Introduction
Pancreatic adenocarcinoma (PAAD) is one of the most
lethal malignancies worldwide, with an increasing incidence, a lack
of specific symptoms, high invasiveness and a 5-year survival rate
of ~10% (1,2). Approximately 60,430 new PAAD cases
and 48,220 deaths of PAAD occurred in the USA in 2021 (2). PAAD has become one of the most
difficult cancer types to treat and is projected to become the
second leading cause of cancer-related death by 2030 worldwide
(3). The efficacy of clinical PAAD
treatments, including surgery, chemotherapy, radiotherapy and
immunotherapy have improved; however, the 5-year overall survival
(OS) rate remains poor (4,5). Therefore, it is necessary to identify
novel prognostic biomarkers and therapeutic targets for PAAD to
guide clinical practice.
Ferroptosis is an iron-dependent form of regulated
cell death that is characterized by overwhelming lipid peroxide
accumulation and redox imbalance (6,7).
Recent studies have reported that ferroptosis is correlated with
the progression and therapeutic response of cancers, and that
activation of the ferroptosis process is a novel strategy for
cancer treatment, especially for malignances resistant to
traditional therapies (8,9). It has been reported that inhibition
of ferroptosis confers sorafenib resistance and is associated with
reduced OS in hepatocellular carcinoma (10,11).
Moreover, ferroptosis may serve an important role in the
development of PAAD. Using pancreatic glutathione peroxidase 4
(GPX4) conditional-knockout mice or high-iron diet models, Dai
et al (12) reported that
ferroptotic damage promotes KRAS-driven PAAD through the
activation of the TMEM173/STING-dependent DNA sensor pathway in
mice. Badgley et al (13)
reported that the import of oxidized cysteine is a critical
dependency of PAAD and that induction of ferroptosis by cysteine
depletion may be a promising target for PAAD treatment. Another
study reported that inhibition of aspartate aminotransaminase
facilitates pancreatic cancer cell death by inducing ferroptosis
(14). Therefore, it was
hypothesized that novel ferroptosis-related biomarkers could have
great potential for predicting the prognosis of patients with PAAD
and identifying therapeutic targets for PAAD.
Long non-coding RNAs (lncRNAs) are RNA transcripts
>200 nucleotides in length that lack a protein-coding capacity.
As a newly reported type of gene regulator, lncRNAs regulate
various important biological processes and participate in the
development and progression of numerous diseases, including PAAD
(15–17). For example, the lncRNA
ENST00000480739 inhibits tumor cell invasion by the regulation of
OS-9 and hypoxia inducible factor-1α in PAAD (18). In another study, upregulation of
LINC01232 was reported to be significantly related to the poor
prognosis of patients with PAAD, and LINC01232 exerted oncogenic
roles in PAAD through the regulation of transmembrane 9 superfamily
member 2 (19). In addition,
lncRNA glutaminase-antisense (GLS-AS) is downregulated in PAAD, and
loss of GLS-AS could impair GLS-mediated metabolism and suppress
cancer progression (20). Several
other lncRNAs, including PVT1 (21), AGAP2-AS1 (22) and PLACT1 (23), have been reported as potential
therapeutic targets or prognostic biomarkers of PAAD. Therefore,
lncRNAs are implicated in the development and progression of PAAD,
and are potential targets for PAAD treatments and biomarkers for
prognosis.
Ferroptosis is an important regulated form of cell
death for cancers that is associated with lncRNAs (24). Numerous lncRNAs participate in the
regulation of ferroptosis and serve important roles in
tumorigenesis and progression through modulation of the
transcription and post-transcriptional modification of
ferroptosis-related genes (25,26).
A recent study reported that the lncRNA NEAT1 improved ferroptosis
sensitivity in non-small cell lung cancer by regulating the
expression of acyl-CoA synthetase long-chain family member 4
(27). Mao et al (28) reported that the lncRNA P53RRA is
downregulated in cancers and promotes ferroptosis through the
nuclear sequestration of p53. Furthermore, it was reported that
LINC00336 regulated the expression of cystathionine-β-synthase to
induce ferroptosis in lung cancer by functioning as a competing
endogenous sponge of microRNA-6852 (29). As these ferroptosis-related lncRNAs
serve important roles in the pathophysiology of malignant diseases,
they may be useful for prognosis prediction and may serve as
therapeutic targets for patients with PAAD; however, the general
role of these lncRNAs in the prognosis prediction remains unknown.
Ferroptosis is rapidly gaining attention in cancer treatment and
cancer genomics databases have been implemented to identify
promising prognostic factors and to explore molecular mechanisms
for multiple types of cancers in recent years. The present study
used lncRNA expression profiles to identify ferroptosis-related
lncRNA signatures (FRLSs), which may serve as novel biomarkers to
predict the prognosis of patients with PAAD and may be utilized as
potential therapeutic targets based on ferroptosis.
Materials and methods
Cell lines and tissue samples
The human pancreatic ductal epithelial cell line
(GENNIO BIO, Guangzhou, China; Cat.no. JNO-089) and human
pancreatic cancer cell lines AsPC-1, BxPC-3, PANC-1 and SW1990
(ATCC, USA) were cultured in complete growth medium with 10% fetal
bovine serum (Gibco, USA), as recommended by the manufacturer. The
human pancreatic ductal epithelial cell line and pancreatic cancer
cell lines at purchase were first passage. Cells were incubated at
37°C in a humidified incubator with 5% CO2.
LINC01133-specific small interfering (si)RNAs (LINC01133-siRNA-1,
sense 5′-AAUGGAUCCAUUCCCUGCAACUG-3′; and LINC01133-siRNA-2, sense
5′-GAAGUGGAAGCAAAGUUCUCCAAAG-3′) and a negative control siRNA
(5′-TTCTCCGAACGTGTCACGT-3′) were purchased from Shanghai GeneChem
Co., Ltd. and transiently transfected using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
A total of 30 paired freshly frozen PAAD tissues and
adjacent nontumor (ANT) tissues were obtained from the Pancreatic
Tumor Bank, Institute of Hepatopancreatobiliary Surgery, Southwest
Hospital, Third Military Medical University (Chongqing, China)
between May and November 2020. The clinicopathological
characteristics of the patients are presented in Table SI. The patient inclusion criteria
were as follows: i) Pathologically diagnosed with pancreatic
adenocarcinoma; ii) no preoperative anti-tumor treatment; and iii)
had complete clinicopathological data. Exclusion criteria were as
follows: i) Previous chemotherapy or other anti-tumor treatment
before surgery; and ii) lacked complete clinical information. The
diagnoses were independently reviewed by two senior pathologists
and classified according to World Health Organization criteria.
PAAD and ANT tissues were stored for total RNA isolation
(snap-frozen in liquid nitrogen and then stored at −80°C). Written
informed consent was obtained from all patients prior to the study.
The present study was approved by the ethical committee of
Southwest Hospital (approval no. 20210312).
Cell Counting Kit-8 (CCK-8)
assays
AsPC-1 and BxPC-3 cells transfected with si-control
and si-LINC01133 were treated with 10 µM erastin for 24 h. Then,
cell viability was assessed using a Cell Counting Kit-8 assay
(Dojindo, Shanghai China Co., Ltd.) according to the manufacturer's
protocols. The data were assessed in at least three independent
experiments. Erastin, a ferroptosis-inducing agent, was purchased
from APExBIO Technology LLC.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the tumor cells or
tissues using an Ultrapure RNA kit (CoWin Biosciences). The RNA
concentration was assessed using a NanoDrop ND-2000
spectrophotometer (Thermo Fisher Scientific, Inc.). RT-qPCR was
performed using a PrimeScript RT reagent kit and SYBR®
Premix Ex Taq™ kit (Takara Biotechnology Co., Ltd.) according to
the manufacturers' protocol. The following thermocycling conditions
were used for qPCR: 95°C for 30 sec, followed by 40 cycles at 95°C
for 5 sec and 60°C for 30 sec. Relative mRNA expression levels were
calculated using the 2−ΔΔCq method (30) and normalized to GAPDH mRNA
expression levels. The primer sequences used were as follows:
LINC01133 forward (F), 5′-TGGATCCATTCCCTGCAACT-3′ and reverse (R),
5′-GTGCTGGGCTCTGGATTCTT-3′; CASC8 F, 5′-GCAGTGAGCCAAGGAGCAAT-3′ and
R, 5′-AACCGCAACACTGGTTGTGT-3′; SLC7A11 F,
5′-TCCCTCTATTCGGACCCATTTA-3′ and R, 5′-TTCTTCTGGTACAACTTCCAGT-3′;
GPX4 F, 5′-CGATACGCTGAGTGTGGTTTGC-3′ and R,
5′-CATTTCCCAGGATGCCCTTG-3′; and GAPDH F, 5′-CAGGAGGCATTGCTGATGAT-3′
and R, 5′-GAAGGCTGGGGCTCATTT-3′.
Collection of data on patients with
PAAD
The Cancer Genome Atlas (TCGA)-PAAD dataset and
clinical information were identified and downloaded from the TCGA
database (portal.gdc.cancer.gov, accessed on 20 September 2021)
using the key words ‘pancreas’, ‘transcriptome profiling’,
‘HTSeq-FPKM’ and ‘TCGA-PAAD’. Data from 181 patients with PAAD were
used as a training cohort. Data on another 90 PAAD samples from the
Pancreatic Cancer-AU (PACA-AU) dataset were downloaded from the
International Cancer Gene Consortium (ICGC) database (https://dcc.icgc.org, accessed on 24 September 2021)
and were used as the validation cohort. The inclusion criteria were
samples with complete follow-up information (survival time >30
days) and complete clinicopathological data. The exclusion
criterion were samples without survival information.
Gene Expression Profiling Interactive
Analysis (GEPIA)
GEPIA (http://gepia.cancer-pku.cn/index.html) is a web server
for large-scale expression analysis and interactive analysis
(31). This database is a highly
cited resource for the comparison of the expression levels of
signature lncRNAs (32–35).
Construction and evaluation of the
prognostic FRLS
The list of ferroptosis-related genes was obtained
from the FerrDb Database (zhounan.org/ferrdb/, accessed on 20
September 2021). The expression matrixes of lncRNAs and mRNAs were
identified using classification and annotation analyses. Potential
ferroptosis-related lncRNAs were identified using Pearson's
correlation analysis (r>0.4; P<0.001). The limma R package
was used to screen the differentially expressed ferroptosis-related
lncRNAs between normal and tumor tissues with a false discovery
rate (FDR) <0.05 and |log2 fold change (FC)|>1. Subsequently,
univariate Cox regression analysis was performed to screen
prognostic ferroptosis-related lncRNAs (threshold, P<0.01).
Next, iterative least absolute shrinkage and selection operator
(LASSO) Cox regression was used with the glmnet R package to
identify the optimal prognostic signature. Consensus genes were
assessed with a frequency >100 within 1,000 LASSO Cox
regressions. Consensus genes were then incorporated into the
multivariate Cox regression analysis to construct the risk score.
Using the median risk score as the cutoff point, patients with PAAD
were divided into a high-risk group (over the median risk score)
and a low-risk group (no more than the median risk score).
