Introduction
Multiple myeloma (MM) involves crosstalk between the
immune and bone systems, and as a blood cancer, homes into the bone
marrow (BM) micro-environment, which is strongly tied to
overexpression of interleukin-6 (IL-6) and bone loss. Like their
normal counterpart, MM cells mainly proliferate and survive within
the bone marrow by physiological and functional interactions with
bone marrow stromal cells (BMSCs) and the surrounding BM micro
environment. The later plays a central role in the pathogenesis of
MM and these interactions have shown to be important in myeloma
cell survival and progression (1–3).
BMSCs produce inflammatory cytokines, such as IL-6, and they
regulate the expression of cell cycle inhibitors-cyclin dependent
kinase inhibitors (CDK) p21WAF1 and p27Kip1,
anti-apoptotic members of the Bcl-2 family and ABC-family drug
transporters in myeloma cell directly via cell receptors and
adhesion molecules. Interleukins are crucial to skeletal
homeostasis and some adhesion molecules, such as cadherins,
facilitate the formation of multicellular structures in the bone
marrow by anchoring to the actin cytoskeleton (4,5).
Epigenetic anomalies have been identified as drivers
in the development of the myeloma (6). DNA methylation is an epigenetic
modification of cytosine catalyzed by DNA methyltransferases
(DNMT1, DNMT3a and DNMT3b). DNA methylation leads to inhibition of
gene expression. Several studies have investigated whether
aberrations in DNA methylation could be expressed as disease
stage-specific, thus changing during disease progression. DNA
hypomethylation was reported as the predominant early change during
myelomagenesis that is gradually transformed to DNA
hypermethylation in relapsed cases and during the disease
progression (7,8). DNA methyltransferase inhibitors,
such as cytidine analogs 5-azacytidine and 5-Aza-2′-deoxycytidine
(Decitabine) (DAC) are currently used to revert aberrant DNA
methylation patterns. The DAC is incorporated only in DNA, whereas
5-azacytidine penetrates in both DNA and RNA, and via incorporation
into newly synthesized RNA, it will interfere with RNA processing,
inhibiting protein synthesis. For these reasons, the 5-azacytidine
is used more often as a demethylation agent for treatment of HMCLs,
though the anti-myeloma activity, supported by enhanced DNA damage,
cell cycle arrest, and induction of myeloma cell apoptosis was
shown in both demethylation agents (9,10).
Histone deacetylases (HDACs) are enzymes responsible
for removing the acetyl group from histones, leading to suppression
of gene expression. This process contributes to the undesired
suppression of tumor suppressor gene expression. HDAC inhibitors
are known to arrest human tumor cell activity in the G1 phase of
the cell cycle and activate CDK inhibitors such as p21WAF1, the
CDKN1A gene of the CIP/KIP family, and p15INK4b
and p16INK4a encoded by the CDKN2B and CDKN2A
genes belonging to the INK4 family of proteins (11,12). Ng et al (13) reported for the first time, the
high incidences of p15INK4b and p16INK4a
alterations in MM patients, not by homozygous deletions or
mutations, but solely by hypermethylation of the 5′CpG islands.
However, the methylation status and transcription of these genes
were subsequently studied in MM-derived cell lines in vitro.
An aberrant methylation of p16INK4a was found in six
analyzed cell lines including RPMI8226 and U266 as well.
Conversely, only the HS-Sultan cell line, but not RPMI8226 and
U266, showed extensive methylation in the 5′upstream region of
p15INK4b (14).
Inhibition of histone deacetylase (HDAC) activity by HDAC
inhibitors on the other hand, results in the accumulation of
acetylated histones leading to altered gene transcription. The HDAC
inhibitor suberohydroxamic acid (SBHA), induces apoptosis,
dependent on the induction of mitochondrial membrane permeability
(15). The SBHA described in
melanoma cell lines (15,16) is able to enhance TRAIL-induced
cell death by the up-regulation of the pro-apoptotic proteins
caspase-8, caspase-3, Bim, Bid, Bak, Bax, while downregulating, at
the same time, the anti-apoptotic proteins Bcl-xL, Mcl-1, and XIAP
(17). The mechanism by which
SBHA regulates such a diverse array of genes involved in apoptosis
is not clear. Likely, SBHA targets HDACs associated with
transcriptional factors that are involved in regulation apoptosis
such as p53 and c-Myc.
The PDLIM4 gene (location 5q.31) encodes PDZ
and LIM protein 4 involved in bone development as was shown in a
study of PDLIM4 gene polymorphisms in the susceptibility to
osteoporotic fracture (18). LIM
protein RIL is an actin associated nuclear protein containing both
PDZ and LIM domains. It belongs to a large family of LIM proteins
(19). PDZ and LIM domain
proteins (PDLIM) are involved in the regulation of a variety of
biological processes, including cytoskeletal organization, and may
act as adaptors between cytoskeleton and intracellular signaling
components (20). The clinical
and prognostic value of the PDLIM family in multiple myeloma is
still unclear. The RIL, PDLIM4 gene, which is frequently
methylated in cancer, has been described as a tumor suppressor. Its
methylation is associated with altered expression in various
cancers, including hematological malignancies (21–23). It is known that RIL re-expression
leads to suppression of cell growth, and it sensitizes cells to
apoptosis (24). In this study,
we examined the methylation and expression patterns of
PDLIM4 as a possible tumor suppressor gene in two human
myeloma cell lines (HMCLs). RPMI8226 and U266 (U266-B1) differ in
both IL-6 expression and p53-functionality. The U266 myeloma cell
line has deletions of chromosome 13 and 17p, which involve the TP53
gene (25). The p53 suppressor
protein is encoded by the TP53 gene located on chromosome 17p13.1,
while deletion of the chromosomal region 17p13-del (17p) is
associated with poor outcome in multiple myeloma (26). The second used RPMI8226 myeloma
cell line has shown deletion of neither chromosome 13, nor
chromosome 17p (25). We also
examined the methylation and expression pattern of another
tumor-suppressor gene, CDKN2B, which is often methylated in
multiple myeloma patients (18,26). The CDKN2B gene belongs to
INK4 family responsible for regulation of cell cycle. Aberrant
methylation of this tumor suppressor gene is considered to be an
important epigenetic change in molecular pathogenesis of multiple
myeloma (13,26,27). Meta-analysis study of Li et
al pointed out the effect of CDKN2B gene methylation on
the prognosis of the disease (28). For this reason, we compared the
CDKN2B gene expression with the expressions of other
cyclin-dependent kinases genes (CDKN2A and CDKN1A).
Our goal was to investigate the in vitro effect of SBHA and
DAC alone, on tumor-suppressor and DNA methyltransferases genes
transcription and the molecular biological behavior in two selected
myeloma cell lines differing in p53-functionality and IL-6
expression.
Materials and methods
Cell lines and cell culture
Cell lines RPMI8226 and U266 represent different
IL-6 expression profile, IL-6 is expressed in U266, but not in
RPMI8226 (7,8). Further, according to Keats Lab
(www.keatslab.org), the U266 cell line has E419X
missense mutation on the RB1 gene and A161T missense mutation on
the TP53 gene, while the RPMI8226 cell line has E285K missense
mutation on the TP53 gene (Table
I). Both human multiple myeloma cell lines were purchased from
ATCC (CCL155™). The cell line RPMI8226 (CCK155TM) was
maintained in RPMI-1640 medium (Sigma-Aldrich) supplemented with
10% fetal bovine serum (FBS), 1% antibiotics
Penicillin-Streptomycin, 1% L-Glutamine and 1% 100 mM Sodium
Pyruvate, while the human multiple myeloma cell line U266 (U266B1)
was cultured in RPMI-1640 medium (Sigma-Aldrich) supplemented with
15% fetal bovine serum (FBS), 1% antibiotics
Penicillin-Streptomycin, 1% L-glutamine and 1% 100 mM sodium
pyruvate. All cells were maintained at 37°C and 5% CO2
atmosphere.
 | Table I.Characteristics of RPMI8226 and U266
myeloma cell lines (www.keatslab.org). |
Table I.
Characteristics of RPMI8226 and U266
myeloma cell lines (www.keatslab.org).
| HMCL | Translocation | TP53 status | p53 expression | IL-6
dependency | (Refs.) |
|---|
| RPMI8226 | t(14,16) | E285K | + | - | (30,57) |
| U266 (U266B1) | t(11,14) | A161T L36M | - | + | (30,58) (36) |
Cell viability assay (MTT assay)
Cell lines were treated with 0.5 and 5 µM DAC
(Sigma-Aldrich) and 10 and 50 µM SBHA (Sigma-Aldrich) for 24 and 48
h. The RPM-I8226 cells were seeded on 96-well plates at 5,000 cells
per well, and the U266 cells at 5,500 cells per well. The cell
viability assay was performed in triplicate using
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide agent
(MTT). Viable cells were detected by measuring absorbance on a
spectrophotometer at a wavelength of 570 nm. The percentage of
viable cells was calculated as follows: average absorbance of
treated cells/average absorbance of control cells ×100.
Cell cycle analysis (FACS)
For fluorescence-activated cell sorting (FACS),
cells were treated for 24 h (10, 50 µM SBHA, 0.5, 5 µM DAC),
harvested, and fixed with 96% ice-cold ethanol overnight at −20°C.
After washing with ice cold PBS + 2% FBS, cells were incubated with
0.2 mg/ml RNAse A + PBS at RT for 30 min. Then, 200 µl of propidium
iodide (PI) was added and analyzed by BD FACSVerse flow cytometry
(BD Biosciences). The results were analyzed using BD FACSuite (BD
Biosciences) software. The percentage of living unaffected cells
was calculated to 100%, then the percentage of live treated cells
was calculated as follows: × = (% of live treated cells determined
by FACSuite software ×100%)/% of live untreated control cells
determined using FACSuite software.
