Introduction
Acute myocardial infarction is an important and
lethal cardiovascular disease characterised by a sharp decline in
the coronary blood supply, and it typically develops into
myocardial fibrosis (1–3). Previous studies on the regulatory
mechanism of myocardial fibrosis following myocardial infarction
showed that myocardial fibrosis is vital to disease progression
(4,5).
Long non-coding RNAs (lncRNAs) are >200
nucleotides in length and do not have protein-coding ability.
LINC00961 encodes a small polypeptide of amino acid response and is
highly expressed in the heart tissue, where it regulates mTORC1
activation (6). Most researchers
have focused on the ability of LINC00961 to inhibit various tumours
(7–9) but recently began studying its
cardiovascular effects. For example, Spencer et al (10) identified that LINC00961 and its
encoded small polypeptide of amino acid response can regulate
endothelial cell adhesion, proliferation and migration, among other
processes. Liu et al (11)
found that LINC00961, via the PI3K-AKT-GSK3β pathway, promoted
myocardial infarction. Wu et al (12) demonstrated that LINC00961
contributed to coronary heart disease by regulating the function of
vascular smooth muscle cells.
Markwald et al (13) first discovered
endothelial-mesenchymal transition (EndMT) through heart formation
developmental studies in 1975. EndMT plays an important role in
pathophysiological processes such as myocardial
ischemia-reperfusion, myocardial infarction, diabetic
cardiomyopathy and fibrosis (14–17). EndMT can be induced by hypoxia and
attenuated by activating AMP-activated protein kinase as well as by
suppressing the mammalian target of rapamycin (mTOR) signalling
pathway in human cardiac microvascular endothelial cells (HCMECs)
(18–20).
Phosphatase and tensin homolog deleted on chromosome
10 (PTEN) is critical for cell growth and affects the survival and
proliferation of tumour cells (21,22). PTEN is also associated with
regenerative processes such as nerve injury recovery (23), cardiac ischemia (24) and wound healing (25). Yang et al (26) revealed that B lymphoma Mo-MLV
insertion region 1 homolog aggravated myocardial fibrosis and
reduced cardiac function after myocardial infarction by inhibiting
the PTEN-PI3K-AKT pathway.
Progress has been made in understanding the
functions of LINC00961. However, whether it regulates EndMT remains
unclear. In the present study, it was examined whether knockdown of
LINC00961 attenuates EndMT and cell injuries by activating the
PTEN-PI3K-AKT pathway.
Materials and methods
Cell culture, cell transfection and
drug treatment
HCMECs were obtained from ScienCell Research
Laboratories, Inc. and cultured in 25-cm2 cell culture
flasks (Corning, Inc.) in a 5% CO2 atmosphere at 37°C.
The culture medium was Roswell Park Memorial Institute-1640
containing 10% fetal bovine serum (both from Gibco; Thermo Fisher
Scientific, Inc.). Endothelial cells were used from passages 4 to
10 for the experiments.
Short hairpin RNA (shRNA) for LINC00961 and negative
control sh-scramble were acquired from Wuhan Aspen Biotechnology
Co., Ltd. The plasmid sequences were as follows: sh-LINC00961-1,
5′-AGTGCCCAGGACTTCTGGACCTTCA-3′; sh-LINC00961-2,
5′-CCUCAGGGAUCCUGUUAU-3′; and sh-LINC00961-3,
5′-GCUUCUUACUUGCUCCUAA-3′. Lentiviruses (LV) carrying negative
control scrambled RNA and shRNA were used for transfection at the
optimal multiplicity of infection value. Transfection of 5 pmol
shRNA was performed using Lipofectamine® RNAiMAX Reagent
(Thermo Fisher Scientific, Inc.) for 5 min at room temperature.
Transfected cells were analyzed following incubation for 48 h at
37°C. The LINC00961 open reading frame (ORF) full-length cDNA clone
vector (containing a restriction site) was obtained from Shanghai
GenePharma Co., Ltd. The LINC00961 overexpression (LV-LINC00961)
vector was prepared by double digestion of the target fragment,
vector fragment acquisition, double digestion of the vector, and
recovery and construction of the recombinant overexpression
vector.
The cells were exposed to 10 ng/ml transforming
growth factor (TGF)-β1 (Aspen Biotechnology Co., Ltd.) in
serum-free medium (27) for 24,
48, 72 and 96 h to induce EndMT. The cells were then randomly
divided into control, TGF-β, TGF-β + sh-LINC00961, TGF-β +
sh-Scramble, TGF-β + LV-LINC00961, TGF-β + LV-Control, and TGF-β +
sh-LINC00961 + VO-Ohpic trihydrate groups. VO-OHpic trihydrate
(VOT, C12H15N2O11), an
inhibitor of PTEN, was purchased from MedChemExpress and diluted in
dimethyl sulfoxide to 35 nmol/l (28). Following the treatments, HCMECs
from each group were collected by centrifugation (1,000 × g at 22°C
for 3 min) for experimental analysis.
Cell Counting Kit-8
A 96-well plate was inoculated with the cell
suspension (1×104 cells/well) and the cells were
precultured at room temperature in an incubator for 24 h. Next, 10
µl of Cell Counting Kit-8 solution (Beyotime Institute of
Biotechnology) was added to each well; the culture plate was
incubated for 2 h at 37°C, after which the absorbance was measured
at 450 nm with a microplate reader (Thermo Fisher Scientific,
Inc.).
Flow cytometric analysis
An Annexin V-FITC apoptosis detection kit (Sungene
Biotech) was used to assess cell apoptosis according to the
manufacturer's protocol. Cells were seeded into six-well plates
(1×105 cells/well) and cultured until reaching 85%
confluence. After digestion, suspension and centrifugation (1,000 ×
g at 22°C for 3 min), the precipitate was obtained, the cells were
resuspended in 300 µl of binding buffer, and 5 µl of Annexin
propidium iodide was added. Annexin propidium iodide was mixed and
incubated for 10 min in the dark before detection with a BD
FACSAriaIII flow cytometer (BD Biosciences).
Immunofluorescence staining
After fixing the cells for 20 min at 4°C in 4%
paraformaldehyde, the cells were washed with phosphate-buffered
saline for 10 min, blocked for 1 h with 5% bovine serum albumin
(Gibco; Thermo Fisher Scientific, Inc.) and incubated overnight at
4°C with the primary antibody α-smooth muscle actin (SMA) (1:5,000;
cat. no. 14395-1-AP; ProteinTech Group, Inc.) or CD31 (1:500; cat.
no. sc-376764, Santa Cruz Biotechnology, Inc.). After incubation
for 1 h with appropriate horseradish peroxidase-conjugated goat
anti-rabbit secondary antibodies (1:10,000; cat. no. AS1107; Aspen
Biotechnology Co., Ltd.) at 22°C, the cells were stained with 5
µg/ml DAPI (Beyotime Institute of Biotechnology) for 2 min at 22°C.
Fluorescence microscopy (BXM1; Olympus Corporation) was performed
to visualise immunofluorescence.
Reverse transcription quantitative
(RT-q) PCR
TRIpure Total RNA Extraction Reagent [ELK (Wuhan)
Biotechnology Co., Ltd.] was used to isolate total RNA from the
HCMECs. All RT-qPCR steps were conducted as previously described
(20,24). Relative expression changes were
calculated using the 2−ΔΔCq method (29), and the selected reference group
was referenced as 1. The primers used in PCR are listed in Table I.
 | Table I.Primer sequences for quantitative
PCR. |
Table I.