Kaplan-Meier survival analysis was performed to evaluate the OS
rates between the two groups with the survival and survminer R
packages. Where two survival curves intersected, the two-stage
procedure (36) was used, which
had good applicability and robustness in different crossing
situations. Receiver operating characteristic (ROC) curve analysis
was performed using the survivalROC R package; P<0.05 was used
as the cutoff criterion. Furthermore, principal component analysis
(PCA) was performed using the Rtsne and ggplot2 packages to assess
the classification ability of the risk signature. Then, the rms
package was used to generate the nomogram for the prediction of the
1, 3 and 5-year OS rates of patients with PAAD. A calibration curve
was constructed to assess the consistency between the actual OS
rates and the nomogram-predicted OS rates. Finally, decision curve
analysis (DCA) was performed to evaluate the net benefits with the
different predictors using the rmda package.
Gene set enrichment analysis
(GSEA)
GSEA (http://www.gsea–-msigdb.org/gsea/downloads.jsp,
version 4.1.0) was performed to evaluate the high and low-risk
groups of the prognostic FRLS and explore the possible cellular
pathways. Gene Ontology (GO) term enrichment and Kyoto Encyclopedia
of Genes and Genomes (KEGG) pathway enrichment analyses were
annotated using the c5.go.v7.4.symbols.gmt and
c2.cp.kegg.v7.4.symbols.gmt gene sets from the GSEA Molecular
Signatures Database. Random assortment times were set to 1,000.
Biological processes GO terms and KEGG pathways were selected on
the sorted samples using default weighted enrichment statistics,
and their enrichment scores and P-values were ranked. Gene clusters
with adjusted P<0.05 were considered as significantly enriched
genes.
Assessment of the correlation of the
risk score with immune infiltration
Tumor IMmune Estimation Resource (TIMER) (37), CIBERSORT (38), CIBERSORT-ABS (38), QUANTISEQ (39), Microenvironment Cell
Populations-counter (MCP-counter) (40), XCELL and Estimating the Proportion
of Immune and Cancer cells (EPIC) (41) algorithms were used to evaluate and
compare the abundance of immune cells between the high- and
low-risk groups based on the FRLS. The immune cell infiltration and
functions were evaluated between the high and the low-risk groups
using the two-sample Wilcoxon test. P<0.05 was used as the
cutoff criterion. Single-sample GSEA (ssGSEA) was used to assess
the immune functions and cells between the high and low-risk groups
using the GSVA package. The correlation of the risk score and
immune checkpoints was evaluated using the difference in gene
expression levels between the two groups.
Statistical analysis
All computational and statistical analyses were
performed using R software (https://www.r–project.org/, version 4.1.0). Fisher's
exact test or the χ2 test were used for analyses of
clinical data. Unpaired or paired two-tailed Student's t-test was
used to compare two groups; one-way ANOVA followed by Tukey's post
hoc test was used for multiple group comparison. The correlation
between ferroptosis-related genes and lncRNAs was assessed using
Pearson's correlation analysis. The ferroptosis-related lncRNA and
gene co-expression network were constructed and visualized using
Cytoscape software (https://cytoscape.org/, version 3.8.2), and the
ggalluvial package was used to generate the Sankey diagram.
Univariate and multivariate Cox regression analyses were performed
to assess whether the prognostic model was independent of other
traditional clinicopathological factors (including age, Sex, tumor
grade and TNM stage) in the prediction of OS of patients with PAAD.
The hazard ratio (HR) and 95% confidence interval (CI) were
evaluated using the survival R package. The Cox model was presented
as the hazard function denoted by h(t). Briefly, the hazard
function could be interpreted as the risk of dying at time t
(42). The Cox regression was
fitted using the covariates as follows: Age, Sex, grade, stage and
risk score. Univariate Cox analyses for all these variables were
first calculated; then multivariate cox analyses were fitted to
describe how these factors jointly impacted on survival. For each
analysis, statistical significance was set at P<0.05.
Results
Identification of prognostic
ferroptosis-related lncRNAs in patients with PAAD
A total of 253 ferroptosis-related genes were
identified using the FerrDb database (43), and expression data were available
for 246 of these genes in the TCGA-PAAD dataset (Table SII). A total of 2,749
ferroptosis-related lncRNAs were identified using Pearson's
correlation analysis (r>0.4; P<0.001). Subsequently, 215
differentially expressed lncRNAs were identified (log2FC>1;
P<0.05; Table SIII).
Furthermore, univariate Cox proportional hazards analysis was
performed and 26 prognostic ferroptosis-related lncRNAs were
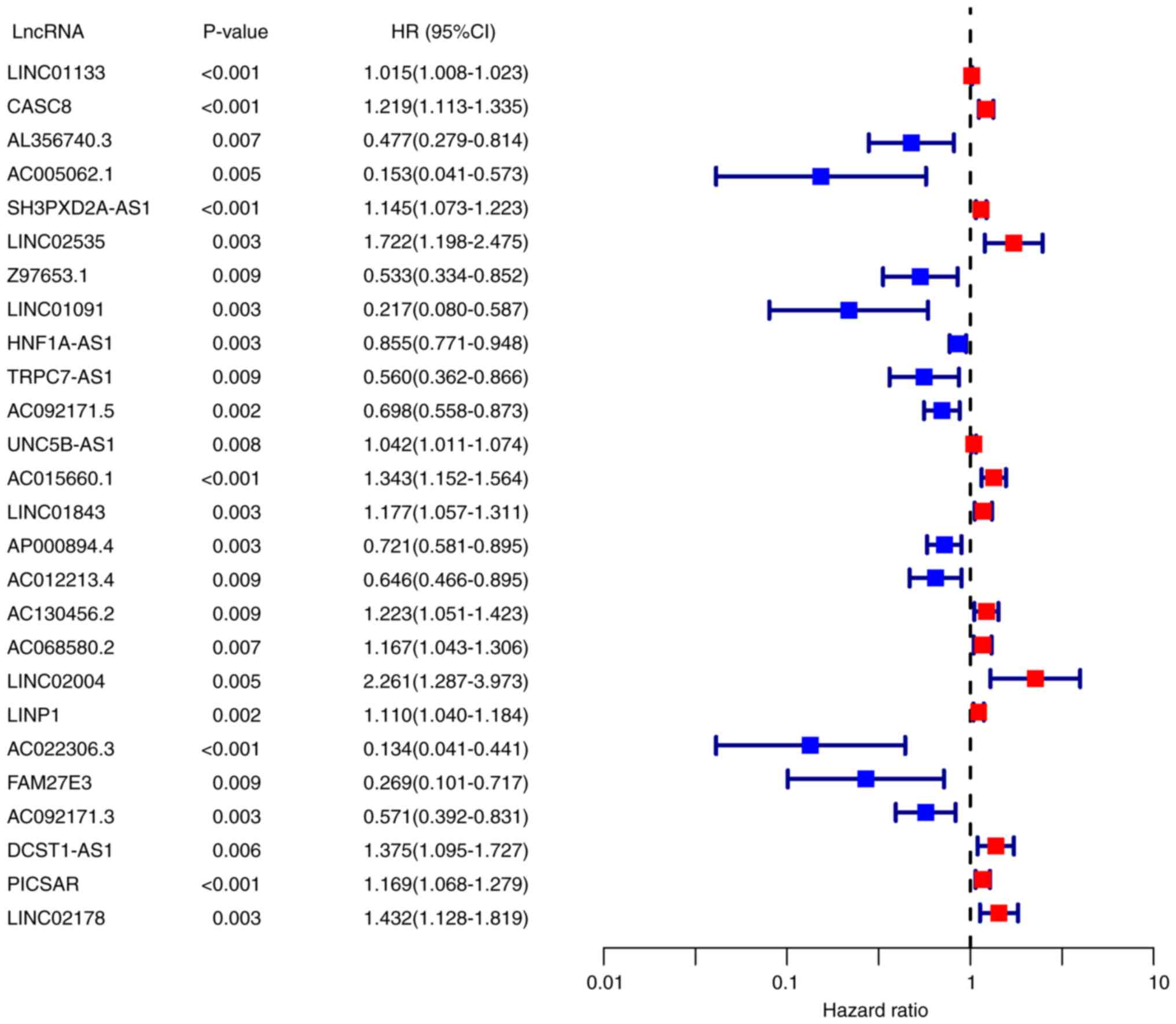
identified (P<0.01; Fig. 1).
Among them, 14 lncRNAs were identified as poor prognostic factors
(HR >1), and 12 were identified as favorable prognostic factors
(HR <1).
Construction and validation of the
prognostic FRLS
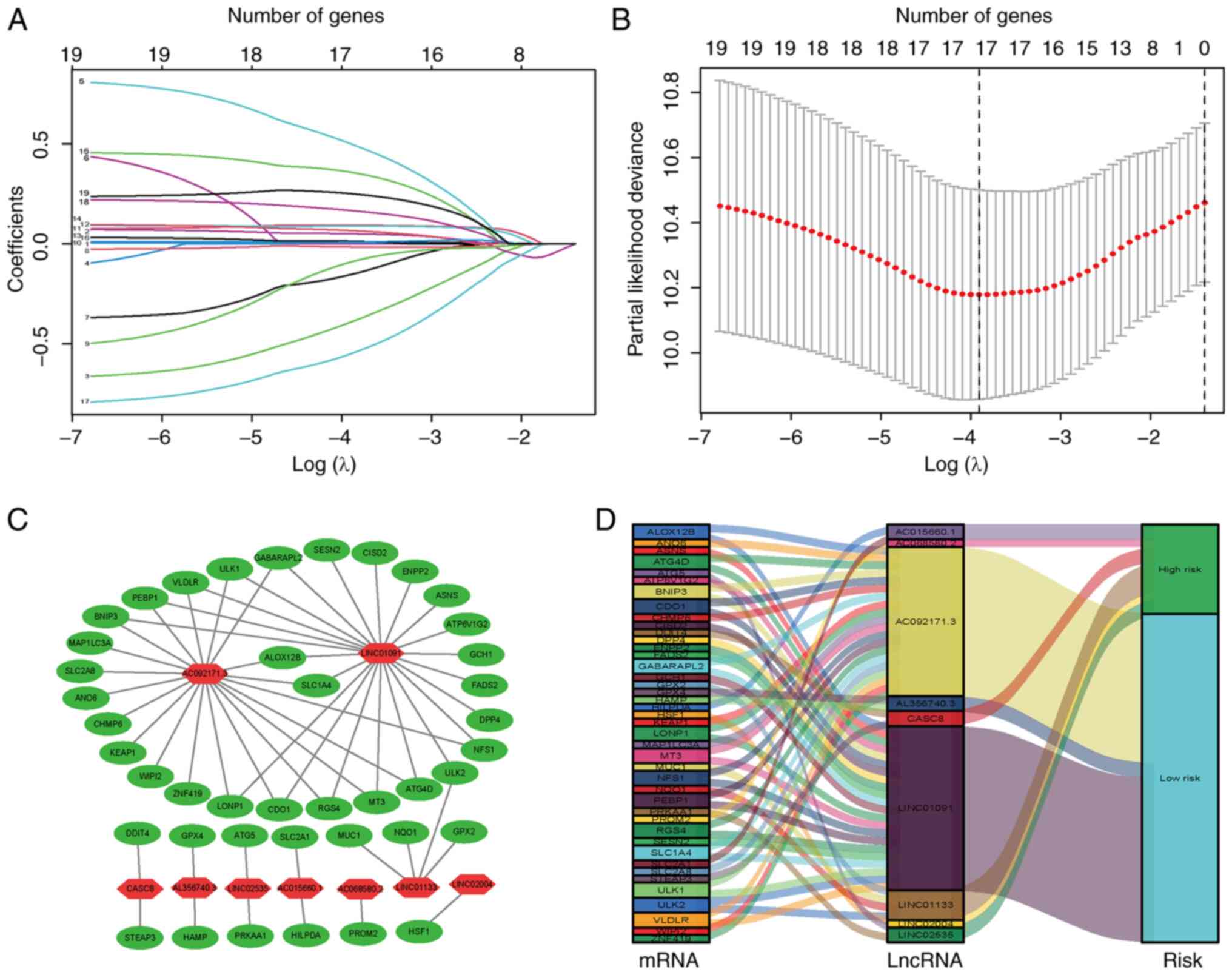
To construct a risk signature, the aforementioned 26
ferroptosis-related lncRNAs were screened using the LASSO
regression algorithm in the TCGA database (Fig. 2A and B); 17 lncRNAs were
obtained(LINC01133, CASC8, AL356740.3, LINC02535, LINC01091,
HNF1A.AS1, TRPC7.AS1, UNC5B.AS1, AC015660.1, LINC01843, AC130456.2,
AC068580.2, LINC02004, LINP1, AC092171.3, DCST1.AS1 and LINC02178).
