Introduction
Colorectal cancer (CRC) is a common tumor. Annually,
there are more than 600,000 CRC deaths worldwide and, at the time
of diagnosis, ~25% of CRC patients have already suffered from
distant metastases (1). With the
advancement of CRC diagnosis and therapy, the 5-year survival rate
of stage I and stage II patients has been improved to >82%
(2). Nonetheless, the prognosis
of CRC patients with advanced disease is still unfavorable and the
5-year survival rate is ≤10% (3).
Hence, it is imperative to clarify the molecular mechanism of CRC
carcinogenesis and progression to seek innovative targets for
diagnosis and therapy.
Circular RNAs (circRNAs) are non-coding (nc) RNA
transcripts that have a stable circular structure and are expressed
in diverse cells. They can work as competing endogenous RNAs
(ceRNA) to down-modulate microRNA (miRNAs/miRs) expression and take
part in the tumorigenesis and development of diverse tumors
(4). Reportedly, circRNAs are
implicated in the tumorigenesis and metastasis of CRC and can be
used as potential targets for CRC diagnosis and therapy, such as
circ_0007031 (5), circRAE1
(6), circ_0007142 (7). In order to explore the novel
circRNAs which participate in CRC progression, the present study
analyzed a microarray dataset (GSE138589) in the Gene Expression
Omnibus (GEO) database and identified a circRNA, circ_0006174,
which is upregulated in CRC tissues. Some previous studies have
reported that circ_0006174 promotes CRC progression and contributes
to the chemoresistance of doxorubicin (8,9).
Nevertheless, its role and mechanism in CRC remains to be
elucidated.
miRNAs are ncRNA transcripts with a length of ~22
nucleotides, which can be used as promising downstream targets of
several ncRNAs such as lncRNAs and circRNAs (10). miRNAs participate in tumorigenesis
by binding to the 3′ UTR of target genes and repressing expression
(10). The bioinformatics
analysis of the present study suggested that miR-1205 had the
potential complementary binding sites with circ_0006174. miR-1205
is reported to take part in the progression of diverse cancers,
including CRC. For instance, miR-1205 modulates the cell cycle and
impedes cell growth of CRC cells by targeting TRIM44 (11). miR-1205 can target CRK such as
proto-oncogene (CRKL) to repress CRC progression (12). calcium-binding epidermal growth
factor domain-containing protein 1 (CCBE1) is vital in the
development of lymphatic vessels. CCBE1 is reported to enhance CRC
metastasis by modulating the TGF-β signaling pathway (13). The bioinformatics analysis of the
present study suggested a potential binding site between miR-1205
and CCBE1 3′UTR. Nonetheless, the miR-1205/CCBE1 axis in CRC is
undefined.
The present study identified a circRNA,
circ_0006174, which is substantially upregulated in CRC tissues and
cell lines. Circ_0006174 was found to adsorb miR-1205 to
up-modulate CCBE1 expression and facilitate CRC progression. This
implies that circ_0006174 is a potential diagnostic biomarker and
therapy target for CRC treatment.
Materials and methods
Participants and tissue cases
Between May 2018 and May 2020, CRC tissues and
para-cancerous tissues (3 cm from the margin of tumor tissues) of
68 Chinese CRC patients (males:females, 31:37; median age at
diagnosis, 60.8 years; age range, 45–72 years) were available from
Guizhou Provincial People's Hospital and all specimens were
determined by pathologists. None of the patients had received
chemotherapy or radiotherapy or other anti-cancer treatments before
surgery. The tissue samples were frozen in liquid nitrogen
immediately after removal. This research was authorized by the
Ethics Committee of Guizhou Provincial People's Hospital (approval
no. 201802004) and complied with the Declaration of Helsinki. Prior
to surgery, each subject completed a written informed consent
form.
Cell culture
Human CRC cell lines (HCT116, SW620, SW480 and HT29)
and the immortalized colonic epithelial cell line NM460 were
procured from ATCC. All cells were cultured in DMEM (Gibco; Thermo
Fisher Scientific, Inc.) containing 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml of
streptomycin at 37°C with 5% CO2.
Oligonucleotide transfection
Guangzhou RiboBio Co., Ltd. synthesized two small
interfering RNA (siRNA) oligonucleotides targeting human
circ_0006174 [si-circ_0006174#1 (50 nM):
5′-GACAGGCAAAATCCTCAATGA-3′ and si-circ_0006174#2 (50 nM):
5′-GCAACTGACAGGCAAAATCCT-3′] and siRNA negative control (50 nM)
(5′-CCATTTACCCGAACGGCAA-3′), circ_0006174 short hairpin RNA
[sh-circ_0006174 (80 nM): 5′-GCATCCATCACTCCAGCATCA-3′] and
corresponding control [sh-NC (80 nM): 5′-TTCTCCGAACGTGTCACGT-3′],
miR-1205 mimics (50 nM) (5′-UCUGCAGGGUUUGCUUUGAG-3′) and miR-1205
inhibitors (50 nM) (5′-CUCAAAGCAAACCCUGCAGA-3′) and miR-NC (50 nM)
(5′-UCACAACCUCCUAGAAAGAGUAGA-3′) and inhibitor NC (50 nM)
(5′-CAGUACUUUUGUGUAGUACAAA-3′). When the cultured cells reached 80%
confluence, the CRC cells were planted into a 6-well plate and the
above oligonucleotides were transfected into the cell line using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Transfection was performed at room temperature
for 6 h. The cells were cultured for 24 h before subsequent
experiments.
Reverse transcription-quantitative
(RT-q) PCR
Cells (5×104/well) were cultured in
6-well plates. According to the manufacturers' protocols, total RNA
was separated from tissues and cell lines using the TRIzol reagent
(Thermo Fisher Scientific, Inc.) and a PrimeScript RT reagent kit
(Takara Biotechnology Co., Ltd.) was utilized for reverse
transcription to synthesize cDNA. Finally, a SYBR Green Premix Ex
Taq kit (Takara Biotechnology Co., Ltd.) was utilized to conduct
RT-qPCR and RNA relative expression was analyzed using the
2−ΔΔCq technique (14)
and GAPDH and U6 were employed as internal controls. The PCR
reaction was as follows: 95°C for 5 min, followed by 40 cycles at
95°C for 30 sec, 60°C for 30 sec and elongation at 72°C for 15 sec.
The following are the primer sequences: circ_0006174,
5′-GCAACTGACAGGCAAAATCC-3′ (forward) and 5′-AGTTGTGGTGGTGGAGGAAG-3′
(reverse); miR-1205, 5′-CTGCAGGGTTTGCTTTGAGG-3′ (forward) and
5′-CTCCAGAACAGGGTTGACAGG-3′ (reverse); CCBE1,
5′-TACCGATATGACCGGGAGAG-3′ (forward) and 5′-AGCTGCCCAAGGTATTGATG-3′
(reverse); U6, 5′-GACTATCATATGCTTACCGT-3′ (forward) and
5′-GGGCAGGAAGAGGGCCTAT-3′ (reverse); GAPDH,
5′-CTTTGGTATCGTGGAAGGACTC-3′ (forward) and
5′-GTAGAGGCAGGGATGATGTTCT-3′ (reverse).
Subcellular fractionation
A PARIS kit (Invitrogen; Thermo Fisher Scientific,
Inc.) was adopted for subcellular fractionation with U6 as a
nuclear control and GAPDH as a cytoplasmic control. RT-qPCR was
then conducted to examine relative expression in the subcellular
fractionation respectively.
