20). It is estimated
that ~90% of all malignant solid tumors are caused by the spread
and distant metastasis of tumor cells (21,22).
The growth of solid tumors is accompanied by the induction of
angiogenesis (23). The present
study aimed to investigate the effect of HMGA2 knockdown and
overexpression on GBC cell invasion, migration and angiogenesis.
The findings of the current article may provide a potential novel
treatment approach for GBC.
Materials and methods
Cell culture and treatment
The human GBC cell line, EH-GB1 (16) was obtained from The Cell Bank of
Type Culture Collection of The Chinese Academy of Sciences. Cells
were cultured in DMEM supplemented with 15% FBS (both from Gibco;
Thermo Fisher Scientific, Inc.), and maintained at 37°C in a
humidified atmosphere containing 5% CO2. HUVECs were
purchased from Clonetics™ (Lonza Group Ltd.) and cultured in DMEM
supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C in
a 5% CO2 incubator. The culture medium of both cultures
was replaced every 1–2 days. Once cells reached 85–90% confluence,
they were passaged at a ratio of 1:3. Untreated cells were used as
the control group.
For the tube formation assays and detection of CD31,
VEGFR2 and VEGFR1 expression, HUVECs between passages two and five
were incubated with DMEM from EH-GB1 cells transfected with
different small interfering RNAs (siRNAs)/overexpression (ov)
plasmids at 37°C for 24 h.
Cell transfection
The ov-HMGA2 and ov-VEGFA pcDNA3.1 plasmids
containing full-length sequences of the genes and the empty plasmid
[used as a negative control (ov-NC)] were obtained from GenScript.
siRNA targeting HMGA2 (siRNA-HMGA2; sense,
5′-CAGCCUGAAUAACUUGAACTT-3′ and antisense,
5′-GUUCAAGUUAUUCAGGCUGTT-3′) and siRNA-NC (sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′) were purchased from Guangzhou RiboBio
Co., Ltd. Cells were seeded into 6-well plates at a density of
2×105 cells/well and transfected with 80 pmol ov-HMGA2,
ov-VEGFA, ov-NC, siRNA-HMGA2 or siRNA-NC using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. At 48 h
post-transfection, the expression levels of HMGA2 and VEGFA were
determined by reverse transcription-quantitative PCR (RT-qPCR) and
western blotting.
Cell Counting Kit 8 (CCK-8) assay
A CCK-8 assay was performed to assess cell
proliferation. Briefly, EH-GB1 cells or transfected cells
(1×104 cells/well) were cultured in 96-well plates for
24, 48 and 72 h at 37°C. At each time point, 10 µl CCK-8 reagent
(Beyotime Institute of Biotechnology) was added into each well and
cells were incubated at 37°C for a further 2 h in the dark. The
optical density of each well was measured at a wavelength of 450 nm
using a microplate reader (Thermo Fisher Scientific, Inc.).
Wound healing assay
For the wound healing assay, 1×105 EH-GB1
cells/well were seeded into 12-well plates. Following incubation at
37°C for 48 h, cells reached 90% confluence, and a scratch was
drawn across the center of each well using a 200 µl plastic pipette
tip to generate an artificial wound. The cells were then washed
three times with fresh serum-free DMEM and cultured in this medium.
Following incubation at 37°C for 24 h, cell migration into the
wound area was visualized using a light microscope (magnification,
×100; Olympus Corporation) and ImageJ v.1.8 software (National
Institutes of Health) was used for quantification.
Transwell assay
Cell invasion was assessed using 24-well Transwell
chambers (Corning, Inc.), which were precoated with Matrigel
(Thermo Fisher Scientific, Inc.) at 37°C for 10 h. Briefly,
2×104 EH-GB1 cells in 200 µl serum-free DMEM were plated
into the upper chamber, while the lower chamber was filled with 500
µl normal DMEM containing 15% FBS. After incubation at 37°C for 20
h, cells were fixed with 4% paraformaldehyde for 20 min and stained
with crystal violet for 20 min both at room temperature. The
invasive cells were counted in three randomly selected fields of
view under a light microscope (magnification, ×100; Nikon
Corporation) and ImageJ v.1.8 software (National Institutes of
Health) was used for quantification.
Western blotting
Total protein was extracted from cells using RIPA
lysis buffer (Beyotime Institute of Biotechnology) supplemented
with protease inhibitors (Roche Diagnostics GmbH). Total protein
was quantified using a BCA assay and the protein samples (30 µg per
lane) were separated via 10% SDS-PAGE. The separated proteins were
subsequently transferred onto PVDF membranes and blocked with 5%
non-fat milk for 2 h at room temperature. The membranes were then
incubated with the following primary antibodies at 4°C overnight:
Anti-HMGA2 (1:1,000; cat. no. ab97276; Abcam), anti-Ki67 (1:1,000;
cat. no. ab16667; Abcam), anti-proliferating cell nuclear antigen
(PCNA; 1:1,000; cat. no. ab18197; Abcam), anti-MMP2 (1:1,000; cat.
no. ab92536; Abcam), anti-MMP9 (1:1,000; cat. no. ab76003; Abcam),
anti-N-cadherin (1:1,000; cat. no. ab76011; Abcam), anti-Slug
(1:1,000; cat. no. ab27568; Abcam), anti-zinc finger E-box-binding
homeobox 1 (ZEB1; 1:1,000; cat. no. ab203829; Abcam), anti-VEGFA
(1:200; cat. no. ab1316; Abcam), anti-CD31 (1:1,000; cat. no.
ab9498; Abcam), anti-VEGFR1 (1:1,000; cat. no. ab32152; Abcam),
anti-VEGFR2 (1:1,000; cat. no. ab134191; Abcam) or anti-GAPDH
(1:1,0000; cat. no. ab181602; Abcam). Following the primary
antibody incubation, the membranes were incubated with a
HRP-conjugated goat anti-rabbit secondary antibody (1:5,000; cat.
no. ab97080; Abcam) or rabbit anti-mouse secondary antibody
(1:5,000; cat. no. ab6728; Abcam) for 2 h at room temperature.
