Introduction
Osteoporosis is a progressive bone disease
characterized by low bone mass and the prevalence of osteoporosis,
based on 343,704 participants from 37 countries, was 19.7%
(1,2). It can result in reduced bone
strength, consequently increasing the risk of fracture.
Osteoporosis is age-related and increasingly recognized as a major
public health concern (3). The
consequences of fracture can be serious, leading to morbidity and
mortality of the patients, and osteoporotic fracture also increases
the risk of subsequent fracture, but the effect of these subsequent
fractures on mortality risk has not been systematically studied
(4). Osteoporosis is a
multifactorial disease caused by endogenous and exogenous factors
such as age, low body mass, diseases disturbing the bone
metabolism, long term medication and lack of physical exercise.
Reduction in endogenous bone marrow only mesenchymal stem cells
(BMSCs), such as proliferation and differentiation, is considered
as one of the causes. The activity of BMSCs and their
microenvironment may be disturbed in patients with osteoporosis
(5,6).
N6-methyl adenosine (m6a) is
an RNA modification method widely found in eukaryotic mRNAs and
long non-coding RNAs (7). The
level of RNA m6a modification is regulated by both
methyltransferase and demethylase. m6a shows dynamic
changes in different tissues and pathological processes (7). It participates in the formation and
development of various diseases (8–10).
For example, Clancy et al (9) found that knocking down the expression
of fat mass and obesity associated (FTO) in adipocytes
significantly increased the expression level of m6a. FTO
was negatively correlated with m6a expression levels.
m6a is involved in precursor cell differentiation by
regulating the runt-related transcription factor (RUNX)1T1 shearing
process. Baylin et al (8)
verified that the AlkB homolog 5, RNA demethylase
(ALKBH5)-m6a-p53 signaling pathway served an important
role in spermatogenesis and apoptosis in mouse testis tissue and
sperm cells; Alkbh5 deficiency leads to aberrant spermatogenesis
and apoptosis in mouse testes. ALKBH5 has been shown to regulate
homeobox protein NANOG (NANOG) protein and mRNA expression in
breast stem cells by m6a modification (10). Next-generation sequencing was used
to elucidate the human epigenome and the dysregulation in diseases
(8); the association between
epigenetic abnormalities and mutations in genes regulating DNA
methylation has been revealed (8).
Epigenetic alterations are novel targets in the development of
specific biomarkers of diseases. However, the effects of
m6a modification on osteoporosis remain to be
elucidated.
Methyltransferase-like 14 (METTL14) is the central
component of the m6a methylated transferase complex,
which is involved in the dynamic reversible process of
m6a modification. METTL14 catalyzes m6a
methylation on mRNA or non-coding RNA to regulate gene expression
and cell phenotypes (11). The
present study found that methyltransferase-like 14 (METTL14) and
m6a modification levels were significantly upregulated
in osteoporosis samples. METTL14 regulated the proliferation of
BMSCs through the m6a/miR-873 signaling pathway.
Therefore, these findings may provide guidance for novel treatment
of osteoporosis.
Materials and methods
Patient tissues and cell cultures
Bone samples were obtained from patients
(male:female=1:1; age range 50–75) who underwent hip replacement in
Tangdu Hospital, Air Force Military Medical University, Xi'an,
Shaanxi; the cohort included 36 patients with osteoporosis and 36
healthy donors. Patients were recruited between September 2018 and
March 2021. There was no significant difference in age and sex
between the two groups. Normal donors with normal bone density had
a T-score ≥-1.0 and patients with osteoporosis had a T score ≤-2.5
(3). Informed consent forms were
signed by the patients and healthy donors, and all research
protocols were reviewed and approved by the Medical Ethics
Committee of Tangdu Hospital (approval no. 202109616). The present
study was performed following the guidelines of the Declaration of
Helsinki. BMSCs were isolated from femoral neck fracture when the
patients or healthy donors underwent hip replacement. Cells were
cultured with 1X mesenchymal stem cell medium (MSCM) + L-glutamine
(Beijing Yuhengfeng Technology Co., Ltd.) supplemented with 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.) and cultured at 37°C in a
5% CO2 incubator. When cells reached 90% confluence,
they were passaged by washing with PBS, followed by dissociation
with 0.25% Trypsin and 0.02% EDTA.