CCK-8 method
Cells (2×103) were inoculated into each
well of a 96-well plate and 10 µl of CCK-8 reagent (Dojindo
Laboratories, Inc.) was added to each well on day 1, 2, 3 and 4 and
then the cells were incubated at 37°C for 1 h. Subsequently, a
microplate reader (Bio-Rad Laboratories, Inc.) was employed to
analyze the absorbance at 450 nm wavelength of each well.
TUNEL assay
Cell apoptosis assay was performed using the TUNEL
Apoptosis Assay kit (Beyotime Institute of Biotechnology). Cells
were cultured in 24-well plates (1×105 cells per well)
and fixed with 4% paraformaldehyde for 15 min at 25°C. TUNEL
solution was added to each well at 37°C for 60 min. After rinsing
with phosphate buffer saline (PBS), the cells were stained with
DAPI for 5 min at 37°C. The slides were mounted using
Fluoromount-G™ (Thermo Fisher Scientific, Inc.). Three
random fields of view were captured with a fluorescence microscope
(Olympus Corporation) and the percentage of TUNEL-positive cells
was calculated.
Transwell assay
Transwell inserts (8-µm pore size; Corning, Inc.)
were used for cell migration and invasion assays. For cell
migration assay 5×104 CRC cells in serum-free medium
were planted in the top compartment of the Transwell compartment.
Subsequently, 600 µl of medium containing 10% FBS was added to the
lower compartment. Following incubation for 24 h at 37°C, the cells
in the top compartment were removed. The cells on the lower surface
of the filter were fixed with 95% ethanol and stained with 0.1%
crystal violet for 15 min at room temperature. Then a light
microscope (Olympus Corporation) was used for counting the cells.
The bottom of the filter was pre-coated with Matrigel (1:10; BD
Biosciences) for cell invasion experiments. The Matrigel-coated
plate was incubated for 30 min at 37°C to polymerize the Matrigel.
The remaining processes were identical to those used in the cell
migration assay.
Dual-luciferase reporter gene
assay
Wild-type and mutant-type sequences of circ_0006174
or CCBE1 3′ UTR containing miR-1205 putative complementary binding
sequences were designed by Promega Corporation, which were
subsequently cloned into psiCHECKTM-2-luciferase reporter plasmid
(Promega Corporation) and co-transfected with miR-1205 mimics
(5′-UCUGCAGGGUUUGCUUUGAG-3′) (Guangzhou RiboBio Co., Ltd.) or
miR-NC (5′-UCACAACCUCCUAGAAAGAGUAGA-3′) (Guangzhou RiboBio Co.,
Ltd.) into CRC cells using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) for 24 h. The relative
luciferase activity of the cells in each group was measured 48 h
later with a dual-luciferase assay system (Promega Corporation).
Renilla luciferase activity was used to normalize the
firefly luciferase activity.
Western blotting
RIPA lysis buffer (Beyotime Institute of
Biotechnology) was utilized to isolate the total protein from the
cells and the concentration of protein was analyzed using the BCA
detection kit (Beyotime Institute of Biotechnology). Sodium dodecyl
sulfate polyacrylamide gel electrophoresis using a 10% gel was
utilized to separate 30 µg protein sample before the protein
samples were transferred to PVDF membranes (MilliporeSigma). The
PVDF membrane was blocked with 5% skimmed milk for 1.5 h at room
temperature and then incubated with anti-CCBE1 (1:1,000; HPA041361;
MilliporeSigma), anti-Ki67 (1:1,000; cat. no. ab16667; Abcam),
anti-c-Myc (1:1,000; cat. no. ab32072; Abcam), anti-cyclin D1
(1:1,000; cat. no. ab134175; Abcam) and anti-GAPDH (1:1,000; cat.
no. ab9485; Abcam) overnight at 4°C and then the membrane was
incubated at room temperature for 1 h with the horseradish
peroxidase-conjugated secondary antibody (1:2,000; cat. no.
ab150077; Abcam). To identify protein bands, the ECL luminescence
reagent (Thermo Fisher Scientific, Inc.) was utilized.
Semi-quantitative analysis was conducted using ImageJ software
(version 1.8.0; National Institutes of Health).
Tumor xenografts and lung metastasis
in mice
Animal experiments were authorized by the Animal
Care Committee of Guizhou Provincial People's Hospital (approval
no. 202109010) and conducted in accordance with the guidelines of
National Health Institute. A total of 10 four-week-old female
BALB/c nude mice (weight, 18–20 g; Guizhou Yikeda Biotechnology Co.
Ltd.) were maintained under standard laboratory conditions at 25°C
with 12-h light/dark cycles, 60% humidity and free access to food
and water. The mice werte randomly divided into two groups. SW620
cells were transfected with sh-NC or sh-circ_0006174 and
resuspended in PBS. Then, the cells were injected into the tail
vein of each nude mice (1×106 cells per mouse). After 30
days, the mice were decapitated and the lungs were removed and
fixed in 4% paraformaldehyde at 4°C for 24 h and embedded in
paraffin. Hematoxylin and eosin (H&E) staining was performed to
evaluate the formation of metastatic nodules. Sections (5-µm thick)
were deparaffinized in xylene, rehydrated in graded alcohol
solution and stained with hematoxylin solution for 30 min, and then
washed with water and counterstained with eosin for 5 min at 25°C.
Images were captured using a microscope (Olympus Corporation).
Bioinformatics
The eligible circRNAs data set (GSE138589) (15) was obtained from the GEO and
included data from 6 CRC tissues and 6 normal tissues. The raw
microarray data were processed using GEO2R. The Circular RNA
Interactome database (https://circinteractome.nia.nih.gov/index.html) was
used to explore the downstream miRNAs of circ_0006174 and the
target genes of miR-1205. Gene Expression Profiling Interactive
Analysis (GEPIA; http://gepia.cancer-pku.cn/) was used to analyze the
relationship between CCBE1 expression and overall survival. The
LinkedOmics (http://www.linkedomics.org/) online platform (16) was used to analyze the signal
pathways associated with CCBE1 in CRC.
Statistical analysis
All the experiments were performed in triplicate and
the data were shown as mean ± standard deviation. Statistical
analysis was executed using SPSS 17.0 data software (SPSS Inc.) and
GraphPad Prism V7.0 (GraphPad Software, Inc.) was adopted for
plotting. Paired (for matched data, such as tumor vs. normal
tissue) and unpaired (all other comparisons between two groups)
Student's t-test was used, while one-way ANOVA was performed to
examine difference among three or more groups, with Turkey's
post-hoc test. Correlation analysis was conducted using Pearson's
correlation analysis. χ2 test was performed to analyze
the association between patients' pathological characteristics and
the expression level of circ_0006174 or CCBE1.