Protein bands were visualized using an ECL reagent (Thermo Fisher
Scientific, Inc.) on a ChemiDoc XRS Imaging system (Bio-Rad
Laboratories, Inc.). ImageJ v.1.8 software (National Institutes of
Health) was used for semi-quantification.
RT-qPCR
Total RNA was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Total RNA was reverse transcribed into cDNA using
PrimeScript Reverse Transcriptase (Takara Bio, Inc.) according to
the manufacturer's protocol. qPCR was subsequently performed using
SYBR Green kit (Beijing Solarbio Science & Technology Co.,
Ltd.) on a StepOnePlus™ Real-Time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The thermocycling conditions were
as follows: 95°C for 15 min, 35 cycles of 94°C for 1 min, 53.6°C
for 30 sec, and 72°C for 30 sec, followed by a final extension at
72°C for 5 min. The primer sequences used for the qPCR are listed
in Table I. GAPDH was used as a
reference gene and the relative gene expression levels were
calculated using the 2−ΔΔCq method (24).
 | Table I.Primer sequences used for reverse
transcription-quantitative PCR. |
Table I.
Primer sequences used for reverse
transcription-quantitative PCR.
| Gene | Primer sequence
(5′→3′) |
|---|
| HMGA2 | F:
GCCAAGAGGCAGACCTAGGAAA |
|
| R:
CATGGCAATACAGAATAAGTGGTCA |
| VEGFA | F:
GCCATCCAATCGAGACCCTG |
|
| R:
ATTAGACAGCAGCGGGCAC |
| CD31 | F:
TGAGTGGTGGGCTCAGATTG |
|
| R:
TGAGTCTAGGTCGGGGAGTG |
| VEGFR1 | F:
CTGGGCAGCAGACAAATCCT |
|
| R:
GCAGTGCTCACCTCTGATTGT |
| VEGFR2 | F:
CGGTCAACAAAGTCGGGAGA |
|
| R:
CAGTGCACCACAAAGACACG |
| GAPDH | F:
CAACAGCCTCAAGATCATCAGC |
|
| R:
TTCTAGACGGCAGGTCAGGTC |
Tube formation assay
In vitro vascular tube formation assay was
performed as previously described (25). Briefly, 96-well plates were
precoated with Matrigel and incubated at 37°C for 1 h. HUVECs were
seeded onto Matrigel-coated 96-well plates at a density of
2×106 cells/well and cultured in the presence of DMEM
from EH-GB1 cells transfected with different siRNAs/ov plasmids at
37°C for 24 h. The tube formation ability of HUVECs was observed
under a phase contrast light microscope (magnification, ×40; MS5;
Leica Microsystems, Ltd.). The number of formed tubes was
quantified using ImageJ v.1.8 software (National Institutes of
Health).
Bioinformatics analysis
The Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING; www.string-db.org) is an online database used for
searching known protein interaction relationships. HMGA2 and VEGFA
were entered together in multiple protein interfaces and Homo
sapiens was chosen as the species, then the results were
automatically displayed.
Statistical analysis
Data are presented as the mean ± SD. All statistical
analyses were performed using GraphPad Prism 8.0 software (GraphPad
Software, Inc.). Statistical comparisons among multiple groups were
performed using a one-way ANOVA followed by a Tukey's post hoc
test. P<0.05 was considered to indicate a statistically
significant difference. All experiments were repeated at least
three times.
Results
HMGA2 knockdown and silencing inhibits
and promotes, respectively, GBC cell proliferation, migration,
invasion and EMT
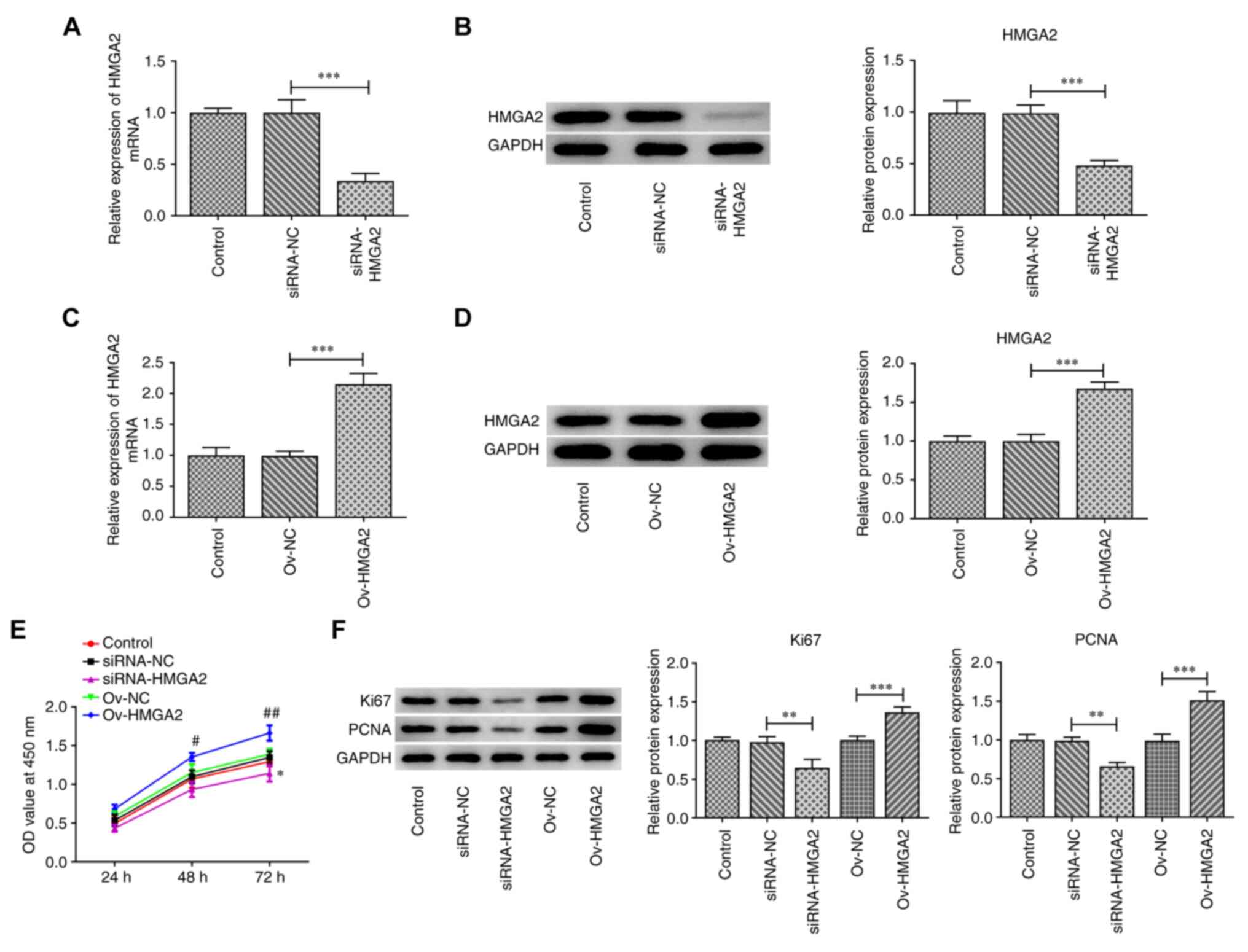
Firstly, HMGA2 was silenced or overexpressed in the
GBC cell line, EH-GB1. As shown in Fig.