Primer sequences for quantitative
PCR.
| Primer name | Primer sequence
(5′-3′) |
|---|
| CD31 | F:
ACCAAGATAGCCTCAAAGTCGG |
|
| R:
TAAGAAATCCTGGGCTGGGAG |
| VE-Cadherin | F:
AAGGACATAACACCACGAAACG |
|
| R:
GAGATGACCACGGGTAGGAAG |
| α-SMA | F:
CTATGCCTCTGGACGCACAAC |
|
| R:
CCCATCAGGCAACTCGTAACTC |
| FSP-1 | F:
GGTGTCCACCTTCCACAAGTAC |
|
| R:
TCCTGGGCTGCTTATCTGG |
| Cyclin D1 | F:
TCCTACTTCAAATGTGTGCAGAAG |
|
| R:
CATCTTAGAGGCCACGAACATG |
| Bcl-2 | F:
AGGATTGTGGCCTTCTTTGAG |
|
| R:
AGCCAGGAGAAATCAAACAGAG |
| Bax | F:
TCTGAGCAGATCATGAAGACAGG |
|
| R:
ATCCTCTGCAGCTCCATGTTAC |
| AKT | F:
TTCTATGGCGCTGAGATTGTGT |
|
| R:
GCCGTAGTCATTGTCCTCCAG |
| PTEN | F:
TGAGAGACATTATGACACCGCC |
|
| R:
TTACAGTGAATTGCTGCAACATG |
| LINC00961 | F:
ATGGAAACGGCAGTGATTGG |
|
| R:
GGCGTCACATGAAGGTCCAG |
| mTOR | F:
AAGCCAAGCCTTGGATTTTG |
|
| R:
GGACGGGTGAGGTAACAGGAT |
| PI3K | F:
GTCCTATTGTCGTGCATGTGG |
|
| R:
TGGGTTCTCCCAATTCAACC |
| GAPDH | F:
CATCATCCCTGCCTCTACTGG |
|
| R:
GTGGGTGTCGCTGTTGAAGTC |
Western blot analysis
Sodium dodecyl sulphate-polyacrylamide gel
electrophoresis (10%) was performed to separate proteins from the
cells and mouse hearts. Western blot analysis was performed as
previously described (20,24).
Details regarding the primary and secondary antibodies used are
provided in Table II.
 | Table II.Information on primary and secondary
antibodies. |
Table II.
Information on primary and secondary
antibodies.
| Antibodies | Species | Supplier | Catalogue
number | Dilution |
|---|
| Primary |
|
|
|
|
|
GAPDH | Rabbit | Abcam | ab37168 | 1:10,000 |
|
PTEN | Rabbit | Abcam | ab267787 | 1:2,000 |
|
p-PI3k | Rabbit | Abcam | ab182651 | 1:500 |
|
PI3k | Rabbit | Cell Signaling
Technology, Inc. | 4292 | 1:3,000 |
|
p-AKT | Rabbit | Cell Signaling
Technology, Inc. | 4060 | 1:1,000 |
|
AKT | Rabbit | Cell Signaling
Technology, Inc. | 9272 | 1:2,000 |
|
SPARR | Rabbit | Cell Signaling
Technology, Inc. | 25823 | 1:1,000 |
|
p-mTOR | Rabbit | Cell Signaling
Technology, Inc. | 5536 | 1:500 |
|
mTOR | Rabbit | Cell Signaling
Technology, Inc. | 2972 | 1:1,000 |
|
CD31 | Mouse | Santa Cruz
Biotechnology, Inc. | sc-376764 | 1:500 |
|
VE-Cadherin | Rabbit | Thermo Fisher
Scientific, Inc. | 36-1900 | 1:500 |
|
α-SMA | Rabbit | ProteinTech Group,
Inc. | 14395-1-AP | 1:5,000 |
|
FSP-1 | Rabbit | ProteinTech Group,
Inc. | 20886-1-AP | 1:1,000 |
| Cyclin
D1 | Rabbit | Cell Signaling
Technology, Inc. | 55506 | 1:1,000 |
|
Bcl-2 | Rabbit | Abcam | ab59348 | 1:1,000 |
|
Bax | Rabbit | Cell Signaling
Technology, Inc. | 2772 | 1:2,000 |
| Secondary |
|
|
|
|
|
HRP-goat anti-rabbit |
| Aspen Biotechnology
Co., Ltd. | AS1107 | 1:10,000 |
|
HRP-goat anti-mouse |
| Aspen Biotechnology
Co., Ltd. | AS1106 | 1:10,000 |
Statistical analysis
All analyses were performed using Prism 9.1.2
(GraphPad Software, Inc.) and SPSS 23.0 software (IBM Corp.).
Unpaired t-tests were used to compare differences between two
groups. The data are presented as the means ± standard deviation;
each experiment was repeated at least three times. P<0.05 was
considered to indicate a statistically significant difference.
Results
Injury and EndMT in HCMECs induced by
TGF-β
As revealed in Fig.
1A, cell viability was reduced over time by TGF-β. After 24 h
of treatment with TGF-β, cell viability in the TGF-β group did not
significantly differ from that in the control group (P>0.05).
However, compared with the control group, the TGF-β group exhibited
significant differences in cell viability after 48, 72, and 96 h of
treatment (P<0.05). Therefore, follow-up experiments were
performed using treatment with TGF-β for 48 h. Treatment with TGF-β
for 48 h increased the apoptotic rate of endothelial cells
(Fig. 1B). It was also revealed
that the mRNA and protein expression levels of α-SMA and fibroblast
specific protein 1 (FSP-1) were elevated, whereas those of CD31 and
VE-cadherin (VE-Cad) were decreased after 48 h of TGF-β treatment
compared with the control group (Fig.
1C and D; P<0.05). Immunofluorescence used to assess the
changes in CD31 and α-SMA expression levels showed the same trends
as RT-qPCR and western blotting (Fig.
1E). These results revealed that the TGF-β-induced injury and
EndMT model in HCMECs was successfully established by treatment
with TGF-β for 48 h.