Results
Circ_0006174 is markedly upregulated
in CRC tissues and cell lines
The GEO database circRNA microarray GSE138589 was
used to analyze the differential circRNA expression in CRC tissues
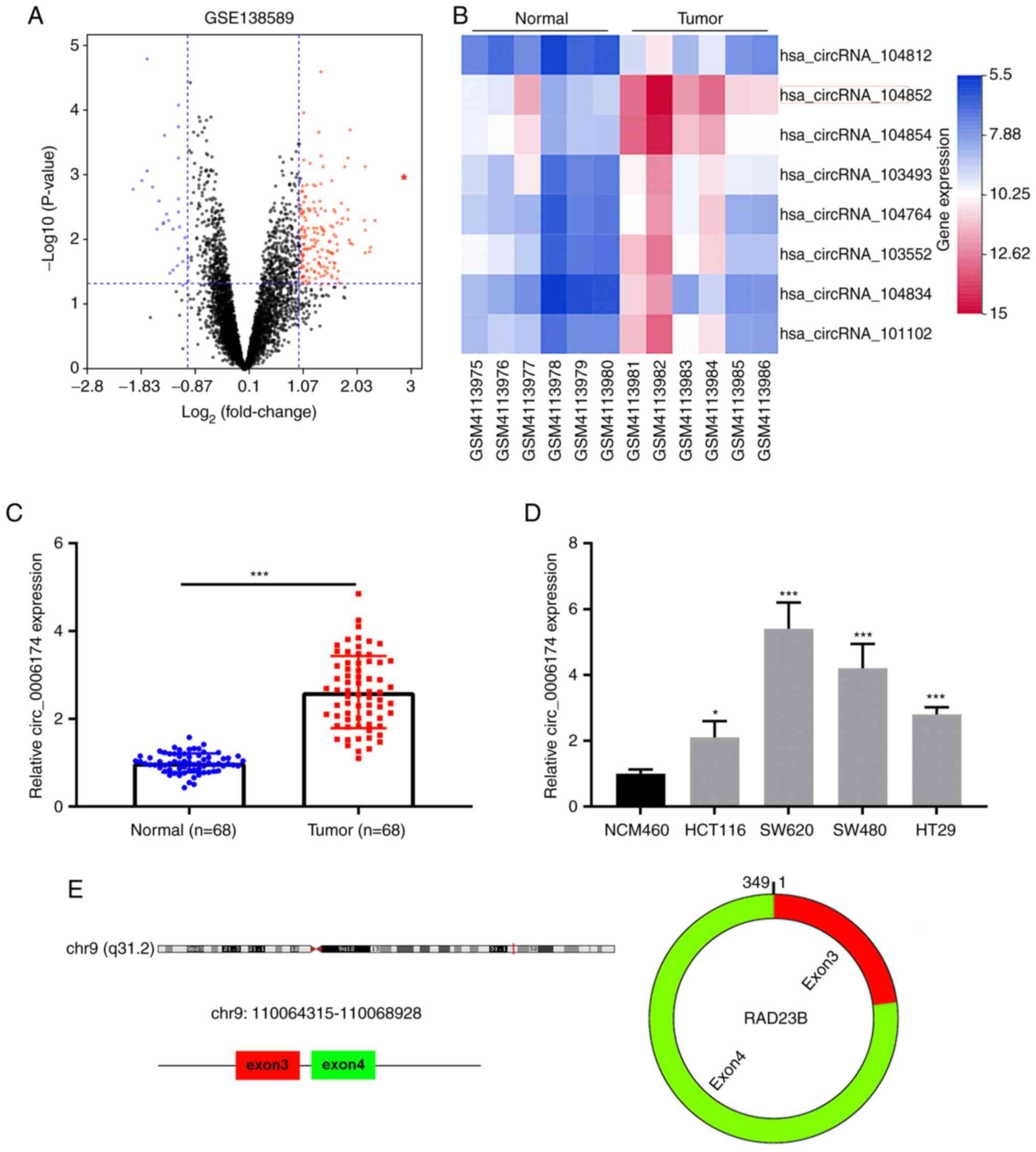
and adjacent tissues using the cut-of criteria
(|log2(fold change)|>1 and P<0.05; Fig. 1A). Fig. 1B displayed the top eight circRNAs
that were markedly upregulated in CRC tissues. Then circ_104852
(circ_0006174) was selected for followed-up analysis. RT-qPCR was
then performed to analyze circ_0006174 expression in 68 pairs of
CRC tissues and paracancerous tissues. The data showed that
circ_0006174 expression in CRC tissues was markedly higher than
that in adjacent tissues (Fig.
1C). Additionally, compared with immortalized human colonic
epithelial cell lines, circ_0006174 expression was notably higher
in CRC cell lines (HCT116, SW620, SW480 and HT29; Fig. 1D). As shown in Fig. 1E, circ_0006174 was a ring
structure composed of the third and fourth exons of RAD23 Homolog B
(RAD23B) gene.
Using the median circ_0006174 expression in CRC
tissues, 68 CRC patients were divided into the overexpression group
and the under-expression group (high expression group: n=34; low
expression group: n=34) to analyze the relationship between
circ_0006174 expression and the clinicopathological characteristics
of CRC patients. It was observed that circ_0006174 overexpression
was associated with larger tumor diameter (P=0.027) and higher T
stage (P=0.014; Table I). The
above data suggested that circ_0006174 was markedly upregulated in
CRC tissues and perhaps implicated in CRC progression as an
oncogene. Given that circ_0006174 had the highest expression in
SW620 and SW480, these two cell lines were chosen for subsequent
experiments.
 | Table I.Correlations between hsa_circ_0006174
expression and clinical features in patients with colorectal
cancer. |
Table I.
Correlations between hsa_circ_0006174
expression and clinical features in patients with colorectal
cancer.
|
|
| circ_0006174 |
|
|---|
|
|
|
|
|
|---|
| Features | n | High (n=34) | Low (n=34) | P-value |
|---|
| Age, years |
|
|
|
|
| ≥60 | 36 | 19 | 17 | 0.808 |
|
<60 | 32 | 15 | 17 |
|
| Sex |
|
|
|
|
|
Male | 31 | 18 | 13 | 0.796 |
|
Female | 37 | 16 | 21 |
|
| Clinical stage |
|
|
|
|
|
T3-T4 | 37 | 24 | 13 | 0.014a |
|
T1-T2 | 31 | 10 | 21 |
|
| Tumor size, cm |
|
|
|
|
| ≥3 | 38 | 24 | 14 | 0.027a |
|
<3 | 30 | 10 | 20 |
|
Knockdown of circ_0006174 markedly
impedes the growth, migration and invasion of CRC cells
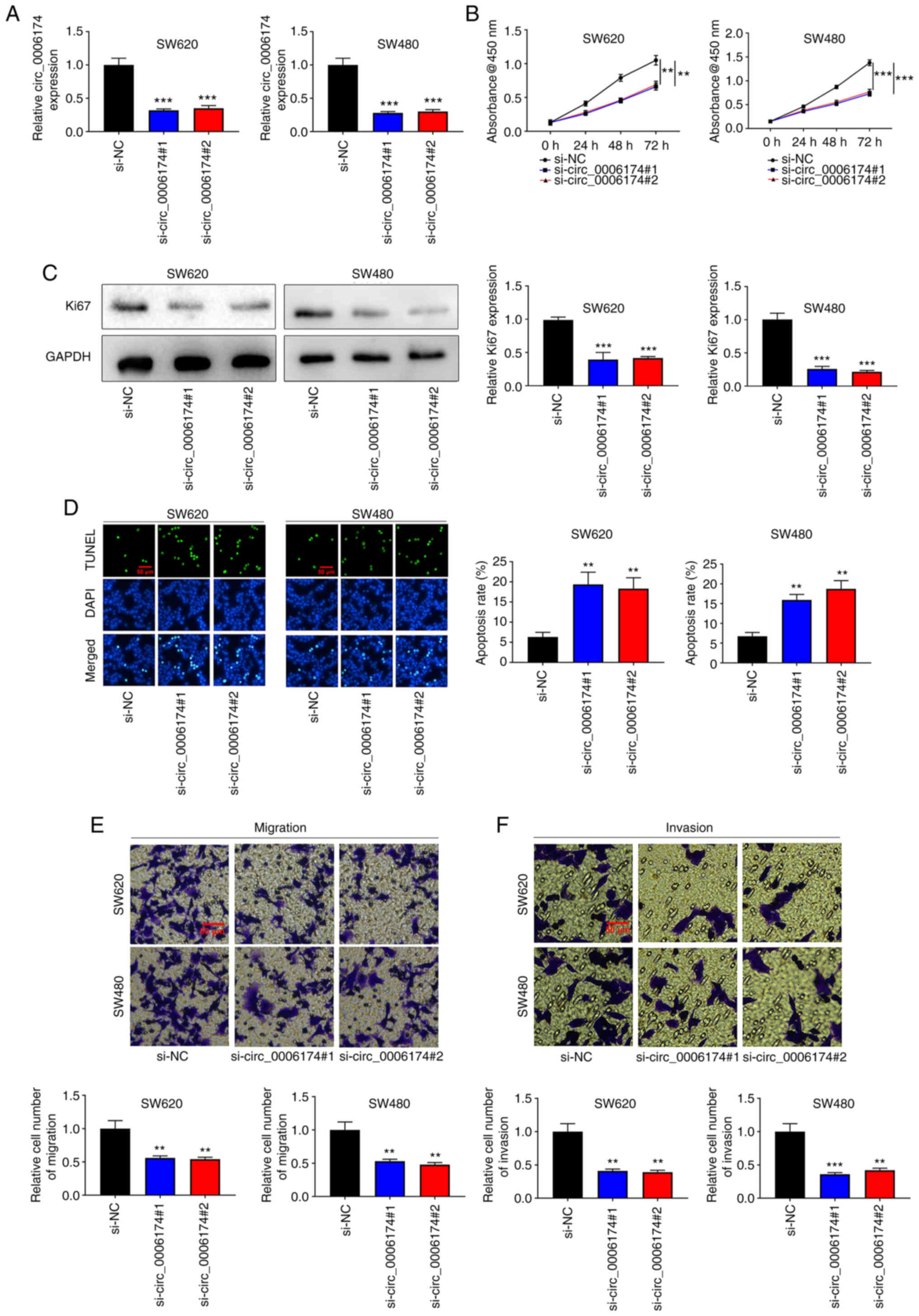
To understand the regulatory effects of circ_0006174
on the biological behavior of CRC cells, two circ_0006174 siRNAs
(si-circ_0006174#1 and si-circ_0006174#2) were transfected into
SW620 and SW480 cell lines and a circ_0006174 low expression model
successfully constructed (Fig.