1A-D, cell transfection with siRNA-HMGA2 or ov-HMGA2
successfully downregulated and upregulated, respectively, the mRNA
and protein expression levels of HMGA2 compared with the siRNA-NC
and ov-NC groups, respectively. Subsequently, a CCK-8 assay was
performed to evaluate cell proliferation. The results showed that
HMGA2 knockdown significantly inhibited cell proliferation at 72 h
post-transfection compared with the siRNA-NC group, while HMGA2
overexpression significantly promoted cell proliferation at 48 and
72 h post-transfection compared with the ov-NC group (Fig. 1E). Consistent with these findings,
the protein expression levels of Ki67 and PCNA were significantly
downregulated following HMGA2 silencing compared with the siRNA-NC
group, while the opposite effect was observed following HMGA2
overexpression compared with the ov-NC group (Fig. 1F).
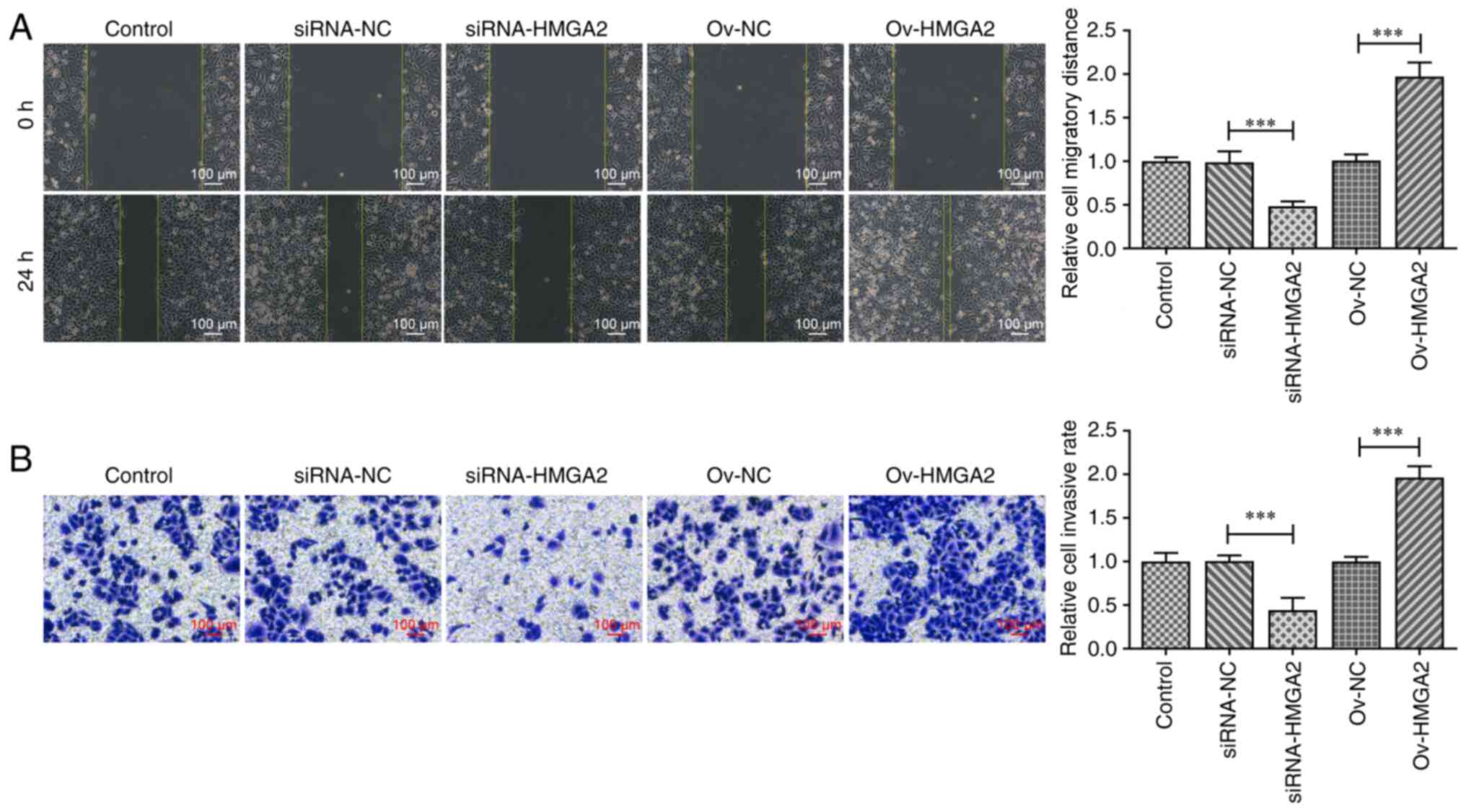
Wound healing and Transwell assays were used to
evaluate the cell migratory and invasive abilities, respectively.