Reverse transcription-quantitative PCR
(RT-qPCR) and cell transfection
Total RNA was extracted from patient samples and
BMSCs (1×106), and cDNA was synthesized using a
PrimeScript 1st strand cDNA Synthesis kit (Takara Bio, Inc.). RNA
extraction, cDNA synthesis and qPCR were performed according to the
manufacturer's protocols. NanoDrop 2000 (Thermo Fisher Scientific,
Inc.) was used to measure the level and quality of RNA. The Power
SYBRGreen PCR Master Mix (Thermo Fisher Scientific, Inc.) was used
for qPCR. The forward and reverse primer pairs were as follows:
Proliferating cell nuclear antigen (PCNA), Forward
5′-TGCTCTGAGGTACCTGAACT-3′ and Reverse 5′-TGCTTCCTCATCTTCAATCT-3′;
osteocalcin, Forward 5′-CGCTACCTGTATCAATGGCTGG-3′ and Reverse
5′-CTCCTGAAAGCCGATGTGGTCA-3′; bone γ-carboxyglutamate protein 2
(Bglap2), Forward 5′-TAGTGAACAGACTCCGGCGCT-3′ and Reverse
5′-TGTAGGCGGTCTTCAAGCCAT-3′; Sp7, Forward
5′-GGCTTTTCTGCGGCAAGAGGTT-3′ and Reverse
5′-CGCTGATGTTTGCTCAAGTGGTC-3′; runt-related transcription factor 2
(Runx2), Forward 5′-TAAAGTGACAGTGGACGGTCCC-3′ and Reverse
5′-TGCGCCCTAAATCACTGAGG-3′; sterol regulatory element-binding
transcription factor 1 (SREBP1), Forward
5′-GACGGGGATCCCTCAGCTCAGAGCCGTGGT-3′ and Reverse
5′-GACGGCAAGCTTTTAGCTTTTGTGAGCGGCATTTC-3′; adiponectin, Forward
5′-CAGGCCGTGATGGCAGAGATG-3′ and Reverse
5′-GGTTTCACCGATGTCTCCCTTAG-3′; CCATT/enhancer binding protein α
(C/EBPα), Forward 5′-CCACGCCTGTCCTTAGAAAG-3′ and Reverse
5′-CAGTTTTTCCAATGTCACCCCTAC-3′; peroxisome proliferator-activated
receptor γ (PPARγ), Forward 5′-AGCCTGCGAAAGCCTTTTGGTG-3′ and
Reverse 5′-GGCTTCACATTCAGCAAACCTGG-3′; METLL14 Forward
5′-CTGAAAGTGCCGACAGCATTGG-3′ and Reverse
5′-CTCTCCTTCATCCAGATACTTACG-3′; METLL3 Forward
5′-AGCCTTCTGAACCAACAGTCC-3′ and Reverse 5′-CCGACCTCGAGAGCGAAAT-3′;
WT1-associated protein, Forward 5′-GCAACAACAGCAGGAGTCTGCA-3′ and
Reverse 5′-CTGCTGGACTTGCTTGAGGTAC-3′; METTL4 Forward
5′-CTTGGTCTGTGGAGGTAGTTGC-3′ and Reverse
5′-CCAGTATAAGACCTTCGTAGGGC-3′; ALKBH5 Forward
5′-GCCTATTCGGGTGTCGGAAC-3′ and Reverse 5′-CTGAGGCCGTATGCAGTGAG-3′;
FTO Forward 5′-ACTTGGCTCCCTTATCTGACC-3′ and Reverse
5′-TGTGCAGTGTGAGAAAGGCTT-3′; YTH m6a RNA-binding protein
2, Forward 5′-GGTTCTGTGCATCAAAAGGATGG-3′ and
5′-CCAAAGAATAGGAAAAGCCAATGG-3′; GAPDH, Forward
5′-CAGTGCCAGCCTCGTCCCGTAGA-3′ and Reverse
5′-CTGCAAATGGCAGCCCTGGTGAC-3′. The 2−ΔΔCq method was
used to calculate all experimental data (11). The program used for qPCR was: 95°C
for 5 min, 45 cycles of 95°C for 15 sec, 60°C for 20 sec and 72°C
for 10 sec.