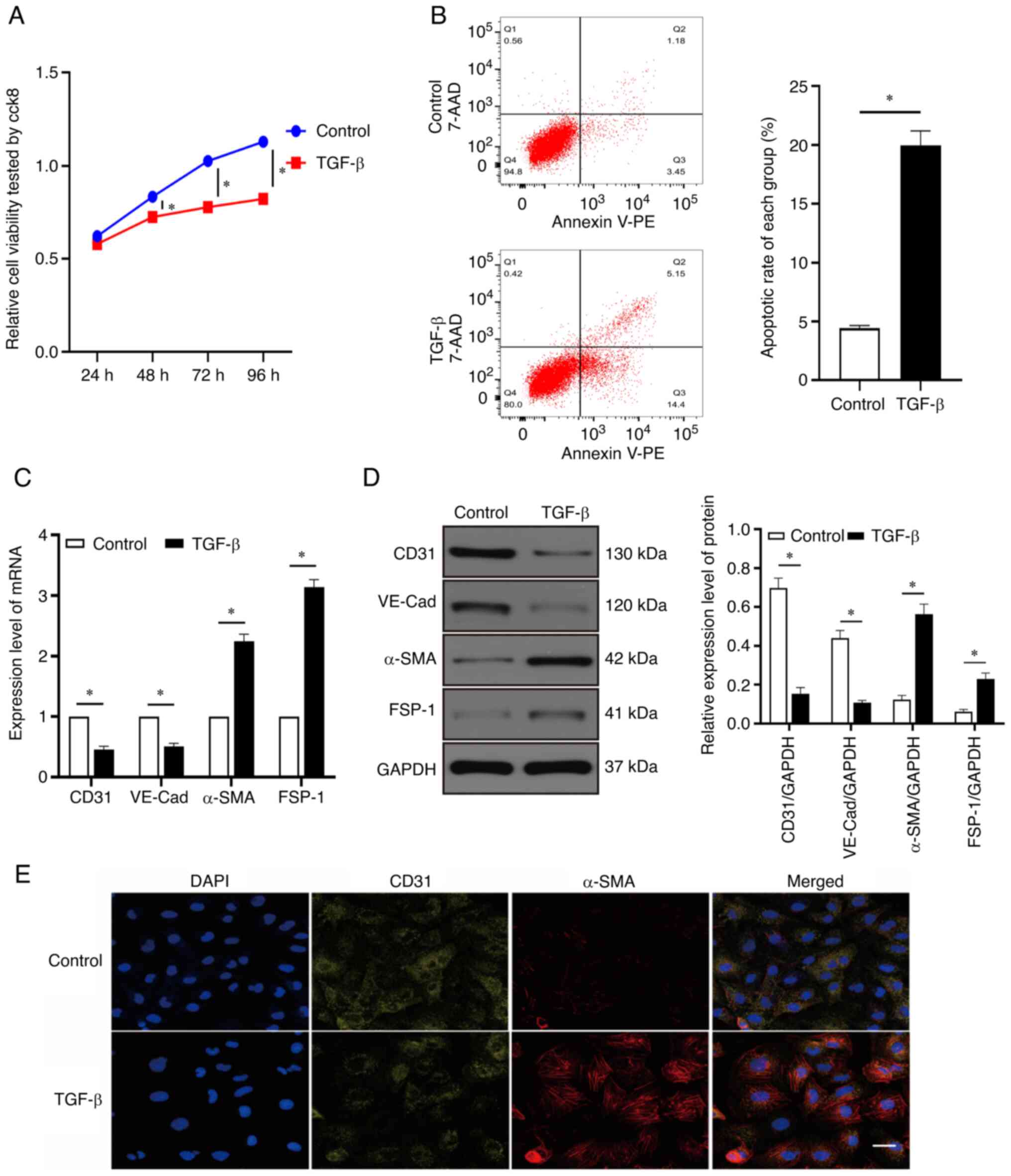 | Figure 1.Injury and EndMT of HCMECs induced by
TGF-β. (A) HCMECs were treated with TGF-β (10 ng/ml) for 24, 48, 72
and 96 h and their viability was detected using a Cell Counting
Kit-8 assay. (B) Rate of cell apoptosis was evaluated using flow
cytometric analysis. (C) Expression of CD31, VE-Cad, α-SMA and
FSP-1 mRNA was evaluated using reverse transcription-quantitative
PCR. (D) Western blotting of CD31, VE-Cad, α-SMA and FSP-1 protein
expression. (E) Immunofluorescence staining was used to identify
CD31+/α-SMA+ cells. α-SMA (red), CD31
(green); scale bars: 50 µm. Each experiment was repeated at least
three times. ns, P>0.05; *P<0.05. n=5 per group. EndMT,
endothelial-mesenchymal transition; HCMECs, human cardiac
microvascular endothelial cells; VE-Cad, VE cadherin; α-SMA,
α-smooth muscle actin; ns, not significant; FSP-1, fibroblast
specific protein 1. |
LINC00961 knockdown attenuates
apoptosis induced by TGF-β in HCMECs
The expression of LINC00961 was suppressed in
endothelial cells after transfection with the sh-LINC00961-1,
sh-LINC00961-2 and sh-LINC00961-3 plasmids; the expression of
LINC00961 was markedly increased using the ORF full-length cDNA
clone vector. LINC00961 levels were remarkably reduced following
transfection of the sh-LINC00961-1 plasmid (Fig. 2A), and thus this plasmid was used
in follow-up experiments. As revealed in Fig. 2B, LINC00961 expression was
significantly increased in the TGF-β group but was significantly
reduced in the sh-LINC00961 group compared with that in control
group (P<0.05). LINC00961 knockdown recovered the cell viability
that had been reduced by TGF-β, whereas LINC00961 overexpression
exacerbated this reduction in cell viability (Fig. 2C; P<0.05). Flow cytometric
analysis was performed to detect the rate of apoptosis, which
showed that LINC00961 overexpression further facilitated
TGF-β-induced apoptosis. However, TGF-β-mediated apoptosis was
reduced by LINC00961 knockdown (Fig.
2D). The mRNA transcription levels of the anti-apoptotic
proteins Bcl-2 and cyclin D1 were decreased by TGF-β but were
recovered when LINC00961 was knocked down, and further reduced when
LINC00961 was overexpressed, whereas the mRNA transcription level
of the proapoptotic protein Bax presented the opposite trend
(Fig. 2E). In addition, western
blotting was performed to assess changes in protein levels of Bax,
Bcl-2 and Cyclin D1. It was identified that the protein levels
exhibited similar trends as those of the mRNA transcription levels
(Fig. 2F). These results
indicated that TGF-β-mediated apoptosis was attenuated by LINC00961
knockdown.