As shown in Fig. 2A and B, cell
migration and invasion were significantly inhibited upon HMGA2
silencing compared with the siRNA-NC group, while both migration
and invasion were significantly increased following HMGA2
overexpression compared with the ov-NC group. Consistent with these
results, the expression levels of cell migration- and
invasion-related proteins, namely MMP2 and MMP9, were significantly
downregulated by HMGA2 silencing compared with the siRNA-NC group.
The opposite effect was observed following HMGA2 overexpression
compared with the ov-NC group (Fig. 3A
and B). The expression levels of the EMT-related proteins,
N-cadherin, Slug and ZEB1, were also significantly upregulated and
downregulated following transfection of EH-GB1 cells with ov-HMGA2
or siRNA-HMGA2, respectively, compared with the respective NCs
(Fig. 3C). The aforementioned
findings suggested that HMGA2 may play a promotive role in the
proliferation, migration, invasion and EMT of GBC cells.
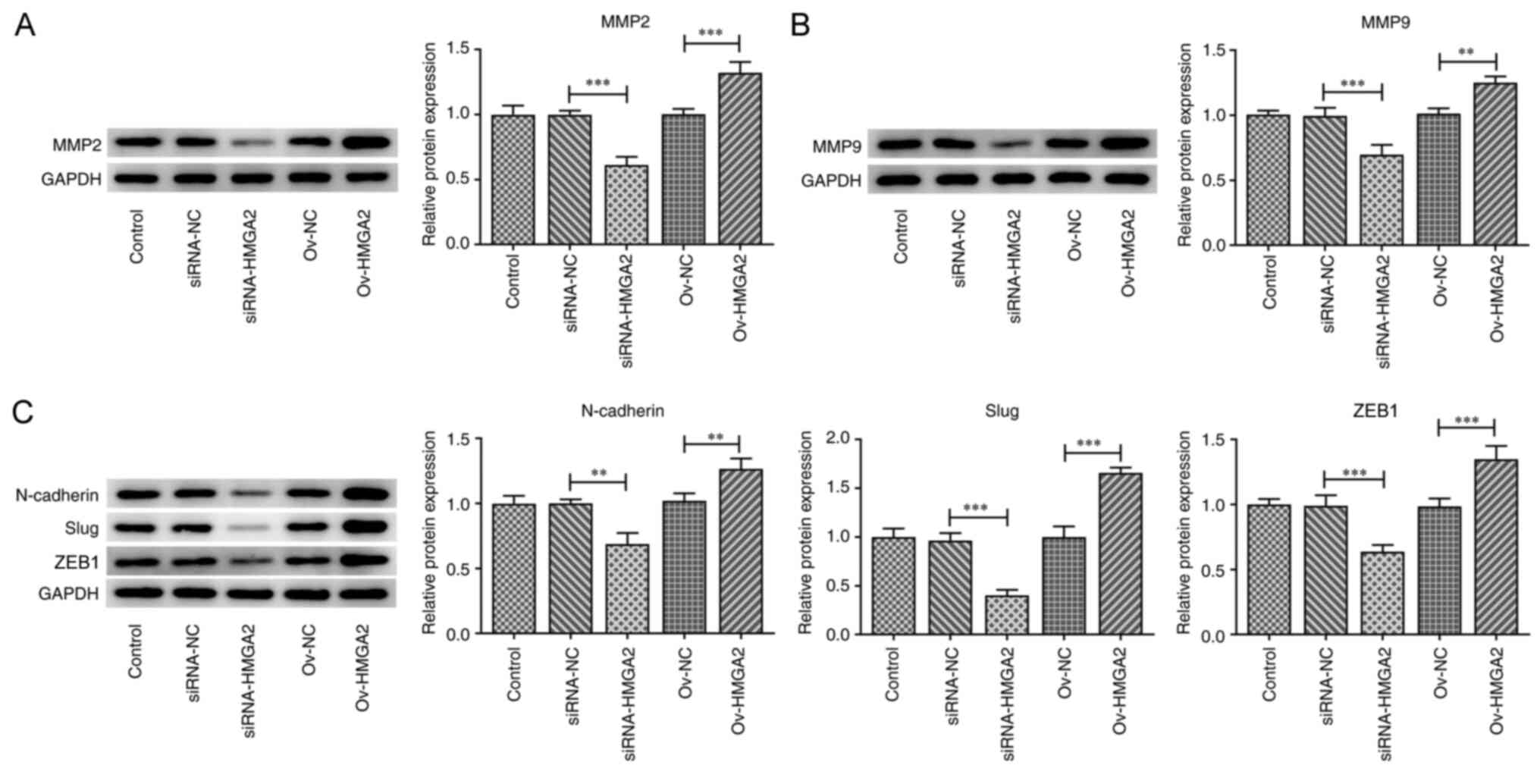 | Figure 3.Effects of HMGA2 overexpression and
knockdown on the epithelial-mesenchymal transition of gallbladder
cancer cells. Protein expression levels of (A) MMP2, (B) MMP9, and
(C) N-cadherin, Slug and ZEB1 in EH-GB1 cells transfected with the
indicated plasmids or siRNAs were detected using western blotting.
