Introduction
At present, root canal therapy and pulpotomy are
important treatments for dental pulp injury (1); however, concomitant complications
and considerable failure rate limit the widespread use of these
treatments in clinical practice (2). Although current root canal filling
materials, such as gutta percha and caprolactone-based points
(2), have good biocompatibility
and effectively seal the apical foramen, treatment often leads to
destruction of dental hard tissue and loss of pulp vitality.
Therefore, retaining biological function of dental pulp is an
urgent requirement in the treatment of pulp disease. Dental pulp
stem cells (DPSCs) have been used as seed cells in reconstruction
of the dental pulp system because of their potential for
multi-directional differentiation, self-renewal and angiogenesis
(3). DPSCs were first isolated
from the pulp of human third permanent molars and subcutaneously
implanted on the back of nude mice to form a pulp-dentin-like
structure in 2000 (4); studies
have confirmed the ability of DPSCs to form hard tissue and dental
cementum (5,6) and to induce osteogenesis in dental
engineering, such as promoting osteoblast differentiation (7,8).
The aforementioned studies indicate that use of DPSCs in
regenerating the pulp-dentin complex and repairing damaged pulp may
become a safe and effective treatment of pulp disease.
Inflammation-mediated tissue repair and regeneration
are key for restoration of damaged dental pulp (9). During the repair and regeneration
process, DPSCs in the microenvironment are recruited, proliferate
and differentiate to repair and regenerate damaged dental pulp
(10). However, the role and
mechanism of DPSCs in pulp restoration is unknown and investigation
is required to understand the involvement of DPSCs in pulp
restoration tissue engineering. Recent studies have demonstrated
that DPSC-derived exosomes are associated with
inflammation-mediated pulp regeneration (11,12). Exosomes are membranous vesicles,
30–150 nm in diameter. The molecular content of exosomes is not
only a fingerprint of the cell phenotype, but can be transferred to
other cells and affect their biological behaviors, including
intercellular communication (13). Specific molecular markers, among
which the most abundant are tetraspanins including CD81, CD63 and
CD9, are detected on the surface of exosomes (14). Exosomes contain proteins, genetic
material and lipids; genetic components including DNA and RNAs
[microRNA (miRNA or miR), long non-coding (lnc)RNA and circular
RNA) are gaining attention (13,14). It has been demonstrated that
dysregulated lncRNAs serve a key role in determining the function
of stem cells, including stem cell pluripotency and differentiation
(14); moreover, lncRNAs also
serve as ‘sponges’ to titrate miRNAs during the differentiation of
stem cells (15).
By contrast, the association between lncRNAs and
miRNAs in DPSCs in dental tissue repair is unclear. To
comprehensively address the aforementioned issue, in the present
study, a variety of in vitro biological experiments based on
an animal model were performed, as well as next-generation
sequencing and bioinformatics analysis, in order to help to broaden
the application prospects of DPSCs, not only in dental tissue
restoration, but also in the field of bone injury repair.
Materials and methods
Animal model
A model of pulpitis was constructed in six-week-old
male Sprague-Dawley rats (weight, 180–200 g), purchased from
Shanghai Laboratory Animal Company. All animals were housed in the
specific-pathogen-free facility in the Institute of Hospital of
Stomatology, Tongji University (Shanghai, China) and were
maintained under a 12/12-h light/dark cycle with free access to
rodent chow and water at room temperature under a controlled
humidity (50±10%). A total of 12 rats were randomly divided into
two groups: Control group and pulp injury model group, with 6 rats
in each group. These animals were placed in a sealed container with
a 4% (vol/vol) isoflurane flow until fully anaesthetized, then
their mandibular incisor labial pulp tissues were resected by an
electrosurgical generator, following which the surgical wounds were
dressed. The diameter of gingival defects was >5 mm, deep to
hard tissue. All pulp tissue was extracted on the 30th day after
modeling and prepared for histomorphometry and statistical
analysis. All animals were sacrificed with CO2
asphyxiation in a chamber (100% CO2, 9.6 l/min, 10 min)
followed by cervical dislocation to confirm death. All animal
experiments were approved by the Institutional Animal Care and Use
Committees of the Hospital of Stomatology, Tongji University
(approval no. 20180606; Jan 1, 2018); studies were performed in
adherence with the international Guide for the Care and Use of
Laboratory Animals.
Primary culture and identification of
DPSCs
Pulp tissue was removed under aseptic conditions,
washed with 0.01 M sterile PBS and cut into small pieces (~1.0
mm3). Following digestion at 37°C for 1 h with 0.3% type
I collagenase and 0.4% dispase, discrete single cell clumps were
pipetted, then the formed single cell suspension was filtered
through a cell sieve (70-µm pore size) and centrifuged at 300 × g
for 5 min at room temperature. After washing the cells with 1X PBS,
the cell precipitate was resuspended in high glucose DMEM
containing 20% FBS (Gibco; Thermo Fisher Scientific, Inc.), and
inoculated into a 5 ml culture flask at a density of
5×104 cells/ml for routine culture at 37°C in a
humidified atmosphere of 5% CO2/95% air. The medium was
changed every 3 days, and cells in the logarithmic growth phase
were collected. The culture supernatant was centrifuged at 2,000 ×
g for 10 min at 4°C, filtered (filter diameter, 0.22 µm) and mixed
with high glucose DMEM containing 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) at a ratio of 1:1 for clonal culture medium. The
first-generation cells in the logarithmic growth phase were diluted
with adaptive medium to 10–15 cells/ml; cells were inoculated in a
96-well plate (100 µl/well) for 12 h at 37°C in a humidified
atmosphere of 5% CO2/95% air and medium was changed
every 5 days. When the cells start to grow, the medium was changed
every 3 days. Identification of DPSCs was performed by
morphological detection using an inverted phase contrast microscope
(CKX53FL; Olympus) and specific marker labeling, including CD34,
CD45, CD29 and CD44, as described previously (16).
Isolation and identification of
DPSC-derived exosomes
To remove cellular debris, medium from DPSCs was
centrifuged at 2,500 × g for 15 min at 4°C and filtered with a 0.22
µm filter. The collected medium containing exosomes was laid on top
of a 30% sucrose/D2O cushion in a sterile UltraClear™
(Beckman Coulter, Inc.) and ultracentrifuged at 100,000 × g for 1 h
at 4°C. The pellets were resuspended in 15 ml PBS and centrifuged
at 4,000 × g at 4°C for 15 min until the volume was concentrated to
~200 µl. The total number of exosomes was determined using CD63
ExoELISA™ (System Biosciences Inc.) according to the
manufacturer's instructions. Exosomes were identified by dynamic
light scattering analysis and transmission electron microscopy
(TEM) according to a previous study (17). Briefly, an enriched exosome
suspension in filtered PBS solution was dispensed on carbon-coated
electron microscopy grids on parafilm and left to absorb for 10 min
at room temperature, then transferred to a drop of
Uranyless® solution (Electron Microscopy Sciences) for 1
min and left to air dry. Excess stain was blotted away. Imaging was
performed with a Jeol JEM-2200FS microscope (Jeol, Ltd.) at 200 kV.
Moreover, the expression of CD63 and CD81 or GM130 and calnexin was
evaluated by western blotting. RNA and proteins were extracted for
further analysis using a Total Exosome RNA & Protein Isolation
kit (Thermo Fisher Scientific, Inc.). To determine the effect of
DPSCs on the migration and osteoblastic differentiation of
mesenchymal stem cell (MSCs), rat MSCs were obtained from Nanjing
Cell Life Biotechnology Co., Ltd. The isolation and purification of
rat MSCs was performed as previously described (16). MSC medium (cat. no. #7501) was
obtained from ScienCell Research Laboratories, Inc. and cultured
MSCs in conditioned medium from DPSCs.
Microarray-based differential
profiling and bioinformatics analysis
Total RNA was isolated from the pulp samples of rats
using TRIzol® (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. The microarray
hybridization was performed using total RNA prepared as
aforementioned. Gene set enrichment analysis (GSEA) tested whether
an a priori defined set of genes shows statistically
significant, concordant differences. The uploaded gene set
consisted of normalized mRNA expression data and was sorted by the
mean log2 signal ratios. Small RNAs of DPSC-derived exosomes were
extracted and used for miRNA sequencing with Illumina HiSeq 2500
platform at Yunxu Co. Ltd. The aggregated distribution of gene
expression levels in pathways was determined by normalized
enrichment score, which represented statistical significance
following enrichment analysis. The KEGG database (http://www.genome.jp/kegg/) was used for pathway
annotation. Pathways that were significantly biased in the control
and model group were identified.
lncRNA-Ankyrin repeat domain (Ankrd)26
and miRNA-150 mimic and inhibitor transfection
Cells were cultured in DMEM supplemented with 10%
FBS (both Gibco; Thermo Fisher Scientific, Inc.). The cells were
cultured at 37°C in a humidified atmosphere of 5% CO2.
Transfection with lncRNA-Ankrd26 and miRNA-150 mimic and inhibitor
were with 30 nM concentration at room temperature performed using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocols. After
48 h of transfection, the cells were harvested for reverse
transcription-quantitative (RT-q)PCR analysis and western blotting.