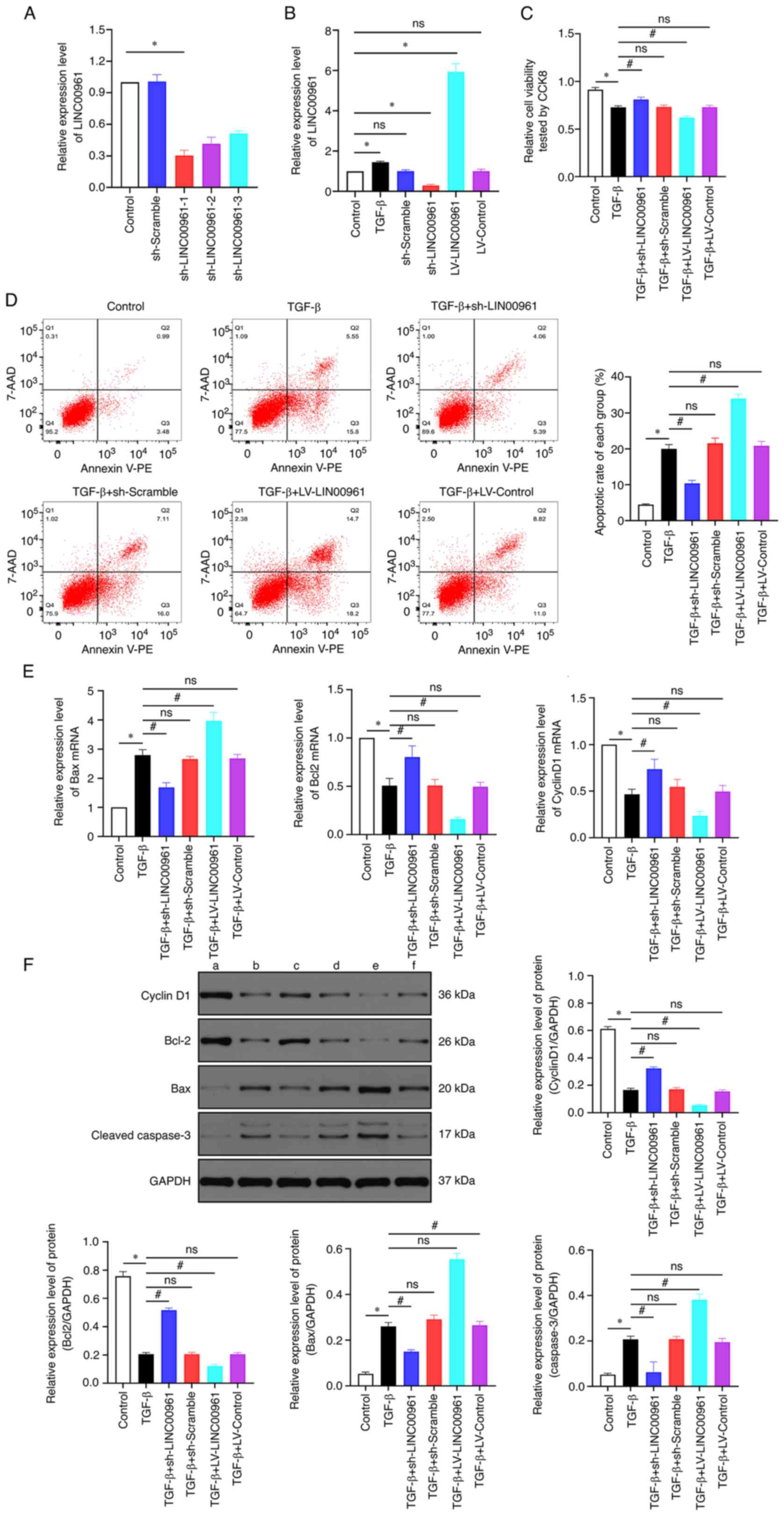 | Figure 2.LINC00961 knockdown attenuates
apoptosis induced by TGF-β in HCMECs. (A and B) Expression level of
LINC00961 was determined using RT-qPCR. (C) Cell viability was
examined using CCK-8 assay. (D) Rate of cell apoptosis was
evaluated using flow cytometric analysis. (E) RT-qPCR was used to
determine expression of apoptosis-related mRNA. (F) Western
blotting to determine expression of apoptosis-related mRNA (a)
Control, (b) TGF-β, (c) TGF-β + sh-LINC00961, (d) TGF-β +
sh-Scramble, (e) TGF-β + LV-LINC00961 and (f) TGF-β + LV-Control.
Each experiment was repeated at least three times. ns, P>0.05;
*P<0.05 and #P<0.05. n=5 per group. HCMECs, human
cardiac microvascular endothelial cells; RT-qPCR, reverse
transcription-quantitative; CCK-8, Cell Counting Kit-8; sh, short
hairpin; LV, lentivirus; ns, not significant. |
LINC00961 knockdown attenuates
TGF-β-induced EndMT
As revealed in Fig.
3, the results of RT-qPCR and western blotting demonstrated
that VE-Cad and CD31 expression was significantly downregulated
after TGF-β treatment compared with the control group (Fig. 3B and C; P<0.05). By contrast,
FSP-1 and α-SMA expression levels were significantly upregulated
(Fig. 3B and C; P<0.05) in the
TGF-β group compared with those in the control group. VE-Cad and
CD31 expression levels were significantly upregulated (P<0.05),
whereas FSP-1 and α-SMA expression levels were significantly
downregulated (P<0.05) in the TGF-β + sh-LINC00961 group
compared with those in the TGF-β group (Fig. 3B and C). The VE-cadherin and CD31
expression levels were significantly downregulated (P<0.05),
whereas FSP-1 and α-SMA levels were significantly upregulated
(P<0.05) in the TGF-β + LV-LINC00961 group compared with those
in the TGF-β group (Fig. 3B and
C). Immunofluorescence used to assess the changes in CD31 and
α-SMA expression levels showed the same trends as RT-qPCR and
western blotting (Fig. 3A). These
data revealed that TGF-β-induced EndMT was attenuated by LINC00961
knockdown.
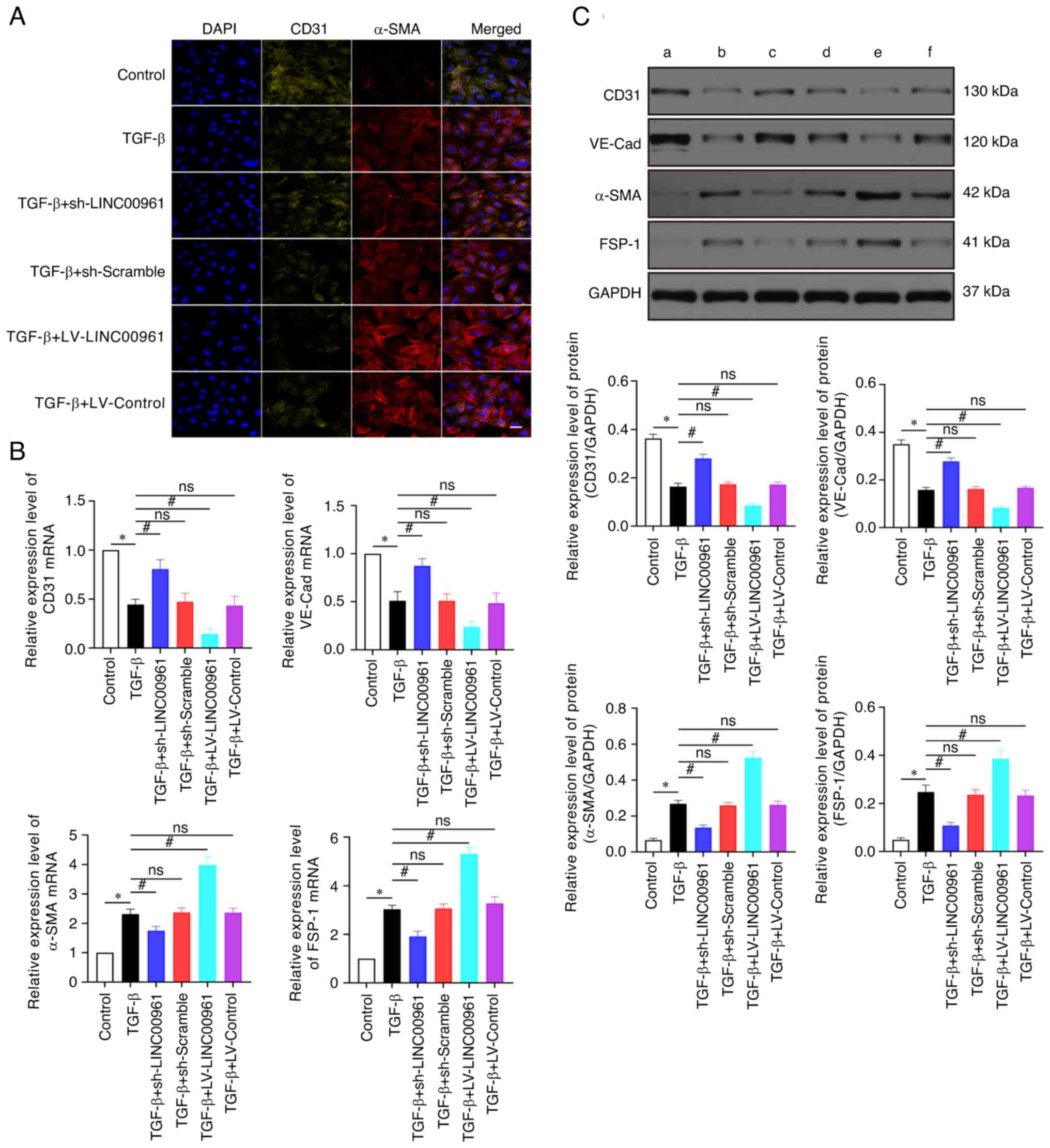 | Figure 3.LINC00961 knockdown attenuates
TGF-β-induced EndMT. (A) Immunofluorescence staining was used to
identify CD31+/α-SMA+ cells. α-SMA (red),
CD31 (green); scale bars: 50 µm. (B) Reverse
transcription-quantitative PCR was used to evaluate EndMT-related
mRNA. (C) Western blotting was used to evaluate EndMT-related
proteins. (a) Control, (b) TGF-β, (c) TGF-β + sh-LINC00961, (d)
TGF-β + sh-Scramble, (e) TGF-β + LV-LINC00961 and (f) TGF-β +
LV-Control. Each experiment was repeated at least three times, ns,
P>0.05; *P<0.05 and #P<0.05. n=5 per group.
