Introduction
Brucella melitensis (B. melitensis) is
a facultatively intracellular bacterium that causes brucellosis,
which is a human zoonosis that can be transmitted though infected
animals or contaminated food products. Brucellosis in humans and
animals can cause spontaneous abortion and diseases, such as
endocarditis, arthritis, meningitis and osteomyelitis. This disease
is a severe threat to human health and ruminant production, and
results in economic loss worldwide (1–5).
Brucella has a small genome, but can invade
the placental trophoblast cells (PTCs) of pregnant animals and
subsequently result in placentitis and even spontaneous abortion
(6). Brucella-infected PTCs
have reportedly adapted to specific pathological changes,
suggesting a unique pathogen-host relationship between B.
melitensis and PTCs (7). PTCs
are a link between mother and fetus and have an important role in
maternal-fetal immunity (8).
Dysfunction of PTCs may result in pregnancy-related diseases and
Brucella interacts with PTCs and can cause infertility and
spontaneous abortion (9). Thus,
PTCs serve a key role in the immune system (10,11).
Bacterial outer membrane proteins (OMPs) can be
easily recognized by the immune system, which produces antibodies
or activates cellular immune responses (12). During infection, OMP25, a surface
protein of Brucella, first establishes contact with host
cells and completes the adhesion process (13). Studies have revealed that the OMP
family is an important virulence factor of Brucella and that
the virulence of Brucella is directly related to the uptake
of iron (14–16); the pathogen efficiently uptakes iron
and increases its virulence (17,18).
Ferritin heavy polypeptide 1 (FTH1) is involved in
cell proliferation, immune response and iron homeostasis. Tsuji
et al (19,20) reported that the upstream region of
FTH1 gene contains an antioxidant response element, which
responds to oxidation reaction and protects cells from oxidative
damage. Oxidative reaction may lead to apoptosis (21).
Trophoblasts are target cells of Brucella and
there have been a number of studies on OMP25 in cell models,
including human monocyte/macrophages, microglial cells and RAW264.7
mouse macrophages (22–24). However, few studies on OMP25 in
human trophoblast cells have been reported. Therefore, the present
study aimed to examine this interaction in HPT-8 cells.
To improve our understanding of the interaction
between OMP25 of B. melitensis 16M strain and host FTH1 in
human trophoblast cell line HPT-8, the present study investigated
the related changes in the biological functions of
Brucella-infected HPT-8 cells. In addition, the mRNA
expression levels of Toll-like receptor (TLR)4, myeloid
differentiation primary response protein MyD88 (MyD88) and
inflammatory factors, including nitric oxide (NO), lactate
dehydrogenase (LDH) and TNF-α, in the OMP25-transfected HPT-8 cells
were analyzed.
Materials and methods
Bacterial strains, plasmids and cell
line
B. melitensis strain 16M strain was procured
from the Chinese Center for Disease Control and Prevention
(Beijing, China). Brucella was cultured in tryptic soy agar
(TSA) or tryptic soy broth (both Sigma-Aldrich; Merck KGaA) without
any antibiotics. Brucella was incubated in an atmosphere
with 5% CO2 at 37°C for 3 days. Subsequently, the cells
were observed and counted. DH5α and BL21 strains of Escherichia
coli (E. coli) were procured from Promega Corporation.
The two strains were grown on Luria-Bertani medium (Beijing
Solarbio Science & Technology Co., Ltd.). The culture medium
was supplemented with antibiotics [100 µg/ml ampicillin or
kanamycin (Invitrogen; Thermo Fisher Scientific, Inc.)] when
necessary. Purification of OMP25 recombinant protein was performed
as previously reported (24).
pGBKT7, a plasmid, was procured from Promega Corporation. pMD18-T
simple vector, another plasmid, was procured from Takara Bio, Inc.
DNA ladder was procured from Takara Bio, Inc. The human trophoblast
HPT-8 cell line was procured from Cell Resource Center (http://m.cellresource.cn/content.aspx)
and was cultured in Dulbecco's modified Eagle's medium (DMEM,
Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal
bovine serum (FBS, Gibco; Thermo Fisher Scientific, Inc.) at 37°C
in an atmosphere with 5% CO2 (vol/vol).
Construction of a cDNA library of
HPT-8 cells infected with B. melitensis 16M strain
A cDNA library of HPT-8 cells infected with B.
melitensis 16M strain was constructed as previously described
(25). Briefly, the number of HPT-8
cells was determined using flow cytometry. Viable B.
melitensis 16M strain was obtained by plating serially diluted
colony-forming units on TSA after a 4 day-incubation at 37°C in an
atmosphere with 5% CO2. The HPT-8 cells were infected
with the 16M strain at a dilution of 100:1 for 4 h at 37°C. The
infected cells were incubated at 37°C in an atmosphere with 5%
CO2.
To validate infection efficiency, genomic DNA was
separately prepared using a number of the infected cells
(1×105/ml) using a DNeasy® Blood & Tissue
kit (Qiagen GmbH), according to the manufacturer's protocols.
OMP25-F and OMP25-R primers (Table
I) were designed for the amplification of the OMP25
gene.
 | Table I.Primers used in the present
study. |
Table I.
Primers used in the present
study.
| Primer | 5′-3′ sequence |
|---|
| OMP25-F | Forward:
GAATTCATGCGCACTCTTAAGTCTCTC |
| OMP25-R | Reverse:
CTGCAGTTAGAACTTGTAGCCGATGCC |
| FTH1-F | Forward:
GCAGGATATAAAGAAACCAGA |
| FTH1-R | Reverse:
TCTCAATGAAGTCACATAAGT |
| 16S rRNA-F | Forward:
CACCCCGACGGCTAACATTCA |
| 16S rRNA-R | Reverse:
AGTGTAGAGGTGAAATTCGTA |
| GAPDH-F | Forward:
GTCGTGGAGTCTACTGGTGT |
| GAPDH-R | Reverse:
TGCTGACAATCTTGAGTGA |
| TLR4-F | Forward:
CTTTAGACCTGTCCCTGAACC |
| TLR4-R | Reverse:
CTAAACCAGCCAGACCTTGA |
| MyD88-F | Forward:
CCTAACCATGTCCCTGAACA |
| MyD88-R | Reverse:
GGTACATAATGGGTCCTTTCC |
To construct a cDNA library of HPT-8 cells, total
RNA was separately prepared from the infected cells using
TRIzol® (Thermo Fisher Scientific, Inc.) and Ultrapure
RNA kit (CoWin Biosciences; cat. no. CW0597S), according to the
manufacturer's protocols. HPT-8 cDNA library construction was
performed according to Make Your Own ‘Mate & Plate’ Library
System Protocol (Clontech Laboratories, Inc.) as previously
described (26). The inserts of
cDNA library were detected by long-distance PCR (LD-PCR) as
previously described (26) with
some modifications. The LD-PCR reaction conditions were as follows:
1 min at 95°C, followed by 40 cycles at 95°C for 15 sec, 68°C for 3
min.
Construction of the pGBKT7-OMP25 bait
system
The open reading frame of OMP25 in the genome of
B. melitensis 16M was amplified using OMP25-F and OMP25-R
primers (Table I). The total PCR
reaction volume was 15 µl, containing ddH2O 13.9 µl,
Up-primer (25 µM) 0.4 µl, Down-primer (25 µM) 0.4 µl, dNTP (2.5
mmol) 0.8 µl, 10× PCR Buffer 2.0 µl, Templet (genome of B.
melitensis 16M) 2.0 µl, Taq DNA Polymerase (5 U/µl) 0.5 µl. The
PCR reaction conditions were as follows: 5 min at 95°C, followed by
30 cycles at 94°C for 30 sec, 55°C for 45 sec and 72°C for 1 min,
and 10 min at 72°C. The PCR product was cloned into pGBKT7 vector
via EcoR I/Xho I sites, and to generate a recombinant
plasmid: pGBKT7-OMP25. The recombinant pGBKT7-OMP25
plasmid was transformed into the Y2HGold yeast strain.
Negative and positive controls of
yeast two-hybrid system
Yeast two-hybrid test was performed, as previously
described (27). Competent Y187
cells that were transformed with pGADT7-T from the
Matchmaker™ Gold Yeast Two-Hybrid System (Clontech
Laboratories, Inc.) was used as a control for the prey system and
pGBKT7-53 and pGBKT7-Lam-transformed yeast Y187 cells were used as
a positive and negative controls, respectively. Then,
pre-two-hybrid experiments were then performed to test the positive
and negative control systems.