Of these, nine lncRNAs were further identified by multivariate Cox
proportional hazards regression analysis (Table I), including LINC01133, CASC8,
AL356740.3, LINC02535, LINC01091, AC068580.2, LINC02004, AC092171.3
and AC015660.1, which were used to construct a prognostic FRLS.
Correlations between the expression levels of these nine lncRNAs
and ferroptosis-related genes are presented (Fig. 2C; Table SIV). Among these candidates,
LINC01091, AC092171.3 and AL356740.3 were predicted to be
protective lncRNAs, whereas the remaining lncRNAs were predicted to
be risk lncRNAs (Fig. 2D). The
risk score of each sample was calculated by taking the sum of the
expression level of each lncRNA and multiplying it by the
corresponding coefficients for each sample.
 | I.Multivariate Cox regression analysis
for the risk model based on nine ferroptosis-related lncRNAs. |
I.
Multivariate Cox regression analysis
for the risk model based on nine ferroptosis-related lncRNAs.
| lncRNA | Coefficient | HR | HR.95L | HR.95H | P-value |
|---|
| LINC01133 | 0.010370471 | 1.01042443 | 1.001045639 | 1.019891091 | 0.029285882 |
| CASC8 | 0.108503871 | 1.114609223 | 0.982588369 | 1.264368436 | 0.091625469 |
| AL356740.3 | -0.454385767 | 0.634837786 | 0.364897336 | 1.104472342 | 0.107778175 |
| LINC02535 | 0.620347204 | 1.859573581 | 1.247702734 | 2.771504628 | 0.002311884 |
| LINC01091 | -0.926940642 | 0.39576264 | 0.116636711 | 1.342871085 | 0.137008934 |
| AC068580.2 | 0.121716427 | 1.129433779 | 0.984743052 | 1.295384273 | 0.081831429 |
| LINC02004 | 0.885501211 | 2.424199123 | 1.138107086 | 5.163610229 | 0.021716368 |
| AC092171.3 | -0.666412005 | 0.513547883 | 0.316408144 | 0.833516562 | 0.006998521 |
| AC015660.1 | 0.176288966 | 1.192782683 | 1.003061085 | 1.418388721 | 0.046092594 |
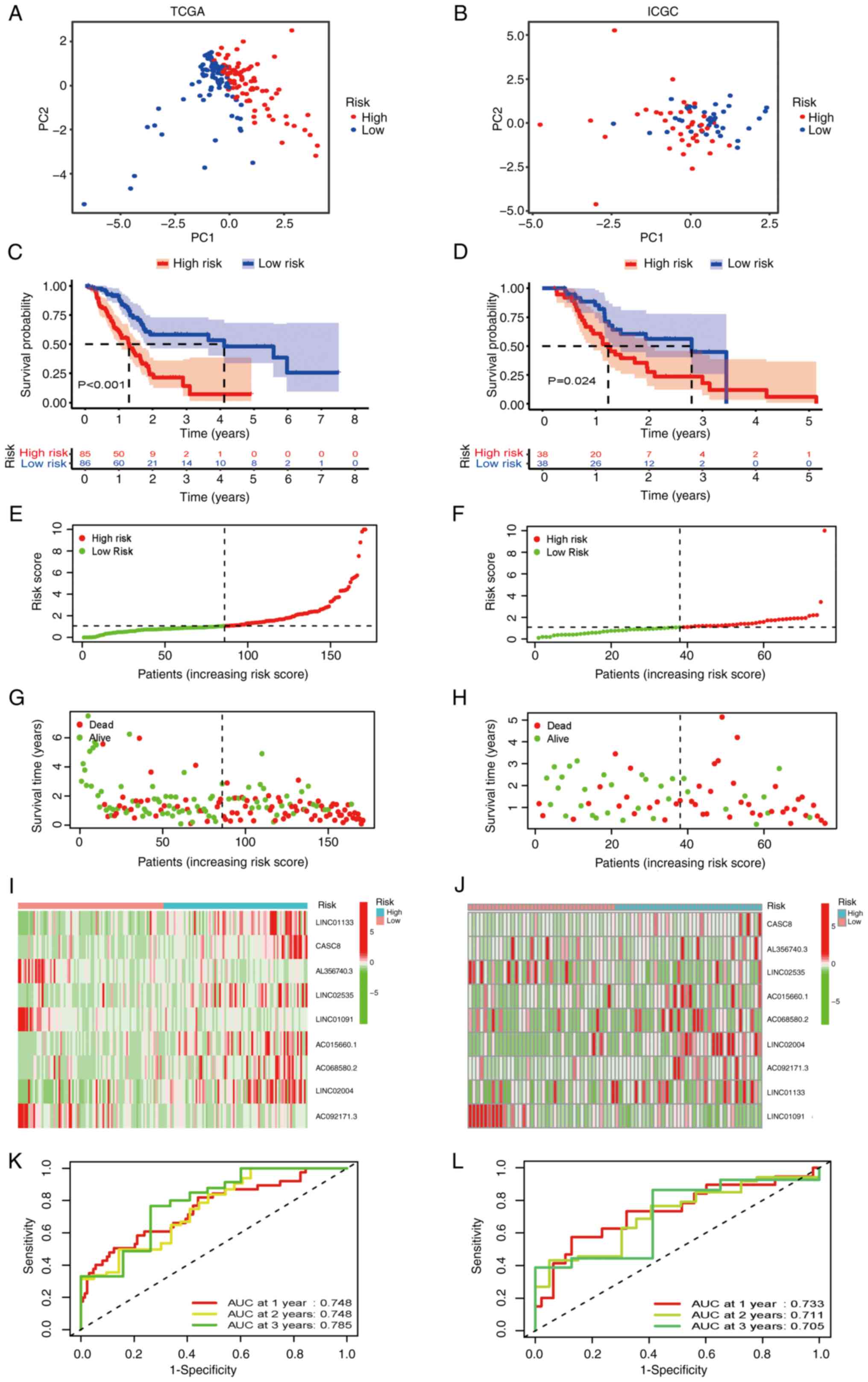
Patients with PAAD were subsequently stratified into
high and low-risk groups using the median risk score as the cutoff
value. The classification ability of the prognostic FRLS was
assessed using PCA in both the TCGA training and ICGC validation
cohorts (Fig. 3A and B,
respectively). The prognostic effectiveness of this model for OS
status of patients with PAAD was assessed using Kaplan-Meier curve
analysis. The results from the TCGA cohort demonstrated that the OS
rate of patients in the high-risk group was significantly lower
(P<0.001; Fig. 3C), which
suggested that the newly developed risk signature may effectively
predict survival. Furthermore, a risk score distribution dot plot
demonstrated that more patients survived in the low-risk group
compared with the high-risk group (Fig. 3E and G). The expression levels of
the nine ferroptosis-related lncRNAs in the risk signature is
presented as a heatmap in Fig. 3I.
Furthermore, the predictive accuracy of the model was evaluated
using time-dependent ROC analysis, with area under the curve (AUC)
values for 1, 2 and 3-year OS of 0.748, 0.748 and 0.785,
respectively (Fig. 3K).
To assess whether the prognostic significance of
FRLS remained in other populations, the same analyses were
performed in the ICGC cohort. In accordance with the results in the
TCGA training cohort, patients in the high-risk group in the ICGC
validation cohort demonstrated a significantly shorter OS compared
with patients in the low-risk group (Fig. 3D). LncRNAs risk distribution,
survival rate and expression profiles in the validation cohort were
shown in Fig. 3F, H and J. The
AUCs at 1, 2 and 3 years OS reached 0.733, 0.711 and 0.705,
respectively (Fig. 3L). All these
results suggested that the prognostic FRLS may accurately and
stably predict the clinical outcome of patients with PAAD.
Correlation of the prognostic FRLS
with clinicopathological factors
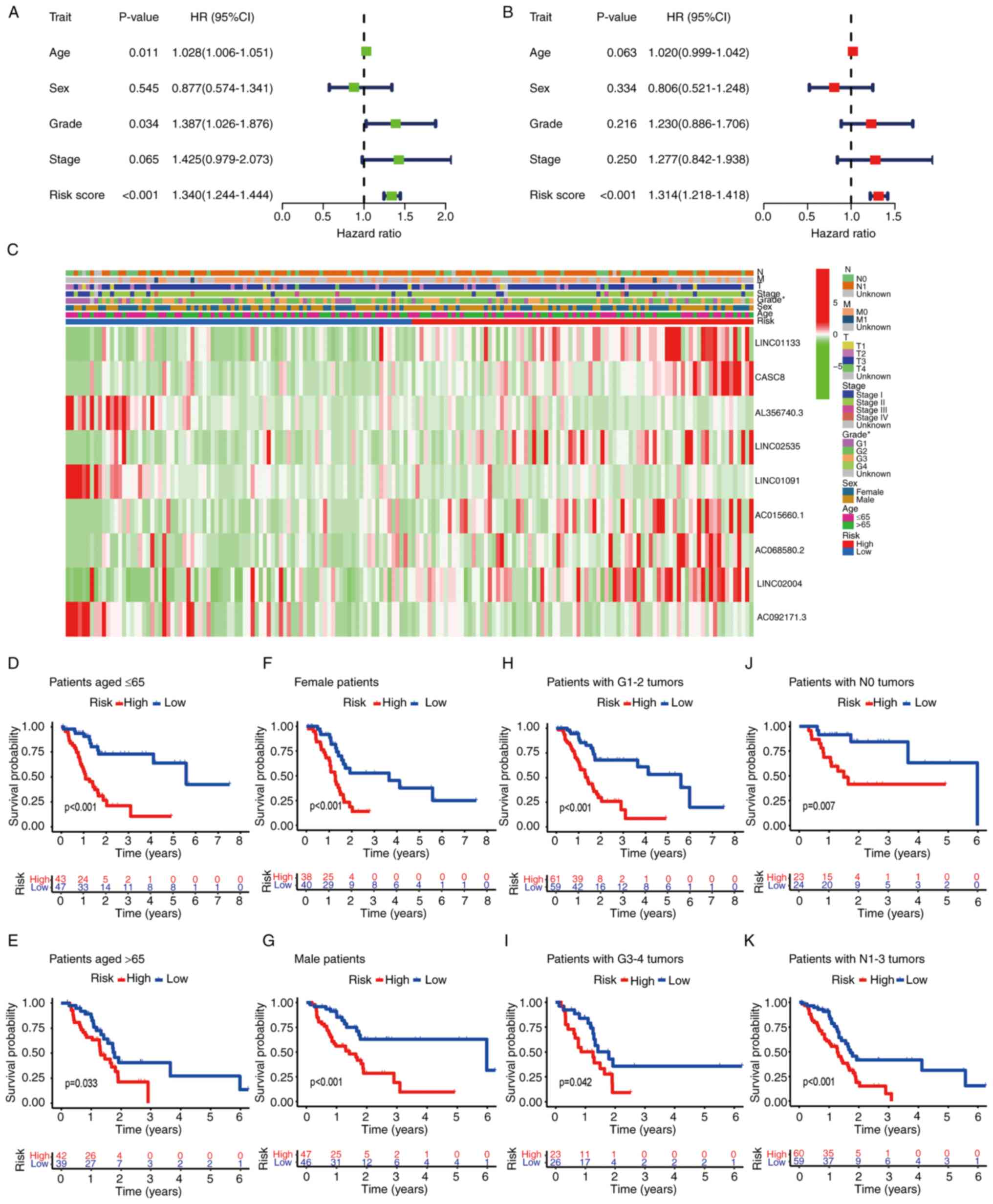
The independent value of the prognostic FRLS was
assessed using univariate and multivariate Cox regression analyses.