EndMT, endothelial-mesenchymal transition; α-SMA, α-smooth muscle
actin; sh-, short hairpin; LV, lentivirus; VE-Cad, VE cadherin;
FSP-1, fibroblast specific protein 1; ns, not significant. |
LINC00961 knockdown attenuates injury
and EndMT of HCMECs by activating the PTEN-PI3K-AKT signalling
pathway
Immunofluorescence staining indicated that α-SMA
expression was significantly increased and CD31 expression was
significantly decreased in the TGF-β + LV-LINC00961 group compared
with that in the TGF-β group. CD31+ was significantly increased,
whereas the α-SMA+ level was decreased significantly in the TGF-β +
sh-LINC00961 group compared with that in the TGF-β group. However,
the expression level was reversed when the TGF-β + sh-LINC00961
group was treated with VOT (Fig.
4A). As revealed in Fig. 4B,
LINC00961 overexpression promoted cell apoptosis that had been
affected by TGF-β. LINC00961 knockdown attenuated the cell
apoptosis rate that had been affected by TGF-β; however, the rate
of cell apoptosis was reversed when the LINC00961 knockdown group
was treated with VOT. The RT-qPCR results demonstrated that PTEN
mRNA transcription levels were significantly reduced (P<0.05)
and the PI3K, AKT and mTOR mRNA transcription levels were
significantly increased (P<0.05) in the TGF-β + LV-LINC00961
group compared with those in the TGF-β group (Fig. 4C). The RT-qPCR results also
indicated that the PETN mRNA transcription level was decreased and
PI3K, AKT and mTOR mRNA transcription levels were reduced
(P<0.05) in the TGF-β + sh-LINC00961 group compared with those
in the TGF-β group. However, these mRNA transcription levels were
reversed when the TGF-β + sh-LINC00961 group was treated with VOT.
In addition, western blotting was performed to examine changes in
protein levels (Fig. 4D).
According to the aforementioned data, LINC00961 knockdown by TGF-β
induced injury and EndMT in HCMECs, possibly through activation of
PTEN and inhibition of PI3K, AKT, and mTOR. The relationships among
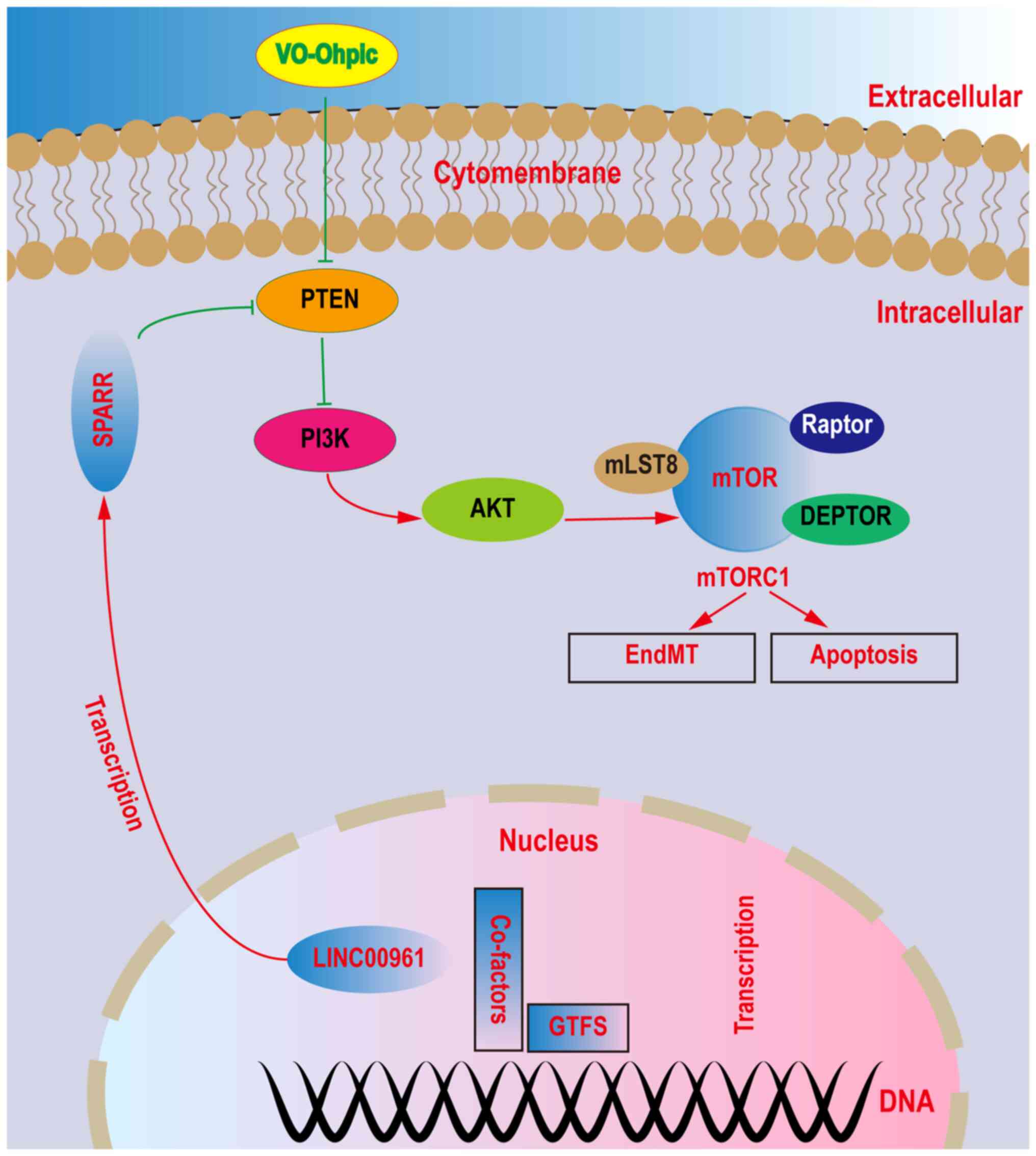
these proteins are schematically represented in Fig. 5.