All transfections were confirmed with appropriate controls,
including mimic negative control or inhibitor negative control. The
transfection efficacy was confirmed (Figs. S1 and S2). The sequence
information is listed in Table
SI.
TLR4 knockdown
The short hairpin RNA (shRNA) carrier was
constructed to target the tlr4 gene by respectively
inserting three different target sequences into the plasmid pLKO.1
(catalog no. 10878; Addgene, Inc.). In brief, lentiviral vectors
pLKO.1 TRC and pWPI.1 were used for constructing recombinant
lentiviruses of short interference RNA (shRNA) constructs and TLR4
shRNA, non-targeting shRNA (shNT). Recombinant lentivirus was
amplified in 293T cells. After transfection, MSCs cells with TLR4
knockdown were screened and obtained for subsequent detection with
western blotting or migration assay.
Migration assay
The migratory capacity of DPSCs with/without
exosomal inhibitor (GW4869) (catalog no. HY-19363; MedChemExpress)
or shTLR4 treatment was tested using a Transwell Boyden Chamber
(6.5-mm; Costar) with polycarbonate membranes (8-µm pore size) on
the bottom of the upper compartment. Cells were seeded in the upper
chamber at a density of 5×104 cells/well with serum-free
high-glucose DMEM; the lower chamber was filled with 600 µl
high-glucose DMEM containing 20% FBS (Gibco; Thermo Fisher
Scientific, Inc.). At the end of the incubation at 37°C for 48 h,
the cells that penetrated through to the lower surface of filter
membranes were fixed with 90% ethanol for 15 min at room
temperature and stained with 0.1% crystal violet solution for 5 min
at room temperature. The stained cells were counted under a light
microscope at ×100 magnification (Olympus Corporation).
Dual-luciferase reporter assay
TargetScan online database (http://www.targetscan.org/vert_71/) was used to
predict target genes for miR-150. The sequences of the TLR4
wild-type (WT) and mutant (Mut) tlr4 gene were cloned and
inserted into the 3′ untranslated region (UTR) of the pEZX-MT01
vector (GeneCopoeia, Inc.). In six-well plates, 293T cells were
cultured to ~70% confluence and co-transfected with WT or Mut
luciferase reporter vector (2 µg) and mimic miRNAs or negative
control (NC; 2 µg) using Lipofectamine 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocols.
After 48 h, luciferase activity was detected using a
Dual-Luciferase® Reporter Assay System (Promega
Corporation) and normalized to Renilla activity. The
sequence information is listed in Table SI.
Ribonucleoprotein immunoprecipitation
(RIP) assay
The RIP assay was performed using the Magna
RIP™ Quad RNA-Binding Protein Immunoprecipitation Kit
(Merck KGaA). The cells transfected with miR-150 mimic or control
were lysed using RIP lysis buffer and then 100 µl of the lysate was
incubated with RIP immunoprecipitation buffer containing magnetic
beads, conjugated with anti-Argonaute-2 (Ago2) G beads (catalog no.
07–590) at 4°C for 90 min. Beads conjugated to human anti-Ago2
antibody or control IgG antibody were centrifugated at 600 × g for
1 min and then washed with RIPA buffer; after being resuspended in
50 mM Tris-HCl (pH 7.0), the beads were finally incubated at 70°C
for 45 min. RNA was extracted using TRIzol (Invitrogen; Thermo
Fisher Sientific, Inc.) following the manufacturer's instructions
and then quantified by RT-qPCR.
RT-qPCR
Total RNA was extracted from the cultured cells and
tissue using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocols. The expression of miRNA
was tested using Mir-X miRNA First Strand Synthesis and Mir-X miRNA
qRT-PCR SYBR kit (Takara Biotechnology Co., Ltd.). The expression
of TLR4 was tested using Transcriptor First Strand cDNA Synthesis
kit and FastStart Universal SYBR Green Master (Roche Diagnostics).
The amplification protocol was performed as follows: 1 cycle of 15
min at 95°C and a further 40 cycles of 15 sec at 95°C and 1 min at
60°C. GAPDH was used as the housekeeping gene for normalization of
the cDNA quantities. All reactions were performed in triplicate.
Data were analyzed according to the 2−ΔΔCq method
(18). The sequence information
is listed in Table SI.
Western blotting
Protein from cells, including DPSCs or MSCs, with a
variety of treatments, and pulp tissue from rats, was extracted
using RIPA buffer containing protease and phosphatase inhibitors
(Beyotime Institute of Biotechnology). The protein content of the
lysate was determined using the BCA protein assay (Beyotime
Institute of Biotechnology). A total of 20 µg protein/lane was
separated by 10% SDS-PAGE; then transferred onto PVDF membranes.
Following blocking with 5% BSA for 1 h at room temperature,
membranes were incubated with primary antibodies (TLR4, 1:1,000;
OCN, 1:1,000; OPN, 1:1,000; RUNX2, 1:1,000; CD36, 1:1,000; CD81,
1:1,000; GM130, 1:1,000; Galnexin, 1:1,000; β-actin, 1:5,000;
GAPDH, 1:1,000) at 4°C overnight followed by incubation with
horseradish peroxidase-conjugated secondary antibodies for 1 h.
Blots were visualized using SuperSignal West Pico Chemiluminescent
Substrate (Thermo Fisher Scientific Inc.). Results were normalized
to β-actin or GAPDH. All experiments were performed three times.
Quantification of western blots was analyzed densitometrically
using QuantityOne software (Bio-Rad Laboratories, Inc.). The
antibody information is listed in Table SII.
Statistical analysis
Statistical analysis was performed using GraphPad
11.0 (GraphPad Software, Inc.). For comparison of quantitative
variables between groups, one-way ANOVA followed by Bonferroni's
post hoc test for multiple comparisons or Mann-Whitney test was
used. For difference in proportions between groups, χ2
test was performed. The association between the various factors was
determined using Pearson's correlation. The independent assay was
performed repeatedly three times. Data are presented as the mean ±
SD. P<0.05 was considered to indicate a statistically
significant difference.
Results
Association between mRNAs, lncRNAs and
TLR signaling pathway during repair and regeneration of pulp
following injury
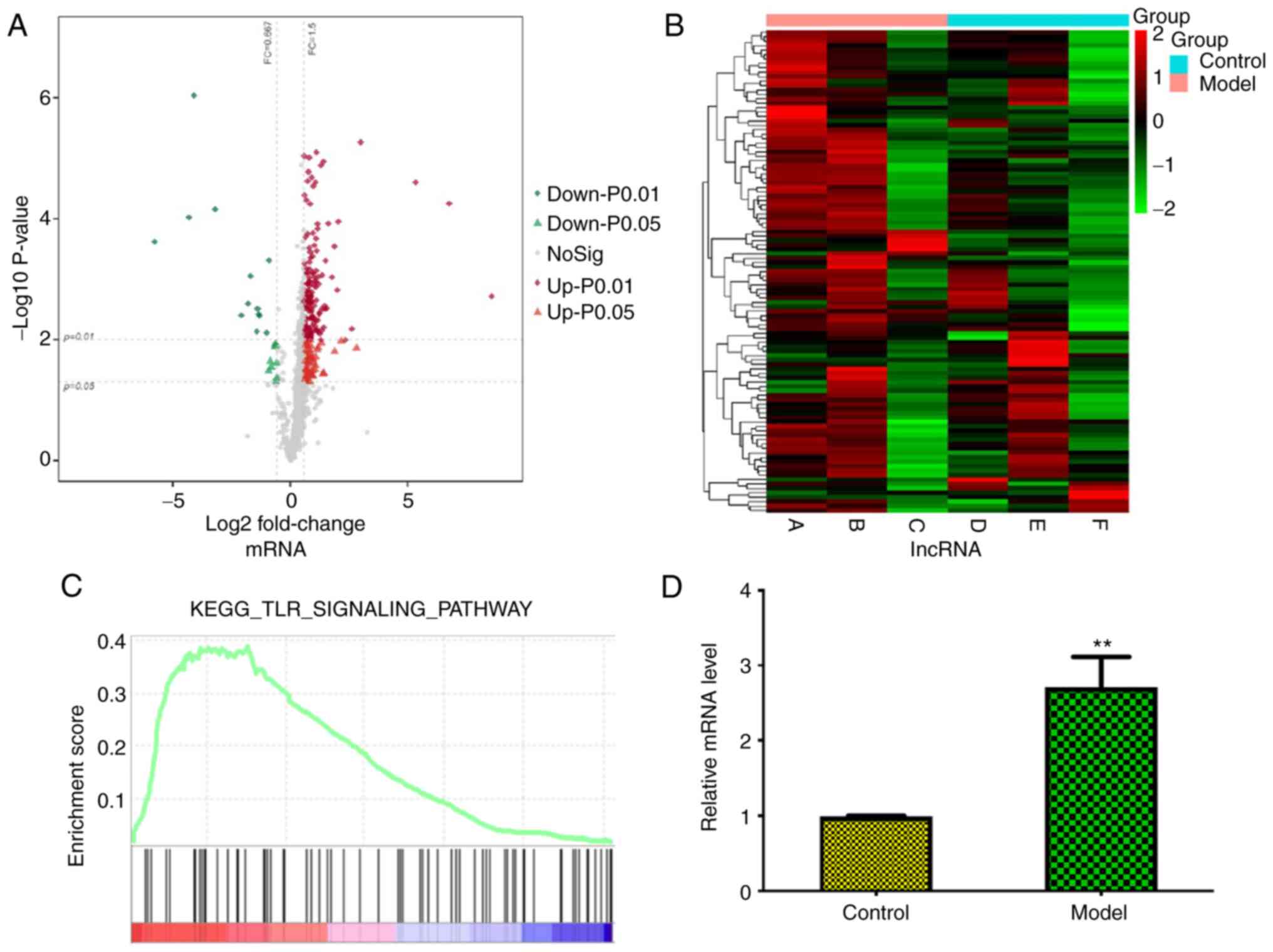
The present study compared next-generation
sequencing mRNA and lncRNA profiles of control and pulp injury
tissue from a gingival repair rat model and identified 10
significantly up- and downregulated mRNAs and lncRNAs ranked by
fold-change value (Fig. 1A and
B), such as AABR07015654, lnc102549726, AABR07026473 and
lncRNA-Ankrd26, among which lncRNA-Ankrd26 represented notably with
gradual increasing expression during the generation of a rat model.