The construction and design of small interfering
(si)RNA targeting METTL14 (si-METTL14; 5′-GCAGCACCUCGGUCAUUUA-3′)
and the negative control (si-NC; 5′-AAGCTTCATAAGGCGCATAGC-3′),
miR-873 mimic (5′-GCAGGAACUUGUGAGUCUCCU-3′) and the control
(5′-UUCUCCGAACGUGUCACGUTT-3′), miR-873 inhibitor
(5′-AGGAGACUCACAAGUUCCUGCTT-3′) and the control
(5′-CAGUACUUUUGUGUAGUACAA-3′) were completed by Shanghai GenePharma
Co., Ltd. Briefly, 25 nM plasmids were used for each transfection.
Transfections of 5×105 cells were performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C for 12 h. At 12 h post-transfection,
culture medium was replenished with fresh culture medium containing
10% FBS. Each experiment was performed three times.
Cell Counting Kit-8 (CCK-8) assay
BMSCs in the logarithmic growth phase
(5×104/well)were seeded into a 96-well culture plate. In
brief, cell viability was determined on days 1, 2, 3 and 4; 10 µl
CCK-8 solution (Dojindo Laboratories, Inc.) was added to the cells,
and the cells were incubated at 37°C for additional 2 h.
Subsequently, the absorbance at 450 nm was detected using a
microplate reader (680; Bio-Rad Laboratories, Inc.).
Western blotting
The total protein samples were extracted from
transfected cells using RIPA buffer (Beyotime Institute of
Biotechnology, Shanghai, China). A BCA kit (Pierce; Thermo Fisher
Scientific, Inc.) was used to determine the protein concentration.
Equal amounts (~30 µg) of samples were separated using 10% SDS-PAGE
gels, and protein were then transferred to nitrocellulose membrane
(MilliporeSigma). The membranes were blocked with PBS containing 5%
non-fat milk for 1 h at room temperature, followed with incubation
with primary antibodies against PCNA (1:1,000; cat. no. ab92552;
Abcam), osteocalcin (1:2,000; cat. no. sc-376835; Santa Cruz),
Bglap (1:1,000; cat. no. MBS3223783; MyBioSource, Inc.), Sp7
(1:2,000; cat. no. ab209484; Abcam), Runx2 (1:2,000; cat. no.
ab92336; Abcam), SREBP1 (1:1,000; cat. no. ab28481; Abcam),
adiponectin (1:2,000; cat. no. ab181281; Abcam), C/EBPα, (1:2,000;
cat. no. ab40761; Abcam), PPARγ (1:2,000; cat. no. ab59256; Abcam)
or β-actin (1:1,000; cat. no. ab8226; Abcam) in a cold room at 4°C
overnight. Subsequently, the membranes were incubated with
horseradish peroxidase-conjugated horse anti-mouse (1:2,000; cat.
no. 7076; Cell Signaling Technology, Inc.) or goat anti-rabbit IgG
(1:2,000; cat. no. 7074; Cell Signaling Technology, Inc.) at room
temperature for 1 h. Protein bands were visualized using an ECL kit
(Pierce; Thermo Fisher Scientific, Inc). The density of bands was
semi-quantified using ImageJ (version 1.44; National Institutes of
Health).
Cell cycle distribution
Cells were seeded onto 6-well plates
(4×105 cells/well). The cell suspension was spun down
using a low-speed centrifugation (120 × g at 4°C for 5 min. Cell
pellets were washed and re-suspended in PBS, fixed with pre-chilled
ethanol (70%) and left at 4°C for two days. Prior to being
subjected to flow cytometry, cells were lysed using ice-cold
methanol, centrifuged at 180 × g at 4°C for 5 min and then
re-suspended in propidium iodide (PI; MilliporeSigma) staining
buffer containing PI (50 µl/ml) and RNase A (250 µl/ml). Cell cycle
distribution was evaluated using a flow cytometer (BD Biosciences),
and the results were analyzed by FlowJo version 7.6 software
(FlowJo LLC).