Interactive screening of Y2HGold
(pGBKT7-OMP25) and Y187 (pGADT7-cDNA) yeast strains
The pGBKT7-OMP25-containing yeast Y2HGold was
inoculated into 50 ml SD/-Trp/Kan (Clontech Laboratories, Inc.)
liquid medium (50 µg/ml) and cultured for 20 h at 30°C with shaking
at 10.48 × g. The bacterial density was adjusted to
>109/ml by a centrifugation at 1,000 × g at 30°C for
10 min. Yeast Y187 cells with a density of >2×107/ml
stored at −80°C were thawed and yeast two-hybrid assays were
performed using Matchmaker™ Gold Yeast Two-Hybrid System
(Clontech Laboratories, Inc.). The resuspended yeast culture was
spread onto 150 mm SD/-Ade/-His/-Leu/-Trp/X-α-Gal (QDO/X) plates
(Clontech Laboratories, Inc.) that were inverted and incubated at
30°C for 72 h. Afterwards, the recombinant plasmids were extracted
from the positive colonies that were identified by
SD/-Ade/-His/-Leu/-Trp screening, PCR amplification and sequencing.
The PCR reaction volume and conditions were described above.
Validation of the yeast two-hybrid
results by co-immunoprecipitation (Co-IP)
The pGBKT7-OMP25 and positive plasmids
identified by pGADT7-cDNA library screening were transcribed and
translated using a TNT T7 polymerase-coupled reticulocyte lysate
system (Promega Corporation). Briefly, the total reaction volume of
TNT T7 transcription and translation was 25 µl, containing TNT
Rabbit Reticulyte Lysate 12.5 µl, TNT Reaction Buffer 1 µl, Amino
Acid Mixture Minus Leucine (1 mM) 0.5 µl, Amino Acid Mixture Minus
Methionine (1 mM) 0.5 µl, Recombinant RNasin Ribonuclease Inhibitor
(40 U/µl) 0.5 µl, DNA template 1.0 µl, Transcend tRNA 0.5 µl,
Nuclease-Free Water 7.0 µl, TNT RNA Polymerase 0.5 µl. In
vitro transcribed and translated bait and prey proteins (10 µl)
were incubated at room temperature for 1 h. Then, 10 µl c-Myc
monoclonal antibody (cat. no. MA1-980; Invitrogen; Thermo Fisher
Scientific, Inc.) was added to the protein mixture and the mixture
was incubated at room temperature for another 1 h. Subsequently,
the above samples were added to a centrifuge tube containing 3 µl
Protein A bead (Matchmaker™ Co-IP kit, cat. no. 630449,
Clontech Laboratories, Inc.) and incubated at room temperature for
1 h. Then 500 µl Buffer 1 was added and centrifuged at 350 × g for
30 sec at room temperature. The supernatant was discarded, 600 µl
Buffer 2 added and centrifuged at 350 × g for 30 sec at room
temperature and the supernatant discarded. The proteins were
electrophoresed via SDS-PAGE on 12% gels and electrotransferred to
a nitro-cellulose (NC) membrane using a Mini Trans-Blot Cell
(Bio-Rad Laboratories, Inc.) at 200 mA for 1 h. The NC membrane was
incubated in blocking solution [5% nonfat milk in Tris-buffered
saline 0.05% Tween-20 (TBST)] for 1 h at room temperature. Then the
membrane was incubated in binding buffer (6 µl Streptavidin-AP in
15 ml TBST) at room temperature for 1 h. After three washes with
TBST, the membrane was stained with Western Blue Stabilized
Substrate (Promega Corporation) at room temperature for 2–6 h. The
signal was detected and densitometry performed by a Micro-Chemi
instrument (GelView 6000Plus, Guangzhou Biolight Biotechnology Co.,
Ltd.).
Reverse transcription-quantitative
(RT-q) PCR
The total RNA was separately prepared from the
infected cells using TRIzol® (Thermo Fisher Scientific,
Inc.) and Ultrapure RNA kit (CoWin Biosciences; cat. no. CW0597S),
according to the manufacturer's protocols. cDNA was synthesized
from the total RNA of the infected cells (1×105) using
reverse transcription kit (cat. no. 639505; Takara Biotechnology
Co., Ltd.) at 42°C for 50 min and at 85°C for 5 min, and was then
used as a PCR template for amplification of FTH1 and
construction of pGM-T-A/B/C/D plasmids. The plasmids were extracted
from the positive clones that were identified using PCR and
considered a reference standard after determining the concentration
of plasmids. Furthermore, the pGM-T-FTH1 plasmid was identified
using EcoR I/Xho I digestion. Genes were amplified as
templates with pGM-T-FTH1 plasmid, 16M, GAPDH and HPT-8 cDNA with
primers (Table I) designed for
FTH1, 16S rRNA and reference gene GAPDH. RT-qPCR was performed
according to Roche LightCycler® 480 (Roche Diagnostics,
Switzerland) protocol; standard curves were generated based on the
standard templates. SYBR Green I Master was used as fluorophore
(Roche Diagnostics). The total RT-qPCR reaction volume was 10 µl,
containing ddH2O 3.6 µl, up-primer (10 µM) 0.2 µl,
down-primer (10 µM) 0.2 µl, template 1.0 µl, SYBR Green I Master
5.0 µl. The thermocycling conditions were as follows: Preincubation
for 5 min at 95°C, and then 40 cycles of amplification (95°C for 30
sec, 60°C for 30 sec and 72°C for 30 sec). The relative
transcriptional levels were determined using the 2−∆∆Cq
method (28). All assays were
performed in triplicate and repeated at least three times.
Construction of pSIREN-siRNA vector
targeting FTH1 gene
The human FTH1 gene (NCBI Reference Sequence:
NM_002032) was submitted to Ambion (Thermo Fisher Scientific,
Inc.); three pairs of positive small interfering (si)RNA fragments
were identified for the FTH1 gene and a pair of siRNA
fragments containing a different gene was considered the negative
control (Table II). siRNA
fragments were synthesized by Sangon Biotech Co., Ltd. pSIREN-siRNA
expression vector with green fluorescent protein was constructed
according to the RNAi-Ready pSIREN-RetroQ ZsGreen Vector kit
(Clontech Laboratories, Inc.) protocol. The diluted siRNA fragment
(antisense strand) concentration was 100 µmol/l and the PCR
conditions were as follows: 95°C for 30 sec, 72°C for 2 min, 37°C
for 2 min and 25°C for 2 min. Synthetic double strands were diluted
to 0.5 µmol/l and 5 µl synthetic double strands were
electrophoresed in 1% agarose gel containing ethidium bromide; the
DNA fragments were ligated using RNAi-Ready pSIREN-RetroQ ZsGreen
Vector at 16°C overnight. Subsequently, the ligated product was
transformed into competent E. coli DH5α cells for 16–18 h
and sequenced by Sangon Biotech Co., Ltd., to identify the positive
plasmids, and digested using Mlu I and named pSIREN-A/B/C/D.
The plasmids were prepared using Endotoxin-Free Plasmid Highpure
kit (Tiangen Biotech Co., Ltd.), according to the manufacturer's
protocols.
 | Table II.Synthesized DNA sequences associated
with siRNA. |
Table II.