GSEA analysis showed that these significantly altered mRNAs were
enriched in ‘Toll-like receptor signaling pathway’ (Fig. 1C); among these mRNAs, TLR4 was
most highly expressed, which was confirmed by qPCR (Fig. 1D). In addition, analysis of the
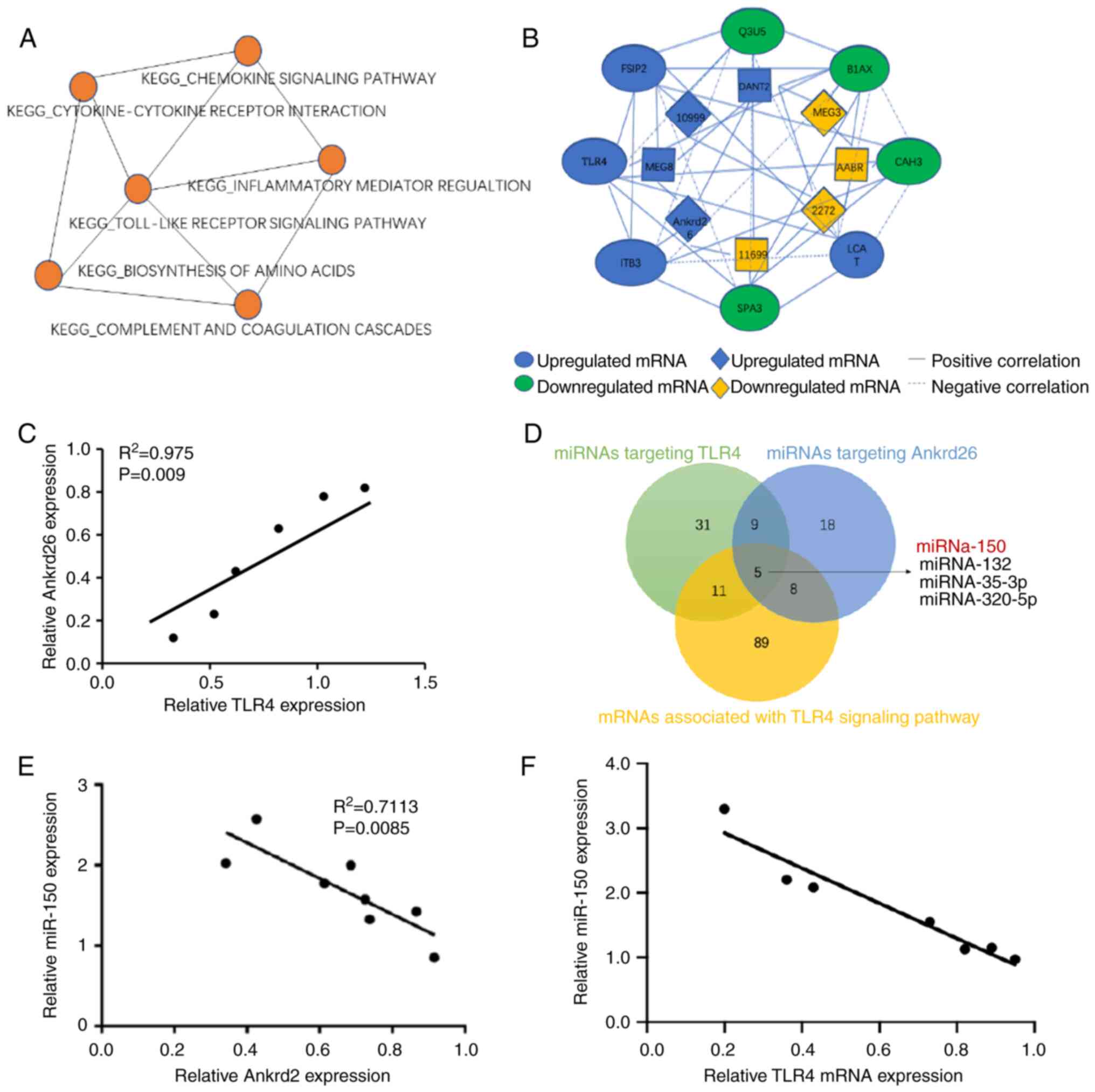
enrichment map showed ‘TLR signaling pathway’ contained genes which
overlapped with six other pathways. The gene co-expression network
displayed an association between lncRNA-Ankrd26 and TLR4 (Fig. 2A and B). Moreover, correlation
analysis showed lncRNA-Ankrd26 was positively linearly associated
with mRNA levels of TLR4 (Fig.
2C). To identify the association between lncRNA-Ankrd26 and
TLR4, potential miRNAs targeting Ankrd26 and TLR4 were
investigated. miR-150 was identified to be involved in the
association between Ankrd26 and TLR4 (Fig. 2D). Expression of miR-150 was
negatively associated with expression of both lncRNA-Ankrd26 and
TLR4 in model pulp tissue (Fig. 2E
and F). The results suggested that the
lncRNA-Ankrd26-miR-150-TLR4 axis could play a crucial role in the
repair and regeneration of injury pulp tissues.
DPSCs promote migration and
osteoblastic differentiation of MSCs via exosomes
The seeded DPSCs began to grow in 24 h. The primary
cells were relatively single, short spindle-like or round. At the
third passage, cell exhibited a spindle-shaped, fibroblast-like
appearance with a spherical or orbicular-ovate nucleus, as well as
rapid proliferation in a whorl-like formation (Fig. 3A). These cells displayed
logarithmic proliferation at 4–6 days and the doubling time was
24.37 h (Fig. 3B). Cell surface
markers CD29 and CD44 were highly expressed and CD34 and CD45 were
minimally expressed in DPSCs (Fig.
3C). To determine the effect of DPSCs on migration and
osteoblastic differentiation of MSCs, MSCs were cultured in
conditioned medium from DPSCs. Morphological alteration of MSCs was
observed after 4 days as well as significantly increased migration
and osteoblastic differentiation with the control (Fig. 3D and E). Due to the association
between lncRNA-Ankrd26, TLR4 and miR-150, levels of lncRNA-Ankrd26,
TLR4 and miR-150 were measured in MSCs cultured alone or in the
conditioned medium from DPSCs. The results showed significantly
higher levels of lncRNA-Ankrd26 and TLR4 and significantly lower
levels of miR-150 in MSCs with conditioned culture than in MSCs
cultured alone (Fig. 3F and G).
When exosomal inhibitor (GW4869) was added to the conditioned
media, migration and differentiation significantly decreased in
MSCs compared with MSCs in conditioned media without GW4869; there
was a significant decrease in lncRNA-Ankrd26 and TLR4 and
significant increase in miR-150 levels in conditioned culture MSCs
with GW4869 compared with those without GW4869 (Fig. 3H-J). These results suggested that
DPSC-derived exosomes could promote the migration and osteoblastic
differentiation of MSCs.