Methyl-specific PCR (MSP-PCR)
Genomic DNA was isolated by the Wizard®
Genomic DNA Purification kit (Promega). Extracted DNA (~400 ng) was
modified with sodium bisulfite using the EpiTect®
Bisulfite kit according to the manufacturer's instructions
(Qiagen). MethPrimer, based on Primer3 (https://www.urogene.org/methprimer/), was used for
designing MSP-PCR primers specific for unmethylated (U) and
methylated (M) DNA sequences (Table
II). PCR products were analyzed by 2% agarose gel
electrophoresis (1 h, 55 V). The percentage of methylation was
subsequently quantified by methyl-specific sequencing
(pyrosequencing).
 | Table II.Primer design for MSP-PCR. |
Table II.
Primer design for MSP-PCR.
| Gene | Forward PCR
primer | Reverse PCR
primer |
|---|
| CDKN2B (U) |
5′-TGAGGATTTTGTGATGTGTTT-3′ |
5′-CATACAATAACCAAACAACCAATCA-3′ |
| CDKN2B (M) |
5′-TGAGGATTTCGCGACGCGTTC-3′ |
5′-CGTACAATAACCGAACGACCGATCG-3′ |
| PDLIM4 (U) |
5′-GATGGGTTGTAGGTGTGTTAGTTG-3′ |
5′-CTTTTAAAATCACTTTTAAA-3′ |
| PDLIM4 (M) |
5′-GATGGGTCGTAGGTGTGTTAGTC-3′ |
5′-CTTTAAAATCGCTTTTAAAAACGAT-3′ |
Methyl-specific sequencing
(Pyrosequncing)
Isolated and modified DNA was used to determine the
percentage of both PDLIM4 and CDKN2B genes promoters
methylation. DNA was isolated from both human myeloma cell lines
(RPMI8226, U266) after 48 h treatment with 10, 50 µM SBHA, 0.5 and
5 µM DAC. The CpG islands identification and PCR primers design for
the following pyrosequencing reaction were designed using PyroMark
Assay Design SW 2.0 (Qiagen) (Table
III). 1 µl of bisulfite treated DNA was added in a 25-µl PCR
reaction mixture containing 12.5 µl 1X PyroMark PCR Master Mix
(Qiagen), 1 µl 25 mM MgCl2, 2.5 µl 1× CoralLoad Concentrate
(Qiagen), 0.2 µl forward primer and 0.2 µl biotinylated reverse
primer. For HotStartTaq Polymerase activation, the PCR reaction
mixture was initially denaturated at 95°C for 15 min followed by 45
cycles of denaturation at 94°C for 30 sec, annealing at 56°C for 30
sec, elongation at 72°C for 30 sec, and the final extension at 72°C
for an additional 10 min after the last cycle. The resulting
biotinylated PCR product was immobilized on Streptavidin
Sepharose® HP (GE Healthcare), precipitated with 70%
ethanol, passed through a denaturation step and then a washing step
using the PyroMark Q96 Vacuum Workstation (Qiagen). The amplicons
were transferred to each well of the PyroMark Q96 plate containing
40 µl of 0.4 µM sequencing primer diluted in annealing buffer
(Qiagen). Control samples (bisulfite modified unmethylated and
methylated DNA; Qiagen) were part of a set of analyzed samples. The
pyrosequencing analysis was performed according to the PyroMark CpG
Software 1.0.11 (Qiagen). The methylation value was quantified in
terms of the methylation level (MtL) as the average percentage of
cytosines methylated per CpG: MtL (%) = (∑% methylated
cytosines)/No. of CpGs analyzed).
 | Table III.Primer design for pyrosequencing
analysis. |
Table III.
Primer design for pyrosequencing
analysis.
| Gene | Forward PCR
primer | Reverse PCR
primer | Sequencing
primer | CpGs |
|---|
| PDLIM4 |
5′-GGGTTTATGAGGA |
5′-biotin-ACACCCCC |
5′-TGTAGATAGTTGG | 9 |
| Promotor (216
bp) |
GGTATTTGAGTTG-3′ | ACTCAACTCTC-3′ | GTTTGG-3′ |
|
| CDKN2B |
5′-AGGAGTTGAGGG |
5′-biotin-TCCCCACC |
5′-GGATATTTAGAGA | 13 |
| Promotor (223
bp) | TAGTGGT-3′ | CCCTTAAACT-3′ |
GTAGTGTAGTTA-3′ |
|
Reverse transcription-quantitative PCR
(RT-qPCR)
Myeloma cells were seeded in 6-well plates at a
concentration of 1×105 cells/well, and after SBHA and
DAC treatments they were harvested after 48 h. Total RNA from both
cell lines was isolated with a High Pure RNA Isolation kit (Roche),
and reverse transcription of 1,000 ng of total RNA was made by
Transcriptor First Strand cDNA Synthesis kit (Roche, Basel,
Switzerland). TRI Reagent® BD (Molecular Research
Centre, Inc.) was used for isolation of total RNA from unsorted BM
aspirate cells. The real-time PCR was carried out using Taq-Man
probes with Xceed qPCR Probe Mix (Institute of Applied
Biotechnologies) using LightCycler® 480 System (Roche).
The cDNA expression of CDKN2A (Hs00923894_m1, CDKN2B
(Hs00793225_m1), CDKN1A (Hs00355782_m1), PDLIM4
(Hs00896441_m1), DNMT1 (Hs01041237_g1), DNMT3A
(Hs01027162_m1), and DNMT3B (Hs00171876_m1) genes was
normalized to the expression of endogenous housekeeping control
GAPDH (Hs01041237_g1). All used probes were provided by Thermo
Fisher Scientific. Untreated cells were used as the calibrator
(control) for the 2−ΔΔCq quantification approach, and
commercially accessible human BM total RNA (Takara Bio USA, Inc.)
for 2−ΔΔCq quantification approach of cDNA prepared from
RNA of unsorted BM patient cells. The experiments were done in
triplicates, repeated in three independent measurements.
Immunocytochemical staining
Immunocytochemical staining method was used for
detection of target p15INK4b and PDZ and LIM protein 4. After 48-h
treatment (10, 50 µM SBHA, 0.5, 5 µM DAC) untreated control (DMSO)
and treated cells (RPMI8226 and U266 cell lines) were spread on a
microscope slide and fixed by mixture of acetone: methanol (1:1).
After antigen recovery in citrate buffer (pH 6.0) for 15 min in
120°C, a block of endogenous peroxidase (5%
H2O2) was performed for 15 min, followed by a
block of non-specific background staining with Protein Block (Dako)
for 10 min. For washing between these steps were used Tris buffer
(pH 7.6). The cells were incubated over night with primary
antibodies (Elabscience), and protein detection was performed using
the EnVision™ Detection System Peroxidase/DAB,
Rabbit/Mouse (Dako). Nuclei of cells were stained with hematoxylin
and samples were dehydrated and coverslipped. Results were
evaluated by ImageJ software by measuring staining intensity of
microscopic photos, taken from at least five different field of
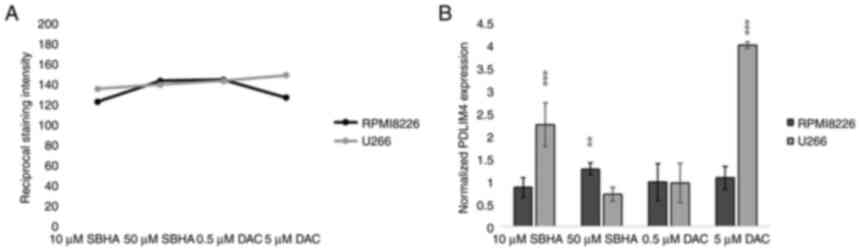
vision. The results were evaluated as reciprocal staining
intensity: reciprocal staining intensity = 255-measured staining
intensity (29) and then
converted to percentages as follows: (reciprocal staining intensity
of treated cells/reciprocal staining intensity of control untreated
cells) ×100.
Statistical analysis
The significance of results was determined using
unpaired Student's t-test with Bonferroni correction. All
statistical analyses were performed using SPSS software version 20
(IBM Corp.) and P<0.05 was considered to indicate a
statistically significant difference.