2A). The CCK-8 method was utilized to appraise cell growth. The
data revealed that cell growth in the knocked down circ_0006174
group was markedly lower compared with the control group (Fig. 2B). Similarly, Ki-67, the
proliferation-related protein, was significantly decreased in the
knocked down circ_0006174 group (Fig.
2C). Additionally, the results of TUNEL assay revealed that the
apoptosis rate of SW620 and SW480 cells was markedly promoted in
the si-circ_0006174#1 and si-circ_0006174#2 group compared with the
si-NC group (Fig. 2D). Transwell
assay was employed to evaluate cell migration and invasion and the
data showed that as opposed to the control group, the cell
migration and invasion of si-circ_0006174#1 and si-circ_0006174#2
groups were markedly repressed (Fig.
2E and F). The above results suggested that circ_0006174
mediated the malignant phenotype of CRC cells.
Circ_0006174 is the molecular sponge
of miR-1205
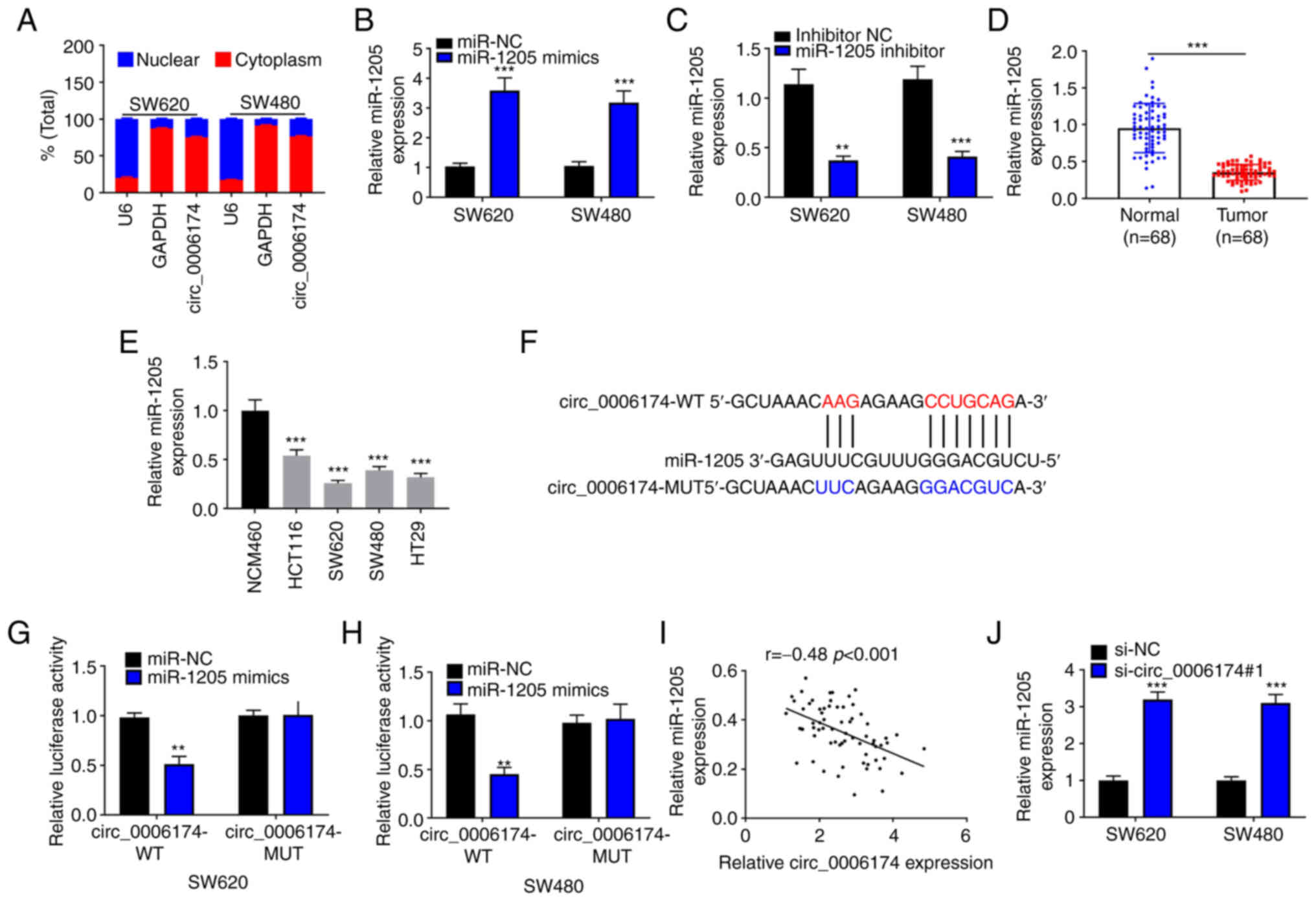
RT-qPCR suggested that circ_0006174 mainly existed
in the cytoplasm of SW620 and SW480 (Fig. 3A). The transfection of miR-1205
mimics could significantly increase the expression of miR-1205 in
SW620 and SW480 cells and the transfection of miR-1205 inhibitor
decreased the expression of miR-1205 in SW620 and SW480 cells
(Fig. 3B and C). Furthermore,
RT-qPCR was performed to determine miR-1205 expression in CRC
tissues and cell lines and the data showed that miR-1205 was
under-expressed in CRC tissues and cell lines (Fig. 3D and E). Hence, circ_0006174 may
be implicated in the modulation of downstream gene expression at
the post-transcriptional level as ceRNAs. In the Circular RNA
Interactome database, it was observed that miR-1205 contained a
binding site complementary to circ_0006174 (Fig. 3F). To confirm the binding
relationship between the two, a dual-luciferase reporter gene assay
was conducted and the data confirmed that miR-1205 markedly
repressed the luciferase activity of the cells in
circ_0006174-wild-type (WT) group, while the luciferase activity of
the cells in circ_0006174-mutant (MUT) group was unchanged
(Fig. 3G and H). Pearson's
correlation analysis showed that miR-1205 expression in CRC tissue
was negatively correlate with circ_0006174 expression (Fig. 3I). miR-1205 expression was