Synthesized DNA sequences associated
with siRNA.
| Primer | 5′-3′ sequence |
|---|
| FTH1-A1 |
GATCCAAACTGATGAAGCTGCAGAACTTCAAGAGAGTTCTGCA |
|
|
GCTTCATCAGTTTTTTTTTACGCGTG |
| FTH1-A2 |
AATTCACGCGTAAAAAAAAACTGATGAAGCTGCAGAACTCTCT |
|
|
TGAAGTTCTGCAGCTTCATCAGTTTG |
| FTH1-B1 |
GATCCAAAGAAACCAGACCGTGATGATTCAAGAGATCATCACG |
|
|
GTCTGGTTTCTTTTTTTTTACGCGTG |
| FTH1-B2 |
AATTCACGCGTAAAAAAAAAGAAACCAGACCGTGATGATCTCT |
|
|
TGAATCATCACGGTCTGGTTTCTTTG |
| FTH1-C1 |
GATCCAATCAGTCACTACTGGAACTGTTCAAGAGACAGTTCCA |
|
|
GTAGTGACTGATTTTTTTTACGCGTG |
| FTH1-C2 |
AATTCACGCGTAAAAAAAATCAGTCACTACTGGAACTCTCTCT |
|
|
TGAACAGTTCCAGTAGTGACTGATTG |
| FTH1-D1 |
GATCCAATTCGAATCTCGCTGACCAGTTCAAGAGACTGGTCAG |
|
|
CGAGATTCGAATTTTTTTTACGCGTG |
| FTH1-D2 |
AATTCACGCGTAAAAAAAATTCGAATCTCGCTGACCAGTCTCT |
|
|
TGAACTGGTCAGCGAGATTCGAATTG |
Detection of pSIREN-siRNA interference
effect
pSIREN-A/B/C/D plasmids were transfected into HTP-8
cells using a Lipofectamine® 2000 Transfection Reagent
kit (cat. no. 11668030; Invitrogen Co., Ltd.), and pSIREN-D was
used as the negative control. The transfected HPT-8 cells were
cultured in an atmosphere with 5% CO2 at 37°C for 48 h
and HPT-8 containing green fluorescent protein were observed under
a LSM510 confocal laser scanning microscope (Zeiss GmbH). Total RNA
was extracted from the transfected HTP-8 cells using Ultrapure RNA
kit (CoWin Biosciences, cat. no. CW0597S), according to the
manufacturer's protocols. cDNA was synthesized using a first-strand
cDNA synthesis kit (CoWin Biosciences, cat. no. CW0741), according
to the manufacturer's protocols. The mRNA expression of FTH1
gene was measured using RT-PCR as described above. The target gene
detection value and the reference gene detection value were
regarded as a correction value in evaluating the effect of each
interfering plasmid and were statistically analyzed before and
after transfection. The FTH1 gene corresponding value/GAPDH
detection value was used as a correction value and interference
efficiency was calculated as follows: Interference efficiency
(%)=(RNAi negative control group-RNAi test group)/RNAi negative
control group ×100% (29).
Determination of OMP25 concentration
by Bradford protein assay
The determination of OMP25 concentration was
performed using a protein assay kit (Shanghai Yeasen Biotechnology
Co., Ltd.).
Expression levels of NO, TNF-α and LDH
in HPT-8 cells transfected with OMP25
The expression levels of NO, TNF-α and LDH were
measured as previously described (30). HPT-8 cells were used to evaluated
the effects of NO, TNF-α and LDH activity. Briefly,
1×106 cells/well were cultured in a 24-well plate for 24
h at 37°C and transfected with OMP25 at low doses (0.25, 0.5 and 1
µg/ml) or high doses (1, 5, 10, 20 and 40 µg/ml). A group without
OMP25 was used as a control. Subsequently, 24 or 48 h
post-transfection, the supernatant was centrifuged at 16,000 × g at
room temperature for 15 min and collected, NO and TNF-α expression
levels were determined using a ELISA kit (NO, cat. no. KGE001;
TNF-α, cat. no. DTA00D; R&D Systems, Inc.) and LDH expression
level was determined using LDH assay kit (cat. no. ab102526; Abcam,
Inc.), according to the manufacturer's protocols.
Expression levels of NO and TNF-α
following pEGFP-OMP25 plasmid and siRNA-c co-transfection
Monolayers of HPT-8 cells at a density of
1×106 cells/well were cultured in a 6-well plate until
the cells achieved ~60% confluence. pEGFP-OMP25 (3 µg) and
siRNA-c (0, 2.5, 5, or 10 µl) were co-transfected into HPT-8 cells
using Lipofectamine® 2000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
At 48 h post-transfection, the supernatant was centrifuged at
16,000 × g at room temperature for 15 min and collected, and the
expression levels of NO and TNF-α were measured as described
above.
RT-qPCR for detection of TLR4 and
MyD88 mRNA levels
To determine the mRNA levels of TLR4 and MyD88, the
genes of TLR4 and MyD88 were cloned into pMD18-T simple vector. The
standard curves of RT-qPCR were constructed using the primers
TLR4-S, TLR4-A, MyD88-S and MyD88-A (Table I). RT-qPCR was performed as
described above to measure TLR4 and MyD88 mRNA levels and the
results were normalized against the standard curves.
Statistical analysis
The statistical differences between the groups were
analyzed using one-way analysis of variance (ANOVA) followed by
Tukey's post hoc test. The expression levels of NO, TNF-α and LDH
are presented as the mean ± standard deviation. The differences
between groups were analyzed using ANOVA in SPSS 17.0 software
(SPSS, Inc.). P<0.05 was considered to indicate a statistically
significant difference. The experiments were repeated three
times.
Results
Detection of infected HPT-8 cells
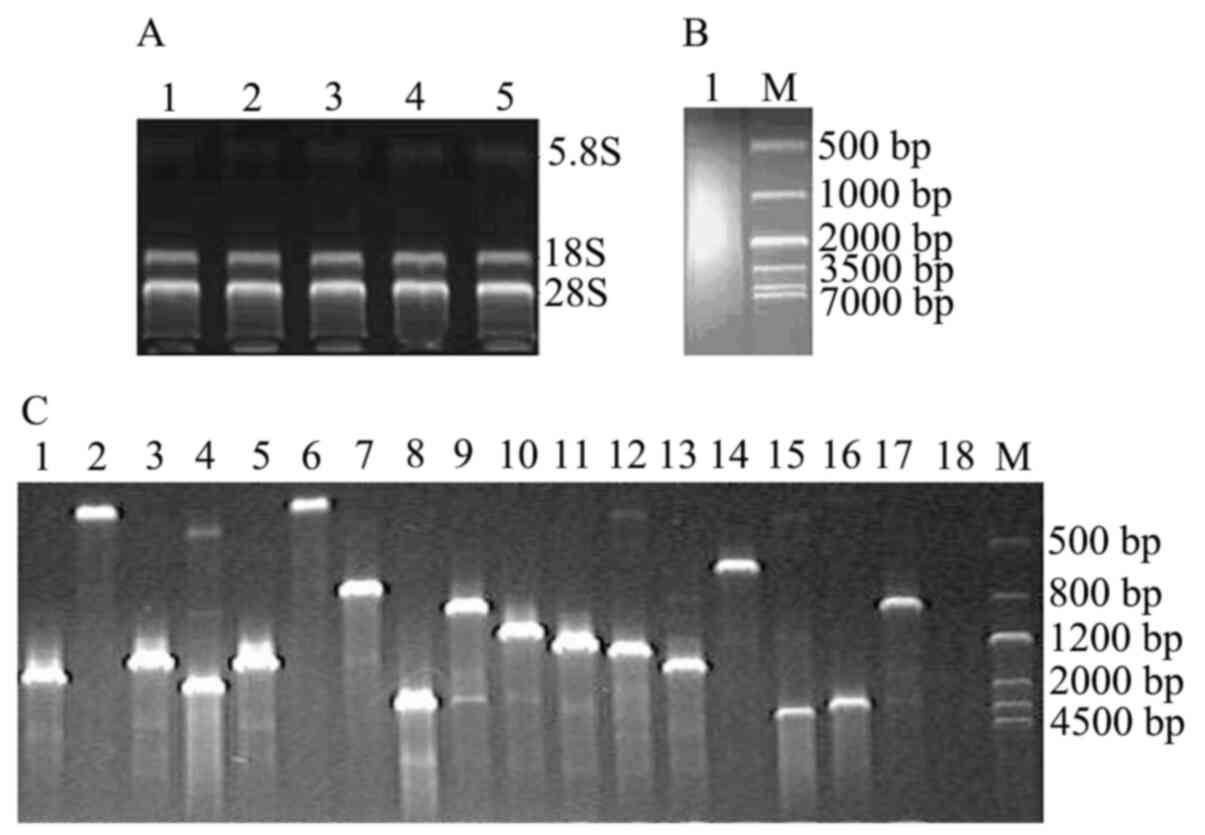
Three bands corresponding to 28S, 18S and 5.8S rRNA
in the total RNA extracts were observed (Fig. 1A). The A260/280 value was 1.90 (20
min post-infection), 2.02 (1 h post-infection), 1.92 (2 h
post-infection), 1.85 (3 h post-infection) and 2.04 (4 h
post-infection), indicating the high purity of the total RNA. The
library capacity of the cDNA was 1.43×106 transformants;
thereby reaching the construction requirements. The cDNA library
was also uncontaminated. The size of the cDNA insert fragments were
mainly in the range of 0.2–5 kb and with the majority near the 1–2
kb region (Fig. 1B and C).