Results
The SBHA anti-proliferative effect in
myeloma cells
The MTT assay and FACS analysis were used as an
assessment of the anti-proliferative effects of both, histone
deacetylase inhibitor SBHA and demethylation agent DAC. The
RPMI8226 cells were more potent in regard to cell death induction
than the U266 cell line, as evidenced by treatment with both tested
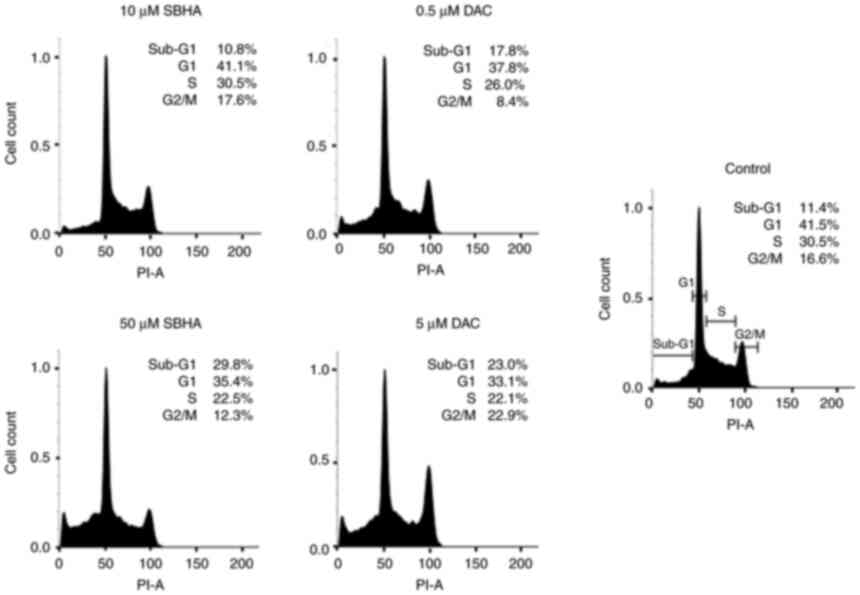
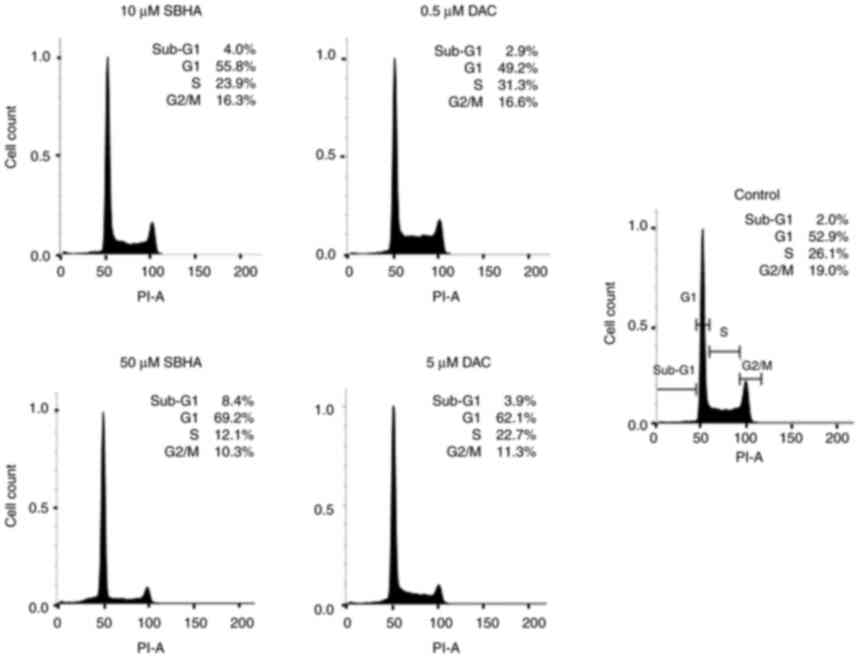
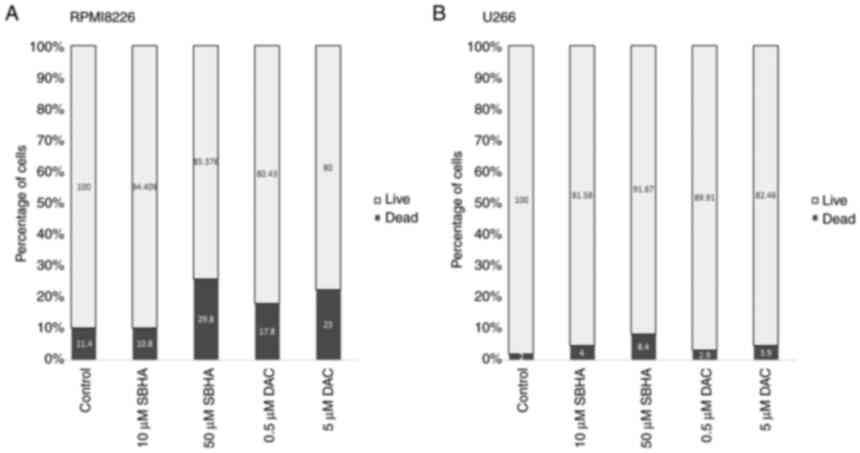
compounds (Fig. 1, Fig. 2, Fig. 3). As depicted in Fig. 2, the U266 cells terminate the cell
cycle predominantly at the G1 checkpoint and the proportion of dead
cells in Sub-G1 population increased in both agents gradually, in a
dose-dependent manner compared to the untreated U266 cell culture.
Fig. 3 shows the differences in
the response of both myeloma lines to used agents, with a more
pronounced effect after 48 h of treatment in RPMI8226 cell line.
The increase in sub-G1 fraction in RPMI8226 cells treated with 50
µM SBHA (29.8%) (Fig. 1)
contrasts with the G1 accumulation observed after treatment of U266
cells with 50 µM SBHA (8.4%) (Fig.
2), which indicates a difference in cell cycle regulation
between these cell lines. For this reason, the cyclin-dependent
kinase inhibitors p15INK4b, p16INK4a,
p21WAF1 cDNA expression was analyzed (Fig. 4).
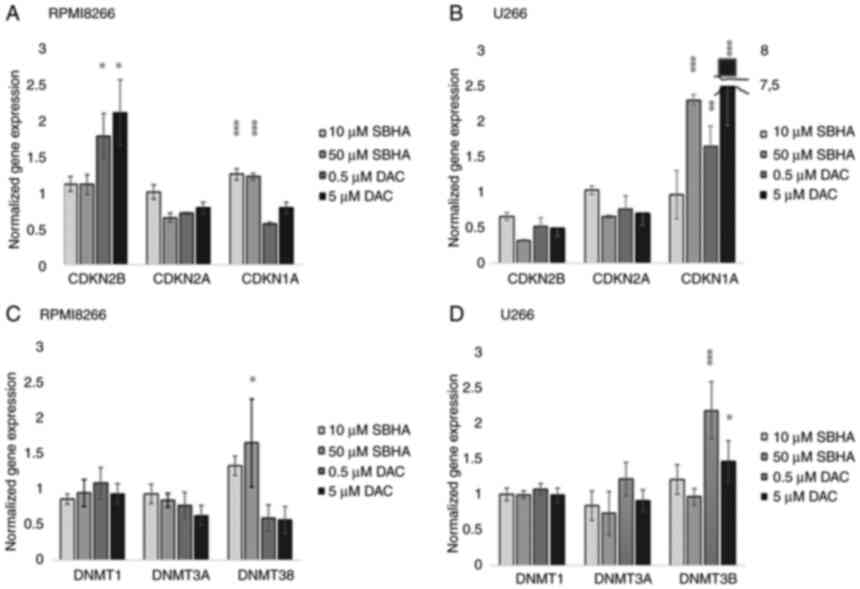 | Figure 4.The cDNA expression profile of
p15INK4b, p16INK4a, p21WAF1 in (A) RPMI8226 and (B) U266 after 48-h
treatment with SBHA (10 and 50 µM) and DAC (0.5 and 5 µM). The cDNA
expression profile of DNMT1, DNMT3A and DNMT3B in (C) RPMI8226 and
(D) U266 cell lines treated with SBHA (10 and 50 µM) and DAC (0.5
and 5 µM) for 48 h. The significance was determined using unpaired
Student's t-test with Bonferroni correction. *P<0.05 vs. Control
(DMSO), **P<0.01 vs. Control (DMSO) and ***P<0.001 vs.
Control (DMSO). The groups were normalized to the control (DMSO)
group, which was set at 1. DAC, 5-Aza-2′-deoxycytidine
(Decitabine); DMSO, dimethyl sulfoxide; MtL, methylation level;
MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
agent; PDLIM4 (RIL), PDZ and LIM domain 4; PI, propidium iodide;
SBHA, suberohydroxamic acid. |
Different CDKs and DNMTs expressions
in myeloma cell lines treated with SBHA and DAC
The significant (P<0.05) increased expression of
CDKN2B gene was detected in RPMI8226 cells after treatment
of both 0.5 and 5 µM DAC concentrations (Fig. 4A; Table IV), while 10 and 50 µM SBHA
treatments resulted to significant (P<0.0001) increase the
CDKN1A expression in U266 cells (Fig. 4B). A similar significant increase
in CDKN1A expression was detected in U266 cells treated with both
used DAC concentrations, and 10 µM SBHA as well (Fig. 4B). Conversely, the CDKN2A
gene expression was not significantly changed, although this gene
is methylated in both of these myeloma cell lines (25,28). An expression analysis of DNA
methyltransferases (DNMT1, DNMT3A and DNMT3B) showed significantly
increased relative DNMT3B (P<0.001) gene expression after
0.5 µM (P<0.001) and 5 µM DAC (P=0.05) treatments in U266 cell
line (Fig. 4D). On the contrary,
in RPMI8226 cells, the significantly increase DNMT3B gene
expression was determined after their treatment with 50 µM SBHA
(P<0.05) (Fig. 4C; Table V).
 | Table IV.Expression profile of CDK genes in
RPMI8226 and U266 cell lines after 48 h treatment with SBHA (10 and
50 µM) and DAC (0.5 and 5 µM). |
Table IV.
Expression profile of CDK genes in
RPMI8226 and U266 cell lines after 48 h treatment with SBHA (10 and
50 µM) and DAC (0.5 and 5 µM).
| Cell
treatments | CDKN2B | CDKN2A | CDKN1A |
|---|
| RPMI8226 |
|
|
|
| 10 µM
SBHA | 1.123±0.10 | 1.010±0.10 |
1.260±0.08c |
| 50 µM
SBHA | 1.121±0.14 | 0.656±0.06 |
1.227±0.04c |
| 0.5 µM
DAC |
1.790±0.32a | 0.715±0.01 | 0.569±0.03 |
| 5 µM
DAC |
2.122±0.46a | 0.791±0.08 | 0.791±0.08 |
| U266 |
|
|
|
| 10 µM
SBHA | 0.662±0.05 | 1.031±0.06 | 0.970±0.34 |
| 50 µM
SBHA | 0.318±0.01 | 0.657±0.02 | 2.750±1.19 |
| 0.5 µM
DAC | 0.522±0.12 | 0.764±0.18 |
1.655±0.28b |
| 5 µM
DAC | 0.494±0.11 | 0.707±0.17 |
7.811±0.55c |
 | Table V.Expression profile of DNMTs genes in
U266 and RPMI8226 cell lines after 48 h treatment with SBHA (10 and
50 µM) and DAC (0.5 and 5 µM). |
Table V.