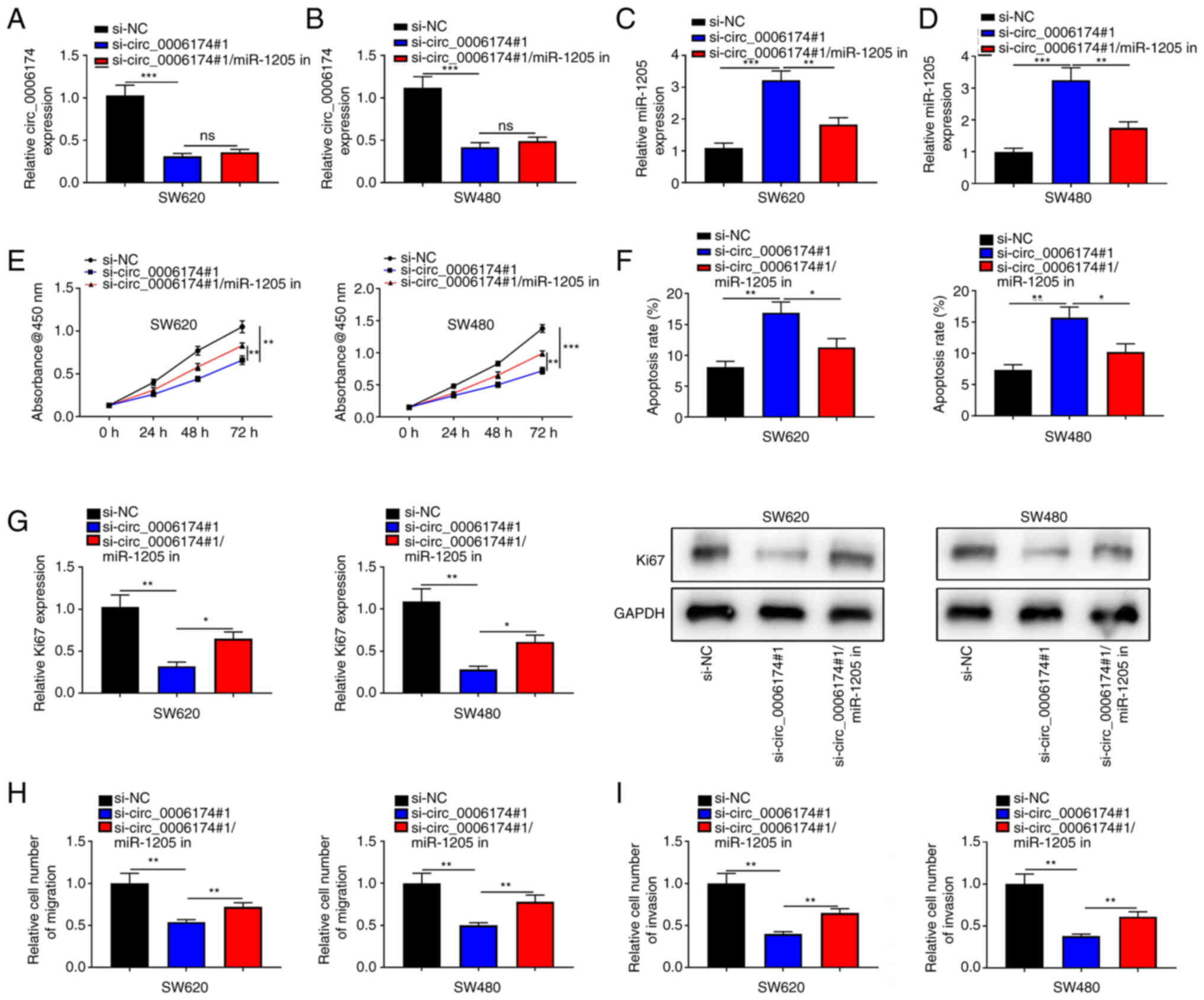
markedly promoted in si-circ_0006174#1 group (Fig. 3J). To validate that circ_0006174
functioned in CRC by adsorbing miR-1205, ‘rescue’ experiments were
performed and the data showed that the co-transfection of miR-1205
inhibitors partly abolished the effects of circ_0006174 depletion
on CRC cell growth, apoptosis, migration and invasion (Fig. 4). The above data implied that
circ_0006174 was the molecular sponge of miR-1205 and circ_0006174
was oncogenic in CRC through regulating the expression of
miR-1205.
Circ_0006174 decoys miR-1205 to
modulate CCBE1 expression
Next, StarBase database was utilized to detect the
downstream target of miR-1205 and it was revealed that CCBE1 was
the most potential target gene (Fig.
5A). Dual-luciferase reporter gene experiments confirmed that
the luciferase activity of CRC cells co-transfected with the
luciferase plasmids psiCHECKTM-2-CCBE1-WT and miR-1205 mimics was
markedly reduced compared with the control group, while no obvious
change in the luciferase activity of the cells was observed in the
cells co-transfected with CCBE1-MUT and miR-1205 mimics (Fig. 5B). This indicated that miR-1205
could bind with CCBE1 3′UTR. GEPIA database suggested that the
survival rate of CRC patients with high CCBE1 expression was worse
(Fig. 5C). The data of RT-qPCR
showed that CCBE1 was markedly upregulated in CRC tissues and cell
lines (Fig. 5D and E). Based on
the expression of CCBE1 in CRC tissues, the 66 CRC patients were
divided into high expression group and low expression group. It was
revealed that CCBE1 expression was not associated with
clinicopathological features of the patients (Table II). Notably, circ_0006174
expression and miR-1205 expression in CRC tissues were negatively
correlated and there was a positive correlation between
circ_0006174 expression and CCBE1 expression (Fig. 5F and G). The findings of RT-qPCR
and western blotting suggested that inhibition of miR-1205 markedly
elevated CCBE1 expression in CRC cell lines (Fig. 5H) and the transfection of miR-1205
inhibitor counteracted the suppressive effect of knocking down
circ_0006174 on CCBE1 expression (Fig. 5I). The above data confirmed that
in CRC cell lines, circ_0006174 positively modulated CCBE1
expression by targeting miR-1205.