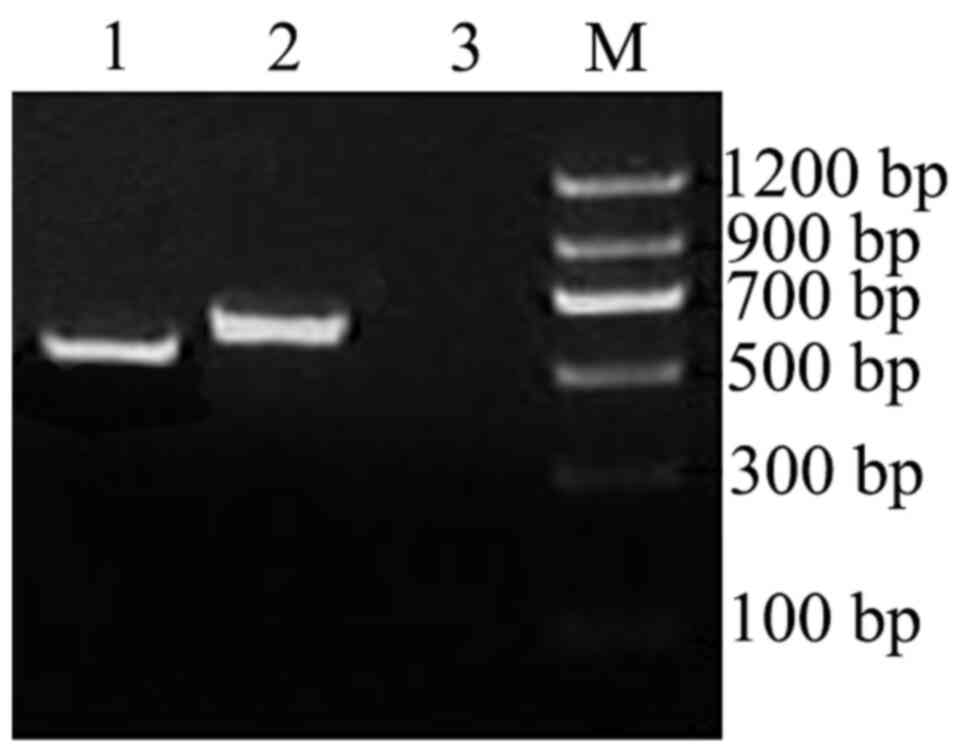
Yeast two-hybrid assay
DNA fragments of 640 bp size obtained using PCR and
restriction enzyme digestion indicated that the target was the
plasmid pGBKT7-OMP25. Y2HGold containing pGBKT7-OMP25
had no self-activation or toxicity for cells (Fig. 2A and B).
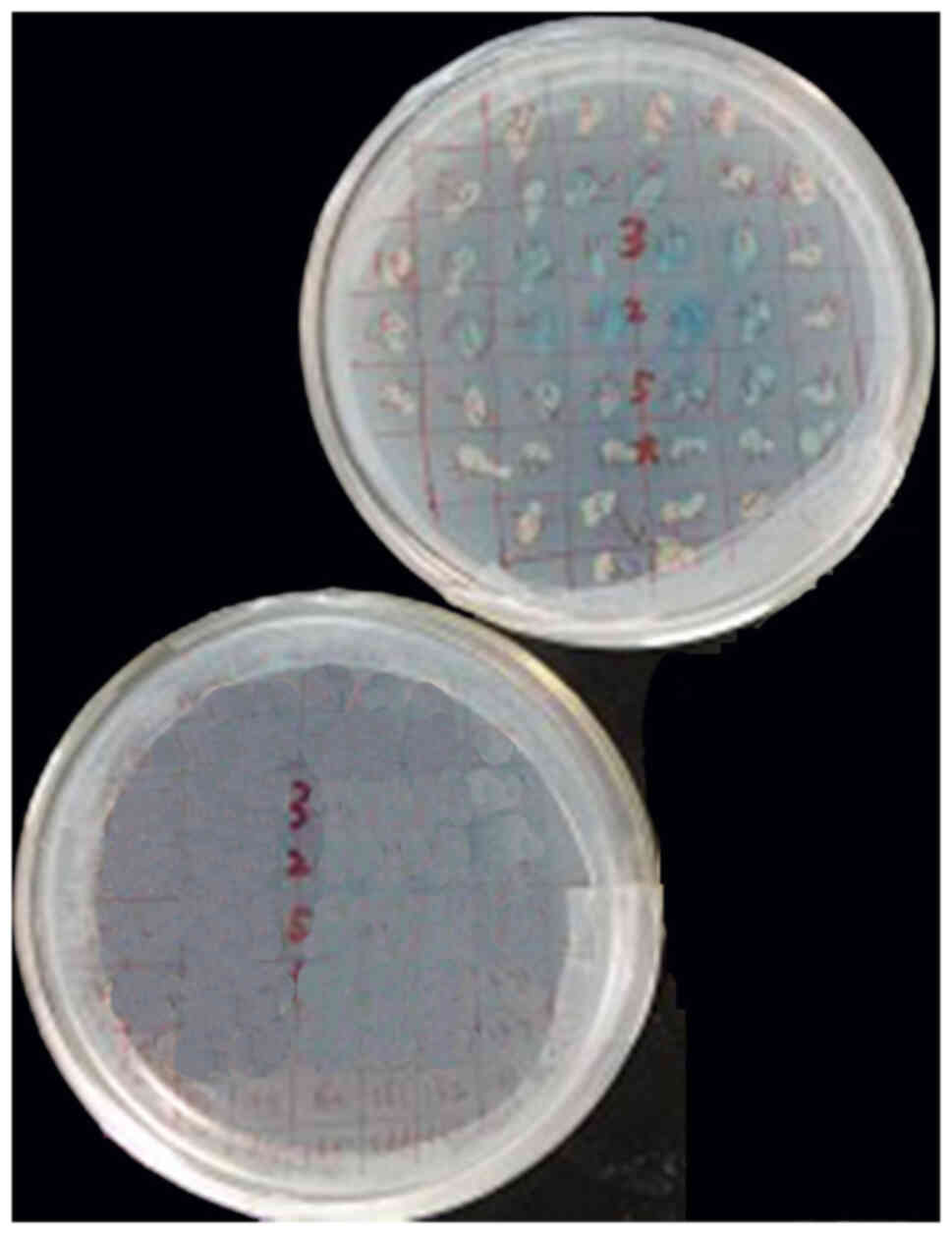
Control assay
Blue colonies (2–3 mm) on the
SD/-Ade/-His/-Leu/-Trp/X-α-Gal plates with Y187 (pGADT7-T) that
matched Y2HGold (pGBKT7-53) as positive control were observed. No
colonies were grown on the SD/-Ade/-His/-Leu/-Trp/X-α-Gal plates
with Y187 (pGADT7-T) that matched Y2HGold (pGBKT7-lam) as negative
control (Fig. 3).