Univariate Cox regression analysis demonstrated that the risk
score, age and tumor grade were associated with the OS of patients
with PAAD (P<0.001; Fig. 4A).
Multivariate Cox regression analysis demonstrated that the risk
score was an independent prognostic factor for the prediction of
the OS of patients with PAAD (P<0.001; Fig. 4B).
To further evaluate the roles of the FRLS in the
development of PAAD, the correlations of the risk score with
clinicopathological features were evaluated. The associations
between the prognostic model based on the nine-gene FRLS and the
clinicopathological factors are presented as a heatmap (Fig. 4C). Stratification analysis was
performed according to the following clinical factors: Age (≤65 and
>65), sex (female and male), tumor grade (G1-2 and G3-4) and N
stage (N0 and N1-3). The risk signature demonstrated significant
differences in prognosis, with high-risk patients having
significantly poorer OS compared with low-risk patients in all
subgroups (Fig. 4D-K). In summary,
these results suggested that the FRLS was closely related to PAAD
progression.
Construction of a predictive
nomogram
To assess the potential clinical practicality of the
prognostic model based on the nine-gene FRLS, a nomogram was
constructed using the risk score and clinicopathological features
to estimate the 1, 3 and 5-year OS rates of patients with PAAD
(Fig. 5A). The calibration curve
of the nomogram demonstrated that the predicted OS rate was close
to the actual OS rate at 1, 3 and 5 years (Fig. 5B). Furthermore, the DCA results
demonstrated that this nomogram based on the nine-gene FRLS had
better clinical practicality for the prognosis prediction of
patients with PAAD (Fig. 5C).
Finally, a 5-year OS time-dependent ROC curve was generated. The
AUC value of the clinical prognostic nomogram was 0.748, which was
markedly higher compared with the AUC values of age, sex, grade and
stage (Fig. 5D), which further
suggested the discriminative ability of the nine-gene FRLS combined
with tumor pathological characteristics to predict the survival
time of patients with PAAD.
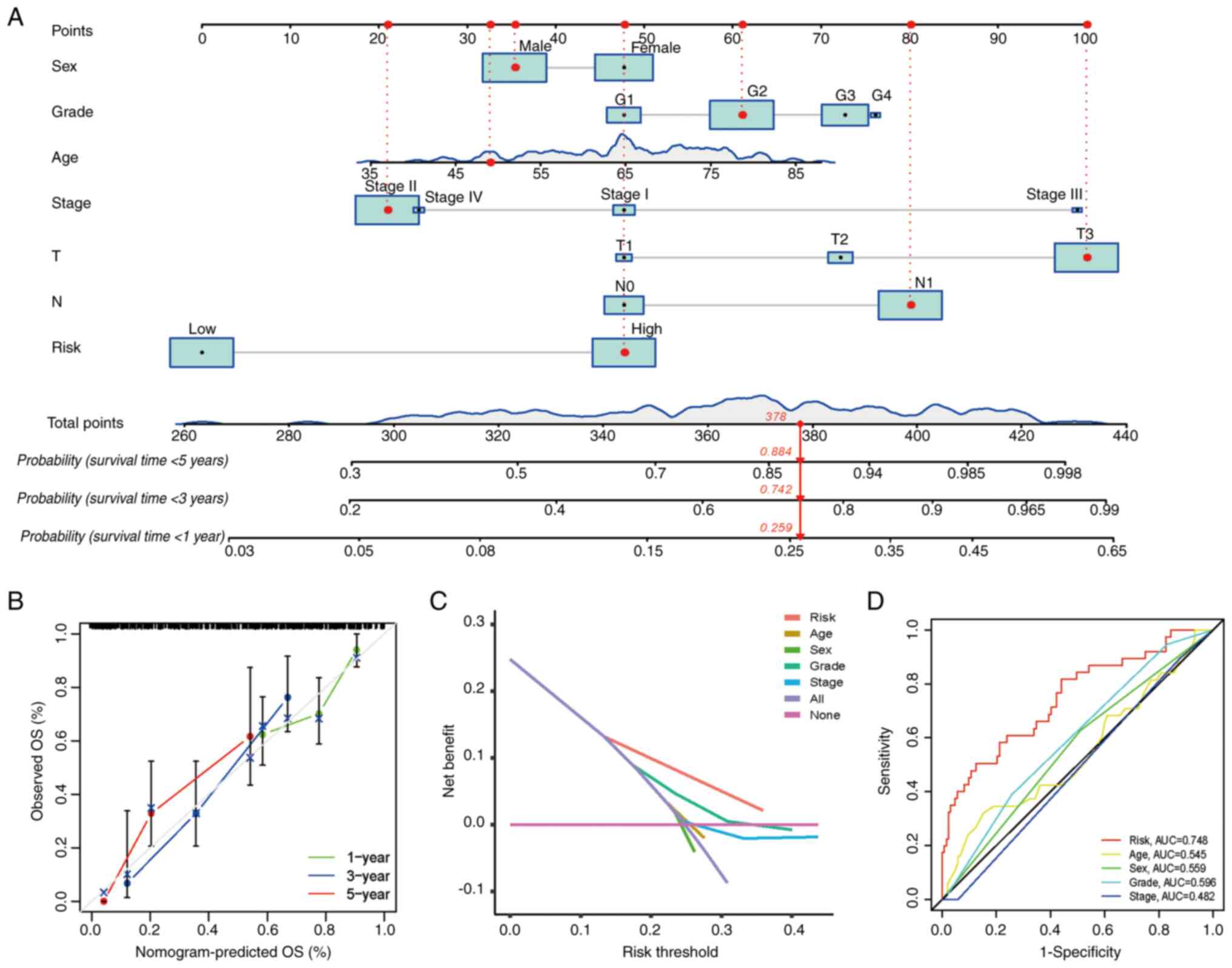 | Figure 5.Clinical prognostic nomogram for
survival prediction. (A) Clinical prognostic nomogram developed to
predict 1, 3 and 5-year OS. (B) Calibration curves presenting
nomogram predictions for 1-year, 3-year and 5-year OS. (C) Decision
curve analysis of the clinical practicality of the nomogram for
prognosis prediction. (D) Time-dependent receiver operating
characteristic curve analyses for predicting OS at 5 years by risk
score, age, sex, grade and stage. AUC, area under the curve; G,
grade; N, node; OS, overall survival; T, tumor. |
Identification of FRLS-associated
biological pathways
To evaluate the significant changes in functional
phenotypes between the high- and low-risk groups of the prognostic
FRLS, GSEA between the two risk groups was used. The GO terms
‘antigen processing and presentation of peptide antigen via MHC
class 1’ [normalized enrichment score (NES)=1.73; P=0.008],
‘antigen processing and presentation of exogenous antigen via MHC
class 1’ (NES=1.66; P=0.02), ‘interleukin-1-mediated signaling
pathway’ (NES=1.59; P=0.002) and ‘innate immune response in mucosa’
(NES=1.62; P=0.002) were significantly enriched in patients with
PAAD with high risk scores. However, ‘voltage-gated cation channel
activity’ (NES=−1.95; P=0.000), ‘voltage-gated potassium channel
activity’ (NES=−1.89; P=0.000) and ‘mast cell activation involved
in the immune response’ (NES=−1.60; P=0.004) were significantly
enriched in patients with PAAD with low risk scores (Fig. 6A-E; Table SV). Notably, several cell
death-related biological processes, such as ‘positive regulation of
cell aging’, ‘regulation of necrotic cell death’ and ‘cell redox
homeostasis’, were enriched in PAAD samples with high risk scores
(Table SV).