**P<0.01, ***P<0.001. HMGA2, high mobility group AT-hook 2;
ZEB1, zinc finger E-box-binding homeobox 1; Slug, snail family
transcriptional repressor 2; siRNA, small interfering RNA; NC,
negative control; ov-, overexpression. |
HMGA2 knockdown downregulates, while
HMGA2 overexpression upregulates, VEGFA expression in GBC
cells
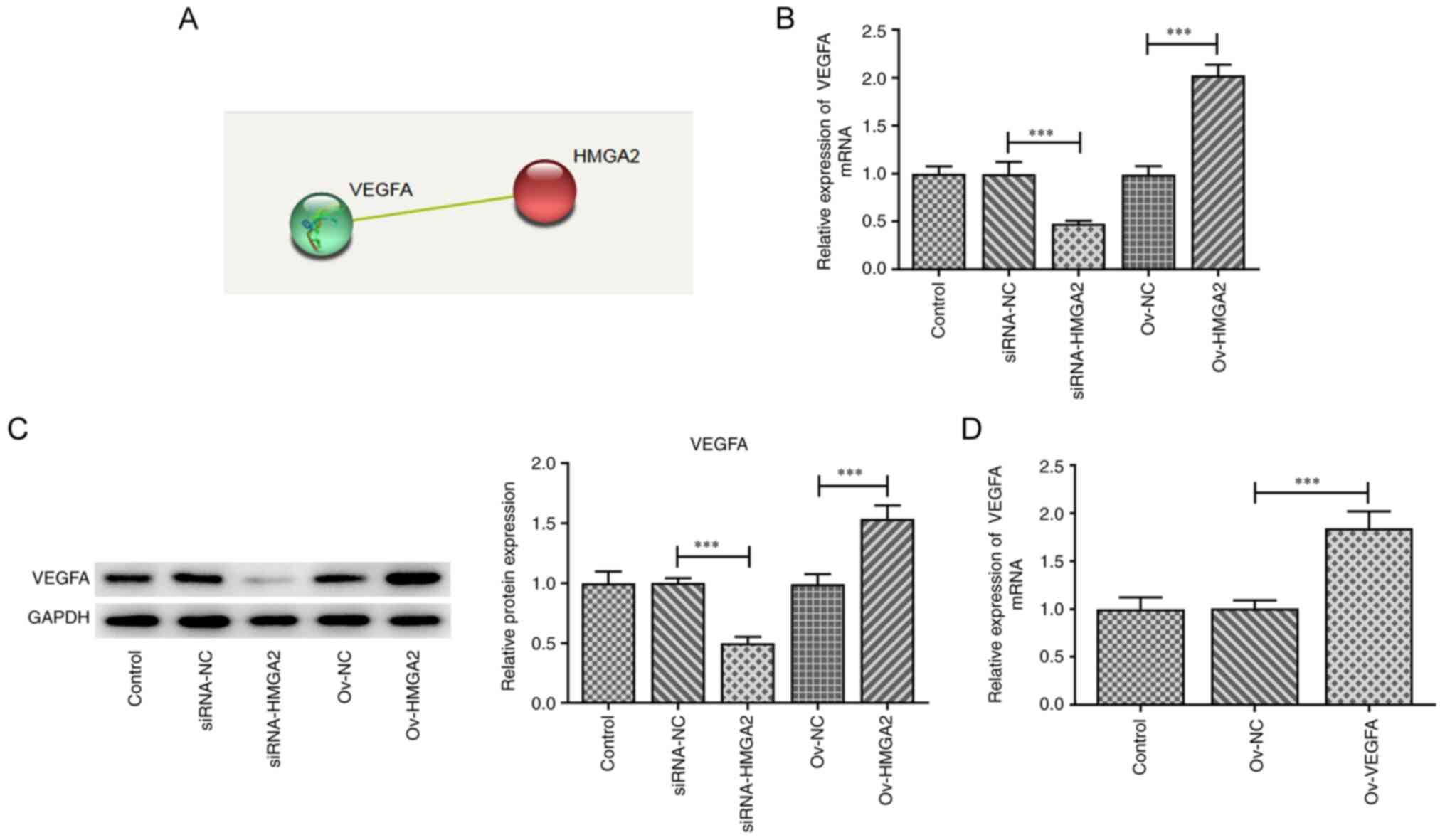
Bioinformatics analysis using the STRING database
predicted that HMGA2 could interact with VEGFA (Fig. 4A). Subsequently, the expression
levels of VEGFA were determined in EH-GB1 cells following HMGA2
knockdown or overexpression. The results showed that both the mRNA
and protein expression levels of VEGFA were downregulated by HMGA2
knockdown compared with the siRNA-NC group, while the opposite
effect was observed upon HMGA2 overexpression compared with the
ov-NC group (Fig. 4B and C). GBC
cells were transfected with ov-VEGFA plasmid, and the results
demonstrated that the expression level VEGFA was upregulated in the
ov-VEGFA group compared with the ov-NC group (Fig. 4D). These data indicated that HMGA2
may positively regulate the expression of VEGFA.
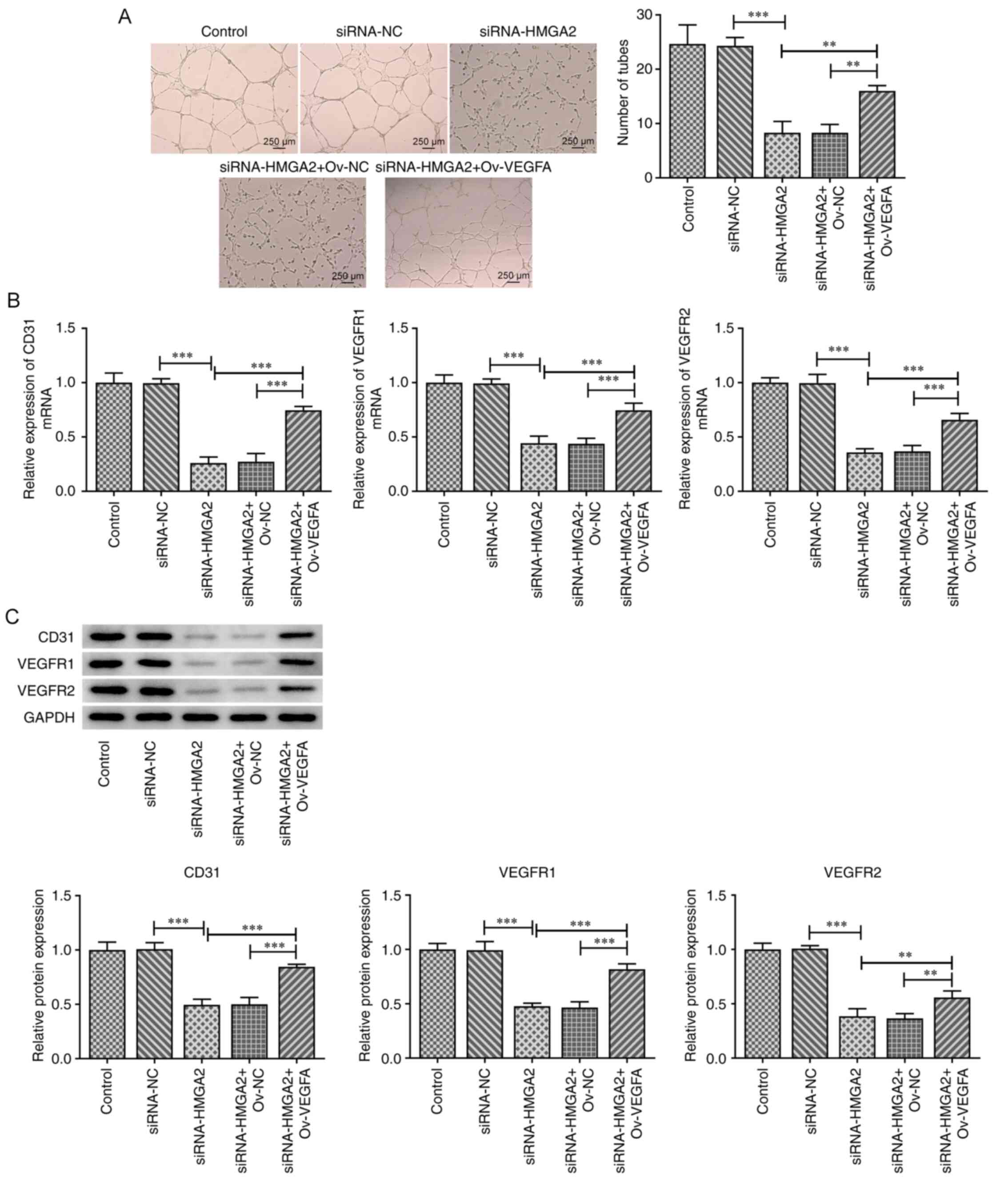
VEGFA overexpression abrogates the
inhibitory effect of HMGA2 silencing on HUVEC tube formation and
expression of CD31, VEGFR1 and VEGFR2
It is well known that VEGFA can induce tumor
angiogenesis (26). Therefore, GBC
cells transfected with siRNA-HMGA2 were transfected with an
ov-VEGFA plasmid and the culture medium was then used to stimulate
HUVECs. Subsequently, the tube formation ability of HUVECs and the
expression levels of CD31, VEGFR1 and VEGFR2 were evaluated. As
shown in Fig. 5A, HMGA2 silencing
significantly attenuated the tube formation ability of HUVECs
compared with the siRNA-NC group. However, the co-transfection with
ov-VEGFA partially rescued the tube formation ability of HUVECs
transfected with siRNA-HMAG2. In addition, both the mRNA and
protein expression levels of CD31, VEGFR1 and VEGFR2 were markedly
downregulated in HUVECs following HMGA2 silencing compared with the
siRNA-NC group. However, the aforementioned effect was partially
rescued by VEGFA overexpression (Fig.
5B and C). These results suggested that HMGA2 knockdown may
inhibit the angiogenesis of HUVECs, possibly via downregulating
VEGFA expression.
Discussion
The HMGA2 protein belongs to the HMGA subfamily of
HMG proteins and encodes a 108 amino acid protein. HMGA2 is a
small, non-histone, chromatin-associated protein with no intrinsic
transcriptional activity (27).
However, it can modulate gene transcription via altering chromatin
architecture, thereby enhancing or suppressing the transcriptional