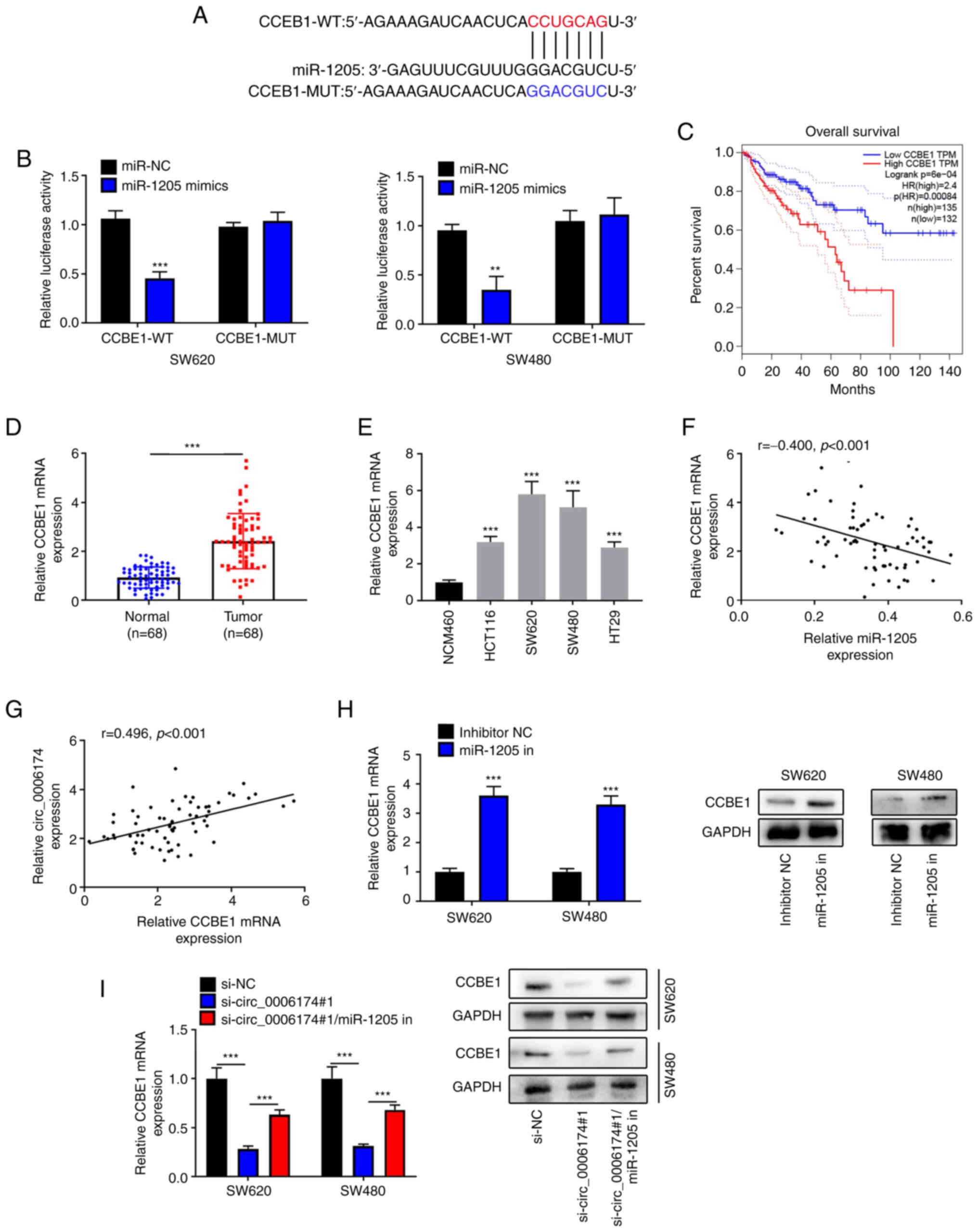 | Figure 5.Circ_0006174 targets miR-1205 to
modulate CCBE1 expression. (A) Bioinformatics was used to predict
the binding site of CCBE1 3′UTR to miR-1205. (B) Dual-luciferase
reporter gene experiment was used to confirm the binding sites
between miR-1205 and CCBE1 3′UTR. (C) GEPIA database analyzed the
relationship between CCBE1 expression and the survival rate of CRC
patients. (D) RT-qPCR was utilized to examine CCBE1 mRNA5
expression in 68 cases of CRC tissues. (E) RT-qPCR was performed to
determine CCBE1 mRNA expression in normal human colonic epithelial
cell line and CRC cell lines (HCT116, SW620, SW480 and HT29).
Pearson's correlation coefficient was used to analyze the
association between (F) miR-1205 expression and CCBE1 expression
and (G) CCBE1 expression and circ_0006174 expression in CRC tissue.
(H) RT-qPCR and western blotting experiments were employed to
determine CCBE1 mRNA and protein expression in CRC cells following
transfection with miR-1205 inhibitor. (I) RT-qPCR and western
blotting were performed to determine CCBE1 mRNA and protein
expression after the cells were co-transfected with circ_0006174
siRNA and miR-1205 inhibitor. **P<0.01 and ***P<0.001. circ,
circular RNA; miR, microRNA; CCBE1, calcium-binding epidermal
growth factor domain-containing protein 1; GEPIA, Gene Expression
Profiling Interactive Analysis; CRC, colorectal cancer; RT-qPCR,
reverse transcription-quantitative PCR; WT, wild-type; MUT, mutant;
NC, negative control. |
 | Table II.Correlations between CCBE1 expression
and clinical features in patients with colorectal cancer. |
Table II.
Correlations between CCBE1 expression
and clinical features in patients with colorectal cancer.
|
|
| CCBE1 |
|
|---|
|
|
|
|
|
|---|
| Features | n | High (n=34) | Low (n=34) | P-value |
|---|
| Age, years |
|
|
|
|
|
≥60 | 36 | 19 | 17 | 0.627 |
|
<60 | 32 | 15 | 17 |
|
| Sex |
|
|
|
|
|
Male | 31 | 16 | 15 | 0.807 |
|
Female | 37 | 18 | 19 |
|
| Clinical stage |
|
|
|
|
|
T3-T4 | 37 | 20 | 17 | 0.465 |
|
T1-T2 | 31 | 14 | 17 |
|
| Tumor size, cm |
|
|
|
|
| ≥3 | 38 | 18 | 20 | 0.625 |
|
<3 | 30 | 16 | 14 |
|
Circ_0006174 upregulates Wnt
pathway-related proteins and promotes lung metastasis in vivo
To further identify the downstream mechanism of
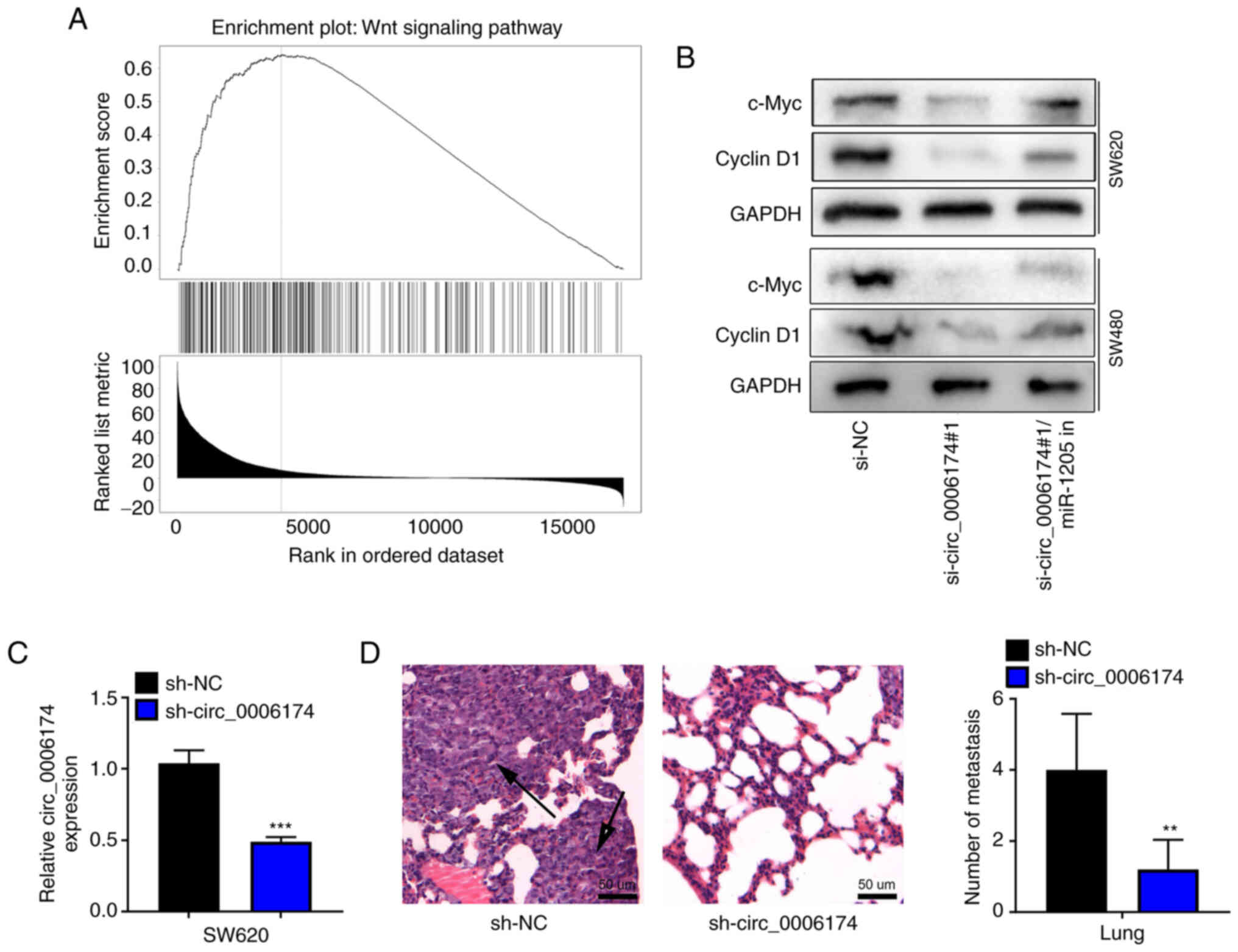
CCBE1 in CRC, gene set enrichment analysis was performed with
LinkedOmics database and the results showed that CCBE1 was
positively associated with the activation of Wnt pathway (Fig. 6A). Western blot assay confirmed
that the expression levels of c-Myc and cyclin D1 were
significantly downregulated in the si-circ_0006174#1 group compared
with the si-NC group and the transfection of miR-1205 inhibitor
reversed the inhibitory effect of circ_0006174 knockdown on the
expression of c-Myc and cyclin D1 in SW620 and SW480 cells
(Fig. 6B). RT-qPCR suggested that
the expression of circ_0006174 in sh-circ_0006174 group was
decreased compared with the sh-NC group (Fig. 6C). H&E staining further showed
that the number of lung metastatic nodules were fewer and the
metastatic nodules were much smaller in sh-circ_0006174#1 group
(Fig. 6D). These results further
supported that circ_0006174 was an oncogenic circRNA in CRC.
Discussion
Reportedly, circRNAs are closely associated with CRC
carcinogenesis and development (17). circ_0006174 is a circRNA which is
transcribed from chromosome chr9:110064315-110068928+. A previous
study reports that circ_0006174 is highly expressed in CRC tissues
and cells that and circ_0006174 can promotes CRC progression via
sponging miR-138-5p and upregulating MACC1 (8). Another study reports that the
exosomal circ_0006174 promotes doxorubicin resistance of CRC cells
(9). In the present study, by
analyzing GEO microarray data, it was found that circ_0006174 was
significantly upregulated in CRC tissues. This result was also
confirmed in the 68 specimens of CRC patients. Moreover,
circ_0006174 expression in CRC cell lines was notably higher than
that in normal human colonic epithelial cell lines. Additionally,
circ_0006174 overexpression was associated with larger tumor
diameter and higher clinical stage of CRC patients. Furthermore,