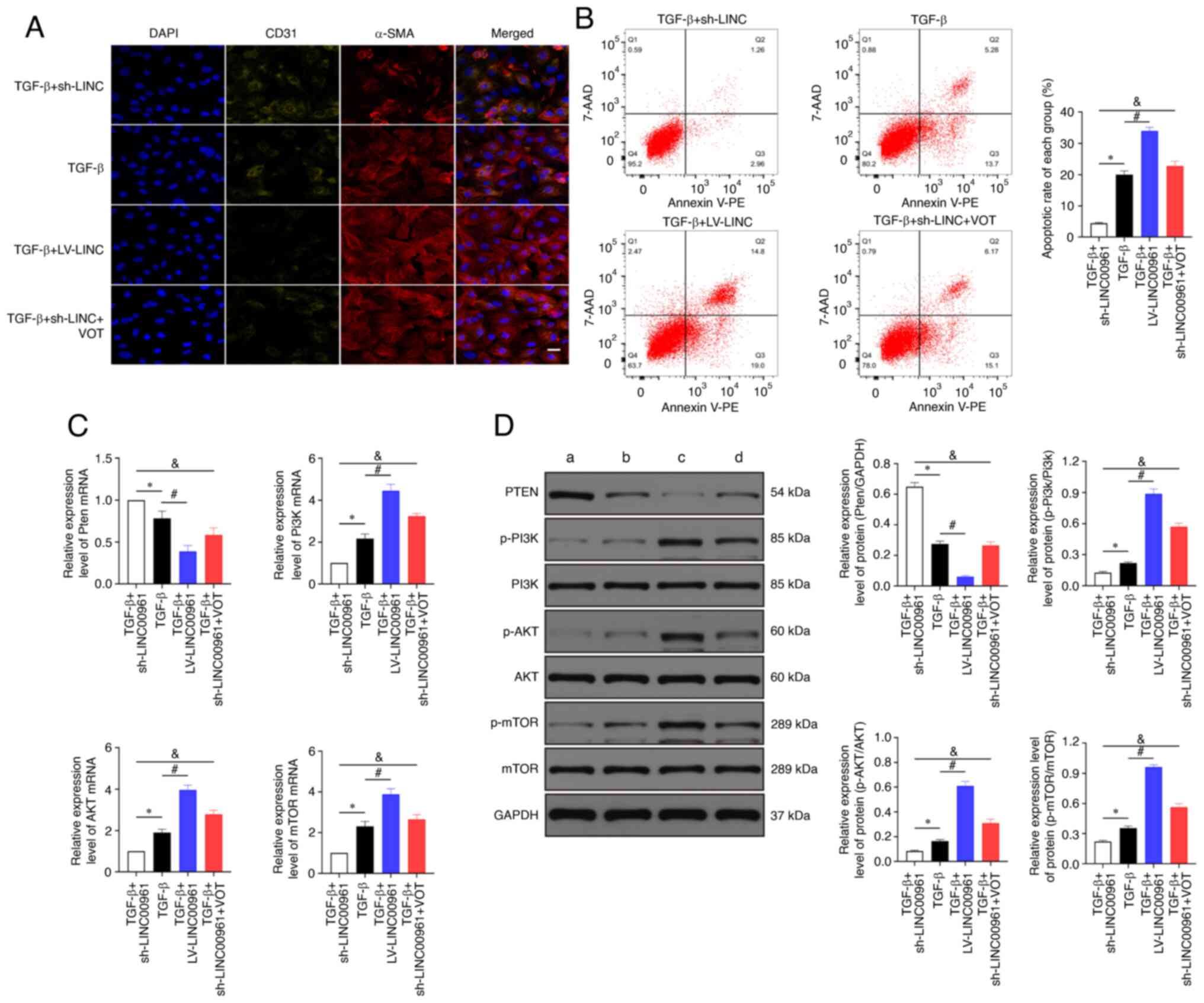 | Figure 4.LINC00961 knockdown attenuates injury
and EndMT in HCMECs by activating the PTEN-PI3K-AKT signalling
pathway. (A) Immunofluorescence staining was used to identify CD31
+ /α-SMA+ cells. α-SMA (red), CD31 (green); scale bars: 50 µm. (B)
Rate of cell apoptosis was evaluated using flow cytometry. (C)
Reverse transcription-quantitative PCR was used to determine the
expression of PTEN, PI3K, AKT and mTOR mRNA. (D) Western blotting
was used to determine the protein expression of PTEN, p-PI3K, PI3K,
p-AKT, AKT, p-mTOR and mTOR. (a) TGF-β + sh-LINC00961, (b) TGF-β,
(c) TGF-β + LV-LINC00961 and (d) TGF-β + sh-LINC00961 + VOT. Each
experiment was repeated at least three times. ns, P>0.05;
*P<0.05, #P<0.05 and &P<0.05.
n=5 per group. EndMT, endothelial-mesenchymal transition; HCMECs,
human cardiac microvascular endothelial cells; PTEN, phosphatase
and tensin homolog deleted on chromosome 10; α-SMA, α-smooth muscle
actin; p-, phosphorylated; sh-, short hairpin; LV, lentivirus; VOT,
VO-OHpic trihydrate; ns, not significant. |
Discussion
HCMECs play crucial physiological roles in
maintaining the normal function of the heart, and their dysfunction
leads to various cardiovascular diseases, such as ischemic
cardiomyopathy, diabetic cardiomyopathy, myocardial infarction and
heart failure (30–32). Human umbilical vein endothelial
cells and human coronary artery endothelial cells have been used to
investigate EndMT (33). To meet
physiological requirements in vivo, HCMECs were used to
evaluate EndMT. EndMT can be induced by TGF-β and hypoxia in
HCMECs, and certain studies (18,34) showed that hypoxia can activate
autophagy; therefore, TGF-β was used to induce EndMT to avoid the
effect of autophagy.
LncRNAs are associated with a range of cellular
biological functions, such as RNA splicing, protein localisation
and chromatin modification (35,36). Previously, Matsumoto et al
(37) reported that LINC00961
generated SPAR polypeptide that acted via the lysosome to suppress
amino-acid-mediated mTORC1 activity, thereby modulating skeletal
muscle regenerative response following injury. Since their
discovery, lncRNAs had reshaped our thinking on the design and
regulatory landscape of genomes in metazoans. LncRNAs bear some of
the hallmarks of mRNAs, as they are transcribed by RNA polymerase
II, spliced, capped, and polyadenylated, yet they have been
initially surmised to have little to no ORF information. LncRNAs
were generally less widely expressed than mRNAs, thus raising the
notion that they actively regulated specific biological processes
(37). The efficacy of LINC00961
in inhibiting tumour growth, invasion and metastasis and regulating
endothelial cell function has been extensively examined (9,38,39). The role of lncRNA LINC00961 has
also gained attention in cardiovascular diseases (12,40,41). Based on the relationship between
LINC00961 and cardiovascular diseases, the effect of this lncRNA on
EndMT was examined in vitro and its underlying mechanisms
were investigated.
During EndMT, the expression of related proteins is
altered, including increased FSP-1 and SMA and decreased CD31 and
VE-Cad (42). It was found that
the protein expression of BAX, caspase, α-SMA and FSP-1 was
increased, whereas that of Bcl-2, cyclin D1, CD31 and VE-cadherin
was decreased in endothelial cells treated with TGF-β. LINC00961
knockdown reversed these alterations.