activity of several human genes, eventually affecting a variety of
biological processes (28). It has
been reported that HMGA2 is upregulated in numerous types of human
cancer, indicating that it may serve a crucial role in cancer
development and carcinogenesis (10). Emerging evidence has suggested that
HMGA2 played an important role in the majority of human cancer
types, including lung (16),
pancreatic (29), colorectal
(15) and breast (30) cancer, via regulating the cell cycle,
apoptosis, DNA damage repair and cell senescence, as well as
promoting EMT and maintaining telomere length (10). The results of the present study
showed that HMGA2 could promote GBC cell proliferation, migration,
invasion and EMT. HMGA2 is normally expressed in mesenchymal cells
and EH-GB1 is a metastatic gallbladder cancer cell line, in which
E-cadherin is expressed at low levels (31). When epithelial cells undergo EMT,
they adopt a mesenchymal cell phenotype to promote metastasis
(32). The results of the present
study revealed that HMGA2 knockdown effectively inhibited the
aforementioned cellular processes in GBC cells. Furthermore, HMGA2
silencing also suppressed angiogenesis in HUVECs via targeting
VEGFA.
Ki67 and PCNA are the most commonly used cell
proliferation markers (33).
Moreover, the increased expression of both markers is associated
with the active proliferation of tumor cells (34). In the present study, HMGA2 knockdown
and overexpression downregulated and upregulated, respectively, the
expression levels of both Ki67 and PCNA, suggesting that HMGA2 may
promote GBC cell proliferation. The invasion and metastasis of
malignant tumors has been discovered to contribute to chemotherapy
failure and death in patients with cancer (35). The results of the wound healing and
Transwell assays in the current study revealed that HMGA2 silencing
attenuated the migratory and invasive abilities of GBC cells,
whereas HMGA2 overexpression exhibited the opposite results.
MMP2 and MMP9 belong to the MMP family and are
responsible for degrading the extracellular matrix, thus
accelerating tumor cell migration, invasion and angiogenesis
(36). Herein, the protein
expression levels of MMP2 and MMP9 were found to be downregulated
following HMGA2 knockdown and upregulated following HMGA2
overexpression. Furthermore, the same results were observed when
the expression levels of N-cadherin, Slug and ZEB1 were determined.
The upregulated expression levels of N-cadherin, Slug and ZEB1 are
typical features of EMT, and represents one of the key steps
required for the invasion and metastasis of malignant tumors of
epithelial origin (37).
Collectively, these data suggested that HMGA2 may promote GBC cell
migration and invasion.