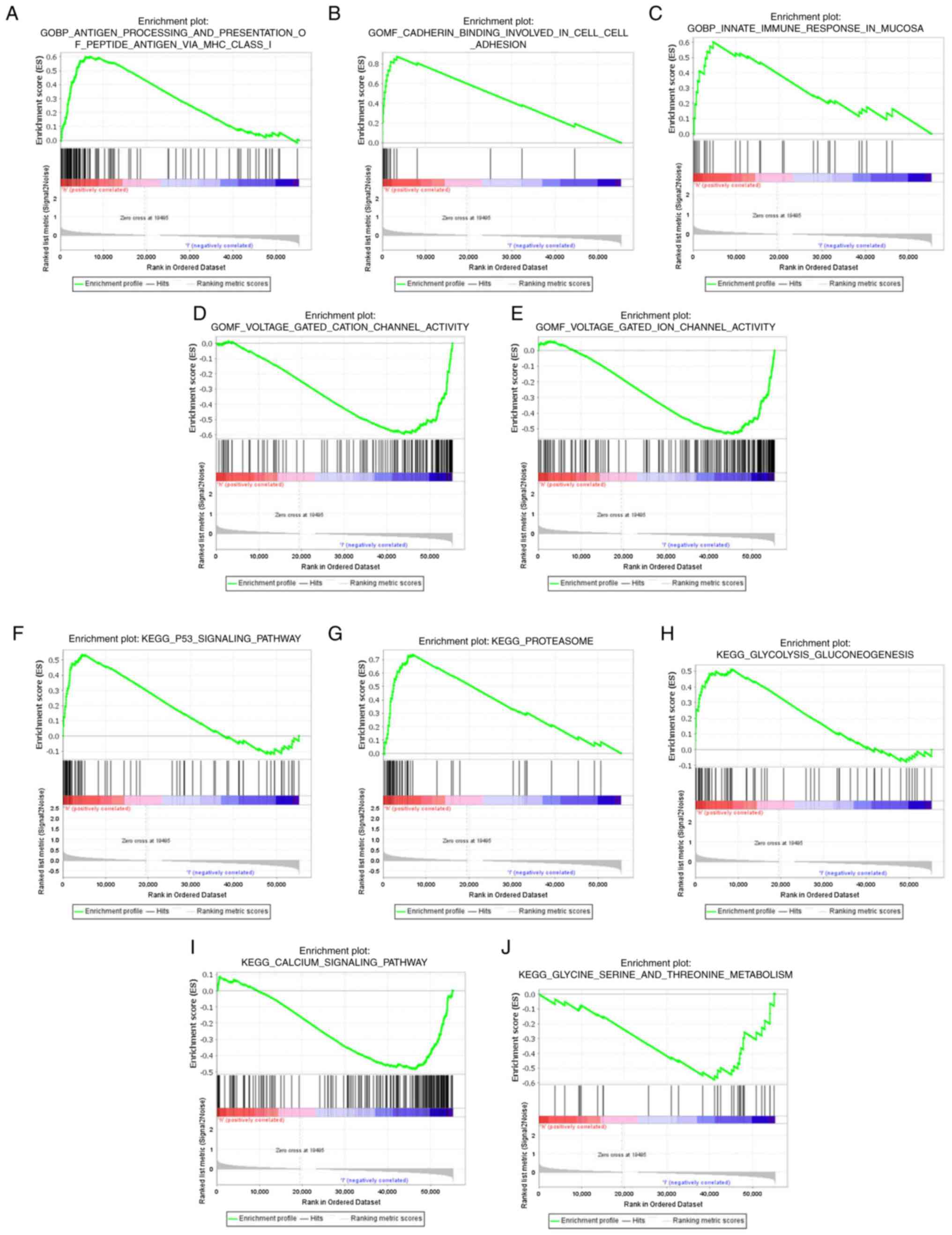 | Figure 6.Functional analysis of the low-risk
and high-risk groups. In the GO enrichment analyses, three GO
items, namely, (A) antigen processing and presentation of peptide
antigen via MHC class 1, (B) cadherin binding involved in cell
adhesion and (C) innate immune response in mucosa, were
significantly enriched in the high-risk group. GO items (D)
voltage-gated cation channel activity and (E) voltage-gated ion
channel activity showed significantly differential enrichment in
the low-risk group. In the KEGG enrichment analyses, three KEGG
items, namely, (F) p53 signaling pathway, (G) proteasome and (H)
glycolysis/gluconeogenesis, were significantly enriched in the
high-risk group. KEGG items (I) calcium signaling pathway and (J)
glycine, serine and threonine metabolism showed significantly
differential enrichment in the low-risk group. GO, Gene Ontology;
KEGG, Kyoto Encyclopedia of Genes and Genomes; MHC, major
histocompatibility complex. |
KEGG pathway analysis demonstrated that the lncRNAs
in the newly developed signature were enriched in classical tumor
pathways and metabolism (Fig.
6F-J; Table SVI). The ‘p53
signaling pathway’ (NES=1.72; P=0.009, ‘proteasome’ (NES=1.75;
P=0.013) and ‘glycolysis/gluconeogenesis’ (NES=1.64; P=0.008) were
significantly enriched in the high-risk group. The ‘calcium
signaling pathway’ (NES=−1.68; P=0.002), ‘glycine, serine and
threonine metabolism’ (NES=−1.66; P=0.010) and ‘tryptophan
metabolism’ (NES=−1.56; P=0.037) were significantly enriched in the
low-risk group. These results indicated that the gene sets were
connected with the tumor immune microenvironment, as well as
carcinogenesis and tumor progression pathways.
Correlation of the FRLS with immune
infiltration
As the GSEA results suggested that the FRLS was
associated with the tumor immune microenvironment, the correlation
of the nine-gene FRLS with immune cell infiltration was assessed.
Heatmaps of immune infiltration were generated using the TIMER,
CIBERSORT, CIBERSORT-ABS, QUANTISEQ, MCP-counter, XCELL and EPIC
algorithms (Fig. 7A). Comparative
analysis of immune functions and immune cells was performed to
assess the differences in cytolytic activity, major
histocompatibility complex (MHC) class I, T cell co-inhibition, T
cell co-stimulation, type I IFN response, type II IFN response, B
cells, CD8+ T cells, mast cells, neutrophils,
plasmacytoid dendritic cells (pDCs), T helper (Th) cells,
follicular helper T cells (Tfhs) and tumor-infiltrating lymphocytes
(TILs) between the two risk groups (P<0.05; Fig. 7B and C). Furthermore, violin plots
were generated based on the CIBERSORT algorithm and demonstrated
the relationship between the risk score and immune cell
infiltration (Fig. 7D). Patients
with PAAD in the low-risk group demonstrated significantly higher
ratios of CD8+ T cells (P<0.001), activated memory
CD4+ T cells (P=0.003) and monocytes (P=0.01) compared
with the high-risk group. However, M0 macrophages (P=0.002) were
significantly positively associated with a high-risk score. In
addition, correlation scatter plots demonstrated the same trends
between the prognostic signature and immune cell infiltration in
CD8+ T cells (r=−0.28; P=0.002), activated memory
CD4+ T cells (r=−0.23; P=0.011), monocytes (r=−0.18;
P=0.045) and M0 macrophages (r=0.25; P=0.005) (Fig. 7E-H).
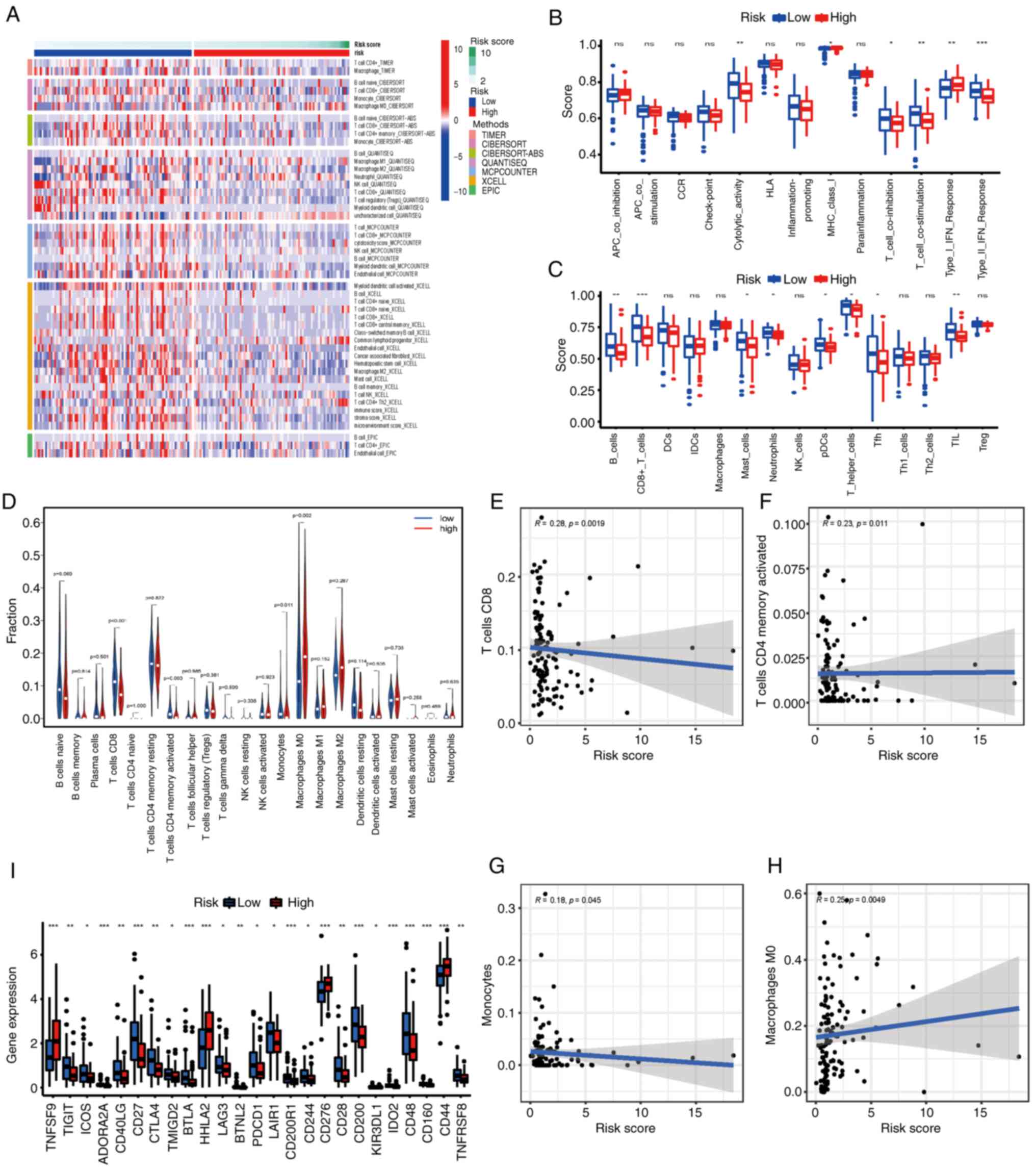 | Figure 7.Correlations of the risk score with
the immune microenvironment and immune checkpoints compared between
the high and low-risk groups. (A) Heatmaps for immune responses
using the TIMER, CIBERSORT, CIBERSORT-ABS, QUANTISEQ, MCP-COUNTER,
XCELL and EPIC algorithms in the two groups. P<0.05 was
considered to indicate a statistically significant difference.
Boxplots presenting the single-sample gene set enrichment analysis
(B) immune function and (C) immune cell scores compared between the
two groups. (D) Violin plot presenting the immune-infiltrating
lymphocytes in the low- and high-risk groups. (E-H) Scatter plots
presenting the correlation between the risk score and immune cell
infiltration. (I) Expression of immune checkpoints compared between
the low- and high-risk groups. The Kruskal-Wallis test was used to
assess differences between the groups. *P<0.05, **P<0.01 and
***P<0.001. APC, antigen presenting cell; CCR, chemokine
receptor; DCs, dendritic cells; HLA, human leukocyte antigen; iDCs,
inflammatory dendritic cells; Tfh, T follicular helper cell; MHC,
major histocompatibility complex; NK, natural killer; ns, not
significant; pDCs, plasmacytoid dendritic cells; TIL,
tumor-infiltrating lymphocytes; Treg, regulatory T cells. |
Given the importance of immune checkpoint blockade
immunotherapy, the relationship between the expression of immune
checkpoints and the two risk groups was evaluated (Fig. 7I). The low-risk group tended to
demonstrate higher mRNA expression levels of 22 immune checkpoint
genes, including TIGIT, inducible T-cell co-stimulator,
adenosine A2a, CD40 ligand, CD27, CTLA4, transmembrane and
immunoglobulin domain containing 2, B and T-lymphocyte attenuator,
lymphocyte activating 3, BTNL2, programmed cell death 1,
leukocyte-associated immunoglobulin-like receptor 1, CD200 receptor
1, CD244, CD28, CD200, KIR3DL1, IDO2, CD48, CD160, CD44 and
TNFRS, whereas the other three immune checkpoint genes
(TNFSF9, HHLA2 and CD276) were highly expressed in
high-risk patients (all P<0.05). These results suggested that
the FRLS may be a potential biomarker for immune checkpoint
inhibitor therapy.
Validation of ferroptosis-related
lncRNA expression in cells and tissues
LINC01133 and CASC8 were selected for validation, as
their LASSO coefficients were the top two among the risk lncRNAs.