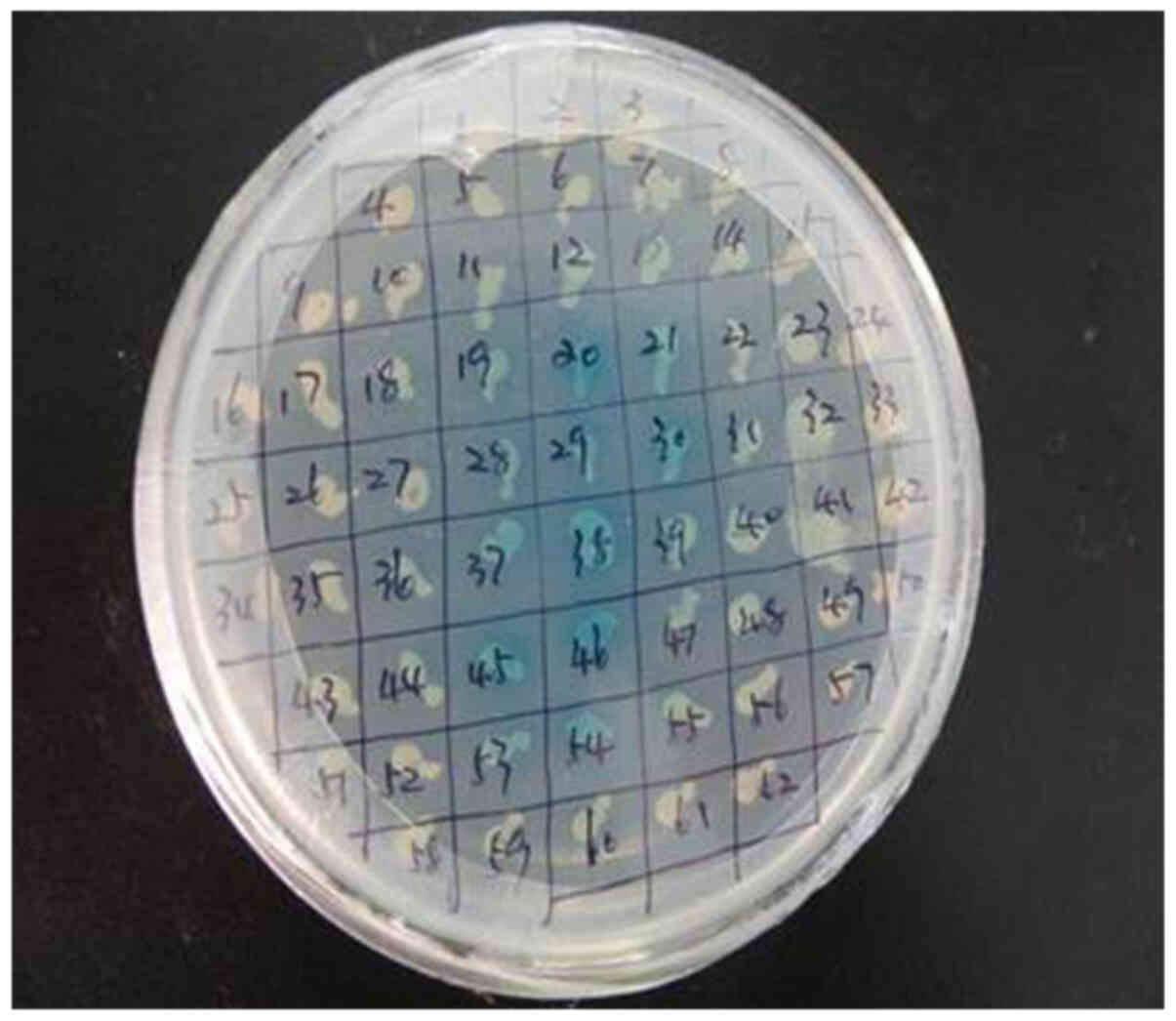
Interaction between prey protein and
OMP25 bait protein
Colonies of 2–3 mm on SD/-Ade/-His/-Leu/-Trp plates
with Y2HGold (pGBKT7-OMP25) matched Y187 (pGADT7-library)
and these colonies switched to SD/-Ade/-His/-Leu/-Trp/X-α-Gal
changed into blue colonies and were considered the positive
colonies (Fig. 4). The OMP25 bait
protein gene trap sequence of the prey protein gene was analyzed
and the results indicated that the pGBKT7-OMP25 bait plasmid
eventually yielded seven positive AD plasmids. Blast analysis
results are shown in Table
III.
 | Table III.Blast analysis result of AD
plasmid. |
Table III.
Blast analysis result of AD
plasmid.
| Gene | GenBank (Accession
no.) |
|---|
| Hypothetical
protein LOC789060 | NM_001110188 |
| NADH dehydrogenase
1 α subcomplex | NM_175826 |
| Ribonucleoprotein,
PTB-binding 1 mRNA | NM_001103108 |
| Eyes absent homolog
2 mRNA | NM_001035464 |
| Histone deacetylase
5 mRNA | NM_001038025 |
| Solute carrier
family 3 | NM_001024488 |
| Similar to Steroid
hormone receptor ERR1 | XM_001790466 |
| Tryptophanyl-tRNA
synthetase mRNA | BC102806 |
| Insulin-like growth
factor binding protein 6 mRNA | NM_001040495 |
| Amidohydrolase
domain containing 2 mRNA | NM_001101104 |
| Actin, β mRNA | BC142413 |
| Ancient ubiquitous
protein 1 mRNA | BC102888 |
| Haloacid
dehalogenase-like hydrolase domain containing | BC102640 |
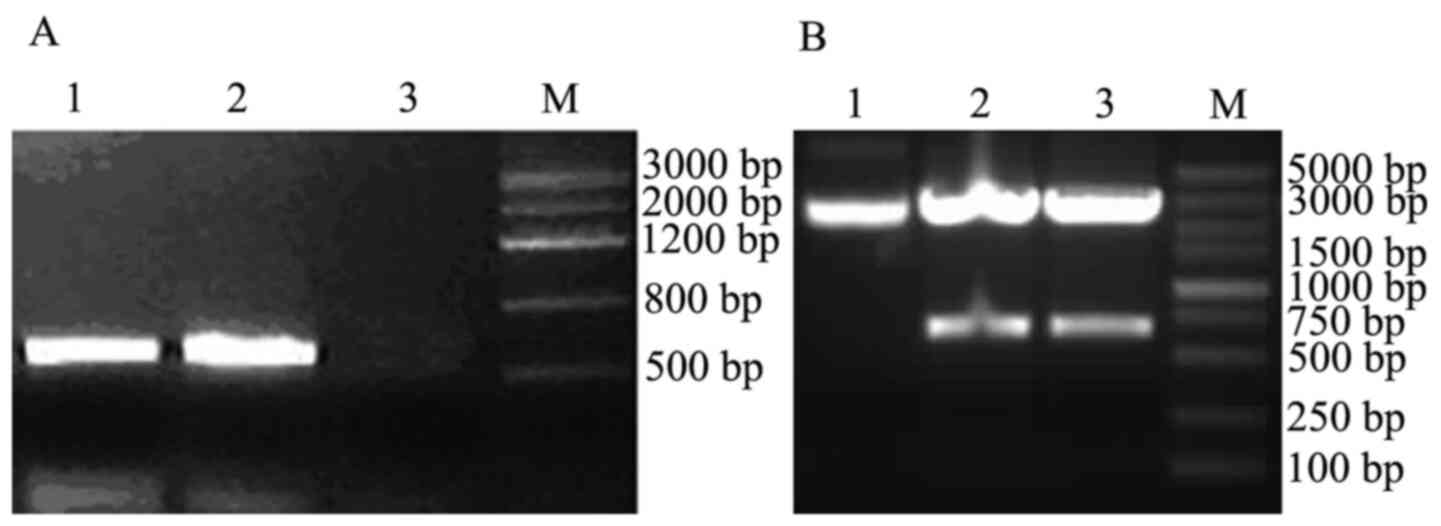
Verification of the interaction
between prey protein and OMP25 by Co-IP
OMP25 carrying c-myc tag and FTH1 carrying HA
tag, were amplified with templates as pGBKT7-OMP25 and
pGADT7-prey (FTH1 gene) using Advantage 2 PCR kit protocol
(Fig. 5). The Co-IP results
indicated that OMP25 interacted with FTH1.