Osteogenic differentiation of
BMSCs
For osteogenic differentiation of BMSCs, when the
cells reached 70% confluence, the culture medium was replenished
with fresh medium containing L-DMEM, 10% FBS, 100 nM dexamethasone,
10 mM β-glycerophosphate and 250 mM L-ascorbic acid (Gibco; Thermo
Fisher Scientific, Inc.). The cells were cultured for 22 days at
37°C and the medium was replenished every two days. Subsequently,
cells were fixed using 4% paraformaldehyde at room temperature for
30 min and stained using 0.1% Alizarin Red S (cat. no. A5533;
MilliporeSigma) at room temperature until obvious calcium deposits
appeared.
Adipogenic differentiation of
BMSCs
BMSCs were seeded in a 6-well plate
(4×105 cells/well). When the cells reached 70%
confluence, they were incubated with adipocyte-inducing medium
comprising L-DMEM, 10% FBS, 1.0 mmol/l dexamethasone, 0.5 mmol/l
isobutyl-methylxanthine and 10 mg/l insulin (all from
MilliporeSigma) at 37°C for 12 days. Cells were then stained with
0.3% Oil Red O (cat. no. O8018; Beijing Solarbio Science &
Technology Co., Ltd.) at room temperature for 1 h.
Co-immunoprecipitation (Co-IP)
A Pierce Co-Immu-noprecipitation kit (Thermo Fisher
Scientific) was used. Cell lysates were incubated with protein A
agarose with gentle shaking at 4°C for 10 min. To remove
non-specific binding, the samples were centrifuged at 4°C at 1,000
× g for 15 min. The lysates with equal amounts (1 mg) of total
protein were incubated with 2 µg rabbit anti-METTL14 antibody (cat.
no. HPA038002; MilliporeSigma) and a negative control rabbit IgG
antibody at 4°C overnight. Subsequently, the samples were incubated
with protein A/G agarose at 4°C for another 2 h. After
centrifugation at 1,000 × g at 4°C for 10 min, the agarose was
rinsed using cold lysis buffer, and proteins were eluted using
Laemmli buffer (Bio-Rad Laboratories, Inc.) and subjected to 10%
SDS-PAGE. The mutual binding between different proteins by the
complexes bound after immunoprecipitation was detected by western
blotting analysis according to the aforementioned protocol.
RNA m6a RIP
Total RNA was extracted from 1×106
isolated BMSCs of osteoporosis group and controls following the
transfection with si-METTL14 or si-NC. m6a -IP-qPCR was
performed following a protocol with minor modifications. Total RNAs
were extracted using TRIzol® (Thermo Fisher Scientific,
Inc.) and purified through GenElute mRNA Miniprep kit
(MilliporeSigma). All RNAs were placed in a sonicator at 30 Hz for
3 min at room temperature and treated with DNase I; the samples
were sonicated for 10 sec. In total, 5 µg of purified mRNAs were
fragmented into 200–300 nt fragments by incubation in RNA
fragmentation reagent (Thermo Fisher Scientific, Inc.) at 94°C for
30 sec and then stopped with stop solution. Of the fragmented
mRNAs, 10% were kept as input. The remaining mRNAs were incubated
with 6.25 µg m6a-specific antibody (cat. no. 202003;
Synaptic Systems GmbH) or rabbit IgG (Beyotime Institute of
Biotechnology) in IP buffer [50 mM Tris-HCl (pH 7.4), 750 mM NaCl,
0.5% Igepal CA-630, 0.4 U/µl RNasin (Promega Corporation)] for 2 h
at 4°C. Then the mixture was incubated with 25 µl Dynabeads Protein
A (Thermo Fisher Scientific, Inc.) for another 2 h at 4°C The
antibodies were bound to the magnetic beads and incubated in 100 µl
RIP Wash buffer (Sigma Aldrich) at room temperature for 30 min.
After extensive washing, the bound mRNAs were eluted with 6.7 mM
N6-methyladenosine (Sigma) in IP buffer and then recovered with
ethanol precipitation. The immunoprecipitated mRNAs and input mRNAs
were processed as in RT-qPCR aforementioned to evaluate the
relative expression levels of primary (pri)-miRNA.