Expression profile of DNMTs genes in
U266 and RPMI8226 cell lines after 48 h treatment with SBHA (10 and
50 µM) and DAC (0.5 and 5 µM).
| Cell
treatments | DNMT1 | DNMT3A | DNMT3B |
|---|
| RPMI8226 |
|
|
|
| 10 µM
SBHA | 0.858±0.07 | 0.929±0.14 | 1.324±0.14 |
| 50 µM
SBHA | 0.943±0.19 | 0.842±0.10 |
1.865±0.66a |
| 0.5 µM
DAC | 1.080±0.22 | 0.772±0.18 | 0.591±0.19 |
| 5 µM
DAC | 0.932±0.14 | 0.624±0.14 | 0.567±0.19 |
| U266 |
|
|
|
| 10 µM
SBHA | 1.005±0.09 | 0.843±0.21 | 1.216±0.21 |
| 50 µM
SBHA | 0.999±0.05 | 0.736±0.31 | 0.968±0.12 |
| 0.5 µM
DAC | 1.080±0.07 | 1.222±0.23 |
2.192±0.4b |
| 5 µM
DAC | 0.992±0.10 | 0.912±0.16 |
1.469±0.29a |
Expression profile of the unmethylated
CDKN2B and the methylated PDLIM4
To determinate the methylation state of promoter
sequences, gene methylation status was performed by MSP-PCR with
primers specific for unmethylated (U) and methylated (M) promoter
sequences. The unmethylated CDKN2B gene promoter sequence
was detected in both untreated myeloma cell lines, RPMI8226 (3%)
and U266 (3%) (Figs. 5 and
6), a methylation was therefore
not determined after epigenetic treatment. Moreover, the 62%
average promoter PDLIM4 gene methylation in RPMI8226
(Figs. 5A and 7B), and 59% in U266 (Figs. 6A and 8B), contrasted with the unmethylated
state of the CDKN2B gene promoter (3%) (Figs. 5B and 6B). The unmethylated CDKN2B
promoter analyzed region in both myeloma cell lines was detected
whereas the PDLIM4 analyzed promoter region was detected as
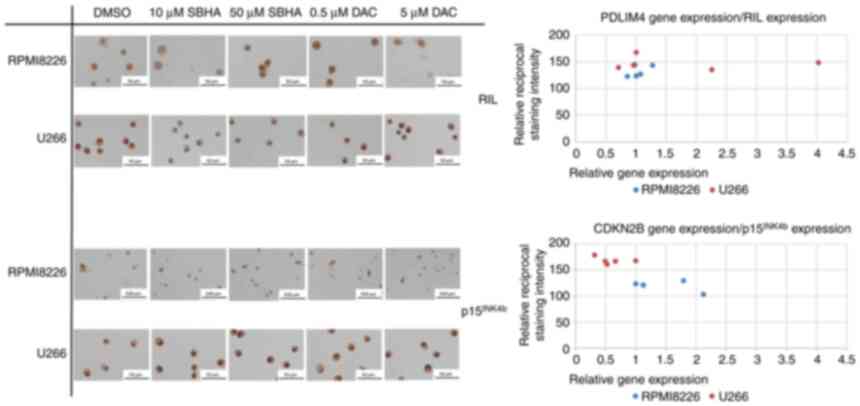
methylated in control cells (DMSO) (Figs. 7 and 8). These results were confirmed
quantitatively by the pyrosequencing, when 13 CpGs of the
CDKN2B and 9 CpGs of the PDLIM4 were analyzed. In
RPMI82226, but not U266, cells, 0.5 µM DAC demethylation treatment
caused significant reduction of PDLIM4 promoter MtL to 44%
in comparison to untreated cells (P-value 0.004; P<0.05)
(Figs. 9 and 10). Although the methylation reduction
was not significantly confirmed by elevated protein level using
immunochemistry staining detection (Fig. 11), U266 cells treated with 10 µM
SBHA and 5 µM DAC showed increased normalized PDLIM4/RIL
expression compared with that in control (DMSO) cells, detected by
both RT-PCR and ICC staining methods, thus both at the cDNA and
protein level (Fig. 12).
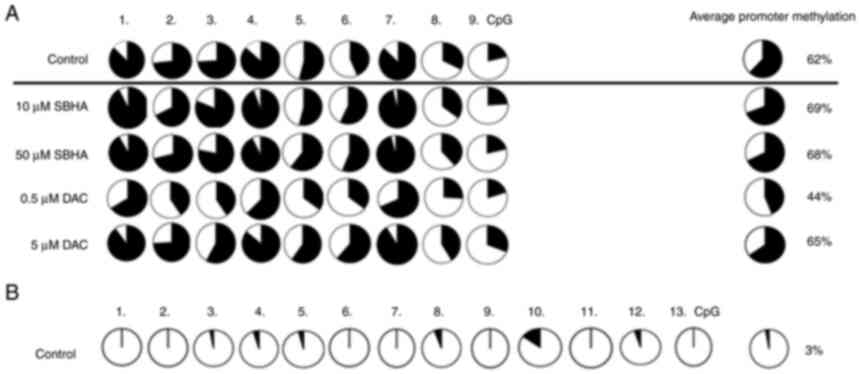 | Figure 5.(A) Comparison of methylation status
at 9 CpGs of the PDLIM4 gene promoter sequence (216 bp) in control
cells and after 48-h of RPMI8226 treatments (10, 10 µM SBHA, 0.5, 5
µM DAC) and (B) 13 CpGs of the CDKN2B gene promoter sequence (233
bp) in control cells performed by bisulfite pyrosequencing. Black
areas in the pie charts represent % of methylated, white areas % of
unmethylated cytosine residues. Pyromarks of sequences obtained
from RPMI8226 cell line. DAC, 5-Aza-2′-deoxycytidine (Decitabine);
DMSO, dimethyl sulfoxide; MtL, methylation level; MTT,
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide agent;
PDLIM4 (RIL), PDZ and LIM domain 4; PI, propidium iodide; SBHA,
suberohydroxamic acid. |
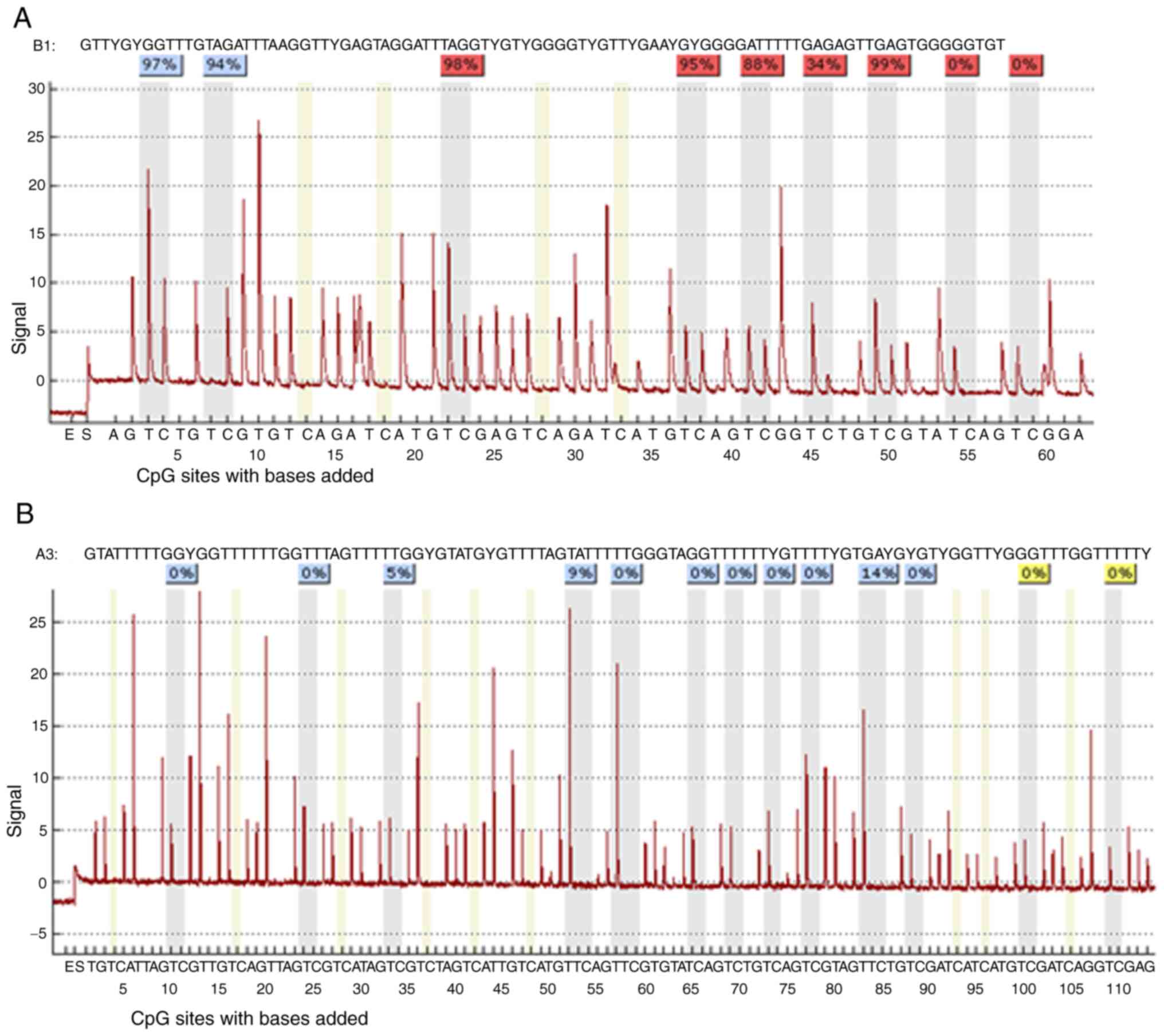 | Figure 6.The sequence analyzed is the (A)
PDLIM4 and (B) CDKN2B genes promoters. The percentages at the top
of the figure indicate the level of methylation of a particular CpG
site, the resulting number for a sample is then calculated by
software as the average methylation of all CpG sites of the
sequence under investigation. DAC, 5-Aza-2′-deoxycytidine
(Decitabine); DMSO, dimethyl sulfoxide; MtL, methylation level;
MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
agent; PDLIM4 (RIL), PDZ and LIM domain 4; PI, propidium iodide;
SBHA, suberohydroxamic acid. |
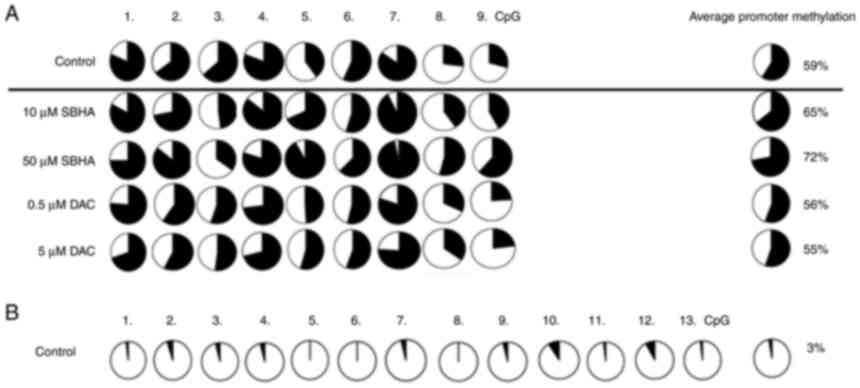 | Figure 7.(A) Comparison of the methylation
status at 9 CpGs of the PDLIM4 gene promoter sequence (216 bp) in
control cells and after 48-h U266 treatments (10, 10 µM SBHA, 0.5,
5 µM DAC) and (B) 13 CpGs of the CDKN2B gene promoter sequence (233
bp) in control cells (B) performed by bisulfite pyrosequencing.