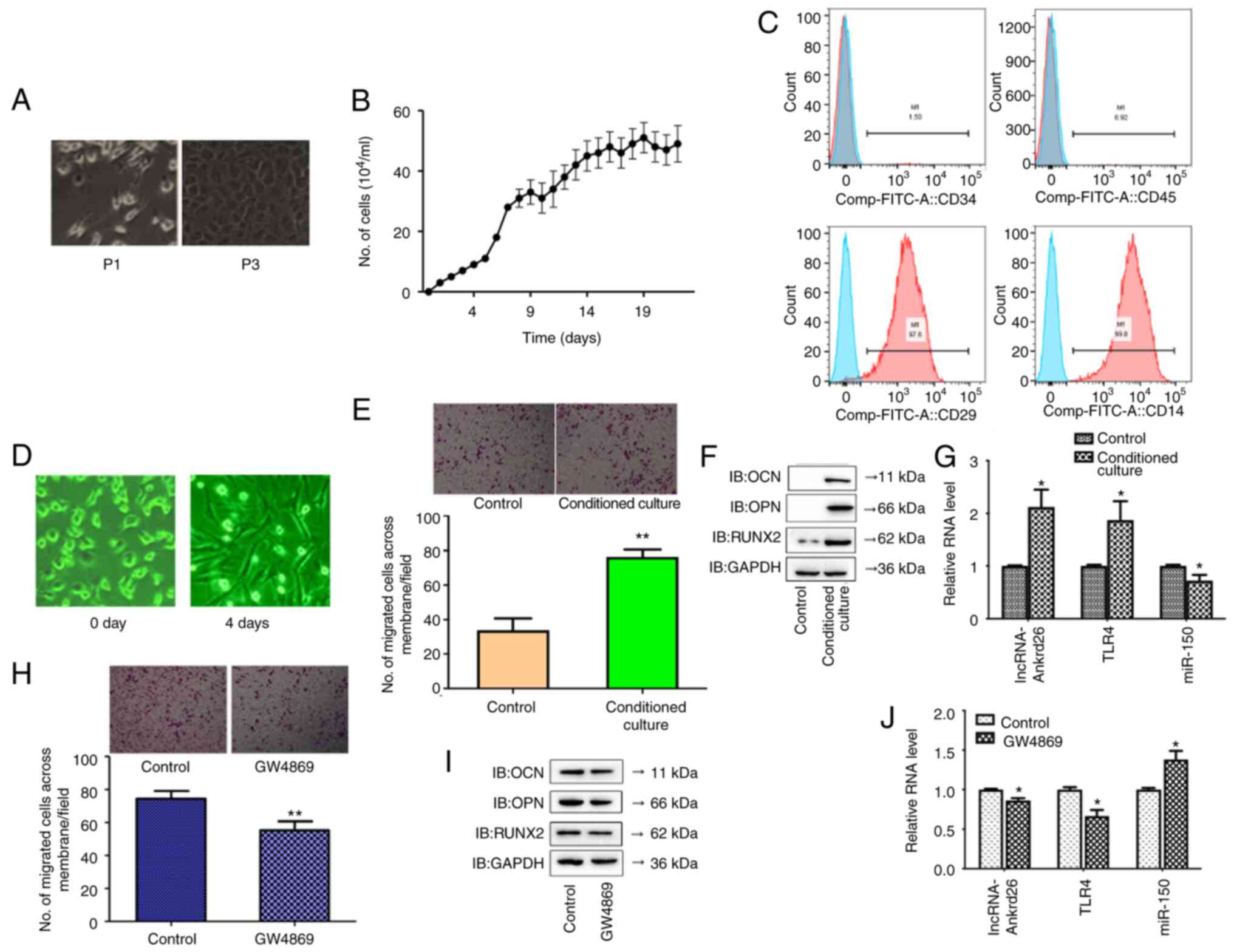 | Figure 3.Morphology, propagation, and
characterization of DPSCs. (A) Morphology of DPSCs from
subcutaneous fat tissue culture in vitro. Magnification,
×100. (B) Growth curves of DPSCs. (C) Expression of surface
antigens (CD29, CD34, CD44, CD45) in DPSCs was detected by flow
cytometry. The same negative control (IgG) is included in all
plots. (D) Morphological alteration of MSCs after 4 days (×200).
(E) Cell migration in MSCs with/without conditioned culture (×100
magnification); **P<0.01 compared with cells without conditioned
culture. (F) Protein levels of osteoblastic differentiation-related
markers, including OCN, OPN and RUNX2, in MSCs with conditioned
culture, compared with control. (G) mRNA levels of lncRNA-Ankrd26,
TLR4 and miR-150 in MSCs with conditioned culture. *P<0.05
compared with control. (H) Cell migration in MSCs with/without
GW4869 treatment (×100 magnification); **P<0.01 compared with
cells without treatment. (I) Protein levels of osteoblastic
differentiation-related markers, including OCN, OPN and RUNX2, in
MSCs treated with GW4869. (J) mRNA levels of lncRNA-Ankrd26, TLR4
and miR-150 in MSCs treated with GW4869. *P<0.05 compared with
control. The results are presented as the mean ± standard
deviation. DPSC, dental pulp stem cell; MSC, mesenchymal stem cell;
lnc, long non-coding; Ankrd, ankyrin repeat domain; TLR, Toll-like
receptor; miR, microRNA; P, passage; IB, immunoblot. |
DPSC-derived exosomal lncRNA-Ankrd26
induces migration and osteoblastic differentiation of MSCs
The present study identified exosomes in medium from
DPSCs by TEM (Fig. 4A), which
demonstrated positive expression of CD63 and CD81 and negative
expression of GM130 and calnexin (Fig. 4B). lncRNA-Ankrd26 expression was
significantly increased in DPSC-derived exosomes with
lncRNA-Ankrd26 transfection compared with cells without
transfection but was markedly inhibited in exosomes when DPSCs were
transfected with siRNA targeting lncRNA-Ankrd26 (Fig. 4C and D). Moreover, migration and
differentiation significantly decreased in MSCs cultured with
conditioned medium from DPSCs with lncRNA-Ankrd26 siRNA
transfection compared with MSCs cultured with conditioned medium
from DPSCs with control siRNA transfection (Fig. 4E and F). Together, these results
suggested that DPSC-mediated promotion of migration and
osteoblastic differentiation of MSCs may result from exosomal
lncRNA-Ankrd26.
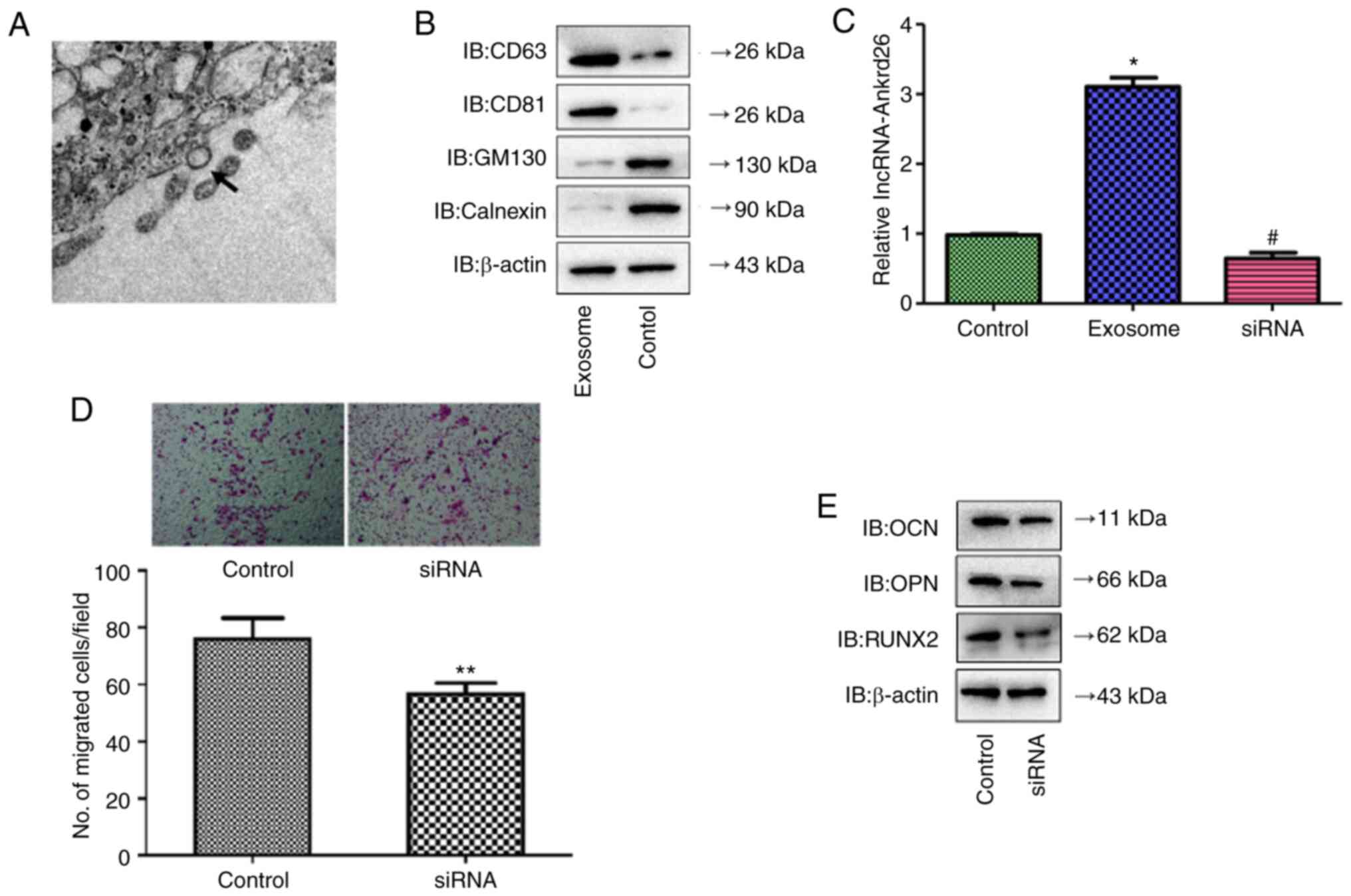 | Figure 4.Identification of DPSC-derived
exosomes. (A) Exosomes extracted from DPSCs were identified by
transmission electron microscopy. Magnification, ×150,000. Arrow
indicates the representative exosome. (B) Protein levels of CD63,
CD81, GM130 and calnexin in DPSC-derived exosomes compared with
DPSCs lysate (control) were determined by western blot analysis.