The PTEN inhibitor VOT was used to explore the
underlying mechanism of LINC00961 in EndMT and cell injuries.
LINC00961 knockdown downregulated the protein levels of p-PI3K,
p-AKT, and p-mTOR and upregulated the protein level of PTEN. When
PTEN was inhibited using VOT, the effects of LINC00961 knockdown on
p-PI3K, p-AKT and p-mTOR were diminished. Based on these results,
LINC00961 knockdown attenuated TGF-β-induced EndMT and cell
injuries by activating the PTEN-PI3K-AKT pathway, which is
consistent with the results of previous studies. For example,
LINC00961 downregulation promotes the proliferation and inhibits
the apoptosis of vascular smooth muscle cells (12). Liu et al (11) revealed that LINC00961 promoted
myocardial infarction in the myocardial cell line H9c2. Considering
the association of the PTEN-PI3K-AKT pathway with EndMT, LINC00961
has been considered as a diagnostic target to promote EndMT and
myocardial fibrosis. In summary, LINC00961 knockdown attenuated
endothelial cell injury and EndMT induced by TGF-β via the
PTEN-PI3K-AKT signalling pathway. To confirm our results, further
studies are needed to investigate the effects of LINC00961 in
animal models and other cell lines (such as cardiomyocytes and
fibroblasts). Additionally, other proteins related to the
PTEN-PI3K-AKT pathway and EndMT and further methods should be used,
including a luciferase assay.
In conclusion, it was revealed that LINC00961
knockdown attenuates injuries and EndMT in HCMECs in vitro
by activating PTEN expression and inhibiting PI3K, AKT and mTOR
expression. Inhibition of LINC00961 expression may prevent the
occurrence of EndMT-related cardiovascular diseases, such as
myocardial fibrosis and heart failure, and shows potential as a
therapeutic target for cardiovascular diseases.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Youth Project of Jiangxi
Provincial Education Department Project (grant no. GJJ190129) and
the National Natural Science Foundation of China.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JXH performed the high-throughput sequencing
experiments and the bioinformatics analysis. BGL, ZQZ, TK, WQ and
SHH performed the histological examination of the kidney, and were
major contributors in writing the manuscript. JXH and BGL confirm
the authenticity of all the raw data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Animal experiments were approved by the Laboratory
Animal Ethics Committee of the First Affiliated Hospital of
Nanchang University (Nanchang, China; approval no. 20210213).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
EndMT
|
endothelial-mesenchymal transition
|
|
HCMEC
|
human cardiac microvascular
endothelial cell
|
|
lncRNA
|
long non-coding RNA
|
|
LV
|
lentivirus
|
|
PTEN
|
phosphatase and tensin homolog deleted
on chromosome 10
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
SMA
|
α-smooth muscle actin
|
|
TGF-β
|
transforming growth factor beta
|
|
VOT
|
VO-OHpic trihydrate
|
References
|
1
|
Talman V and Ruskoaho H: Cardiac fibrosis
in myocardial infarction-from repair and remodeling to
regeneration. Cell Tissue Res. 365:563–581. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vaskova E, Ikeda G, Tada Y, Wahlquist C,
Mercola M and Yang PC: Sacubitril/valsartan improves cardiac
function and decreases myocardial fibrosis via downregulation of
exosomal miR-181a in a rodent chronic myocardial infarction model.
J Am Heart Assoc. 9:e0156402020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu W, Sun J, Guo Y, Liu N, Ding X, Zhang
X, Chi J, Kang N, Liu Y and Yin X: Calhex231 ameliorates myocardial
fibrosis post myocardial infarction in rats through the
autophagy-NLRP3 inflammasome pathway in macrophages. J Cell Mol
Med. 24:13440–13453. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chi YC, Shi CL, Zhou M, Liu Y, Zhang G and
Hou SA: Selective cyclooxygenase-2 inhibitor NS-398 attenuates
myocardial fibrosis in mice after myocardial infarction via snail
signaling pathway. Eur Rev Med Pharmacol Sci. 21:5805–5812.
2017.PubMed/NCBI
|
|
5
|
Shibamoto M, Higo T, Naito AT, Nakagawa A,
Sumida T, Okada K, Sakai T, Kuramoto Y, Yamaguchi T, Ito M, et al:
Activation of DNA damage response and cellular senescence in
cardiac fibroblasts limit cardiac fibrosis after myocardial
infarction. Int Heart J. 60:944–957. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tajbakhsh S: lncRNA-encoded polypeptide
SPAR(s) with mTORC1 to regulate skeletal muscle regeneration. Cell
Stem Cell. 20:428–430. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mu X, Mou KH, Ge R, Han D, Zhou Y and Wang
LJ: Linc00961 inhibits the proliferation and invasion of skin
melanoma by targeting the miR367/PTEN axis. Int J Oncol.
55:708–720. 2019.PubMed/NCBI
|
|
8
|
Wu H, Dai Y, Zhang D, Zhang X, He Z, Xie X
and Cai C: LINC00961 inhibits the migration and invasion of colon
cancer cells by sponging miR-223-3p and targeting SOX11. Cancer
Med. 9:2514–2523. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang L, Shao L and Hu Y: Long noncoding
RNA LINC00961 inhibited cell proliferation and invasion through
regulating the Wnt/beta-catenin signaling pathway in tongue
squamous cell carcinoma. J Cell Biochem. 120:12429–12435. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Spencer HL, Sanders R, Boulberdaa M,
Meloni M, Cochrane A, Spiroski AM, Mountford J, Emanueli C,
Caporali A, Brittan M, et al: The LINC00961 transcript and its
encoded micropeptide SPAAR regulate endothelial cell function.
Cardiovasc Res. 116:1981–1994. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu S, He Y, Shi J, Liu L, Ma H, He L and
Guo Y: STAT1-avtiviated LINC00961 regulates myocardial infarction
by the PI3K/AKT/GSK3β signaling pathway. J Cell Biochem.
120:13226–13236. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu CT, Liu S and Tang M: Downregulation of
linc00961 contributes to promote proliferation and inhibit
apoptosis of vascular smooth muscle cell by sponging miR-367 in
patients with coronary heart disease. Eur Rev Med Pharmacol Sci.
23:8540–8550. 2019.PubMed/NCBI
|
|
13
|
Markwald RR, Fitzharris TP and Smith WN:
Sturctural analysis of endocardial cytodifferentiation. Dev Biol.
42:160–180. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu X, Mujahid H, Rong B, Lu QH, Zhang W,
Li P, Li N, Liang ES, Wang Q, Tang DQ, et al: Irisin inhibits high
glucose-induced endothelial-to-mesenchymal transition and exerts a
dose-dependent bidirectional effect on diabetic cardiomyopathy. J
Cell Mol Med. 22:808–822. 2018.PubMed/NCBI
|
|
15
|
Feng B, Cao Y, Chen S, Chu X, Chu Y and
Chakrabarti S: miR-200b mediates endothelial-to-mesenchymal
transition in diabetic cardiomyopathy. Diabetes. 65:768–779. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yin Y, Zhang Q, Zhao Q, Ding G, Wei C,
Chang L, Li H, Bei H, Wang H, Liang J and Jia Z: Tongxinluo
attenuates myocardiac fibrosis after acute myocardial infarction in
rats via inhibition of endothelial-to-mesenchymal transition.