Biotin-labeled pri-let-7e probe was generated by
in vitro transcription followed by biotin-labeling (Thermo
Fisher Scientific, Inc.). A total of 1 pmol of biotin-labeled RNA
probes was incubated with increasing concentrations of purified
recombinant protein expressed in E. coli in reaction buffer
[20 mM HEPES-KOH (pH 8.0), 150 mM KCl, 1.5 mM MgCl2, 0.2
mM EDTA, 0.1% (w/v) Triton X-100] at 4°C for 1 h. After the
reaction, the samples mixed with loading buffer [10 mM Tris-HCl, 3%
(w/v) sucrose and dyes] were loaded on 8% (w/v) native gel and run
at a constant voltage of 100 V. After electrophoresis, the probe
was transferred onto hybond-N+ membrane and detected using
Chemiluminescent Nucleic Acid Detection Module Kit (Thermo Fisher
Scientific, Inc.).
cDNA was synthesized using a PrimeScript 1st strand
cDNA Synthesis kit (Takara Bio, Inc.). RNA extraction, cDNA
synthesis and qPCR were performed according to the manufacturer's
protocols. NanoDrop 2000 (Thermo Fisher Scientific, Inc.) was used
to measure the level and quality of RNA. The Power SYBRGreen PCR
Master Mix (Thermo Fisher Scientific, Inc.) was used for qPCR. The
forward and reverse primer pairs were as follows: miR-873, Forward
5′-GCAGGAACUUGUGAGUCUCCU-3′ and Reverse
5′-AGGAGACUCACAAGUUCCUGC-3′; let-7e Forward
5-′GGGTGAGGTAGGAGGTTGT-3′ and Reverse 5′-CAGTGCGTGTCGTGGAGT-3′;
GAPDH, Forward 5′-CAGTGCCAGCCTCGTCCCGTAGA-3′ and Reverse
5′-CTGCAAATGGCAGCCCTGGTGAC-3′. The 2-ΔΔCq method was
used to calculate all experimental data (12). The program used for qPCR was: 95°C
for 5 min, 45 cycles of 95°C for 15 sec, 60°C for 20 sec and 72°C
for 10 sec.
Statistical analysis
SPSS v15.0 statistical software (SPSS, Inc.) was
used to analyze the experimental data. All data are expressed as
the mean ± SD. Differences between and among groups were
respectively analyzed using t-test or one-way ANOVA followed by
Tukey's post-hoc test. To investigate the linear correlations
between two variables, Pearson's correlation test was performed.
P<0.05 was considered to indicate a statistically significant
difference.
Results
m6a modification levels are
significantly upregulated in osteoporosis samples and downregulated
METTL14 significantly inhibits m6a modification levels
in BMSCs
The present study compared the m6a
modification levels in osteoporotic and healthy bone samples using
RT-qPCR, and the results indicated that the relative levels of
m6a modification in osteoporosis tissues were
significantly higher compared with the controls (Fig. 1A). To further explore the cause of
the higher levels of m6a modification, the relative
levels of methyltransferases and demethylases in the two groups
were compared. The data revealed that the relative content of
METTL14 in osteoporosis samples was notably higher than other
enzymes such as METTL3, WTAP, METTL4,ALKBH5, FTO and YTHDF2
compared with normal bone samples (Fig. 1B). Furthermore, the relative
expression levels of m6a and METTL14 in BMSCs were
significantly increased in osteoporosis BMSCs (Fig. 1C and D). In addition, the protein
levels of METTL14 were increased in osteoporosis BMSCs (Fig. 1E). The m6a content in
total RNA and expression of METTL14 was reduced in cells treated
with si-METTL14 (Fig. 1F and G).
Co-IP experiments were used to further verify the relationship
between METTL14 and m6a. The results showed that METTL14
could bind to the DiGeorge syndrome critical region 8 (DGCR8)
protein in BMSCs. Following treatment with Rnase, the binding
effects between METTL14 and DGCR8 was inhibited (Fig. 1H). Silencing of METTL14
significantly reduced the binding effects of METTL14 and DGCR8 and
the level of m6a modification (Fig. 1I).