the loss-of-function experiments results implied that circ_0006174
could enhance cells proliferation, metastasis and inhibit cell
apoptosis rate in vitro and promote lung metastasis in
vivo. These findings revealed that circ_0006174 served as an
oncogenic factor to participant in CRC progression.
The ceRNA hypothesis provides a new mechanism of RNA
interaction. circRNA can function as ceRNA to modulate gene
expression by targeting miRNA (18). The present study revealed that
circ_0006174 was mainly distributed in the cytoplasm, so it is
hypothesized that circ_0006174 serves as a ceRNA. Bioinformatics
and dual-luciferase reporter gene assay showed that miR-1205 could
directly and specifically bind with circ_0006174. As a tumor
suppressor, miR-1205 restrains the development of diverse
malignancies. For instance, miR-1205 represses NSCLC progression by
downregulating KRAS expression (19). In gliomas, miR-1205 targets BATF3
to restrain cell growth and invasion (20). In ovarian cancer, the
miR-1205/SH2D3A axis modulates cell growth, migration and invasion
(21). Reportedly, miR-1205 is
significantly downregulated in CRC tissues and it represses the
growth and metastasis of CRC cells by targeting MYO6 (22). The findings of the present study
showed that miR-1205 was markedly downregulated in CRC tissues and
cell lines and was negatively correlated with circ_0006174
expression; additionally, circ_0006174 knockdown in CRC cell lines
markedly enhanced miR-1205 expression; besides, circ_0006174
expression and miR-1205 expression in CRC tissues were negatively
correlated; miR-1205 inhibitor counteracted the suppressing effect
of knocking down circ_0006174 on the growth, migration and invasion
of CRC cells. Hence, it was validated that circ_0006174 was the
molecular sponge of miR-1205 and circ_0006174 works in CRC cells by
modulating miR-1205.
Reportedly, CCBE1 can not only modulate
extracellular matrix remodeling, but also are crucial in
angiogenesis and lymphangiogenesis (23). Accumulating research has
demonstrated that CCBE1 is vital in cancer biology. CCBE1 is
identified as a target gene of miR-330-3p, which is related to the
aggressive phenotype of breast cancer (24). In gastrointestinal stromal tumor
patients, the patients with CCBE1 overexpression have worse overall
survival and relapse-free survival compared with patients with
CCBE1 under-expression (25). In
CRC, CCBE1 overexpression is reported to be markedly associated
with tumor differentiation, lymph node metastasis, vascular
invasion, liver metastasis and TNM stage of CRC patients (26). Notably, the present study observed
that there was no significant association between CCBE1 expression
level and the characteristics of the patients and this is probably
due to the heterogeneity of the tissue samples. CCBE1 is
demonstrated to facilitate lymphangiogenesis and lymphatic
metastasis in CRC (13). In the
present study, CCBE1 was identified as the target gene of miR-1205.
Bioinformatics analysis revealed that CCBE1 was overexpressed in
CRC tissues and was associated with the unfavorable prognosis of
CRC patients. Furthermore, it was also revealed that CCBE1 was
markedly upregulated in CRC tissues and cell lines and circ_0006174
upregulated CCBE1 expression by inhibiting miR-1205. The present
study partly explains the mechanism of CCBE1 dysregulation in
CRC.
As one of the most important intracellular signal
pathways, Wnt/β-catenin signaling is associated with diverse
cellular processes, such as cell proliferation, differentiation,
migration, survival and metastasis. Dysregulated Wnt pathway has
been reported to promote CRC progression. For instance, lncRNA
SLCO4A1-AS1 promotes CRC cell proliferation, migration, invasion
and epithelial-mesenchymal transition via the Wnt/β-catenin pathway
(27). SNHG16 is regulated by the
Wnt pathway and plays an oncogenic role in CRC (28). In the present study, it was
confirmed that CCBE1 was associated with the activation of Wnt
pathway in CRC. Knockdown of circ_0006174 could decrease
Wnt-related proteins by regulating miR-1205 in CRC cells. However,
how circ_0006174/miR-1205/CCBE1 axis modulates the activation of
Wnt signaling requires further investigation.