RT-qPCR was performed to evaluate the expression levels of
LINC01133 and CASC8 in pancreatic cancer tissues, and the results
demonstrated that LINC01133 and CASC8 were significantly increased
compared with ANT tissues (Fig. 8A and
B). Furthermore, the mRNA expression levels of SLC7A11 and
GPX4, two essential ferroptosis-related genes (43), were significantly increased in
pancreatic cancer tissues compared with ANT tissues (Fig. 8C and D). The SLC7A11/GPX4 axis
serves as the canonical defense against ferroptosis by facilitating
intracellular glutathione biosynthesis and alleviating lipid
peroxidation (44). Next, the
GEPIA (31) database was used to
verify the expression levels of ferroptosis-related lncRNAs in
pancreatic cancer tissues, ANT tissues and normal pancreatic
tissues. The RNA expression levels of LINC01133 and CASC8 were
significantly upregulated in pancreatic cancer tissues compared
with normal tissues (Fig. 8E and
F). Moreover, Kaplan-Meier survival analysis demonstrated that
upregulation of these two lncRNAs were associated with shorter
overall survival in the GEPIA database (Fig. 8G and H). The expression levels of
LINC01133 and CASC8 were assessed in pancreatic cancer cells and
pancreatic ductal epithelial cell lines. The results demonstrated
that LINC01133 and CASC8 expression levels were significantly
upregulated in pancreatic cancer cells compared with pancreatic
ductal epithelial cells (Fig. 8I and
J). To evaluate the role of LINC01133 in the ferroptosis
process of pancreatic cancer, LINC01133 expression was then knocked
down in AsPC-1 and BxPC-3 cells, both of which have high levels of
LINC01133 (Fig. 8K). The results
demonstrated that downregulation of LINC01133 significantly
decreased the survival rate of AsPC-1 and BxPC-3 cells after
ferroptosis induction using erastin (Fig. 8L), which suggested that LINC01133
may have been involved in the ferroptosis of pancreatic cancer
cells.
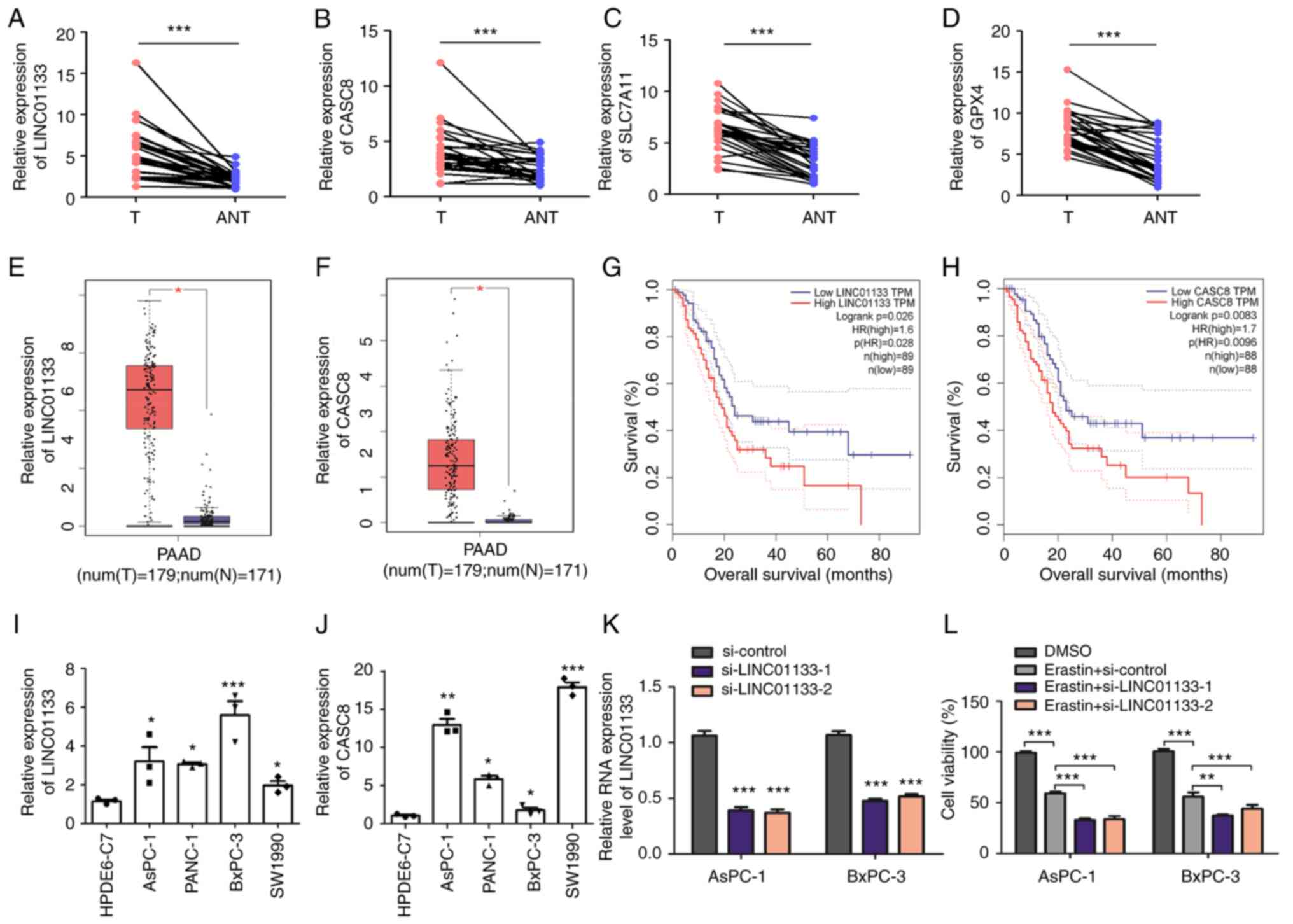 | Figure 8.Evaluation of ferroptosis-related
lncRNA expression in cells and tissues. RT-qPCR analysis of (A)
LINC01133, (B) CASC8, (C) SLC7A11 and (D) GPX4 expression levels in
30 paired pancreatic cancer tissues and ANT tissues. Comparison of
the differential expression of (E) LINC01133 and (F) CASC8 in the
Gene Expression Profiling Interactive Analysis database.
Kaplan-Meier survival curves for the ferroptosis-related lncRNAs
(G) LINC01133 and (H) CASC8. *P<0.05, **P<0.01 and
***P<0.001. (I and J) RT-qPCR analysis of LINC01133 and CASC8
expression levels in pancreatic cancer and HPDE normal pancreatic
ductal epithelial cell lines. *P<0.05, **P<0.01 and
***P<0.001 vs HPDE6-C7. (K) AsPC-1 and BxPC-3 cells were
transiently transfected with si-control and si-LINC01133 using
Lipofectamine. LINC01133 expression was confirmed to be knocked
down 24 h after transfection using RT-qPCR. ***P<0.001 vs
si-control. (L) AsPC-1 and BxPC-3 cells transfected with si-control
and si-LINC01133 were treated with 10 µM erastin for 24 h. Then,
cell viability was assessed using a Cell Counting Kit-8 assay.
Unpaired or paired two-tailed Student's t-test was used to compare
two groups; one-way ANOVA followed by Tukey's post hoc test was
used for multiple group comparison. lncRNA, long non-coding RNA;
RT-qPCR, reverse-transcription quantitative PCR; T, pancreatic
cancer tissues; ANT, adjacent non-tumor tissues; PAAD, pancreatic
adenocarcinoma; si, small interfering RNAs; HR, hazard ratio. |
Discussion
LncRNAs serve an important role in the ferroptotic
process in cancers (24). In the
present study, the relationship between lncRNAs and
ferroptosis-related genes in PAAD, and the expression of
ferroptosis-related lncRNAs and their relationship with the
prognosis of patients with PAAD were assessed. A novel prognostic
FRLS containing nine ferroptosis-related lncRNAs was then
constructed to predict the prognosis of patients with PAAD. In both
the training cohort from the TCGA database and the validation
cohort from the ICGC database, the FRLS demonstrated robust
prognostic predictive ability for patients with PAAD. Univariate
and multivariate Cox regression analyses demonstrated that the FRLS
was an independent prognostic factor for PAAD. Furthermore,
combining the prognostic FRLS with clinicopathological factors, a
nomogram with improved predictive ability for OS was constructed.
Calibration curves demonstrated that the nomogram predictions for
1-year, 3-year and 5-year survival were close to the actual OS
rates. Functional enrichment analyses demonstrated the difference
in immune-related biological processes and classical tumor pathways
between the high and low-risk groups. The differential immune
infiltration between the two risk groups was further analyzed by
comparison of the abundance of immune cells and immune functions
and the mRNA expression levels of immune checkpoint genes. These
results demonstrated that the nine-gene FRLS was a novel prognostic
biomarker and potential therapeutic target for PAAD.
The prognostic signature constructed in the present
study was derived from nine ferroptosis-related lncRNAs. Among
them, LINC01133 had been previously reported to serve important
roles in gastric cancer (45),
ovarian cancer (46), breast
cancer (47) and cervical cancer
(48,49). In PAAD, LINC01133 was reported to
be upregulated, and loss of LINC01133 was reported to suppress the
development of PAAD cells via the Wnt signaling pathway (50). In another study, Huang et al
(51) reported that overexpression
of LINC01133 was associated with poorer prognosis in patients with
PAAD and that the C/EBPβ-LINC01133 axis served an oncogenic
function through upregulation of CCNG1. CASC8 was reported to be
correlated with the progression of non-small cell lung cancer
(52), retinoblastoma (53) and several other cancers (54,55).
Upregulation of CASC8 was reported to be associated with the poor
prognosis of patients with PAAD (56). In the prognostic signature
identified in the present study, LINC01133 and CASC8 were
upregulated and associated with poor prognosis in PAAD, which was
consistent with the findings of the aforementioned studies.
Furthermore, LINC02535 was demonstrated to regulate DNA damage
repair in cervical cancer (57)
and to promote cell growth in poorly differentiated gastric cancer
(58). However, the role and
prognostic value of the remaining ferroptosis-related lncRNAs in
cancer were unknown. The specific mechanisms of ferroptosis-related
lncRNAs and their contributions to ferroptosis in PAAD are complex
and still obscure, and require further study.
The results of KEGG enrichment analyses demonstrated
that several pathways were enriched when compared between the high
and low-risk groups, including the p53, proteasome, glycolysis,
calcium and chemokine signaling pathways. Previous studies reported
that p53 signaling pathway was closely related to ferroptosis
(59,60). Ferroptosis can be regulated by p53
signaling pathway through transcriptional or post-translational
mechanisms. p53 can facilitate ferroptosis through the inhibition
of SLC7A11 expression or by the induction of spermidine/spermine N
(1)-acetyltransferase 1 and
glutaminase 2 activity. However, p53 can also inhibit ferroptosis
by the inhibition of dipeptidyl peptidase 4 or through the
enhancement of cyclin dependent kinase inhibitor 1 A (61). It has been reported that proteasome
function is often inhibited during ferroptosis (62). Numerous metabolic and degradation
pathways, including the ubiquitin-proteasome system, orchestrate
the complex ferroptotic response through the regulation of iron
accumulation or lipid peroxidation (63). Recent studies reported that
cellular energy metabolism activities such as glycolysis, pentose
phosphate pathway and the tricarboxylic acid cycle are associated
with ferroptosis by the regulation of essential ferroptosis
molecules, including nicotinamide adenine dinucleotide phosphate,
reactive oxygen species and glutathione (64). It was also reported that
calcium-iron crosstalk regulates iron-induced ferroptosis (65). Martin-Sanchez et al
(66) reported that ferrostatin-1,
an inhibitor of ferroptosis, could prevent the upregulation of
IL-33 and other chemokines and cytokines. In general, KEGG analyses
suggested the probable mechanisms of the high-risk and low-risk
groups, and these signaling pathways were generally related to
ferroptosis. However, the specific mechanisms of
ferroptosis-related lncRNAs in PAAD are complex and still unclear
and require further evaluation in future work.