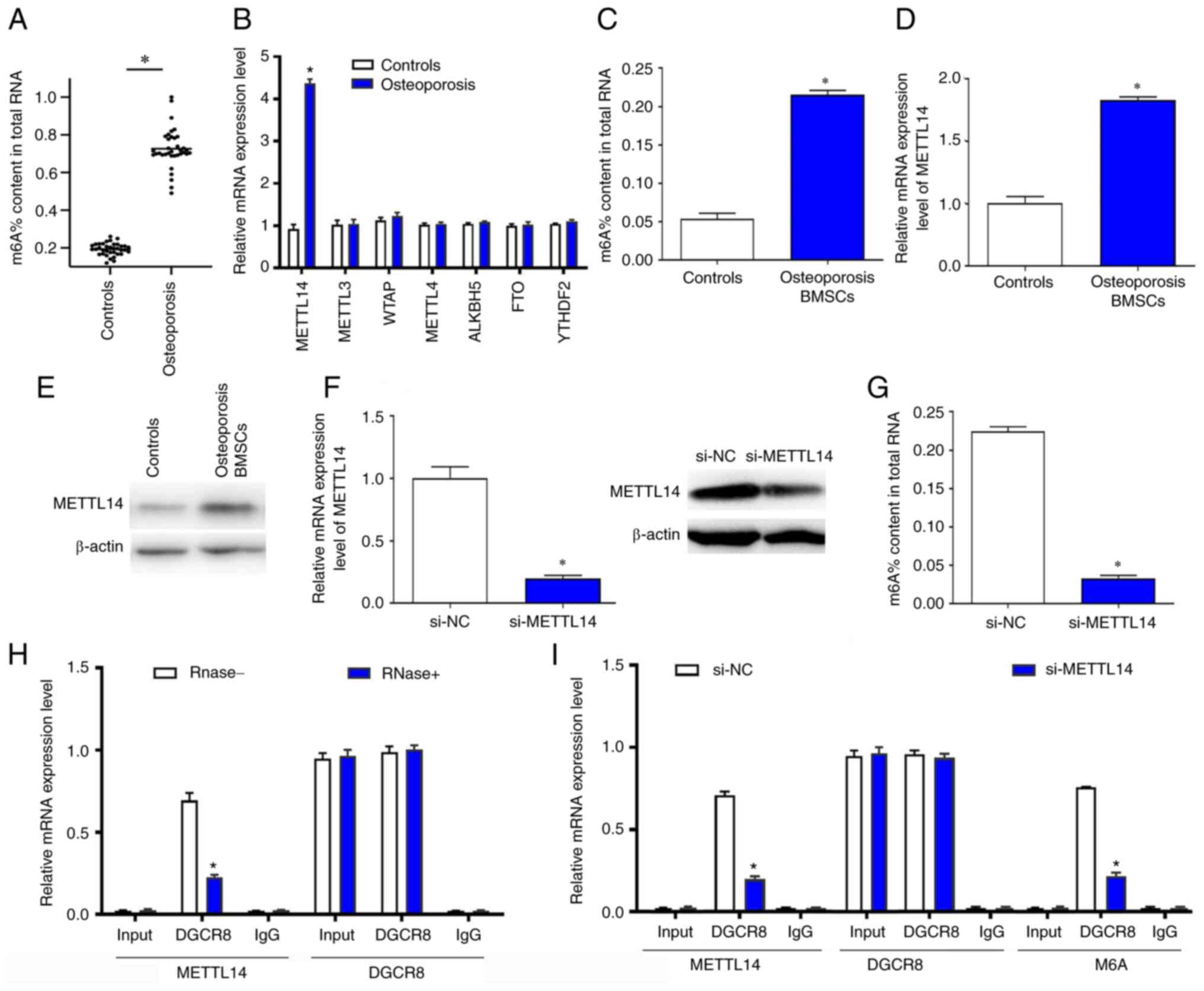 | Figure 1.m6a modification levels
were significantly overexpressed in patients with osteoporosis and
downregulated METTL14 significantly inhibited m6a
modification levels in BMSCs. (A) m6a content in healthy
controls and patients with osteoporosis was detected by RT-qPCR
assay. (B) METTL14 and other methyltransferase and demethylase mRNA
expression levels in the two experimental groups were measured
using RT-qPCR. (C) m6a content in total RNA of
osteoporosis BMSCs after transfection with si-NC and si-METTL14
were detected. (D) METTL14 relative expression levels were
evaluated in BMSCs. (E) The protein expression levels of METTL14
was evaluated in osteoporosis BMSCs by western blotting. (F) The
mRNA levels of METTL14 was decreased in cells transfected with
si-METTL14. (G) The protein levels of METTL14 and m6a%
content in total RNA was reduced in cells treated with si-METTL14.