(C) lncRNA-Ankrd26 expression was significantly higher in
DPSC-derived exosomes with lncRNA-Ankrd26 transfection compared
with untransfected cells (control) but was markedly inhibited in
exosomes when DPSCs were transfected with siRNA targeting
lncRNA-Ankrd26. *P<0.05 compared with control;
#P<0.05 compared with exosome. (D) Migration and
differentiation significantly decreased in MSCs cultured with
conditioned medium from DPSCs with lncRNA-Ankrd26 siRNA
transfection compared with MSCs cultured with conditioned medium
from DPSCs with control siRNA transfection. **P<0.01 compared
with control. (E) Protein levels of osteoblastic
differentiation-related markers, including OCN, OPN and RUNX2, in
MSCs were determined by western blotting. DPSC, dental pulp stem
cell; MSC, mesenchymal stem cell; lnc, long non-coding; Ankrd,
ankyrin repeat domain; si, small interfering; IB, immunoblot. |
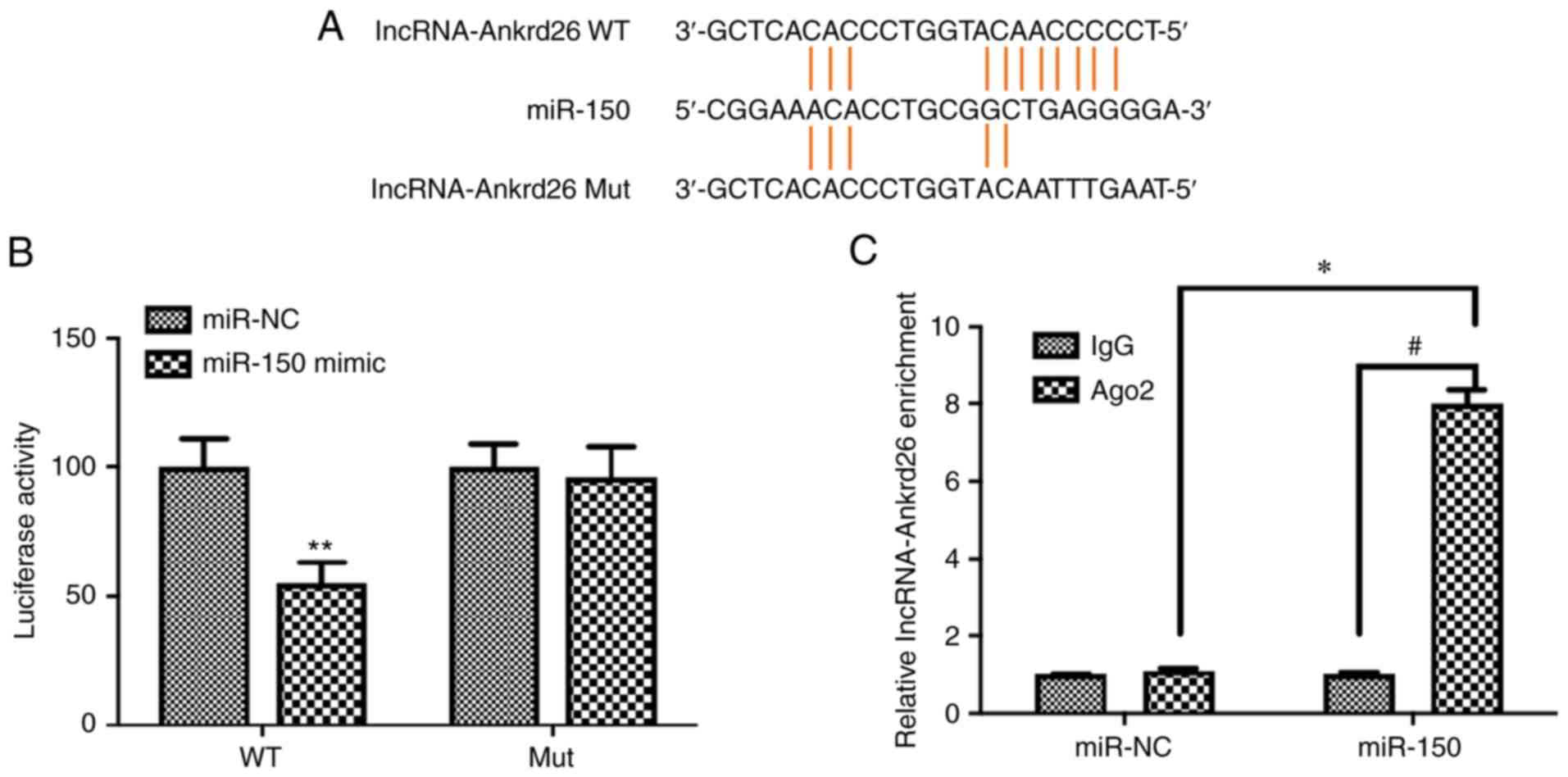
lncRNA-Ankrd26 directly regulates
miR-150
A putative binding site of lncRNA-Ankrd26 to miR-150
was mutated and co-transfected with miR-150 mimic into 293T cells
(Fig. 5A). Luciferase activity
assay demonstrated that, for 293T cells with WT lncRNA-Ankrd26
binding site construct, luciferase reporter activity significantly
decreased when cells were transfected with miR-150 mimic compared
with cells with miR-NC; Mut binding site construct-transfected
cells did not show the significant alteration in the activity
regardless of transfection with miR-150 mimic or miR-NC (Fig. 5B). To confirm the role of
lncRNA-Ankrd26 in regulating miR-150 expression, anti-AGO2
ribonucleoprotein immunoprecipitation assay was performed using
293T cells transfected with miR-NC or miR-150 mimic and
lncRNA-Ankrd26 levels were evaluated by RT-qPCR. The level of
lncRNA-Ankrd26 in cells transfected with miR-150 mimic was
significantly increased (Fig.
5C). The results suggested that there was a direct interaction
between lncRNA-Ankrd26 and miR-150.
Role of exosomal lncRNA-Ankrd26 is
dependent on miR-150/TLR4 signaling
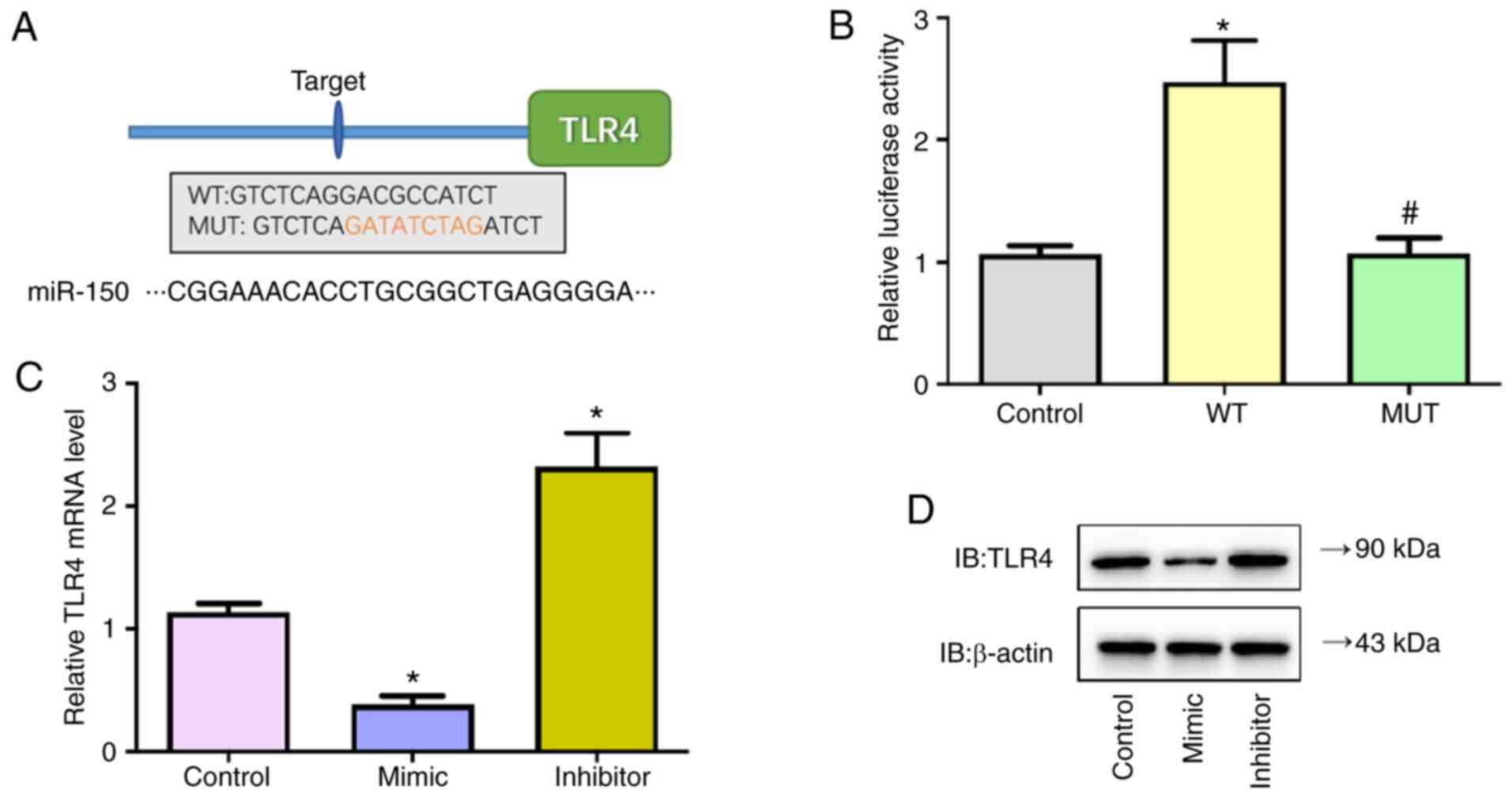
The present study confirmed direct binding of
miR-150 on the 3′-UTR of TLR4 by dual-luciferase reporter assay.