Bio-function of the interaction
between FTH1 and OMP25 during brucellosis
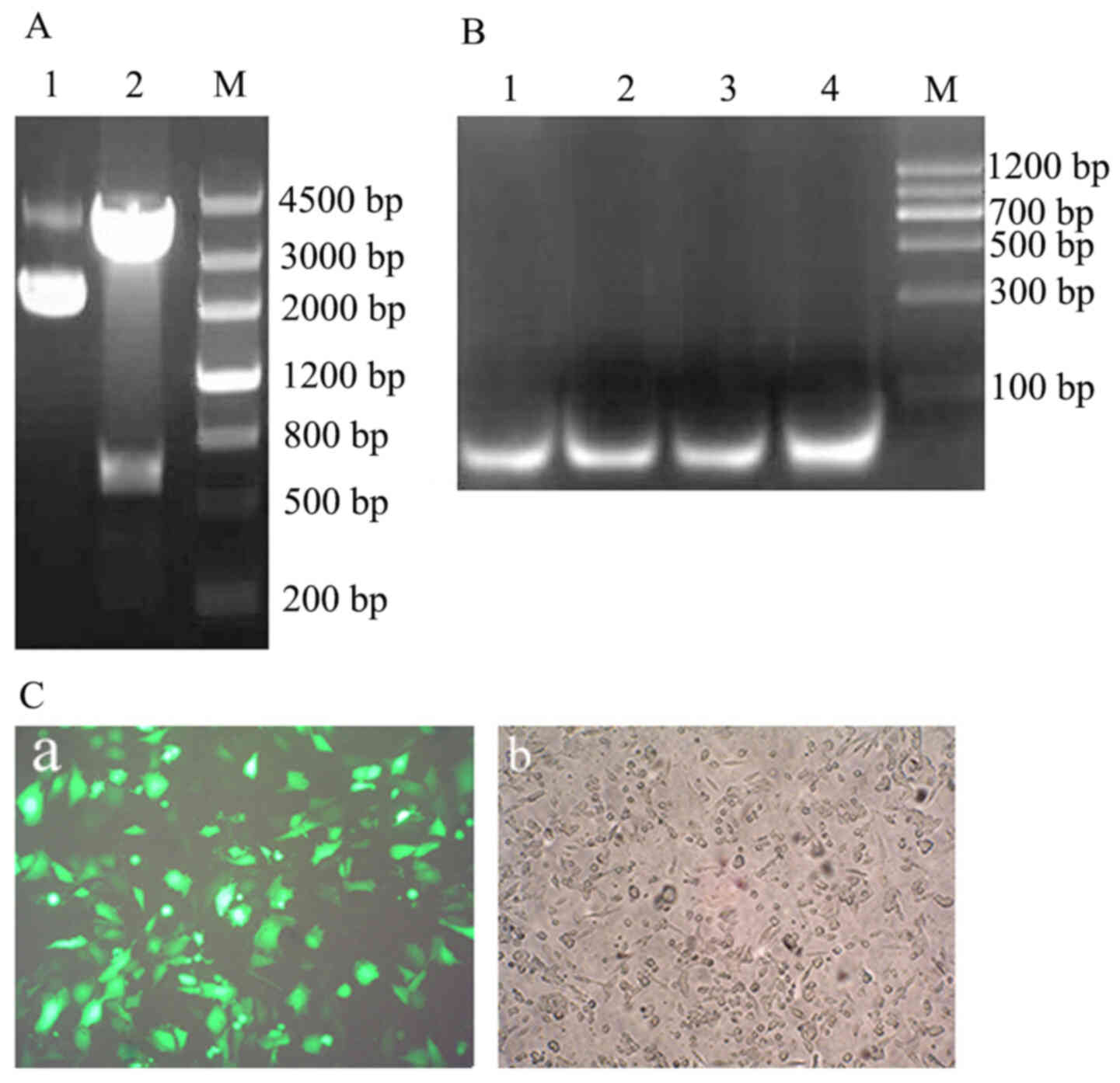
The obtained FTH1 gene ligated into pGM-T
vector generated a positive plasmid: pGM-FTH1; which was
identified using EcoR I/Xho I digestion and target
fragment indicated a 550 bp band (Fig.
6A).
The results of the green fluorescent protein
expression vector pSIREN-siRNA transfection indicated that a 69 bp
positive fragment was inserted into Linearized pSIREN Vector and
was sequenced (Fig. 6B); the
sequencing results indicated that four small fragments of DNA
sequences were identical with sequences in the original
sequence.
The RT-qPCR detection results of Brucella
survival ability in the transfected cells indicated that the
relative expression of 16S rRNA decreased by ~84% in HPT-8 cells
containing interference plasmid vs. empty HPT-8 cells infected with
Brucella; the viability of brucella was decreased in
the host cells transfected with FTH1 interference
fragment.
HPT-8 cells were transfected and subsequently
observed under a LSM 510 Laser scanning confocal microscope 48 h
post-transfection. The results indicated green fluorescence protein
expression in HPT-8 cells and a ~100% plasmid transfection
efficiency was achieved (Fig. 5C-a and
C-b).
The interference effect of four RNAi plasmids,
namely pSIREN-A, pSIREN-B, pSIREN-C and pSIREN-D on the FTH1
gene were detected, respectively. pSIREN-A, pSIREN-B and pSIREN-C
interference plasmids exerted interference effects and an
interference efficiency of ≤98% was achieved for pSIREN-C (Table IV).
 | Table IV.Optimal siRNA plasmid screening. |
Table IV.
Optimal siRNA plasmid screening.
| Detection
value | pSIREN-A | pSIREN-B | pSIREN-C | Negative |
|---|
| GAPDH average Cq
value (x) | 22.92 | 26.90 | 25.36 | 30.38 |
| GAPDH copies
(10y) | 11,081,537.00 | 1,475,707.00 | 3,219,586.00 | 253,162.90 |
| FTH1 average Cq
value (x) | 24.69 | 30.38 | 31.90 | 29.50 |
| FTH1 copies
(10y) | 3,939,671.00 | 120,759.10 | 47,599.24 | 207,014.10 |
| Inhibition ratio
for FTH1 (%) a | 57% | 89% | 98% | - |
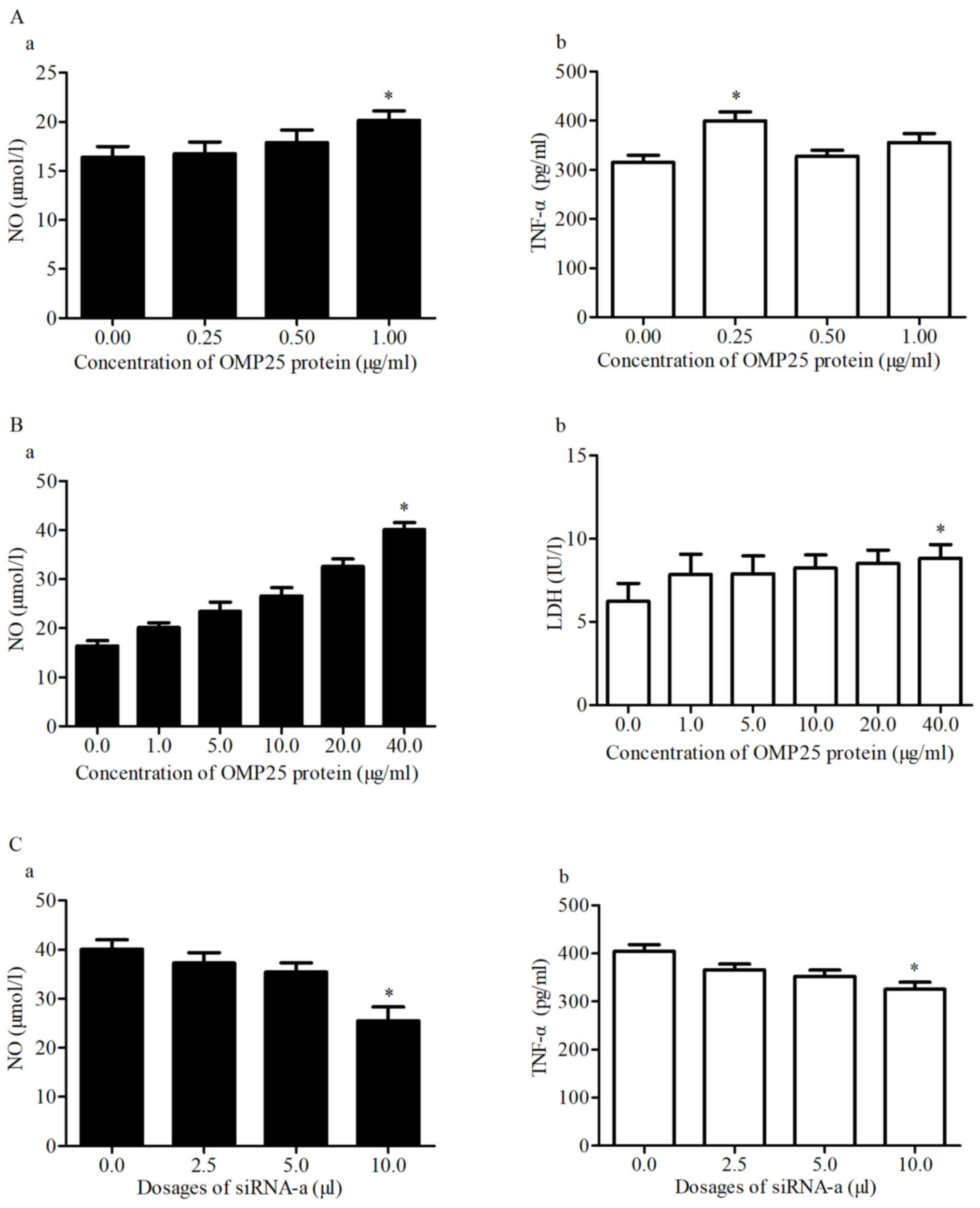
NO, TNF-α and LDH activity in HPT-8
cells treated with OMP25 and siRNA-a
Low doses of OMP25 affected HPT-8 cells after 24 h
and the levels of NO and TNF-α were positively associated with the
concentration of OMP25 (Fig. 7A-a and
-b). High doses of OMP25 affected HPT-8 cells after 24 h and
the levels of NO and LDH were increased with the concentration of
OMP25 (Fig. 7B-a and -b). Different
doses of siRNA-a were transfected into HPT-8 cells and the levels
of NO and LDH were decreased with the concentration of siRNA-a
(Fig. 7C-a and C-b).