(H) The relationship between METTL14 and DGCR8 was verified by
Co-IP assay. (I) Co-IP was used to explore the relationship between
METTL14 and m6a modification. The data are presented as the mean ±
SD; *P<0.05 vs. control BMSCs or si-NC. ALKBH5, AlkB homolog 5,
RNA demethylase; BMSCs, bone marrow mesenchymal stem cells; Co-IP,
co-immunoprecipitation; DGCR8, DiGeorge syndrome critical region 8;
FTO, fat mass and obesity associated; m6a,
n6-methyl-adenosine; METTL14, methyltransferase-like 14; NC,
negative control; RT-qPCR, reverse transcription-quantitative PCR;
si, small interfering RNA. |
Silencing of METTL14 promotes the
proliferation and differentiation of BMSCs
To explore the effects of METTL14 silencing, BMSCs
were transfected with si-METTL14 and cultured for 3 days. As a
result, the proliferation of BMSCs was significantly improved
compared with si-NC group (Fig.
2A). The mRNA and protein expression levels of cell
proliferation marker PCNA were examined by RT-qPCR and western
blotting, respectively, and the data show that the expression of
PCNA in BMSCs was significantly increased following transfection
with si-METTL14 (Fig. 2B and C).
Flow cytometry was used to detect the effects of METTL14 on the
cell cycle of BMSCs. Following the transfection with si-METTL14,
the proportion of cells at S phase was increased significantly in
BMSCs compared with sh-NC, whereas the percentage of cells at the
G0/G1 phase were markedly decreased (Fig. 2D-F). In addition, the results of
Alizarin Red S and Oil Red O staining indicated that knockdown of
METTL14 promoted the differentiation of BMSCs (Fig. 2G and I). Consistent with these
findings, the expression levels of corresponding differentiation
markers osteocalcin, Bglap2, Sp7, Runx2, SREBP1, Adiponectin,