Immune cells that infiltrate tumors exert diverse
effects on PAAD progression (67).
The number and proportion of infiltrating immune cells serve
important roles in regulation of the immunotherapy response and
affect cancer progression (68).
However, the relationship between the ferroptotic process and
immune infiltration levels in PAAD remains unclear. In the present
study, the numbers of B cells, CD8+ T cells, mast cells,
neutrophils, pDCs, Th cells, Tfhs and TILs were significantly
associated with risk score, which suggested that these immune cells
may serve a substantial role in the development of PAAD. Compared
with patients with PAAD in the low-risk group, those in the
high-risk group tended to have fewer infiltrated tumor-killing
immune cells, such as B cells, CD8+ T cells, Th cells
and TILs, which suggested that the antitumor ability of the
high-risk group was weaker. Therefore, we hypothesized that
ferroptosis is markedly associated with the number and proportion
of tumor-infiltrating immune cells in PAAD. Recently, certain
studies have begun to report the effect of combining cancer
immunotherapy with ferroptosis inducers. Wang et al
(69) first reported that
immunotherapy-activated CD8+ T cells could promote tumor
cell lipid peroxidation and ferroptosis through the release of IFNγ
to decrease the expression of SLC3A2 and SLC7A11. Given the
potential role of the ferroptosis-related lncRNAs in the signature
identified in the present study in the process of immune
infiltration, the underlying mechanism of these lncRNAs in the
regulation of the immune response of PAAD should be evaluated in
further studies. Checkpoint blockade immunotherapy has been
reported to have improved the prognosis of patients multiple
advanced malignancies; however, not all patients respond to this
treatment (70). The results of
the present study demonstrated that patients with PAAD in the
high-risk group, based on the FRLS, tended to have lower mRNA
expression levels of immune checkpoints. Significant differences in
these molecules between the two groups indicated differences in
sensitivity to immunotherapies. However, further studies are needed
to confirm the feasibility and effectiveness of this combined
regimen of ferroptosis inducers and immune checkpoint inhibitors
for the treatment of PAAD.
There have been numerous systematic bioinformatic
analyses of PAAD; however, the present study proposed a novel
nine-gene FRLS to predict prognosis for patients with PAAD.
Furthermore, a prognostic nomogram including the FRLS was
constructed that demonstrated improved accuracy for survival risk
stratification. Importantly, the FRLS indicated in the present
study may be involved in immune infiltration and have an effect on
cancer immunotherapy. In addition, the AUC values for the training
and test groups were 0.785 and 0.733, respectively. In previous
studies, several prognostic models have also been constructed based
on lncRNAs to assess the prognosis of patients with PAAD. Chen
et al (71) constructed an
evaluation model based on ferroptosis-related lncRNAs, and the AUC
values were 0.70 at 1 year, 0.71 at 3 years and 0.75 at 5 years,
which were lower than those for the model developed in the present
study. In another study, Shi et al (72) reported a prognostic three lncRNA
signature, and the AUC of this model was 0.716. Wei et al
(73) built a nine lncRNA
signature model for PAAD, and the AUC for this model for the
prediction of the 2-year survival rate was 0.703. These results
suggested that the prognostic model based on nine
ferroptosis-related lncRNAs constructed in our study may improve
the prognostic power for PAAD. The AUC values at 2 and 3 years
slightly decreased compared with 1 year. Several other prognostic
models also reported a decrease in the AUC values at 2 and 3 years
(71,74,75).
Numerous factors affect the AUC value and one of the important
factors is the uncertainty of the sample. The so-called uncertainty
of the sample refers to whether the corresponding label is
uncertain for the same sample; that is, the samples with the same
feature values. In the present study, the ICGC cohort had only six
patients with a survival time of >3 years; however, five of them
had a survival status of death, which may have affected the AUC
value for 3-year overall survival. Furthermore, in a finite sample,
the probability is commonly obtained by estimating the frequency of
the sample. This estimate gradually approaches the true value as
the sample size increases. As the ICGC cohort only had 76 PAAD
samples (survival time >30 days), this may have also affected
the AUC value for overall survival.
There were several limitations to the present study.
First, the FRLS was only tested and validated in the TCGA and ICGC
cohorts. If possible, it should be verified in other, larger
cohorts to improve the accuracy and robustness of the prognostic
prediction model. Furthermore, this was a retrospective study and
it would be more convincing to evaluate the clinical application of
the signature in prospective cohorts. Finally, LINC01133 and CASC8
were highly expressed in PAAD cells and tissues; however, they were
not confirmed targets in PAAD treatment. Additional in vitro
and in vivo experiments are required to fully characterize
the role and mechanism of these lncRNAs in PAAD ferroptosis. In
short, the prognostic model constructed in the present study
requires further validation.
In conclusion, the present study constructed an FRLS
for PAAD to predict prognosis. The prognostic signature and nine
ferroptosis-related lncRNAs may be molecular biomarkers and
therapeutic targets for patients with PAAD.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from The Postdoctoral
Program of Chongqing (grant no. 2021XM2017).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JL, BN and YL designed the study. JL and WL acquired
and analyzed the data. HW collected clinical samples and revised
the manuscript. BN and YL wrote and revised the manuscript. All
authors reviewed and approved the final manuscript. BN and YL
confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
The experimental procedures for human tissues were
approved by the ethics committee of Southwest Hospital (Chongqing,
China; approval no. 20210312).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mizrahi JD, Surana R, Valle JW and Shroff
RT: Pancreatic cancer. Lancet. 395:2008–2020. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2021. CA Cancer J Clin. 71:7–33. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rahib L, Smith BD, Aizenberg R, Rosenzweig
AB, Fleshman JM and Matrisian LM: Projecting cancer incidence and
deaths to 2030: The unexpected burden of thyroid, liver, and
pancreas cancers in the United States. Cancer Res. 74:2913–2921.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bliss LA, Witkowski ER, Yang CJ and Tseng
JF: Outcomes in operative management of pancreatic cancer. J Surg
Oncol. 110:592–598. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dixon SJ, Lemberg KM, Lamprecht MR, Skouta
R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS,
et al: Ferroptosis: An iron-dependent form of nonapoptotic cell
death. Cell. 149:1060–1072. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cao JY and Dixon SJ: Mechanisms of
ferroptosis. Cell Mol Life Sci. 73:2195–2209. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liang C, Zhang X, Yang M and Dong X:
Recent progress in ferroptosis inducers for cancer therapy. Adv
Mater. 31:e19041972019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hassannia B, Vandenabeele P and Berghe TV:
Targeting ferroptosis to iron out cancer. Cancer Cell. 35:830–849.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun X, Niu X, Chen R, He W, Chen D, Kang R
and Tang D: Metallothionein-1G facilitates sorafenib resistance
through inhibition of ferroptosis. Hepatology. 64:488–500. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Houessinon A, Francois C, Sauzay C,
Louandre C, Mongelard G, Godin C, Bodeau S, Takahashi S, Saidak Z,
Gutierrez L, et al: Metallothionein-1 as a biomarker of altered
redox metabolism in hepatocellular carcinoma cells exposed to
sorafenib. Mol Cancer. 15:382016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dai E, Han L, Liu J, Xie Y, Zeh HJ, Kang
R, Bai L and Tang D: Ferroptotic damage promotes pancreatic
tumorigenesis through a TMEM173/STING-dependent DNA sensor pathway.
Nat Commun. 11:63392020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Badgley MA, Kremer DM, Maurer HC,
DelGiorno KE, Lee HJ, Purohit V, Sagalovskiy IR, Ma A, Kapilian J,
Fir CEM, et al: Cysteine depletion induces pancreatic tumor
ferroptosis in mice. Science. 368:85–89. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kremer DM, Nelson BS, Lin L, Yarosz EL,
Halbrook CJ, Kerk SA, Sajjakulnukit P, Myers A, Thurston G, Hou SW,
et al: GOT1 inhibition promotes pancreatic cancer cell death by
ferroptosis. Nat Commun. 12:48602021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu SJ, Dang HX, Lim DA, Feng FY and Maher
CA: Long noncoding RNAs in cancer metastasis. Nat Rev Cancer.
21:446–460. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Goodall GJ and Wickramasinghe VO: RNA in
cancer. Nat Rev Cancer. 21:22–36. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sun YW, Chen YF, Li J, Huo YM, Liu DJ, Hua
R, Zhang JF, Liu W, Yang JY, Fu XL, et al: A novel long non-coding
RNA ENST00000480739 suppresses tumour cell invasion by regulating
OS-9 and HIF-1α in pancreatic ductal adenocarcinoma. Br J Cancer.
111:2131–2141. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li Q, Lei C, Lu C, Wang J, Gao M and Gao
W: LINC01232 exerts oncogenic activities in pancreatic
adenocarcinoma via regulation of TM9SF2. Cell Death Dis.
10:6982019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Deng SJ, Chen HY, Zeng Z, Deng S, Zhu S,
Ye Z, He C, Liu ML, Huang K, Zhong JX, et al: Nutrient
stress-dysregulated antisense lncRNA GLS-AS impairs GLS-mediated
metabolism and represses pancreatic cancer progression. Cancer Res.