The growth of solid tumors is accompanied by the
induction of angiogenesis and VEGFA is considered as a key
regulator of this processes (38).
Angiogenesis is a biological process that leads to the formation of
new blood vessels from pre-existing blood vessels (39). Tumor cells produce or cause the
microenvironment to generate pro-angiogenic signals, which can
recruit and expand endothelial cells (40). In addition, a retrospective review
reported that VEGFA was expressed in ~80% of GBCs, and 56.3% of the
84 patients had high expression levels of VEGFA, which has been
found to be an independent prognostic factor for survival in GBC
(41). Notably, the current study
demonstrated that HMGA2 could bind with VEGFA to positively
regulate its expression. Therefore, it was hypothesized that HMGA2
may modulate angiogenesis via targeting VEGFA. The present study
co-transfected GBC cells with siRNA-HMGA2 and ov-VEGFA and the CM
was collected to stimulate HUVECs. Consistent with the
aforementioned hypothesis, HMGA2 silencing significantly attenuated
the tube formation ability of HUVECs. Consistently, the mRNA and
protein expression levels of CD31, VEGFR1 and VEGFR2 were also
downregulated following HMGA2 knockdown. The expression of CD31,
also known as platelet endothelial cell adhesion molecule 1, is
often used to evaluate tumor angiogenesis (42). VEGFA is activated after its binding
to VEGFR1 and VEGFR2, two receptors involved in
angiogenesis-related signaling pathways (43). Herein, the results suggested that
HMGA2 may be involved in the angiogenesis of GBC cells.
Furthermore, the overexpression of VEGFA partially abrogated the
inhibitory effect of HMGA2 silencing on angiogenesis, thus
verifying that HMGA2 may regulate angiogenesis via modulating VEGFA
expression. However, the analysis of MMP activation in the present
study was limited to western blotting, and gelatin zymography and
RT-qPCR are required for a more comprehensive analysis of MMP
activation in the future. In addition, the present study only used
in vitro cell models to generate the results; therefore,
future studies should focus on verifying the results of the current
study using in vivo models to determine the potential
mechanisms involved in the effects of HMGA2.
In conclusion, the results of the present study
suggested that the overexpression of HMGA2 in GBC cells may promote
cancer progression by inducing cell migration, invasion and
angiogenesis. Therefore, HMGA2 may serve as a predictive factor in
GBC and targeting HMGA2 could be considered as a potential
therapeutic approach for GBC.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Author's contributions
JY and YZ conceived and designed the present study,
and JY was involved in drafting the manuscript. JY, PD and XQ
conducted the experiments. XQ and YH performed the data analysis.
YZ performed the bioinformatics analysis and provided critical
comments on the revision of the manuscript. All authors read and
approved the final manuscript. JY and YZ confirm the authenticity
of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhou F, Zhang Y, Sun J and Yang X:
Characteristics of a novel cell line ZJU-0430 established from
human gallbladder carcinoma. Cancer Cell Int. 19:1902019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ramachandran A, Srivastava DN and
Madhusudhan KS: Gallbladder cancer revisited: The evolving role of
a radiologist. Br J Radiol. 94:202007262021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hickman L and Contreras C: Gallbladder
cancer: Diagnosis, surgical management, and adjuvant therapies.
Surg Clin North Am. 99:337–355. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brandi G, Venturi M, Pantaleo MA and
Ercolani G: Cholangiocarcinoma: Current opinion on clinical
practice diagnostic and therapeutic algorithms: A review of the
literature and a long-standing experience of a referral center.
Digestive Liver Dis. 48:231–241. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Siegel RL, Fedewa SA, Miller KD,
Goding-Sauer A, Pinheiro PS, Martinez-Tyson D and Jemal A: Cancer
statistics for Hispanics/Latinos, 2015. CA Cancer J Clin.
65:457–480. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Goetze TO: Gallbladder carcinoma:
Prognostic factors and therapeutic options. World J Gastroenterol.
21:12211–12217. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang W, Zhao B, Li Y, Qi D and Wang D:
Modification of the 8th American joint committee on cancer staging
system for gallbladder carcinoma to improve prognostic precision.
BMC Cancer. 20:11292020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sharma A, Sharma KL, Gupta A, Yadav A and
Kumar A: Gallbladder cancer epidemiology, pathogenesis and
molecular genetics: Recent update. World J Gastroenterol.
23:3978–3998. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sternby Eilard M, Lundgren L, Cahlin C,
Strandell A, Svanberg T and Sandström P: Surgical treatment for
gallbladder cancer-a systematic literature review. Scand J
Gastroenterol. 52:505–514. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang S, Mo Q and Wang X: Oncological role
of HMGA2 (Review). Int J Oncol. 55:775–788. 2019.PubMed/NCBI
|
|
11
|
Li Y, Zhao Z, Xu C, Zhou Z, Zhu Z and You
T: HMGA2 induces transcription factor Slug expression to promote
epithelial-to-mesenchymal transition and contributes to colon
cancer progression. Cancer Lett. 355:130–140. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lovnicki J, Gan Y, Feng T, Li Y, Xie N, Ho
CH, Lee AR, Chen X, Nappi L, Han B, et al: LIN28B promotes the
development of neuroendocrine prostate cancer. J Clin Invest.
130:5338–5348. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun J, Sun B, Sun R, Zhu D, Zhao X, Zhang
Y, Dong X, Che N, Li J, Liu F, et al: HMGA2 promotes vasculogenic
mimicry and tumor aggressiveness by upregulating Twist1 in gastric
carcinoma. Sci Rep. 7:22292017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fedele M, Palmieri D and Fusco A: HMGA2: A
pituitary tumour subtype-specific oncogene? Mol Cell Endocrinol.