The present study has some limitations. For example,
whether CCBE1 promotes the proliferation and metastasis of CRC
cells through Wnt pathway needs to be verified. The other potential
target genes of miR-1205 in CRC remains largely unknown. which will
be investigated in the future.
In summary, the present study elaborated on the
biological role of circ_0006174 in CRC cells and revealed the role
of the circ_0006174/miR-1205/CCBE1 axis in CRC progression. Hence,
circ_0000326 is likely to be a novel target for CRC therapy.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Cultivation Fund of
National Natural Science Foundation (grant no.
qiankehe2018-5764-11).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XZ, DC and BH designed the experiments and wrote the
manuscript. FY performed the bioinformatics analysis. XZ, FY, LY
and MZ performed the experiments. XZ and BH confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
This research was authorized by the Ethics Committee
of Guizhou Provincial People's Hospital (approval no. 201802004)
and complied with the Declaration of Helsinki. Prior to surgery,
each subject completed a written informed consent form.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Miller KD, Nogueira L, Mariotto AB,
Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL and Siegel
RL: Cancer treatment and survivorship statistics, 2019. CA Cancer J
Clin. 69:363–385. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Van Cutsem E, Nordlinger B and Cervantes
A; ESMO Guidelines Working Group, : Advanced colorectal cancer:
ESMO clinical practice guidelines for treatment. Ann Oncol. 21
(Suppl 5):v93–v97. 2010. View Article : Google Scholar
|
|
4
|
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P
and Wu M: CircRNA: Functions and properties of a novel potential
biomarker for cancer. Mol Cancer. 16:942017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang Y, Wang H, Zhang J, Chu Z, Liu P,
Zhang X, Li C and Gu X: Circ_0007031 serves as a sponge of miR-760
to regulate the growth and chemoradiotherapy resistance of
colorectal cancer via regulating DCP1A. Cancer Manag Res.
12:8465–8479. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Du J, Xu J, Chen J, Liu W, Wang P and Ye
K: circRAE1 promotes colorectal cancer cell migration and invasion
by modulating miR-338-3p/TYRO3 axis. Cancer Cell Int. 20:4302020.
View Article : Google Scholar
|
|
7
|
Wen T, Wu H, Zhang L, Li K, Xiao X, Zhang
L and Zhang Y: Circular RNA circ_0007142 regulates cell
proliferation, apoptosis, migration and invasion via
miR-455-5p/SGK1 axis in colorectal cancer. Anticancer Drugs.
32:22–33. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wei J, Lin Y, Wang Z, Liu Y and Guo W:
Circ_0006174 accelerates colorectal cancer progression through
regulating miR-138-5p/MACC1 axis. Cancer Manag Res. 13:1673–1686.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang Y, Tan X and Lu Y: Exosomal transfer
of circ_0006174 contributes to the chemoresistance of doxorubicin
in colorectal cancer by depending on the miR-1205/CCND2 axis. J
Physiol Biochem. 79:39–50. 2022. View Article : Google Scholar
|
|
10
|
Jorgensen BG and Ro S: MicroRNAs and
‘sponging’ competitive endogenous RNAs dysregulated in colorectal
cancer: Potential as noninvasive biomarkers and therapeutic
targets. Int J Mol Sci. 23:21662022. View Article : Google Scholar
|
|
11
|
Han B, Wang X and Yin X: Knockdown of
circRAD23B exerts antitumor response in colorectal cancer via the
regulation of miR-1205/TRIM44 axis. Dig Dis Sci. 67:504–515. 2022.
View Article : Google Scholar
|
|
12
|
Jiang Y, Liu G, Ye W, Xie J, Shao C, Wang
X and Li X: ZEB2-AS1 accelerates epithelial/mesenchymal transition
through miR-1205/CRKL pathway in colorectal cancer. Cancer Biother
Radiopharm. 35:153–162. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Song J, Chen W, Cui X, Huang Z, Wen D,
Yang Y, Yu W, Cui L and Liu CY: CCBE1 promotes tumor
lymphangiogenesis and is negatively regulated by TGFβ signaling in
colorectal cancer. Theranostics. 10:2327–2341. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46:D956–D963. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Artemaki PI, Scorilas A and Kontos CK:
Circular RNAs: A new piece in the colorectal cancer puzzle. Cancers
(Basel). 12:24642020. View Article : Google Scholar
|
|
18
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yan H, Chen X, Li Y, Fan L, Tai Y, Zhou Y,
Chen Y, Qi X, Huang R and Ren J: MiR-1205 functions as a tumor
suppressor by disconnecting the synergy between KRAS and MDM4/E2F1
in non-small cell lung cancer. Am J Cancer Res. 9:312–329.
2019.PubMed/NCBI
|
|
20
|
Yi C, Li H, Li D, Qin X, Wang J, Liu Y,
Liu Z and Zhang J: Upregulation of circular RNA circ_0034642
indicates unfavorable prognosis in glioma and facilitates cell
proliferation and invasion via the miR-1205/BATF3 axis. J Cell
Biochem. 120:13737–13744. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang G, Zhang H and Li P: Upregulation of
hsa_circRNA_102958 indicates poor prognosis and promotes ovarian
cancer progression through miR-1205/SH2D3A axis. Cancer Manag Res.
12:4045–4053. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Luan Y, Li X, Luan Y, Zhao R, Li Y, Liu L,
Hao Y, Oleg Vladimir B and Jia L: Circulating lncRNA UCA1 promotes
malignancy of colorectal cancer via the miR-143/MYO6 axis. Mol Ther
Nucleic Acids. 19:790–803. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hogan BM, Bos FL, Bussmann J, Witte M, Chi
NC, Duckers HJ and Schulte-Merker S: Ccbe1 is required for
embryonic lymphangiogenesis and venous sprouting. Nat Genet.
41:396–398. 2009. View
Article : Google Scholar
|
|
24
|
Mesci A, Huang X, Taeb S, Jahangiri S, Kim
Y, Fokas E, Bruce J, Leong HS and Liu SK: Targeting of CCBE1 by
miR-330-3p in human breast cancer promotes metastasis. Br J Cancer.
116:1350–1357. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tian GA, Zhu CC, Zhang XX, Zhu L, Yang XM,
Jiang SH, Li RK, Tu L, Wang Y, Zhuang C, et al: CCBE1 promotes GIST
development through enhancing angiogenesis and mediating resistance
to imatinib. Sci Rep. 6:310712016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao YR, Liu H, Xiao LM, Jin CG, Zhang ZP
and Yang CG: The clinical significance of CCBE1 expression in human
colorectal cancer. Cancer Manag Res. 10:6581–6590. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yu J, Han Z, Sun Z, Wang Y, Zheng M and
Song C: LncRNA SLCO4A1-AS1 facilitates growth and metastasis of
colorectal cancer through β-catenin-dependent Wnt pathway. J Exp
Clin Cancer Res. 37:2222018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Christensen LL, True K, Hamilton MP,
Nielsen MM, Damas ND, Damgaard CK, Ongen H, Dermitzakis E, Bramsen
JB, Pedersen JS, et al: SNHG16 is regulated by the Wnt pathway in
colorectal cancer and affects genes involved in lipid metabolism.
Mol Oncol. 10:1266–1282. 2016. View Article : Google Scholar
|