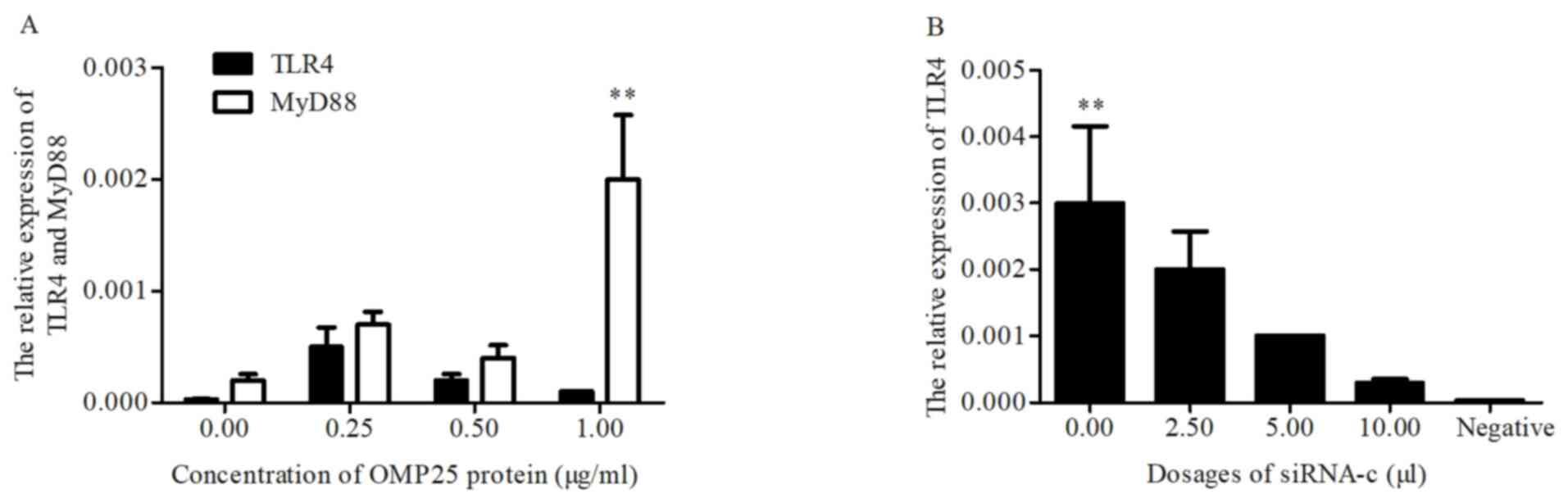
TLR4 and MyD88 mRNAs levels in HPT-8
cells treated with OMP25 protein and siRNA-c
Low doses of OMP25 affected HPT-8 cells and the
relative expression levels of TLR4 and MyD88 were associated with
the concentration of OMP25 (Fig.
8A). No significant differences observed for TLR4 (Fig. 8A).
siRNA-c and pGEFP-OMP25 were co-transfected
into HPT-8 cells and the relative expression levels of TLR4 were
decreased with the increase of the concentration of siRNA-c
(Fig. 8B).
Discussion
B. melitensis can efficiently invade
trophoblast cells in mammals and rapidly multiply within the
placenta, which can result in spontaneous abortion and placentitis
(24,25). Trophoblast cells are therefore the
primary target cells of B. melitensis in the infected host;
however, the infection of trophoblast cells remain poorly
understood. Spontaneous abortion in pregnant women resulting from
brucellosis may indicate that if Brucella can reach the
placenta and enter trophoblasts (27,30),
they may also become an important cellular niche. In vitro
studies regarding intracellular trafficking are few. The present
study successfully obtained a cDNA library from HPT-8 cell lines
infected with B. melitensis strain 16M and demonstrated that
the protein OMP25 directly interacts with eukaryotic proteins such
as FTH1, suggesting that OMP25 is a functional protein involved in
the alteration of host cell processes. The interactions between
B. melitensis protein and host proteins in vivo
further indicated that Brucella protein OMP25 serves an
important role in the modulation of host cells via protein-protein
or protein-DNA interactions.
B. melitensis OMP25 is a virulence protein
involved in intracellular replication. LDH, an intracellular
enzyme, is released into the extracellular matrix when cells are
damaged or undergo death; this results in an increase in LDH
activity in cell supernatants. In addition, LDH is considered an
index for cell injury (31). OMP25
at concentrations >5 µg/ml resulted in increased amounts of LDH
in HPT-8 cells.
TLR is expressed on the surface of epithelial cells
and is important to bacterial infection. The present study found
that TLR4 mRNA was lowly expressed in the HPT-8 cells, whereas a
low concentration of OMP25 upregulated TLR4 and inflammatory
signaling molecules, including TNF-α and NO, in the HPT-8 cells.
The activation of TLR4, MyD88, TNF-α and NO resulted in the
activation of the nuclear transcription factor NF-κB, which may
serve a key regulatory role in a variety of inflammatory responses
in host cells.
In a previous study (32), we found that OMP25 serves an
important role in the activation of the MAPK signaling pathway in
HPT-8 cells infected with Brucella. 2308ΔOmp25 mutant strain
could not activate p38 phosphorylation protein, ERK and JNK
branches in HPT-8 cells. However, in the present study, it was
found that OMP25 interacts with FTH1 in HPT-8 cells. The present
study provided a theoretical basis regarding the new functions of
OMP25.
The present study focused on the interaction between
OMP25 and key molecules of trophoblast cells. Further studies,
including on cell and animal experiments, are required to confirm
whether OMP25 can help in the infection of trophoblast cells. The
present study did not prove that OMP25 can help trophoblast
infection and this is one of its limitations.
In conclusion, the results of the present study
indicated that OMP25 interacts with FTH1. The mRNA expression
levels of TLR4, MyD88 and inflammatory factors, including NO, LDH
and TNF-α significantly increased after the interaction of OMP25
and FTH1 in the HPT-8 cells. OMP25-FTH1 interaction may contribute
to the exacerbation of the intracellular replication of
Brucella and the likelihood of a subsequent abortion or
stillbirth of the infected fetus. Further research is required to
elucidate the mechanisms underlying the molecular interactions of
OMP25 with the host, such that other Brucella effector
proteins and host cell targets can be identified to further
elucidate and define the molecular mechanisms of brucellosis.