Black areas in the pie charts represent % of methylated, white
areas % of unmethylated cytosine residues. Pyromarks of sequences
obtained from U266 cell line. DAC, 5-Aza-2′-deoxycytidine
(Decitabine); DMSO, dimethyl sulfoxide; MtL, methylation level;
MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
agent; PDLIM4 (RIL), PDZ and LIM domain 4; PI, propidium iodide;
SBHA, suberohydroxamic acid. |
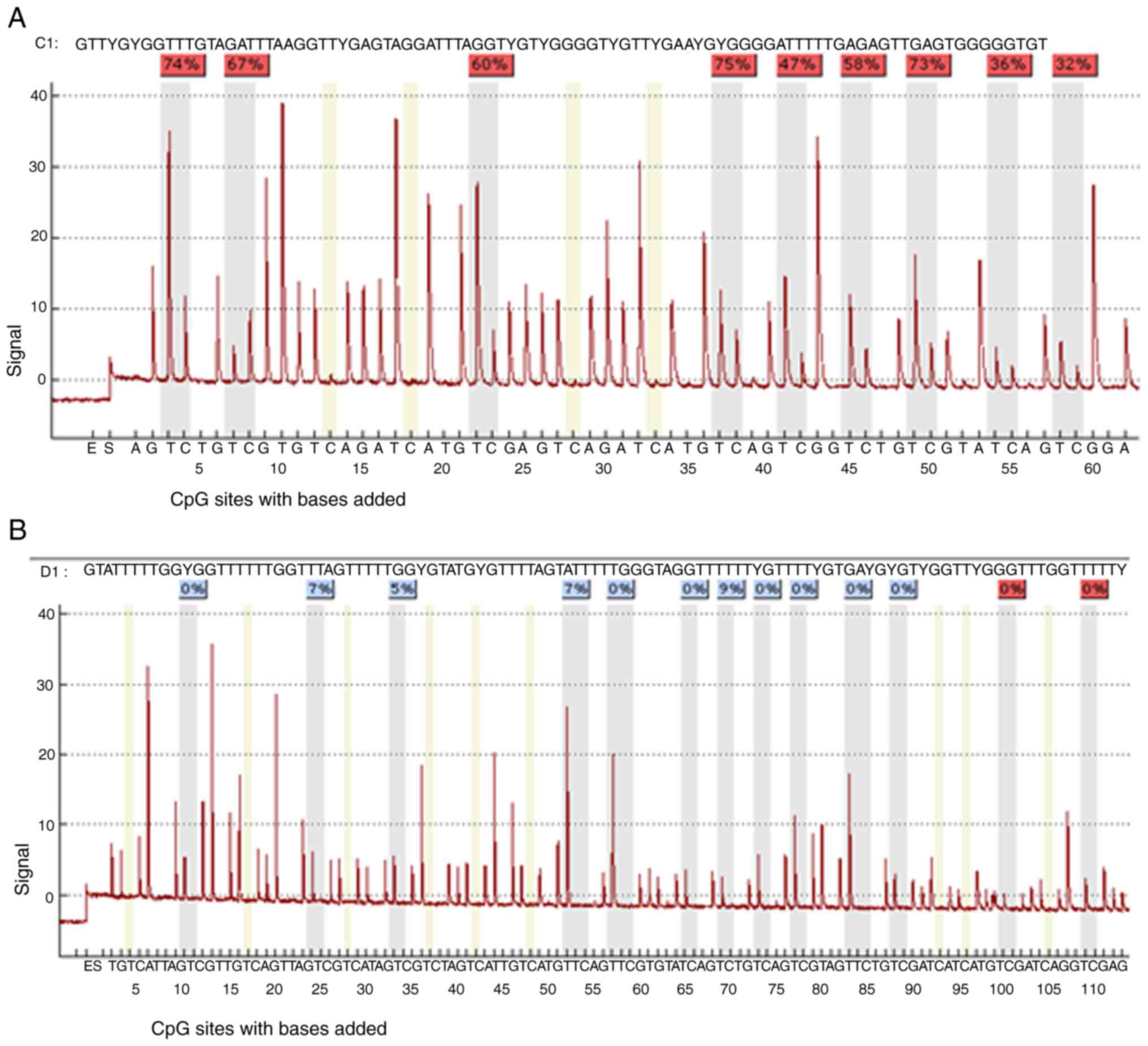 | Figure 8.The sequence analyzed is the (A)
PDLIM4 and (B) CDKN2B genes promoter. The percentages at the top of
the figures indicate the level of methylation of a particular CpG
site, the resulting number for a sample is then calculated by
software as the average methylation of all CpG sites of the
sequence under investigation. DAC, 5-Aza-2′-deoxycytidine
(Decitabine); DMSO, dimethyl sulfoxide; MtL, methylation level;
MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
agent; PDLIM4 (RIL), PDZ and LIM domain 4; PI, propidium iodide;
SBHA, suberohydroxamic acid. |
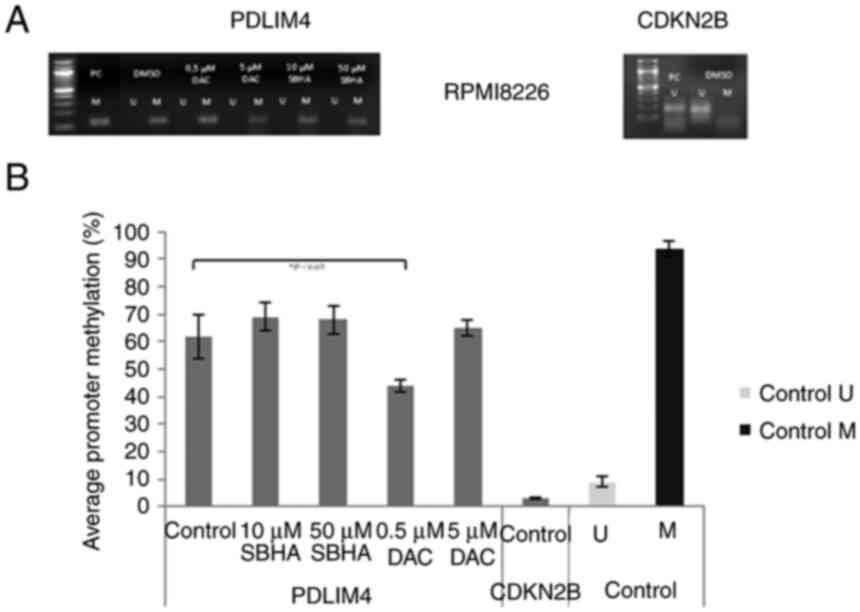 | Figure 9.(A) Methylation status of PDLIM4 and
CDKN2B genes after 48-h treatment of the RPMI8226 cell line
detected by MSP-PCR. (B) Average PDLIM4 gene promoter methylation
in treated RPMI8226 cells in comparison to average CDKN2B gene
promotor methylation, and to unmethylated (U) (9%) and methylated
(M) (94%) control DNA detected by pyrosequencing. The significance
was determined using unpaired Student's t-test and Bonferroni
correction. *P<0.05 vs. PDLIM4 Control (DMSO). DAC,
5-Aza-2′-deoxycytidine (Decitabine); DMSO, dimethyl sulfoxide; MtL,
methylation level; MTT,
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide agent;
PDLIM4 (RIL), PDZ and LIM domain 4; PI, propidium iodide; SBHA,
suberohydroxamic acid. |
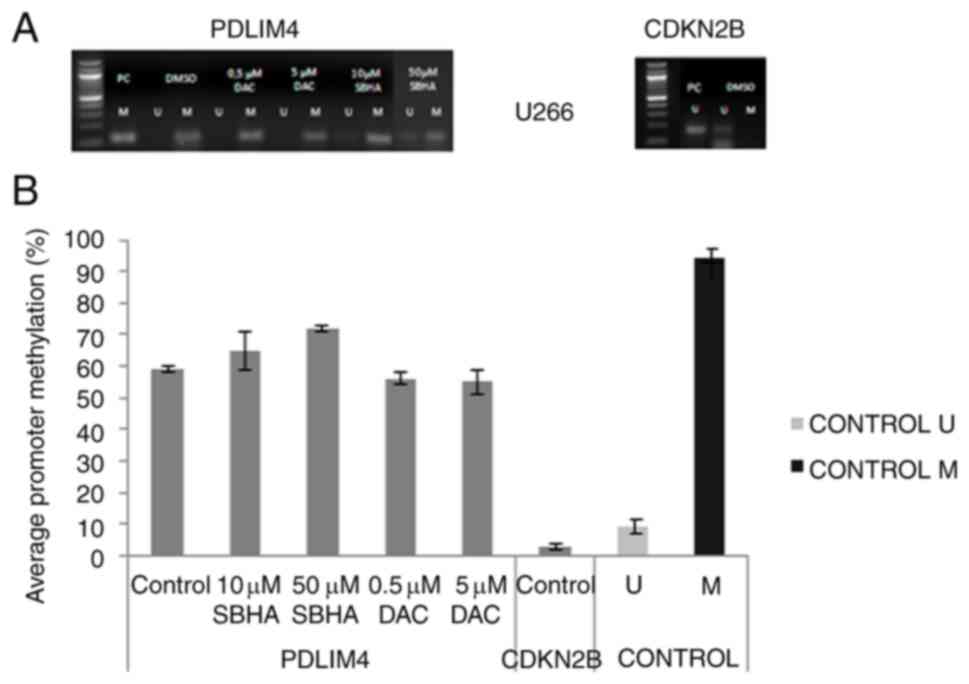 | Figure 10.(A) Methylation status of PDLIM4 and
CDKN2B genes after 48-h treatment of the U266 cell line detected by
MSP-PCR. (B) Average PDLIM4 gene promoter methylation in treated
U266 cells in comparison to average CDKN2B gene promotor
methylation, and to unmethylated (U) (9%) and methylated (M) (94%)
control DNA detected by pyrosequencing. The significance was
determined using unpaired Student's t-test and Bonferroni
correction. DAC, 5-Aza-2′-deoxycytidine (Decitabine); DMSO,
dimethyl sulfoxide; MtL, methylation level; MTT,
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide agent;
PDLIM4 (RIL), PDZ and LIM domain 4; PI, propidium iodide; SBHA,
suberohydroxamic acid. |
Discussion
Two human myeloma cell lines, differing in
p53-functionality and IL-6 expression, RPMI8226 and U266, were used
in the study (30,31). The IL-6 has been shown to regulate
DNA methylation by inducing expression of FLI-1, a transcription
factor of DNMT1 (32).