BioMed Res Int. 16:65954372019.PubMed/NCBI
|
|
17
|
Zheng X, Peng M, Li Y, Wang X, Lu W, Wang
X, Shan Y, Li R, Gao L and Qiu C: Cathelicidin-related
antimicrobial peptide protects against cardiac fibrosis in diabetic
mice heart by regulating endothelial-mesenchymal transition. Int J
Biol Sci. 15:2393–2407. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zou J, Liu Y, Li B, Zheng Z, Ke X, Hao Y,
Li X, Li X, Liu F and Zhang Z: Autophagy attenuates
endothelial-to-mesenchymal transition by promoting snail
degradation in human cardiac microvascular endothelial cells.
Biosci Rep. 37:BSR201710492017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu Y, Zou J, Li B, Wang Y, Wang D, Hao Y,
Ke X and Li X: RUNX3 modulates hypoxia-induced
endothelial-to-mesenchymal transition of human cardiac
microvascular endothelial cells. Int J Mol Med. 40:65–74. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hu J, Zheng Z, Li X, Li B, Lai X, Li N and
Lei S: Metformin attenuates hypoxia-induced endothelial cell injury
by activating the AMP-activated protein kinase pathway. J
Cardiovasc Pharmacol. 77:862–874. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao X, Qin T, Mao J, Zhang J, Fan S, Lu Y,
Sun Z, Zhang Q, Song B and Li L: PTENP1/miR-20a/PTEN axis
contributes to breast cancer progression by regulating PTEN via
PI3K/AKT pathway. J Exp Clin Cancer Res. 38:2562019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chai C, Song LJ, Han SY, Li XQ and Li M:
MicroRNA-21 promotes glioma cell proliferation and inhibits
senescence and apoptosis by targeting SPRY1 via the PTEN/PI3K/AKT
signaling pathway. CNS Neurosci Ther. 24:369–380. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hervera A, De Virgiliis F, Palmisano I,
Zhou L, Tantardini E, Kong G, Hutson T, Danzi MC, Perry RB, Santos
CXC, et al: Reactive oxygen species regulate axonal regeneration
through the release of exosomal NADPH oxidase 2 complexes into
injured axons. Nat Cell Biol. 20:307–319. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cheng S, Zhang X, Feng Q, Chen J, Shen L,
Yu P, Yang L, Chen D, Zhang H, Sun W and Chen X: Astragaloside IV
exerts angiogenesis and cardioprotection after myocardial
infarction via regulating PTEN/PI3K/Akt signaling pathway. Life
Sci. 227:82–93. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cao L, Graue-Hernandez EO, Tran V, Reid B,
Pu J, Mannis MJ and Zhao M: Downregulation of PTEN at corneal wound
sites accelerates wound healing through increased cell migration.
Invest Ophthalmol Vis Sci. 52:2272–2278. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang W, Wu Z, Yang K, Han Y, Chen Y, Zhao
W, Huang F, Jin Y and Jin W: BMI1 promotes cardiac fibrosis in
ischemia-induced heart failure via the PTEN-PI3K/Akt-mTOR signaling
pathway. Am J Physiol Heart Circ Physiol. 316:H61–H69. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang M, Weng H and Zheng J: NAD(+)
repletion inhibits the endothelial-to-mesenchymal transition
induced by TGF-β in endothelial cells through improving
mitochondrial unfolded protein response. Int J Biochem Cell Biol.
117:1056352019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zonghai C, Tao L, Pengjiao M, Liang G,
Rongchuan Z, Xinyan W, Wenyi N, Wei L, Yi W and Lang B:
Mycobacterium tuberculosis ESAT6 modulates host innate immunity by
downregulating miR-222-3p target PTEN. Biochim Biophys Acta Mol
Basis Dis. 1868:1662922022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lei W, Li J, Li C, Chen L, Huang F, Xiao
D, Zhang J, Zhao J, Li G, Qu T, et al: MARCH5 restores endothelial
cell function against ischaemic/hypoxia injury via Akt/eNOS
pathway. J Cell Mol Med. 25:3182–3193. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Feng B, Chen S, Gordon AD and Chakrabarti
S: miR-146a mediates inflammatory changes and fibrosis in the heart
in diabetes. J Mol Cell Cardiol. 105:70–76. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Quan X, Liu X, Qin X, Wang Y, Sun T, Li Z,
Zhu L, Chen J, Zhou Y, Singh S, et al: The role of LR-TIMAP/PP1c
complex in the occurrence and development of no-reflow.
EBioMedicine. 65:1032512021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tian J, Zhang M, Suo M, Liu D, Wang X, Liu
M, Pan J, Jin T and An F: Dapagliflozin alleviates cardiac fibrosis
through suppressing EndMT and fibroblast activation via
AMPKα/TGF-β/Smad signalling in type 2 diabetic rats. J Cell Mol
Med. 25:7642–7659. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Qureshi-Baig K, Kuhn D, Viry E, Pozdeev
VI, Schmitz M, Rodriguez F, Ullmann P, Koncina E, Nurmik M,
Frasquilho S, et al: Hypoxia-induced autophagy drives colorectal
cancer initiation and progression by activating the PRKC/PKC-EZR
(ezrin) pathway. Autophagy. 16:1436–1452. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Geisler S and Coller J: RNA in unexpected
places: Long non-coding RNA functions in diverse cellular contexts.
Nat Rev Mol Cell Biol. 14:699–712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Matkovich SJ, Edwards JR, Grossenheider
TC, de Guzman Strong C and Dorn GW II: Epigenetic coordination of
embryonic heart transcription by dynamically regulated long
noncoding RNAs. Proc Natl Acad Sci USA. 111:12264–12269. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Matsumoto A, Pasut A, Matsumoto M,
Yamashita R, Fung J, Monteleone E, Saghatelian A, Nakayama KI,
Clohessy JG and Pandolfi PP: mTORC1 and muscle regeneration are
regulated by the LINC00961-encoded SPAR polypeptide. Nature.
541:228–232. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pan LN and Sun YR: LINC00961 suppresses
cell proliferation and induces cell apoptosis in oral squamous cell
carcinoma. Eur Rev Med Pharmacol Sci. 23:3358–3365. 2019.PubMed/NCBI
|
|
39
|
Chen D, Zhu M, Su H, Chen J, Xu X and Cao
C: LINC00961 restrains cancer progression via modulating
epithelial-mesenchymal transition in renal cell carcinoma. J Cell
Physiol. 234:7257–7265. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Spiroski AM, Sanders R, Meloni M,
McCracken IR, Thomson A, Brittan M, Gray GA and Baker AH: The
influence of the LINC00961/SPAAR locus loss on murine development,
myocardial dynamics, and cardiac response to myocardial infarction.
Int J Mol Sci. 22:9692021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yin J, Liu Q, Chen C and Liu W: Small
regulatory polypeptide of amino acid response negatively relates to
poor prognosis and controls hepatocellular carcinoma progression
via regulating microRNA-5581-3p/human cardiolipin synthase 1. J
Cell Physiol. 234:17589–17599. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Piera-Velazquez S, Li Z and Jimenez SA:
Role of endothelial-mesenchymal transition (EndMT) in the
pathogenesis of fibrotic disorders. Am J Pathol. 179:1074–1080.
2011. View Article : Google Scholar : PubMed/NCBI
|