79:1398–1412. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou C, Yi C, Yi Y, Qin W, Yan Y, Dong X,
Zhang X, Huang Y, Zhang R, Wei J, et al: LncRNA PVT1 promotes
gemcitabine resistance of pancreatic cancer via activating
Wnt/beta-catenin and autophagy pathway through modulating the
miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes. Mol Cancer. 19:1182020.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hui B, Ji H, Xu Y, Wang J, Ma Z, Zhang C,
Wang K and Zhou Y: RREB1-induced upregulation of the lncRNA
AGAP2-AS1 regulates the proliferation and migration of pancreatic
cancer partly through suppressing ANKRD1 and ANGPTL4. Cell Death
Dis. 10:2072019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ren X, Chen C, Luo Y, Liu M, Li Y, Zheng
S, Ye H, Fu Z, Li M, Li Z and Chen R: lncRNA-PLACT1 sustains
activation of NF-κB pathway through a positive feedback loop with
IkappaBalpha/E2F1 axis in pancreatic cancer. Mol Cancer. 19:352020.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jiang N, Zhang X, Gu X, Li X and Shang L:
Progress in understanding the role of lncRNA in programmed cell
death. Cell Death Discov. 7:302021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu Y, Zhang S, Gong X, Tam S, Xiao D, Liu
S and Tao Y: The epigenetic regulators and metabolic changes in
ferroptosis-associated cancer progression. Mol Cancer. 19:392020.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gai C, Liu C, Wu X, Yu M, Zheng J, Zhang
W, Lv S and Li W: MT1DP loaded by folate-modified liposomes
sensitizes erastin-induced ferroptosis via regulating
miR-365a-3p/NRF2 axis in non-small cell lung cancer cells. Cell
Death Dis. 11:7512020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu H and Liu A: Long non-coding RNA NEAT1
regulates ferroptosis sensitivity in non-small-cell lung cancer. J
Int Med Res. 49:3000605219961832021.PubMed/NCBI
|
|
28
|
Mao C, Wang X, Liu Y, Wang M, Yan B, Jiang
Y, Shi Y, Shen Y, Liu X, Lai W, et al: A G3BP1-interacting lncRNA
promotes ferroptosis and apoptosis in cancer via nuclear
sequestration of p53. Cancer Res. 78:3484–3496. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang M, Mao C, Ouyang L, Liu Y, Lai W, Liu
N, Shi Y, Chen L, Xiao D, Yu F, et al: Long noncoding RNA LINC00336
inhibits ferroptosis in lung cancer by functioning as a competing
endogenous RNA. Cell Death Differ. 26:2329–2343. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47:W556–W560. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ma W, Yao Y, Xu G, Wu X, Li J, Wang G,
Chen X, Wang K, Chen Y, Guo Y, et al: Identification of a
seven-long non-coding RNA signature associated with Jab1/CSN5 in
predicting hepatocellular carcinoma. Cell Death Discov. 7:1782021.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Su Y, Zhang T, Tang J, Zhang L, Fan S,
Zhou J and Liang C: Construction of competitive endogenous RNA
network and verification of 3-Key LncRNA signature associated with
distant metastasis and poor prognosis in patients with clear cell
renal cell carcinoma. Front Oncol. 11:6401502021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hu J, Xu L, Shou T and Chen Q: Systematic
analysis identifies three-lncRNA signature as a potentially
prognostic biomarker for lung squamous cell carcinoma using
bioinformatics strategy. Transl Lung Cancer Res. 8:614–635. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhou Z, Yang Z, Cui Y, Lu S, Huang Y, Che
X, Yang L and Zhang Y: Identification and validation of a
ferroptosis-related long non-coding RNA (FRlncRNA) signature to
predict survival outcomes and the immune microenvironment in
patients with clear cell renal cell carcinoma. Front Genet.
13:7878842022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li H, Han D, Hou Y, Chen H and Chen Z:
Statistical inference methods for two crossing survival curves: A
comparison of methods. PLoS One. 10:e01167742015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48:W509–W514. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Finotello F, Mayer C, Plattner C,
Laschober G, Rieder D, Hackl H, Krogsdam A, Loncova Z, Posch W,
Wilflingseder D, et al: Molecular and pharmacological modulators of
the tumor immune contexture revealed by deconvolution of RNA-seq
data. Genome Med. 11:342019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Becht E, Giraldo NA, Lacroix L, Buttard B,
Elarouci N, Petitprez F, Selves J, Laurent-Puig P, Sautès-Fridman
C, Fridman WH and de Reyniès A: Estimating the population abundance
of tissue-infiltrating immune and stromal cell populations using
gene expression. Genome Biol. 17:2182016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Racle J, de Jonge K, Baumgaertner P,
Speiser DE and Gfeller D: Simultaneous enumeration of cancer and
immune cell types from bulk tumor gene expression data. Elife.
6:e264762017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bradburn MJ, Clark TG, Love SB and Altman
DG: Survival analysis part II: Multivariate data analysis-an
introduction to concepts and methods. Br J Cancer. 89:431–436.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhou N and Bao J: FerrDb: A manually
curated resource for regulators and markers of ferroptosis and
ferroptosis-disease associations. Database (Oxford).
2020:baaa0212020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Koppula P, Zhuang L and Gan B: Cystine
transporter SLC7A11/xCT in cancer: Ferroptosis, nutrient
dependency, and cancer therapy. Protein Cell. 12:599–620. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yang XZ, Cheng TT, He QJ, Lei ZY, Chi J,
Tang Z, Liao QX, Zhang H, Zeng LS and Cui SZ: LINC01133 as ceRNA
inhibits gastric cancer progression by sponging miR-106a-3p to
regulate APC expression and the Wnt/β-catenin pathway. Mol Cancer.
17:1262018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu S and Xi X: LINC01133 contribute to
epithelial ovarian cancer metastasis by regulating miR-495-3p/TPD52
axis. Biochem Biophys Res Commun. 533:1088–1094. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Song Z, Zhang X, Lin Y, Wei Y, Liang S and
Dong C: LINC01133 inhibits breast cancer invasion and metastasis by
negatively regulating SOX4 expression through EZH2. J Cell Mol Med.
23:7554–7565. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Feng Y, Qu L, Wang X and Liu C: LINC01133
promotes the progression of cervical cancer by sponging miR-4784 to
up-regulate AHDC1. Cancer Biol Ther. 20:1453–1461. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang D, Zhang Y and Sun X: LINC01133
promotes the progression of cervical cancer via regulating
miR-30a-5p/FOXD1. Asia Pac J Clin Oncol. 17:253–263. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Weng YC, Ma J, Zhang J and Wang JC: Long
non-coding RNA LINC01133 silencing exerts antioncogenic effect in
pancreatic cancer through the methylation of DKK1 promoter and the
activation of Wnt signaling pathway. Cancer Biol Ther. 20:368–380.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Huang CS, Chu J, Zhu XX, Li JH, Huang XT,
Cai JP, Zhao W and Yin XY: The C/EBPβ-LINC01133 axis promotes cell
proliferation in pancreatic ductal adenocarcinoma through
upregulation of CCNG1. Cancer Lett. 421:63–72. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Jiang X, Guan J, Xu Y, Ren H, Jiang J,
Wudu M, Wang Q, Su H, Zhang Y, Zhang B, et al: Silencing of CASC8
inhibits non-small cell lung cancer cells function and promotes
sensitivity to osimertinib via FOXM1. J Cancer. 12:387–396. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yang B, Gu B, Zhang J, Xu L and Sun Y:
CASC8 lncRNA promotes the proliferation of retinoblastoma cells
through downregulating miR34a methylation. Cancer Manag Res.
12:13461–13467. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Cui Z, Gao M, Yin Z, Yan L and Cui L:
Association between lncRNA CASC8 polymorphisms and the risk of
cancer: A meta-analysis. Cancer Manag Res. 10:3141–3148. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sang Y, Gu H, Chen Y, Shi Y, Liu C, Lv L,
Sun Y and Zhang Y: Long non-coding RNA CASC8 polymorphisms are
associated with the risk of esophageal cancer in a Chinese
population. Thorac Cancer. 11:2852–2857. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang Y, Yang Y, Wang Y, Li X, Xiao Y and
Wang W: High cancer susceptibility candidate 8 expression is
associated with poor prognosis of pancreatic adenocarcinoma:
Validated analysis based on four cancer databases. Front Cell Dev
Biol. 8:3922020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wen D, Huang Z, Li Z, Tang X, Wen X, Liu J
and Li M: LINC02535 co-functions with PCBP2 to regulate DNA damage
repair in cervical cancer by stabilizing RRM1 mRNA. J Cell Physiol.
235:7592–7603. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wu J, Gao L, Chen H, Zhou X, Lu X and Mao
Z: LINC02535 promotes cell growth in poorly differentiated gastric
cancer. J Clin Lab Anal. 35:e238772021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Jiang L, Kon N, Li T, Wang SJ, Su T,
Hibshoosh H, Baer R and Gu W: Ferroptosis as a p53-mediated
activity during tumour suppression. Nature. 520:57–62. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Liu J, Zhang C, Wang J, Hu W and Feng Z:
The regulation of ferroptosis by tumor suppressor p53 and its
pathway. Int J Mol Sci. 21:83872020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Kang R, Kroemer G and Tang D: The tumor
suppressor protein p53 and the ferroptosis network. Free Radic Biol
Med. 133:162–168. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kotschi S, Jung A, Willemsen N, Ofoghi A,
Proneth B, Conrad M and Bartelt A: NFE2L1-mediated proteasome
function protects from ferroptosis. Mol Metab. 57:1014362022.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chen X, Li J, Kang R, Klionsky DJ and Tang
D: Ferroptosis: Machinery and regulation. Autophagy. 17:2054–2081.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yao X, Li W, Fang D, Xiao C, Wu X, Li M
and Luo Z: Emerging roles of energy metabolism in ferroptosis
regulation of tumor cells. Adv Sci (Weinh). 8:e21009972021.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Gleitze S, Paula-Lima A, Nunez MT and
Hidalgo C: The calcium-iron connection in ferroptosis-mediated
neuronal death. Free Radic Biol Med. 175:28–41. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Martin-Sanchez D, Ruiz-Andres O, Poveda J,
Carrasco S, Cannata-Ortiz P, Sanchez-Niño MD, Ortega MR, Egido J,
Linkermann A, Ortiz A and Sanz AB: Ferroptosis, but not
necroptosis, is important in nephrotoxic folic acid-induced AKI. J
Am Soc Nephrol. 28:218–229. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Morrison AH, Byrne KT and Vonderheide RH:
Immunotherapy and prevention of pancreatic cancer. Trends Cancer.
4:418–428. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Gajewski TF, Schreiber H and Fu YX: Innate
and adaptive immune cells in the tumor microenvironment. Nat
Immunol. 14:1014–1022. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wang W, Green M, Choi JE, Gijón M, Kennedy
PD, Johnson JK, Liao P, Lang X, Kryczek I, Sell A, et al: CD8(+) T
cells regulate tumour ferroptosis during cancer immunotherapy.
Nature. 569:270–274. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Cristescu R, Mogg R, Ayers M, Albright A,
Murphy E, Yearley J, Sher X, Liu XQ, Lu H, Nebozhyn M, et al:
Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based
immunotherapy. Science. 362:eaar35932018. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Chen D, Gao W, Zang L, Zhang X, Li Z, Zhu
H and Yu X: Ferroptosis-related IncRNAs are prognostic biomarker of
overall survival in pancreatic cancer patients. Front Cell Dev
Biol. 10:8197242022. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Shi X, Zhao Y, He R, Zhou M, Pan S, Yu S,
Xie Y, Li X, Wang M, Guo X and Qin R: Three-lncRNA signature is a
potential prognostic biomarker for pancreatic adenocarcinoma.
Oncotarget. 9:24248–24259. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wei C, Liang Q, Li X, Li H, Liu Y, Huang
X, Chen X, Guo Y and Li J: Bioinformatics profiling utilized a nine
immune-related long noncoding RNA signature as a prognostic target
for pancreatic cancer. J Cell Biochem. 120:14916–14927. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhang Z, Zeng X, Wu Y, Liu Y, Zhang X and
Song Z: Cuproptosis-related risk score predicts prognosis and
characterizes the tumor microenvironment in hepatocellular
carcinoma. Front Immunol. 13:9256182022. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Guo Y, Qu Z, Li D, Bai F, Xing J, Ding Q,
Zhou J, Yao L and Xu Q: Identification of a prognostic
ferroptosis-related lncRNA signature in the tumor microenvironment
of lung adenocarcinoma. Cell Death Discov. 7:1902021. View Article : Google Scholar : PubMed/NCBI
|