Furthermore, IL-6 signal enhances DNMT1, which promotes the
methylation with subsequent deactivation of p53 enabling cells to
escape cell cycle checkpoint (33). Moreover, although the molecular
mechanism by which DAC induces cancer cell death is not fully
understood, the cytotoxic effect of DAC may be mediated primarily
through Dnmt3a and Dnmt3b, as described in null mutant embryonic
stem (ES) cells (34).
Human myeloma cell lines are widely used for their
representation of primary myeloma cells because they cover patient
diversity (35,36). However, the HMCLs harbor the 14q32
abnormality, which occurs early at the MGUS stage, and display
frequent mutations in NRAS and KRAS genes (37,38). Tessoulin et al performed
whole-exon sequencing of 33 HMCLs, and recurrent bi-allelic losses
were found in genes involved in cell cycle regulation (RB1,
CDKN2C), the NF-κB pathway (TRAF, BIRC2), and the p53 pathway
(TP53, CDKN2A) (36). The
U266 cell line, used in our study, contained 378 missense mutations
including two ones with TP53 gene (Table I), and 231 mutations leading to
different gene silencing. In addition to mutually exclusive and
associated mutations/deletions in genes involved in the MAPK and
p53 pathways, were identified in epigenetic regulator/modifier
genes, such as histone methyltransferases and DNA methylation
modifiers (35,39). Above these findings, up to 53% of
MM patients may present with mutated histone acetylation-, DNA
methylation-, and chromatin remodeling-related genes (40).
Since the identification of IL-6 was used as a main
growth factor for myeloma cells in the past, there is a large
cohort of HMCLs, culturing primary myeloma cells from patients with
extramedullary proliferation with IL-6 (41). Subsequent evaluation on a limited
number of HMCLs lacking IL-6 type HMCLs showed that HMCLs did not
reflect their genetic diversity and chromosomal abnormalities
(42,43). As Moreaux et al described,
TP53 gene abnormalities were found in 65% of HMCLs culturing
without adding exogenous IL-6, while HMCLs depending on the
addition of IL-6, had a trend to have less TP53 abnormalities than
HMCL without IL-6, 58% vs. 81% (37). Thus, HMCLs are heterogenous in
term of how they were obtained, IL-6-dependence, phenotype, and
gene abnormalities. In our study, the significantly increased
DNMT3B expression in the DAC treated U266 cells (Fig. 4D) was determined in an
IL-6-expressing U266 cell line, while the expression of the DNMT1
and DNMT3A was not affected (Fig.
4). Although this finding has not yet been published, and we
may assume that the increased DNMT3B expression is due to the
interaction between DAC and IL-6, it is important to mention the
non-negligible effect of the large number of missense/nonsense
mutations detected in the U266 IL-6-expressing cell line. Further,
epigenetic silencing via DNA methylation is an alternative
mechanism that can result in silencing of genes. On the contrary,
acetylation of histones is associated to gene activation (44).
In the RPMI8226 cell line, the 0.5 µM
5-Aza-2′-deoxy-cytidine treatment caused a reduced level of the
PDLIM4 gene promoter methylation enabling the increase of
gene transcription (Figs. 5 and
6), whereas in IL-6-expressing
U266 cells, we found no demethylation changes after either DAC or
SBHA agents (Figs. 7 and 8). It is very likely that the DAC impact
leading to an increased DNMT3B expression in the IL-6-expressing
cell line may be due to a repressive transcriptional mechanism
including interaction between DNMT3B and IL-6. However, although
the mechanisms of transcriptional repression are not fully
understood yet, they probably involve interactions of methylated
DNA with regulatory proteins, such as binding of methylated DNA
binding proteins (MBPs). In addition, the methylated cytosines in
the promoter region can bind methyl-CpG binding protein 2 (MeCP2),
which form complexes with corepressors that include HDACs (45).
Bone remodeling is a balance between bone resorption
and bone apposition controlled by two cell types, osteoclasts, and
osteoblasts (46). Osteoblasts
are responsible for bone apposition whereas osteoclasts are
specialized for bone resorption. Osteoclastic precursors
differentiate into mature osteoclasts after interaction with
osteoblastic/stromal cells. The cell-cell interactions are
necessary as well as the production of soluble factors by
osteoblasts. Thus, inflammatory cytokines, such as IL-6, can
modulate skeletal homeostasis and osteoclast differentiation.
IL-6-type cytokines utilize the transducing receptor β-subunit
gp130 as a part of a multimeric receptor complex (47). Ligand-induced oligomerization of
receptor subunits activates Janus protein-tyrosine kinases (JAKs),
which allows activation of the signal transducer and activator of
transcription (STATs, predominantly STAT3) (46). Other signaling cascades known to
be activated by IL-6 are phosphoinositide-3-kinase (PI3K)/AKT, and
PKCδ (48,49). However, the activation of STAT3 is
necessary for osteoblast differentiation and bone formation induced
by IL-6 (50). IL-6 can reduce
proliferation of various osteoblastic cells through activation of
STAT3 and enhanced expression of p21WAF1 (51). Then, IL-6-type cytokines can then
protect osteoblastic cells from apoptosis induced by serum
depletion or tumor necrosis factor α (TNFα) (52). Therefore, in our study, increased
p21WAF1 expression detected as result of the action of
both SBHA and DAC agents may not be the desirable therapeutic
outcome in IL-6-expressing U266 myeloma cells.
DAC has a significant therapeutic value for
treatment of patients with myelodysplastic syndrome (MDS), acute
myeloid leukemia (AML), chronic myeloid leukemia (CML), and is
clinically used as a DNA methylation inhibitor for the treatment of
MDS and AML (53). In MM, phase 1
trial of DAC has been performed to study its biological and
clinical effectiveness as monotherapy or combined with lenalidomide
or dexamethasone (54). However,
its therapeutic efficacy has not been well established. In AML
cells, for example, the DAC treatment resulted in the induction of
p21WAF1, which correlated with the arrest of AML cells
in the G1 cell cycle checkpoint (55). Our analyzes reveal that the DAC
treatment induced the DNA methyltransferase 3B enhancement in
IL-6-expressing U266 cells.
In the current study, the cell cycle and viability
of RPMI8226 and U266 cell lines were analyzed, and the increase in
sub-G1 phase was found to tend to increase in the RPMI8226 cells
following SBHA or DAC treatment (Fig.
1). An increase in sub-G1 and distinctly in G1 arrest were
found to have a tendency to increase in U266 cells following SBHA
treatment (Fig. 2). Although,
these findings do not correspond to U266 cell viability by SBHA or
DAC treatment, a significant reduction in the RPMI8226 cell
viability following 48 h DAC treatment may be due to a later onset
of DAC (Fig. 3).
Moreover, the enhanced level of p21WAF1
was detected after U266 cell treatment with DAC, while in RPMI8226
cells, the DAC caused a significant increase in p15INK4B
expression. As was reported in several studies (56–58), the p21WAF1 expression
is regulated through both p53-dependent and p53-independent
mechanism. We found that the 50 µM SBHA treatment caused a
significant increase in p21WAF1 expression in the
p53-deleted U266 cell line (Fig.
4). This finding is in accordance with the previously described
SBHA inhibiting effect of terminating the cell cycle predominantly
at the G1 checkpoint (11,12),
which we observed in p53-deleted U266 cells (Fig. 2), but no in RPMI8226 cells with
functional p53 (Fig. 1).