Firstly, by screening targets of miR-150 using Targetscan, TLR4 was
identified as a potential target of miR-150 (Fig. 6A). Next, predicted WT or Mut
full-length 3′-UTR of TLR4 gene was cloned into a dual-luciferase
reporter plasmid and then co-transfected with miR-150 mimic into
293T cells. Luciferase activity decreased following miR-150 mimic
co-transfection in cells with WT constructs, whereas the activity
was not altered in cells co-transfected with Mut constructs
(Fig. 6B). In addition, the
transcriptional and protein levels of TLR4 in MSC cells transfected
with miR-150 mimic, inhibitors or control were detected; miR-150
mimic significantly decreased mRNA and protein levels of TLR4 while
miR-150 inhibitors significantly increased levels of TLR4 (Fig. 6C and D). These findings verified
that miR-150 negatively regulated TLR4 expression by directly
targeting TLR4 in MSC cells.
To investigate whether the role of DPSC-derived
exosomal lncRNA-Ankrd26 in inducing migration and osteoblastic
differentiation of MSCs required miR-150/TLR4 signaling, TLR4 shRNA
was transfected into MSCs. Following transfection, TLR4 mRNA and
protein levels were significantly decreased (Fig. 7A and B). TLR4 knockdown notably
decreased osteoblastic differentiation markers, including
osteocalcin (OCN), osteopontin (OPN) and RUNX2, as well as cell
migration (Fig. 7C and D).
TLR4-knockdown MSCs cells were cultured in conditioned medium
containing DPSC-derived exosomal lncRNA-Ankrd26. When cultured in
conditioned medium containing DPSC-derived exosomal lncRNA-Ankrd26,
migration and OCN, OPN, RUNX2 expression of MSC cells transfected
with control shRNA were significantly increased, but TLR4-knockdown
MSC cells displayed no significant change (Fig. 7E and F).
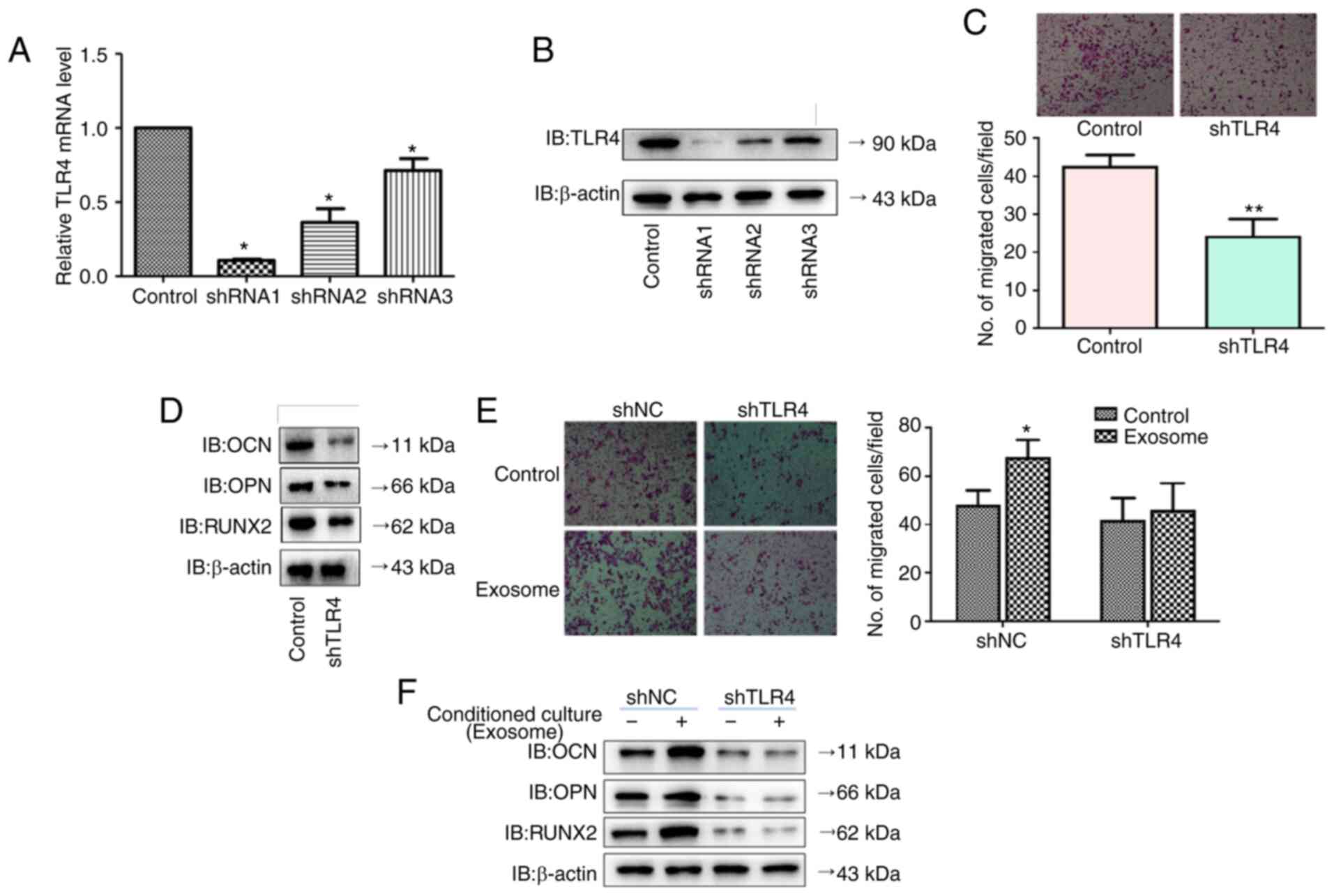 | Figure 7.Effects of DPSC-derived exosomal
lncRNA-Ankrd26 on MSCs requires the involvement of miR-150/TLR4
signaling. (A) mRNA and (B) protein levels of TLR4 were detected
following transfection of TLR4 shRNA into MSCs. *P<0.05 compared
with control. (C) Migration (×100 magnification) and (D)
osteoblastic differentiation markers, including OCN, OPN and RUNX2,
were detected following TLR4 knockdown. **P<0.01 compared with
control. (E) Migration (×100 magnification) and (F) osteoblastic
differentiation markers, including OCN, OPN and RUNX2, were
detected following culture in conditioned medium containing
DPSC-derived exosomal lncRNA-Ankrd26. *P<0.05 compared with
control. The results are presented as the mean ± standard
deviation. DPSC, dental pulp stem cell; MSC, mesenchymal stem cell;
lnc, long non-coding; Ankrd, ankyrin repeat domain; sh, short
hairpin; IB, immunoblot; miR, microRNA; TLR, Toll-like receptor;
OCN, osteocalcin; OPN, osteopontin; NC, negative control. |
Discussion
DPSCs have been used as key seed cells for pulp
regeneration and restoration and exosomes derived from DPSCs have
become a research hotspot in pulp regeneration and restoration
(19,20). A number of studies have shown that
stem cell-derived exosomes are associated with pulp regeneration
and inflammation (21–23), suggesting that stem cell-derived
exosomes have potential application value in pulp repair and
regeneration. Notably, DPSCs hold some advantages, including the
fact they are easy to obtain, and their strong proliferation
ability and neurotropism (24,25). To the best of our knowledge,
however, there are few studies on DPSC-derived exosomes in pulp
injury and repair processing. The study demonstrated that
DPSC-derived exosomes promoted migration and osteoblastic
differentiation of MSCs, which is an important step in the process
of dental pulp regeneration and repair. Mechanistically,
lncRNA-Ankrd26 served as a competitive endogenous miR-150,
regulated differentiation of MSCs via TLR4 signaling and
participated in dental pulp regeneration and repair.