Furthermore, the present study clarified the pathogenic mechanism
of brucellosis and may yield novel insights for the development of
therapeutic options.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the National
Natural Science Foundation of China (grant no. 31860691), the Key
Scientific Research Project of Colleges and Universities in Henan
Province (grant no. 21A230015), the National Innovation Training
Program for College Students of China (grant no. 202010483007), the
Foundation of the Technology Department of Henan Province (grant
no. 212102310746), the International Science and Technology
Cooperation Promotion Plan (grant no. GJHZ201709), the Training
Program for Excellent Young Teachers Colleges and Universities of
Corps (grant no. CZ027202), the Youth Science and technology
innovation leading talent program of Corps (grant no. 2017CB002)
and the Science and Technology Research and Development Program
(grant no. RCZX201403).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HZ, ZL, CC, CW and JL contributed to the conception
and design of the experiment. HZ, XW, ZL, JZ, YZ and YW performed
all experiments and verified the analytical data. JZ, YZ and YW
contributed to the statistical analysis and helped interpret the
results. HZ supervised the experiments in discussion with ZL. JZ,
YZ and YW wrote the manuscript. JL and HZ confirm the authenticity
of all the raw data. All authors discussed the final results and
all authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Purwar S, Metgud SC, Karadesai SG,
Nagamoti MB, Darshan A and Tiwari S: Triad of infective
endocarditis, splenic abscess, and septicemia caused by Brucella
melitensis. J Lab Physicians. 9:340–342. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Asmare K: Neospora caninum versus
Brucella spp. exposure among dairy cattle in Ethiopia: A
case control study. Trop Anim Health Prod. 46:961–966. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cash-Goldwasser S, Maze MJ, Rubach MP,
Biggs HM, Stoddard RA, Sharples KJ, Halliday JEB, Cleaveland S,
Shand MC, Mmbaga BT, et al: Risk factors for human brucellosis in
Northern Tanzania. Am J Trop Med Hyg. 98:598–606. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bayasgalan C, Chultemdorj T, Roth F,
Zinsstag J, Hattendorf J, Badmaa B, Argamjav B and Schelling E:
Risk factors of brucellosis seropositivity in bactrian camels of
Mongolia. BMC Vet Res. 14:3422018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
El-Sayed A and Awad W: Brucellosis:
Evolution and expected comeback. Int J Vet Sci Med. 6 (Suppl
1):S31–S35. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Meador VP and Deyoe BL: Intracellular
localization of Brucella abortus in bovine placenta. Vet
Pathol. 26:513–515. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang J, Li M, Li Z, Shi J, Zhang Y, Deng
X, Liu L, Wang Z, Qi Y and Zhang H: Deletion of the type IV
secretion system effector VceA promotes autophagy and inhibits
apoptosis in Brucella-Infected human trophoblast cells. Curr
Microbiol. 76:510–519. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
von Bargen K, Gorvel JP and Salcedo SP:
Internal affairs: Investigating the Brucella intracellular
lifestyle. FEMS Microbiol Rev. 36:533–562. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Anderson TD and Cheville NF:
Ultrastructural morphometric analysis of Brucella
abortus-infected trophoblasts in experimental placentitis.
Bacterial replication occurs in rough endoplasmic reticulum. Am J
Pathol. 124:226–237. 1986.PubMed/NCBI
|
|
10
|
Sidhu-Muñoz RS, Sancho P and Vizcaíno N:
Evaluation of human trophoblasts and ovine testis cell lines for
the study of the intracellular pathogen Brucella ovis. FEMS
Microbiol Lett; 365. 2018, PubMed/NCBI
|
|
11
|
Watanabe K, Tachibana M, Tanaka S, Furuoka
H, Horiuchi M, Suzuki H and Watarai M: Heat shock cognate protein
70 contributes to Brucella invasion into trophoblast giant
cells that cause infectious abortion. BMC Microbiol. 8:2122008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vassen V, Valotteau C, Feuillie C,
Formosa-Dague C, Dufrêne YF and De Bolle X: Localized incorporation
of outer membrane components in the pathogen Brucella
abortus. EMBO J. 38:e1003232019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Degos C, Hysenaj L, Gonzalez-Espinoza G,
Arce-Gorvel V, Gagnaire A, Papadopoulos A, Pasquevich KA, Méresse
S, Cassataro J, Mémet S and Gorvel JP: Omp25-dependent engagement
of SLAMF1 by Brucella abortus in dendritic cells limits
acute inflammation and favours bacterial persistence in vivo. Cell
Microbiol. 22:e131642020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cloeckaert A, Jacques I, Grilló MJ, Marín
CM, Grayon M, Blasco JM and Verger JM: Development and evaluation
as vaccines in mice of Brucella melitensis Rev.1 single and
double deletion mutants of the bp26 and omp31 genes coding for
antigens of diagnostic significance in ovine brucellosis. Vaccine.
22:2827–2835. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Paul S, Peddayelachagiri BV, Nagaraj S,
Kingston JJ and Batra HV: Recombinant outer membrane protein 25c
from Brucella abortus induces Th1 and Th2 mediated
protection against Brucella abortus infection in mouse
model. Mol Immunol. 99:9–18. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Basaraba RJ, Bielefeldt-Ohmann H,
Eschelbach EK, Reisenhauer C, Tolnay AE, Taraba LC, Shanley CA,
Smith EA, Bedwell CL, Chlipala EA and Orme IM: Increased expression
of host iron-binding proteins precedes iron accumulation and
calcification of primary lung lesions in experimental tuberculosis
in the guinea pig. Tuberculosis (Edinb). 88:69–79. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rossi MS, Fetherston JD, Letoffe S,
Carniel E, Perry RD and Ghigo JM: Identification and
characterization of the hemophore-dependent heme acquisition system
of Yersinia pestis. Infect Immun. 69:6707–6717. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hop HT, Arayan LT, Huy TXN, Reyes AWB,
Baek EJ, Min W, Lee HJ, Rhee MH, Watanabe K, Chang HH and Kim S:
Lipocalin 2 (Lcn2) interferes with iron uptake by Brucella
abortus and dampens immunoregulation during infection of RAW 264.7
macrophages. Cell Microbiol. 20:2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tsuji Y: JunD activates transcription of
the human ferritin H gene through an antioxidant response element
during oxidative stress. Oncogene. 24:7567–7578. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tsuji Y, Moran E, Torti SV and Torti FM:
Transcriptional regulation of the mouse ferritin H gene.
Involvement of p300/CBP adaptor proteins in FER-1 enhancer
activity. J Biol Chem. 274:7501–7507. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qian ZM, Li H, Sun H and Ho K: Targeted
drug delivery via the transferrin receptor-mediated endocytosis
pathway. Pharmacol Rev. 54:561–587. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cui B, Liu W, Wang X, Chen Y, Du Q, Zhao
X, Zhang H, Liu SL, Tong D and Huang Y: Brucella Omp25
upregulates miR-155, miR-21-5p, and miR-23b to inhibit
interleukin-12 production via modulation of programmed death-1
signaling in human monocyte/macrophages. Front Immunol. 8:7082017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ma QL, Liu AC, Ma XJ, Wang YB, Hou YT and
Wang ZH: Brucella outer membrane protein Omp25 induces
microglial cells in vitro to secrete inflammatory cytokines and
inhibit apoptosis. Int J Clin Exp Med. 8:17530–17535.
2015.PubMed/NCBI
|
|
24
|
Zhang J, Guo F, Huang X, Chen C, Liu R,
Zhang H, Wang Y, Yin S and Li Z: A novel Omp25-binding peptide
screened by phage display can inhibit Brucella abortus 2308
infection in vitro and in vivo. J Med Microbiol. 63:780–787. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jiang H, Dong H, Peng X, Feng Y, Zhu L,
Niu K, Peng Y, Fan H and Ding J: Transcriptome analysis of gene
expression profiling of infected macrophages between
Brucella suis 1330 and live attenuated vaccine strain S2
displays mechanistic implication for regulation of virulence.
Microb Pathog. 119:241–247. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang YJ, Liu ZS, Lu SY, Li C, Hu P, Li YS,
Liu NN, Tang F, Xu YM, Zhang JH, et al: Molecular cloning,
expression and characterization of programmed cell death 10 from
sheep (ovis aries). Gene. 558:65–74. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Berta P, Bourg G, Hanna N, Saadeh B,
Armengaud J, Patey G and O'Callaghan D: The Brucella suis
IbpA heat-shock chaperone is not required for virulence or for
expression of the VirB type IV secretion system VirB8 protein. Lett
Appl Microbiol. 58:564–568. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Guo F, Wang Y, Chen C, Zhang H, Qiao J,
Ren Y, Zhang J and Li Z: Interaction between VirB5 of
Brucella type IV secretion system (TFSS) and ferritin heavy
polypeptide 1 (FTH1) in murine macrophage. J Anim Vet Adv.
11:2623–2629. 2012. View Article : Google Scholar
|
|
30
|
Zhang Y, Li T, Zhang J, Li Z, Zhang Y,
Wang Z, Feng H, Wang Y, Chen C and Zhang H: The Brucella
melitensis M5–90 phosphoglucomutase (PGM) mutant is attenuated
and confers protection against wild-type challenge in BALB/c mice.
World J Microbiol. 32:582016. View Article : Google Scholar
|
|
31
|
Meng L, Ma H, Meng J, Li T, Zhu Y and Zhao
Q: Costunolide attenuates oxygen-glucose
deprivation/reperfusion-induced mitochondrial-mediated apoptosis in
PC12 cells. Mol Med Rep. 23:4112021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang J, Zhang Y, Li Z, Liu J, Shao X, Wu
C, Wang Y, Wang K, Li T, Liu L, et al: Outer membrane protein 25 of
Brucella activates mitogen-activated protein kinase signal
pathway in human trophoblast cells. Front Vet Sci. 4:1972017.
View Article : Google Scholar : PubMed/NCBI
|