Moreover, SBHA was able to induce Notch1 intracellular domain
levels, coupled with increase in p53 and p21WAF1, and
may have anti-tumor functions via regulating Notch1/p53 (59). Owing to the in vitro
detected cyclin-dependent kinase inhibitor 2B (CDKN2B)
(p15INK4B) unmethylated promoter regions in both cell
lines (Figs. 5A, B, 7A, B, 9A and 10A) the unmethylated CDKN2B gene
may be included in the SBHA-activated cell cycle inhibiting
signaling pathway in a p53-dependent manner as was detected in the
RPMI8226 cell line (Fig. 4A).
However, unlike studied myeloma cell lines, in MM patients in
vivo, the methylated state of the CDKN2B gene has been
confirmed methylated in several studies (27,60,61). On the other hand, the results of
this study correspond to our previous results, indicating a low
level of CDKN2B gene methylation and higher level of
PDLIM4 gene methylation in patients with MM (62). Additionally, the frequency of
p16INK4a (CDKN2A) hypermethylation increases with
the progression of MM (18,63,64). Hence, during MM development, the
methylation status of the CDKN2B gene can vary in individual
stages of the disease, which may be further accompanied by
increased frequency of CDKN2B gene methylation.
In conclusion, the DAC treatment induces the DNMT3B
enhancement in IL-6-expressing U266 cells, which indicates the
controversial role of DAC treatment as a demethylation agent in
multiple myeloma patients. From the obtained data we assume that
analysing of the IL-6 pathway in multiple myeloma may be a
promising therapeutic target of multiple myeloma since the effect
of blocking IL-6 may act at least in part through regulation of
cell cycle gene expression. Moreover, in IL-6-expressing cells, the
increased expression of cyclin dependent kinase inhibitor
p21WAF may not be indicative of the desired multiple
myeloma cell apoptosis.
Acknowledgements
The authors would like to acknowledge Mrs. Eva
Pimrova (Department of Clinical and Molecular Pathology, Palacky
University Olomouc, Czech Republic) for her technical assistance
with processing of myeloma cell lines.
Funding
This study was funded in part by NV18-03-00500 from the Ministry
of Health of the Czech Republic, the European Regional Development
Fund-Project ENOCH (grant no. CZ.02.1.01/0.0/0.0/16_019/0000868)
and LF_2021_005 from Palacky University Olomouc.
Availability of data and materials
All data generated or analyzed during this study
are included in this published article.
Authors' contributions
KST, PL and JM designed the study, and wrote and
revised the manuscript; PL, DWD, LJ, IF and DP performed the
experiments; KC, JG and MH analysed the dataset. All authors read
and approved the final manuscript. KST and JM confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
AML
|
acute myelogenous leukemia
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
CDK6
|
cyclin-dependent kinase 6
|
|
CDKN2B (p15INK4b)
|
cyclin-dependent kinase inhibitor
2B
|
|
CML
|
chronic myelogenous leukemia
|
|
DAC
|
5-Aza-2′-deoxycytidine
(Decitabine)
|
|
DMSO
|
dimethyl sulfoxide
|
|
MDS
|
myelodysplastic syndrome
|
|
MtL
|
methylation level
|
|
MTT
|
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
agent
|
|
PDLIM4 (RIL)
|
PDZ and LIM domain 4
|
|
PI
|
propidium iodide
|
|
SBHA
|
suberohydroxamic acid
|
References
|
1
|
Bianchi G and Munshi NC: Pathogenesis
beyond the cancer clone(s) in multiple myeloma. Blood.
125:3049–3058. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Abe M: Targeting the interplay between
myeloma cells and the bone marrow microenvironment in myeloma. Int
J Hematol. 94:334–343. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Meads MB, Hazlehurst LA and Dalton WS: The
bone marrow microenvironment as a tumor sanctuary and contributor
to drug resistance. Clin Cancer Res. 14:2519–2526. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Furukawa Y and Kikuchi J: Molecular
pathogenesis of multiple myeloma. Int J Clin Oncol. 20:413–422.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Harmer D, Falank C and Reagan MR:
Interleukin-6 interweaves the bone marrow microenvironment, bone
loss, and multiple myeloma. Front Endocrinol (Lausanne). 9:7882019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Choudhury SR, Ashby C, Tytarenko R, Bauer
M, Wang Y, Deshpande S, Den J, Schinke C, Zangari M, Thanendrarajan
S, et al: The functional epigenetic landscape of aberrant gene
expression in molecular subgroups of newly diagnosed multiple
myeloma. J Hematol Oncol. 13:1082020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Heuck CJ, Mehta J, Bhagat T, Gundabolu K,
Yu Y, Khan S, Chrysofakis G, Schinke C, Tariman J, Vickrey E, et
al: Myeloma is characterized by stage-specific alterations in DNA
methylation that occur early during myelomagenesis. J Immunol.
190:2966–2975. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Walker BA, Wardell CP, Chiecchio L, Smith
EM, Boyd KD, Neri A, Davies FE, Ross FM and Morgan GJ: Aberrant
global methylation patterns affect the molecular pathogenesis and
prognosis of multiple myeloma. Blood. 117:553–562. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Maes K, De Smedt E, Lemaire M, De Raeve H,
Menu E, Van Valckenborgh E, McClue S, Vanderkerken K and De Bruyne
E: The role of DNA damage and repair in decitabine-mediated
apoptosis in multiple myeloma. Oncotarget. 5:3115–3129. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kiziltepe T, Hideshima T, Catley L, Raje
N, Yasui H, Shiraishi N, Okawa Y, Ikeda H, Vallet S, Pozzi S, et
al: 5-Azacytidine, a DNA methyltransferase inhibitor, induces
ATR-mediated DNA double-strand break responses, apoptosis, and
synergistic cytotoxicity with doxorubicin and bortezomib against
multiple myeloma cells. Mol Cancer Ther. 6:1718–1727. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Carew JS, Giles FJ and Nawrocki ST:
Histone deacetylase inhibitors: Mechanisms of cell death and
promise in combination cancer therapy. Cancer Lett. 269:7–17. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bolden JE, Peart MJ and Johnstone RW:
Anticancer activities of histone deacetylase inhibitors. Nat Rev
Drug Discov. 5:769–784. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ng MH, Chung YF, Lo KW, Wickham NW, Lee JC
and Huang DP: Frequent hypermethylation of p16 and p15 genes in
multiple myeloma. Blood. 89:2500–2506. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wong IH, Ng MH, Lee JC, Lo KW, Chung YF
and Huang DP: Transcriptional silencing of the p16 gene in human
myeloma-derived cell lines by hypermethylation. Br J Haematol.
103:168–175. 1998.PubMed/NCBI
|
|
15
|
Zhang XD, Gillespie SK, Borrow JM and
Hersey P: The histone deacetylase inhibitor suberic bishydroxamate
regulates the expression of multiple apoptotic mediators and
induces mitochondria-dependent apoptosis of melanoma cells. Mol
Cancer Ther. 3:425–435. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gillespie S, Borrow J, Zhang XD and Hersey
P: Bim plays a crucial role in synergistic induction of apoptosis
by the histone deacetylase inhibitor SBHA and TRAIL in melanoma
cells. Apoptosis. 11:2251–2265. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Elmallah MIY and Micheau O: Epigenetic
regulation of TRAIL signaling: Implication for cancer therapy.
Cancers (Basel). 11:8502019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen J, Hong Z, Zhao C, Bi Q and Qiu B:
Associations between polymorphisms of the PDLIM4 gene and
susceptibility to osteoporotic fracture in an elderly population of
Han Chinese. Biosci Rep. 39:BSR201815052019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kadrmas JL and Beckerle MC: The LIM
domain: From the cytoskeleton to the nucleus. Nat Rev Mol Cell
Biol. 5:920–931. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ono R, Kaisho T and Tanaka T: PDLIM1
inhibits NF-κB-mediated inflammatory signaling by sequestering the
p65 subunit of NF-κB in the cytoplasm. Sci Rep. 5:183272015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kravchenko DS, Ivanova AE, Podshivalova ES
and Chumakov SP: PDLIM4/RIL-mediated regulation of Src and
malignant properties of breast cancer cells. Oncotarget. 11:22–30.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Y, Qian J, Lin J, Qian W, Yang J, Chai
HY, Wang CZ, Deng ZQ, Yao DM, Chen Q and Ma JC: Reduced expression
of PDLIM4 gene correlates with good prognosis in acute myeloid
leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 21:1111–1115.
2013.PubMed/NCBI
|
|
23
|
Vanaja DK, Ballman KV, Morlan BW, Cheville
JC, Neumann RM, Lieber MM, Tindall DJ and Young CY: PDLIM4
repression by hypermethylation as a potential biomarker for
prostate cancer. Clin Cancer Res. 12:1128–1136. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Boumber YA, Kondo Y, Chen X, Shen L,
Gharibyan V, Konishi K, Estey E, Kantarjian H, Garcia-Manero G and
Issa JP: RIL, a LIM gene on 5q31, is silenced by methylation in
cancer and sensitizes cancer cells to apoptosis. Cancer Res.
67:1997–2005. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fernando RC, de Carvalho F, Mazzotti DR,
Evangelista AF, Braga WMT, de Lourdes Chauffaille M, Leme AFP and
Colleoni GWB: Multiple myeloma cell lines and primary tumors
proteoma: Protein biosynthesis and immune system as potential
therapeutic targets. Genes Cancer. 6:462–471. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lodé L, Eveillard M, Trichet V, Soussi T,
Wuillème S, Richebourg S, Magrangeas F, Ifrah N, Campion L, Traullé
C, et al: Mutations in TP53 are exclusively associated with
del(17p) in multiple myeloma. Haematologica. 95:1973–1976. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wei B, Yang S, Zhang B and Feng Y:
Clinicopathological significance of p15 promoter hypermethylation
in multiple myeloma: A meta-analysis. Onco Targets Ther.