326:19–24. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang X, Wang J and Wu J: Emerging roles
for HMGA2 in colorectal cancer. Transl Oncol. 14:1008942021.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gao X, Dai M, Li Q, Wang Z, Lu Y and Song
Z: HMGA2 regulates lung cancer proliferation and metastasis. Thorac
Cancer. 8:501–510. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hombach-Klonisch S, Kalantari F, Medapati
MR, Natarajan S, Krishnan SN, Kumar-Kanojia A, Thanasupawat T,
Begum F, Xu FY, Hatch GM, et al: HMGA2 as a functional antagonist
of PARP1 inhibitors in tumor cells. Mol Oncol. 13:153–170. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zou Q, Xiong L, Yang Z, Lv F, Yang L and
Miao X: Expression levels of HMGA2 and CD9 and its
clinicopathological significances in the benign and malignant
lesions of the gallbladder. World J Surg Oncol. 10:922012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou H, Guo W, Zhao Y, Wang Y, Zha R, Ding
J, Liang L, Hu J, Shen H, Chen Z, et al: MicroRNA-26a acts as a
tumor suppressor inhibiting gallbladder cancer cell proliferation
by directly targeting HMGA2. Int J Oncol. 44:2050–2058. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yuan L, Guo F, Wang L and Zou Q:
Prediction of tumor metastasis from sequencing data in the era of
genome sequencing. Brief Funct Genomics. 18:412–418. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Scully OJ, Bay BH, Yip G and Yu Y: Breast
cancer metastasis. Cancer Genomics Proteomics. 9:311–320.
2012.PubMed/NCBI
|
|
22
|
Steeg PS: Targeting metastasis. Nat Rev
Cancer. 16:201–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Viallard C and Larrivée B: Tumor
angiogenesis and vascular normalization: Alternative therapeutic
targets. Angiogenesis. 20:409–426. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang JT, Fan YZ, Chen CQ, Zhao ZM and Sun
W: Norcantharidin: A potential antiangiogenic agent for gallbladder
cancers in vitro and in vivo. Int J Oncol. 40:1501–1514.
2012.PubMed/NCBI
|
|
26
|
Li X, Hu Z, Shi H, Wang C, Lei J and Cheng
Y: Inhibition of VEGFA increases the sensitivity of ovarian cancer
cells to chemotherapy by suppressing VEGFA-mediated autophagy. Onco
Targets Ther. 13:8161–8171. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Su L, Deng Z and Leng F: The mammalian
high mobility group protein AT-Hook 2 (HMGA2): Biochemical and
biophysical properties, and its association with adipogenesis. Int
J Mol Sci. 21:37102020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu J and Wei JJ: HMGA2 and high-grade
serous ovarian carcinoma. J Mol Med (Berl). 91:1155–1165. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chiou SH, Dorsch M, Kusch E, Naranjo S,
Kozak MM, Koong AC, Winslow MM and Grüner BM: Hmga2 is dispensable
for pancreatic cancer development, metastasis, and therapy
resistance. Sci Rep. 8:140082018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao W, Geng D, Li S, Chen Z and Sun M:
LncRNA HOTAIR influences cell growth, migration, invasion, and
apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer
Med. 7:842–855. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hao J, Yang Z, Wang L, Zhang Y, Shu Y,
Jiang L, Hu Y, Lv W, Dong P and Liu Y: Downregulation of BRD4
inhibits gallbladder cancer proliferation and metastasis and
induces apoptosis via PI3K/AKT pathway. Int J Oncol. 51:823–831.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shintani Y, Maeda M, Chaika N, Johnson KR
and Wheelock MJ: Collagen I promotes epithelial-to-mesenchymal
transition in lung cancer cells via transforming growth factor-beta
signaling. Am J Respir Cell Mol Biol. 38:95–104. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ruan Y, Wang L and Lu Y: HDAC6 inhibitor,
ACY1215 suppress the proliferation and induce apoptosis of
gallbladder cancer cells and increased the chemotherapy effect of
gemcitabine and oxaliplatin. Drug Dev Res. 82:598–604. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Juríková M, Danihel Ľ, Polák Š and Varga
I: Ki67, PCNA, and MCM proteins: Markers of proliferation in the
diagnosis of breast cancer. Acta Histochem. 118:544–552. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nowakowska A and Tarasiuk J: Invasion and
metastasis of tumour cells resistant to chemotherapy. Postepy Hig
Med Dosw (Online). 71:380–397. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pittayapruek P, Meephansan J, Prapapan O,
Komine M and Ohtsuki M: Role of matrix metalloproteinases in
photoaging and photocarcinogenesis. Int J Mol Sci. 17:6682016.
View Article : Google Scholar
|
|
37
|
Zhou P, Wang C, Hu Z, Chen W, Qi W and Li
A: Genistein induces apoptosis of colon cancer cells by reversal of
epithelial-to-mesenchymal via a Notch1/NF-κB/slug/E-cadherin
pathway. BMC Cancer. 17:8132017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Claesson-Welsh L and Welsh M: VEGFA and
tumour angiogenesis. J Intern Med. 273:114–127. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Simone V, Brunetti O, Lupo L, Testini M,
Maiorano E, Simone M, Longo V, Rolfo C, Peeters M, Scarpa A, et al:
Targeting angiogenesis in biliary tract cancers: An open option.
Int J Mol Sci. 18:4182017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bergers G and Benjamin LE: Tumorigenesis
and the angiogenic switch. Nat Rev Cancer. 3:401–410. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sun XN, Cao WG, Wang X, Wang Q, Gu BX,
Yang QC, Hu JB, Liu H and Zheng S: Prognostic impact of vascular
endothelial growth factor-A expression in resected gallbladder
carcinoma. Tumour Biol. 32:1183–1190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lertkiatmongkol P, Liao D, Mei H, Hu Y and
Newman PJ: Endothelial functions of platelet/endothelial cell
adhesion molecule-1 (CD31). Curr Opin Hematol. 23:253–259. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Freire Valls A, Knipper K, Giannakouri E,
Sarachaga V, Hinterkopf S, Wuehrl M, Shen Y, Radhakrishnan P, Klose
J, Ulrich A, et al: VEGFR1(+) metastasis-associated macrophages
contribute to metastatic angiogenesis and influence colorectal
cancer patient outcome. Clin Cancer Res. 25:5674–5685. 2019.
View Article : Google Scholar : PubMed/NCBI
|