As DPSCs have the characteristics of
multidirectional differentiation and angiogenesis, the use of DPSCs
to regenerate the pulp-dentin complex and repair damaged pulp is
expected to become a safe and effective treatment of pulp disease
(26,27). Success of periodontal cell-based
tissue engineering requires appropriate progenitor cells with the
capacity to differentiate into the required mature tissue-forming
phenotypes and appropriate signals to modulate cellular
differentiation and tissue neogenesis (28). Dental MSCs have the capacity of
differentiation into cells that present some characteristics
associated with osteoblasts, chondrocytes and adipocytes, thus
contributing to tooth growth and repair; easy accessibility of MSCs
provides a tractable model system to study their function and
properties in vivo (27,29). Therefore, MSC-based therapies are
being investigated in bone engineering. However, the association
between DPSCs and MSCs is still unclear. It has recently been
recognized that DPSCs express MSC surface markers, such as CD29,
CD44, CD59, CD73, CD90 and CD146, but do not express hematopoietic
stem cell markers, such as CD14, CD34, CD45 and CD11b, based on
multi-omics analysis (30,31).
In the present study, the DPSCs were purified and identified using
specific markers. Moreover, the present study used a conditioned
culture system and demonstrated that DPSCs promoted migration and
osteoblastic differentiation of MSCs, suggesting the importance in
understanding the crosstalk between different dental cell
populations.
Exosomes serve a key role in regulating cell-cell
interaction (32), which confers
the possibility of the application of exosomes in the clinical
practice of dental disease management. For example, exosomes from
DPSCs rescue human dopaminergic neurons from
6-hydroxy-dopamine-induced apoptosis (33) and suppress carrageenan-induced
acute inflammation in mice (34),
harbor stronger immune-modulating activity (35) and trigger regeneration of dental
pulp-like tissue (30).
Consistently, DPSC promotion of migration and osteoblastic
differentiation of MSCs is mediated by exosomes. To the best of our
knowledge, the present study is the first to report the association
between DPSC-derived exosomes and MSC differentiation.
To facilitate intercellular communication, exosomes
contain RNA and proteins (36).
Previous studies have demonstrated that exosomes participate in
epithelium-mesenchyme crosstalk in tooth morphogenesis and
differentiation by transferring RNA to recipient cells (37–39). Moreover, the present study
demonstrated that DPSC-derived exosomal lncRNA-Ankrd26 induced
migration and osteoblastic differentiation of MSCs by regulating
miR-150/TLR4 signaling, as shown by analysis dysregulated mRNAs,
lncRNAs and TLR signaling pathway during the repair and
regeneration of damaged dental pulp. The present study verified
that lncRNA-Ankrd26 directly regulated miR-150 in MSCs. To the best
of our knowledge, the present study is the first to clarify the
association between lncRNA Ankrd26 and miR-150 in osteoblast
differentiation and tissue restoration. The present study confirmed
the hypothesis that DPSC-derived exosomal lncRNA-Ankrd26 promotion
of migration and osteoblastic differentiation in MSCs was dependent
on of miR-150/TLR4 signaling. To the best of our knowledge, the
present study is the first to demonstrate the association between
lncRNA-Ankrd26 and dental pulp repair. ANKRD26 gene silencing and
variation are associated with regulation of pro-inflammatory
factors (40), platelet
aggregation (41) and
adipogenesis (42). TLR signaling
is a key mediator for inflammatory pathways and tissue response to
both pathogen- and damage-associated molecular pattern factors
(43). TLR4, a key component of
TLR family, serves a role in wound healing (44). TLR4 activation in MSCs mediates
production of multiple cytokines, chemokines and inflammatory
mediators, thus contributing to osteoclastogenesis (45). Enhancement of TLR4 activity
increases osteoblast viability (46) and differentiation of
adipose-derived stem cells (47).
These aforementioned studies suggested that TLR4 modulates
inflammation-induced healing response in different clinical
settings.
There are certain limitations in the present study.
Cell-derived exosomes contain a variety of components including
inflammatory and growth factors, genetic material and lipids
(13,14). Although the present study
demonstrated the role of DPSC-derived exosomal
lncRNA-Ankrd26-miR-150-TLR4 signaling in regulating migration and
osteoblastic differentiation of MSCs in pulp regeneration and
restoration, other exosomal content may serve a role in crosstalk
between DPSCs and MSC. Secondly, the present study used cells from
model animals, which may be not accurately represent human disease.
Moreover, due to the limitations in clinical feasibility, pulp
tissue was used as control; however, control pulp tissue was not
active in pulp restoration. Patient-derived xenografts or more
appropriate cell models in vitro will be used in future. In
addition, the present study only investigated the role of
miR-150/TLR4 signaling in terms of the effects of DPSC exosomal
lncRNA-Ankrd26 on inducing migration and osteoblastic
differentiation of MSCs. Further studies are required to
characterize the mechanism by which exosomes control the migration
and osteoblastic differentiation of MSCs in pulp regeneration and
restoration. Nevertheless, the present results support an important
potential role for DPSC-derived exosomal lncRNA-Ankrd26 in
promoting migration and osteoblastic differentiation of MSCs.
Moreover, considering previous advances in miRNA-mediated therapy
in tissue regeneration and restoration (48–50), lncRNA-Ankrd26 may be a potential
target for patients with dental pulp inflammation.
In summary, the present study investigated the role
of lncRNA/Ankrd26/miR-150-TLR4 signaling in differentiation of stem
cells, which will provide understanding of the molecular mechanism
in dental pulp regeneration and restoration.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like thank Dr Yang Zhang (Fudan
University) for help in depositing the original data to a public
database.
Funding
The present study was supported by the Fund of Stomatology
Hospital Affiliated to Tongji University (grant no. 20183001).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the NCBI BIOSAMPLE repository
(ncbi.nlm.nih.gov/sra; accession no. PRJNA767485).
Authors' contributions
JG conceived and designed the study. LL and JG
analyzed the data, performed experiments and drafted the
manuscript. Both authors have read and approved the final
manuscript. LL and JG confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
All animal studies were approved by the
Institutional Animal Care and Use Committees of the Hospital of
Stomatology, Tongji University (approval no. 20180606; Jan 1,
2018), in compliance with the Basel Declaration and institutional
guidelines for the care and use of animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yamada Y, Nakamura-Yamada S, Kusano K and
Baba S: Clinical potential and current progress of dental pulp stem
cells for various systemic diseases in regenerative medicine: A
concise review. Int J Mol Sci. 20:11322019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chauhan R, Rasaratnam L, Alani A and
Djemal S: Adult dental trauma: What should the dental practitioner
know? Prim Dent J. 5:70–81. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mead B, Logan A, Berry M, Leadbeater W and
Scheven BA: Concise review: Dental pulp stem cells: A novel cell
therapy for retinal and central nervous system repair. Stem Cells.
35:61–67. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gronthos S, Mankani M, Brahim J, Robey PG
and Shi S: Postnatal human dental pulp stem cells (DPSCs) in vitro
and in vivo. Proc Natl Acad Sci USA. 97:13625–13630. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang R, Liu Y, Yu T, Liu D, Shi S and Zhou
Y and Zhou Y: Hydrogen sulfide maintains dental pulp stem cell
function via TRPV1-mediated calcium influx. Cell Death Discov.
4:692018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Munévar JC, Gutiérrez N, Jiménez NT and
Lafaurie GI: Evaluation of two human dental pulp stem cell
cryopreservation methods. Acta Odontol Latinoam. 28:114–121.
2015.PubMed/NCBI
|
|
7
|
Tatsuhiro F, Seiko T, Yusuke T, Reiko TT
and Kazuhito S: Dental pulp stem cell-derived, scaffold-free
constructs for bone regeneration. Int J Mol Sci. 19:18462018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang J, Ma H, Jin X, Hu J, Liu X, Ni L and
Ma PX: The effect of scaffold architecture on odontogenic
differentiation of human dental pulp stem cells. Biomaterials.
32:7822–7830. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kichenbrand C, Velot E, Menu P and Moby V:
Dental pulp stem cell-derived conditioned medium: An attractive
alternative for regenerative therapy. Tissue Eng Part B Rev.
25:78–88. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Morsczeck C and Reichert TE: Dental stem
cells in tooth regeneration and repair in the future. Expert Opin
Biol Ther. 18:187–196. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Iezzi I, Pagella P, Mattioli-Belmonte M
and Mitsiadis TA: The effects of ageing on dental pulp stem cells,
the tooth longevity elixir. Eur Cell Mater. 37:175–185. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schuh CMAP, Benso B and Aguayo S:
Potential Novel Strategies for the treatment of dental pulp-derived
pain: Pharmacological approaches and beyond. Front Pharmacol.