9:4015–4022. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li J, Bi L, Lin Y, Lu Z and Hou G:
Clinicopathological significance and potential drug target of
p15INK4B in multiple myeloma. Drug Des Devel Ther. 8:2129–2136.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nguen DH, Zhou T, Shu J and Mao JH:
Quantifying chromogen intensity in immunohistochemistry via
reciprocal intensity. Cancer InCytes. 2:1–4. 2013.
|
|
30
|
Ingersoll SB, Thoni ND, Ahmed F, Monahan
KA, Caballero L, Batista A, Ahmad S and Edwards JR: Role of the
IL-6 pathway to multiple myeloma cell growth and its implications
in target gene hypermethylation. Blood. 110:47692007. View Article : Google Scholar
|
|
31
|
Ingersoll SB, Ahmad S, Thoni ND, Ahmed FH,
Monahan KA and Edwards JR: Targeting the IL-6 pathway in multiple
myeloma and its implications in cancer-associated gene
hypermethylation. Med Chem. 7:473–479. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hodge DR, Li D, Qi SM and Farrar WL: IL-6
induces expression of the Fli-1 proto-oncogene via STAT3. Biochem
Biophys Res Commun. 292:287–291. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hodge DR, Peng B, Cherry JC, Hurt EM, Fox
SD, Kelley JA, Munroe DJ and Farrar WL: Interleukin 6 supports the
maintenance of p53 tumor suppressor gene promoter methylation.
Cancer Res. 65:4673–4682. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Oka M, Meacham AM, Hamazaki T, Rodić N,
Chang LJ and Terada N: De novo DNA methyltransferases Dnmt3a and
Dnmt3b primarily mediate the cytotoxic effect of
5-aza-2′-deoxycytidine. Oncogene. 24:3091–3099. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fu HY, Shen JZ, Wu Y, Shen SF, Zhou HR and
Fan LP: Arsenic trioxide inhibits DNA methyltransferase and
restores expression of methylation-silenced CDKN2B/CDKN2A genes in
human hematologic malignant cells. Oncol Rep. 24:335–343.
2010.PubMed/NCBI
|
|
36
|
Tessoulin B, Moreau-Aubry A, Descamps G,
Gomez-Bougie P, Maïga S, Gaignard A, Chiron D, Ménoret E, Le Gouill
S, Moreau P, et al: Whole-exon sequencing of human myeloma cell
lines shows mutations related to myeloma patients at relapse with
major hits in the DNA regulation and repair pathways. J Hematol
Oncol. 11:1372018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Moreaux J, Klein B, Bataille R, Descamps
G, Maïga S, Hose D, Goldschmidt H, Jauch A, Rème T, Jourdan M, et
al: A high-risk signature for patients with multiple myeloma
established from the molecular classification of human myeloma cell
lines. Haematologica. 96:574–582. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Manier S, Salem KZ, Park J, Landau DA,
Getz G and Ghobrial IM: Genomic complexity of multiple myeloma and
its clinical implications. Nat Rev Clin Oncol. 14:100–113. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pawlyn C, Kaiser MF, Heuck C, Melchor L,
Wardell CP, Murison A, Chavan SS, Johnson DC, Begum DB, Dahir NM,
et al: The spectrum and clinical impact of epigenetic modifier
mutations in myeloma. Clin Cancer Res. 22:5783–5794. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Caprio C, Sacco A, Giustini V and Roccaro
AM: Epigenetic aberrations in multiple myeloma. Cancers (Basel).
12:29962020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang XG, Gaillard JP, Robillard N, Lu ZY,
Gu ZJ, Jourdan M, Boiron JM, Bataille R and Klein B: Reproducible
obtaining of human myeloma cell lines as a model for tumor stem
cell study in human multiple myeloma. Blood. 83:3654–3663. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fabris S, Agnelli L, Mattioli M, Baldini
L, Ronchetti D, Morabito F, Verdelli D, Nobili L, Intini D, Callea
V, et al: Characterization of oncogene dysregulation in multiple
myeloma by combined FISH and DNA microarray analyses. Genes
Chromosomes Cancer. 42:117–127. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhan F, Hardin J, Kordsmeier B, Bumm K,
Zheng M, Tian E, Sanderson R, Yang Y, Wilson C, Zangari M, et al:
Global gene expression profiling of multiple myeloma, monoclonal
gammopathy of undetermined significance, and normal bone marrow
plasma cells. Blood. 99:1745–1757. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Claus R and Lübbert M: Epigenetic targets
in hematopoietic malignancies. Oncogene. 22:6489–6496. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Nan X, Cross S and Bird A: Gene silencing
by methyl-CpG-binding proteins. Novartis Found Symp. 214:6–21.
46–50. 1998.PubMed/NCBI
|
|
46
|
Theoleyre S, Wittrant Y, Tat SK, Fortun Y,
Redini F and Heymann D: The molecular triad OPG/RANK/RANKL:
Involvement in the orchestration of pathophysiological bone
remodeling. Cytokine Growth Factor Rev. 15:457–475. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Burger R, Günther A, Klausz K, Staudinger
M, Peipp M, Penas EM, Rose-John S, Wijdenes J and Gramatzki M: Due
to interleukin-6 type cytokine redundancy only glycoprotein 130
receptor blockade efficiently inhibits myeloma growth.
Haematologica. 102:381–390. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Heinrich PC, Behrmann I, Haan S, Hermanns
HM, Müller-Newen G and Schaper F: Principles of interleukin
(IL)-6-type cytokine signalling and its regulation. Biochem J.
374:1–20. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chipoy C, Berreur M, Couillaud S, Pradal
G, Vallette F, Colombeix C, Rédini F, Heymann D and Blanchard F:
Downregulation of osteoblast markers and induction of the glial
fibrillary acidic protein by oncostatin M in osteosarcoma cells
require PKCdelta and STAT3. J Bone Miner Res. 19:1850–1861. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Blanchard F, Duplomb L, Baud'huin M and
Brounais B: The dual role of IL-6-type cytokines on bone remodeling
and bone tumors. Cytokine Growth Factor Rev. 20:19–28. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bellido T, O'Brien CA, Roberson PK and
Manolagas SC: Transcriptional activation of the p21(WAF1,CIP1,SDI1)
gene by interleukin-6 type cytokines. A prerequisite for their
pro-differentiating and anti-apoptotic effects on human
osteoblastic cells. J Biol Chem. 273:21137–21144. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Jilka RL, Weinstein RS, Bellido T, Parfitt
AM and Manolagas SC: Osteoblast programmed cell death (apoptosis):
Modulation by growth factors and cytokines. J Bone Miner Res.
13:793–802. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hollenbach PW, Nguyen AN, Brady H,
Williams M, Ning Y, Richard N, Krushel L, Aukerman SL, Heise C and
MacBeth KJ: A comparison of azacitidine and decitabine activities
in acute myeloid leukemia cell lines. PLoS One. 5:e90012010.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Maes K, Menu E, Van Valckenborgh E, Van
Riet I, Vanderkerken K and De Bruyne E: Epigenetic modulating
agents as a new therapeutic approach in multiple myeloma. Cancers
(Basel). 5:430–461. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Schmelz K, Sattler N, Wagner M, Lübbert M,
Dörken B and Tamm I: Induction of gene expression by
5-Aza-2′-deoxycytidine in acute myeloid leukemia (AML) and
myelodysplastic syndrome (MDS) but not epithelial cells by
DNA-methylation-dependent and -independent mechanisms. Leukemia.
19:103–111. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ocker M and Schneider-Stock R: Histone
deacetylase inhibitors: Signalling towards p21cip1/waf1. Int J
Biochem Cell Biol. 39:1367–1374. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lagger G, Doetzlhofer A, Schuettengruber
B, Haidweger E, Simboeck E, Tischler J, Chiocca S, Suske G,
Rotheneder H, Wintersberger E and Seiser C: The tumor suppressor
p53 and histone deacetylase 1 are antagonistic regulators of the
cyclin-dependent kinase inhibitor p21/WAF1/CIP1 gene. Mol Cell
Biol. 23:2669–2679. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Richon VM, Sandhoff TW, Rifkind RA and
Marks PA: Histone deacetylase inhibitor selectively induces p21WAF1
expression and gene-associated histone acetylation. Proc Natl Acad
Sci USA. 97:10014–10019. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Li J, Zheng X, Gao M, Zhao J, Li Y, Meng
X, Qian B and Li J: Suberoyl bis-hydroxamic acid activates Notch1
signaling and induces apoptosis in anaplastic thyroid carcinoma
through p53. Oncol Rep. 37:458–464. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wang X, Zhu YB, Cui HP and Yu TT: Aberrant
promoter methylation of p15 (INK4b) and p16 (INK4a) genes may
contribute to the pathogenesis of multiple myeloma: A
meta-analysis. Tumour Biol. 35:9035–9043. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Stanganelli C, Arbelbide J, Fantl DB,
Corrado C and Slavutsky I: DNA methylation analysis of tumor
suppressor genes in monoclonal gammopathy of undetermined
significance. Ann Hematol. 89:191–199. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Luzna P, Flodrova P, Janovska L,
Zapletalova J, Minarik J, Kolar Z and Trtkova KS: Different gene
methylation status of the CDKN2B and/or PDLIM4 as the result of
comparative analysis to the global DNA methylation in unsorted cell
population of multiple myeloma patients. Ann Hematol Oncol.
6:12572019.
|
|
63
|
Yuregir OO, Yurtcu E, Kizilkilic E, Kocer
NE, Ozdogu H and Sahin FI: Detecting methylation patterns of p16,
MGMT, DAPK and E-cadherin genes in multiple myeloma patients. Int J
Lab Hematol. 32:142–149. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Gonzalez-Paz N, Chng WJ, McClure RF, Blood
E, Oken MM, Van Ness B, James CD, Kurtin PJ, Henderson K, Ahmann
GJ, et al: Tumor suppressor p16 methylation in multiple myeloma:
Biological and clinical implications. Blood. 109:1228–1232. 2007.
View Article : Google Scholar : PubMed/NCBI
|