10:10682019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen T, Moscvin M and Bianchi G: Exosomes
in the pathogenesis and treatment of multiple myeloma in the
context of the bone marrow microenvironment. Front Oncol.
10:6088152020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li Y, Yin Z, Fan J, Zhang S and Yang W:
The roles of exosomal miRNAs and lncRNAs in lung diseases. Signal
Transduct Target Ther. 4:472019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Asgarpour K, Shojaei Z, Amiri F, Ai J,
Mahjoubin-Tehran M, Ghasemi F, ArefNezhad R, Hamblin MR and Mirzaei
H: Exosomal microRNAs derived from mesenchymal stem cells:
Cell-to-cell messages. Cell Commun Signal. 18:1492020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu Y, Xiao Q, Tian H, Zhang L and Zhang G:
Biological effects of the extracellular matrix on rat bone marrow
mesenchymal stem cells. Chin J Curr Adv Gen Surg. 10:26–29.
2007.(In Chinese).
|
|
17
|
Shu S, Yang Y, Allen CL, Maguire O,
Minderman H, Sen A, Ciesielski MJ, Collins KA, Bush PJ, Singh P, et
al: Metabolic reprogramming of stromal fibroblasts by melanoma
exosome microRNA favours a pre-metastatic microenvironment. Sci
Rep. 8:129052018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang CC, Narayanan R, Alapati S and
Ravindran S: Exosomes as biomimetic tools for stem cell
differentiation: Applications in dental pulp tissue regeneration.
Biomaterials. 111:103–115. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hu X, Zhong Y, Kong Y, Chen Y, Feng J and
Zheng J: Lineage-specific exosomes promote the odontogenic
differentiation of human dental pulp stem cells (DPSCs) through
TGFβ1/smads signaling pathway via transfer of microRNAs. Stem Cell
Res Ther. 10:1702019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu B, Zhang X and Li X: Exosomes derived
from mesenchymal stem cells. Int J Mol Sci. 15:4142–4157. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hao ZC, Lu J, Wang SZ, Wu H, Zhang YT and
Xu SG: Stem cell-derived exosomes: A promising strategy for
fracture healing. Cell Prolif. 50:e123592017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mendt M, Rezvani K and Shpall E:
Mesenchymal stem cell-derived exosomes for clinical use. Bone
Marrow Transplant. 54 (Suppl 2):S789–S792. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sharma A: Role of stem cell derived
exosomes in tumor biology. Int J Cancer. 142:1086–1092. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Harrell CR, Jovicic N, Djonov V,
Arsenijevic N and Volarevic V: Mesenchymal stem cell-derived
exosomes and other extracellular vesicles as new remedies in the
therapy of inflammatory diseases. Cells. 8:16052019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhai Q, Dong Z, Wang W, Li B and Jin Y:
Dental stem cell and dental tissue regeneration. Front Med.
13:152–159. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hu L, Liu Y and Wang S: Stem cell-based
tooth and periodontal regeneration. Oral Dis. 24:696–705. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Han J, Menicanin D, Gronthos S and Bartold
PM: Stem cells, tissue engineering and periodontal regeneration.
Aust Dent J. 59 (Suppl 1):S117–S130. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sharpe PT: Dental mesenchymal stem cells.
Development. 143:2273–2280. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Stanko P, Altanerova U, Jakubechova J,
Repiska V and Altaner C: Dental mesenchymal stem/stromal cells and
their exosomes. Stem Cells Int. 2018:89736132018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cui D, Li H, Wan M, Peng Y, Xu X, Zhou X
and Zheng L: The origin and identification of mesenchymal stem
cells in teeth: From odontogenic to non-odontogenic. Curr Stem Cell
Res Ther. 13:39–45. 2018.PubMed/NCBI
|
|
32
|
Mathieu M, Martin-Jaular L, Lavieu G and
Théry C: Specificities of secretion and uptake of exosomes and
other extracellular vesicles for cell-to-cell communication. Nat
Cell Biol. 21:9–17. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jarmalavičiūtė A, Tunaitis V, Pivoraitė U,
Venalis A and Pivoriūnas A: Exosomes from dental pulp stem cells
rescue human dopaminergic neurons from 6-hydroxy-dopamine-induced
apoptosis. Cytotherapy. 17:932–939. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pivoraitė U, Jarmalavičiūtė A, Tunaitis V,
Ramanauskaitė G, Vaitkuvienė A, Kašėta V, Biziulevičienė G, Venalis
A and Pivoriūnas A: Exosomes from human dental pulp stem cells
suppress carrageenan-induced acute inflammation in mice.
Inflammation. 38:1933–1941. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ji L, Bao L, Gu Z, Zhou Q, Liang Y, Zheng
Y, Xu Y, Zhang X and Feng X: Comparison of immunomodulatory
properties of exosomes derived from bone marrow mesenchymal stem
cells and dental pulp stem cells. Immunol Res. 67:432–442. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
van der Grein SG and Nolte-'t Hoen EN:
‘Small Talk’ in the Innate immune system via RNA-containing
extracellular vesicles. Front Immunol. 5:5422014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Colombo M, Raposo G and Théry C:
Biogenesis, secretion, and intercellular interactions of exosomes
and other extracellular vesicles. Annu Rev Cell Dev Biol.
30:255–289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jiang N, Xiang L, He L, Yang G, Zheng J,
Wang C, Zhang Y, Wang S, Zhou Y, Sheu TJ, et al: Exosomes mediate
epithelium-mesenchyme crosstalk in organ development. ACS Nano.
11:7736–7746. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nakao Y, Fukuda T, Zhang Q, Sanui T,
Shinjo T, Kou X, Chen C, Liu D, Watanabe Y, Hayashi C, et al:
Exosomes from TNF-α-treated human gingiva-derived MSCs enhance M2
macrophage polarization and inhibit periodontal bone loss. Acta
Biomater. 122:306–324. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Desiderio A, Longo M, Parrillo L,
Campitelli M, Cacace G, de Simone S, Spinelli R, Zatterale F,
Cabaro S, Dolce P, et al: Epigenetic silencing of the ANKRD26 gene
correlates to the pro-inflammatory profile and increased
cardio-metabolic risk factors in human obesity. Clin Epigenetics.
11:1812019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen MH, Yanek LR, Backman JD, Eicher JD,
Huffman JE, Ben-Shlomo Y, Beswick AD, Yerges-Armstrong LM,
Shuldiner AR, O'Connell JR, et al: Exome-chip meta-analysis
identifies association between variation in ANKRD26 and platelet
aggregation. Platelets. 30:164–173. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fei Z, Bera TK, Liu X, Xiang L and Pastan
I: Ankrd26 gene disruption enhances adipogenesis of mouse embryonic
fibroblasts. J Biol Chem. 286:27761–27768. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
McKeown-Longo PJ and Higgins PJ:
Integration of canonical and noncanonical pathways in TLR4
signaling: Complex regulation of the wound repair program. Adv
Wound Care (New Rochelle). 6:320–329. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bhattacharyya S and Varga J: Endogenous
ligands of TLR4 promote unresolving tissue fibrosis: Implications
for systemic sclerosis and its targeted therapy. Immunol Lett.
195:9–17. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Alonso-Pérez A, Franco-Trepat E,
Guillán-Fresco M, Jorge-Mora A, López V, Pino J, Gualillo O and
Gómez R: Role of toll-like receptor 4 on osteoblast metabolism and
function. Front Physiol. 9:5042018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zheng L, Shen X, Ye J, Xie Y and Yan S:
Metformin alleviates hyperglycemia-induced apoptosis and
differentiation suppression in osteoblasts through inhibiting the
TLR4 signaling pathway. Life Sci. 216:29–38. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yu L, Qu H, Yu Y, Li W, Zhao Y and Qiu G:
LncRNA-PCAT1 targeting miR-145-5p promotes TLR4-associated
osteogenic differentiation of adipose-derived stem cells. J Cell
Mol Med. 22:6134–6147. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Olsen I, Singhrao SK and Osmundsen H:
Periodontitis, pathogenesis and progression: miRNA-mediated
cellular responses to porphyromonas gingivalis. J Oral Microbiol.
9:13333962017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Irhimeh MR, Hamed M, Barthelmes D,
Gladbach Y, Helms V, Shen W and Gillies MC: Identification of novel
diabetes impaired miRNA-transcription factor co-regulatory networks
in bone marrow-derived Lin-/VEGF-R2+ endothelial progenitor cells.
PLoS One. 13:e02001942018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kadota T, Fujita Y, Araya J, Watanabe N,
Fujimoto S, Kawamoto H, Minagawa S, Hara H, Ohtsuka T, Yamamoto Y,
et al: Human bronchial epithelial cell-derived extracellular
vesicle therapy for pulmonary fibrosis via inhibition of TGF-β-WNT
crosstalk. J Extracell Vesicles. 10:e121242021. View Article : Google Scholar : PubMed/NCBI
|