C/EBPa and PPARr were also increased in cells treated with
si-METTL14 (Fig. 2H and J).
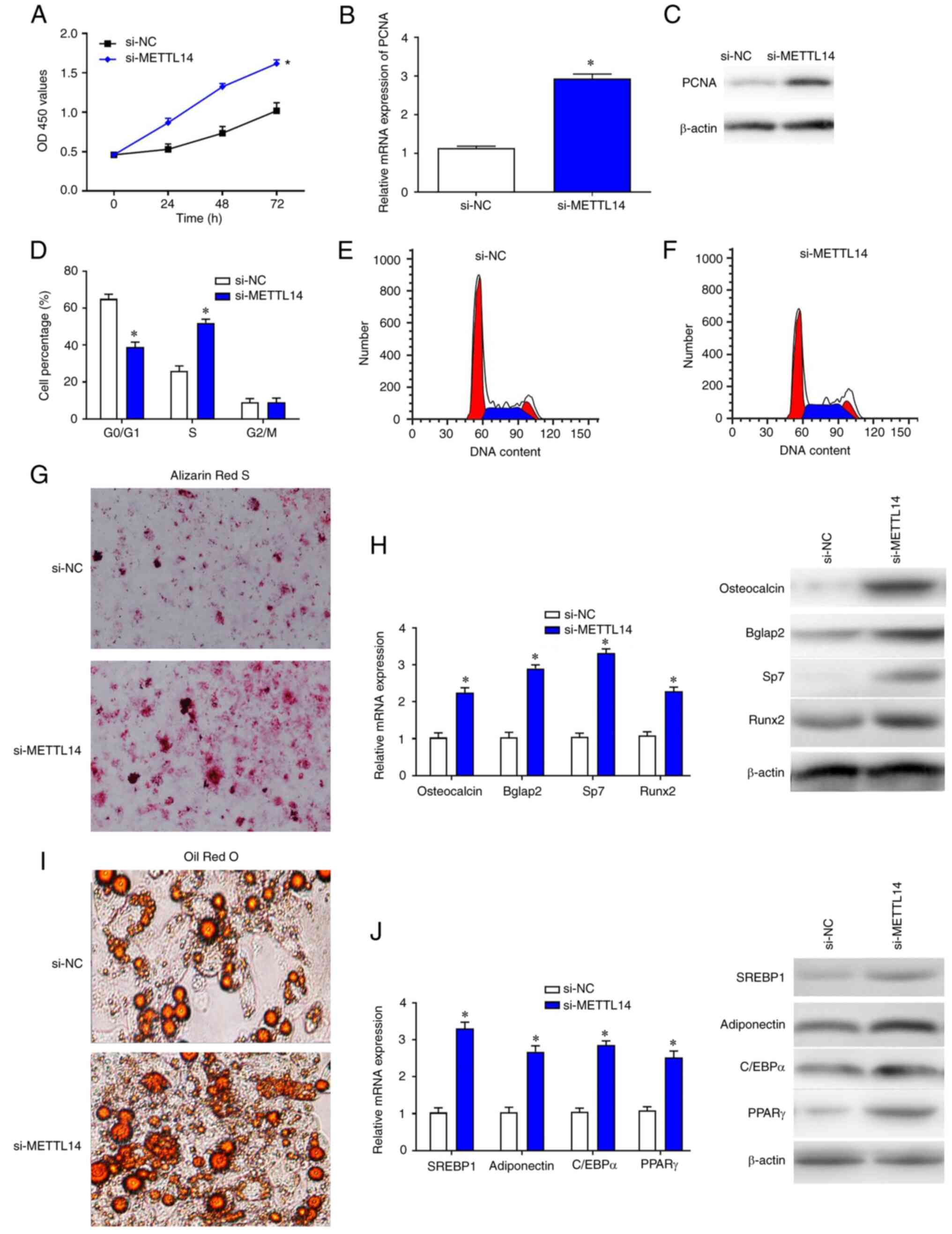 | Figure 2.METTL14 downregulation promotes BMSC
proliferation. (A) Cell Counting Kit-8 assay was used to examine
the proliferative ability of BMSCs after transfection with si-NC
and si-METTL14. (B) Reverse transcription-quantitative PCR and (C)
western blotting were used to measure the expression levels of PCNA
in transfected cells. (D-F) Flow cytometry was used to detect the
percentage of BMSCs at different phases of the cell cycle. (G and
I) The differentiation of cells was enhanced after the treatment
with si-METTL14 (magnification, ×200). (H and I) The expression of
differential-associated markers were increased in cells transfected
with si_METTL14. The data are presented as the mean ± SD;
*P<0.05 vs. si-NC. BMSCs, bone marrow mesenchymal stem cells;
C/EBPα, CCATT/enhancer binding protein α; METTL14,
methyltransferase-like 14; NC, negative control; PCNA,
proliferating cell nuclear antigen; PPARγ, peroxisome
proliferator-activated receptor γ; RunX, runt-related transcription
factor; si, small interfering RNA; SREBP1, sterol regulatory
element binding protein 1. |
METTL14/m6a modification
promotes processing of pri-miR-873 by binding to DGCR8 in
BMSCs
A study has shown that m6a modification
is involved in the processing of pri-miRNA and promotes the
conversion of pri-miRNA into mature miRNA (8). A previous study reported that,
miR-873-3p targets HDAC4 to stimulate matrix metalloproteinase-13
expression upon parathyroid hormone exposure in rat osteoblasts
(13). Furthermore, miR-873 could
affect the proliferation and migration of different types of cells
(14–16). The results of the present study
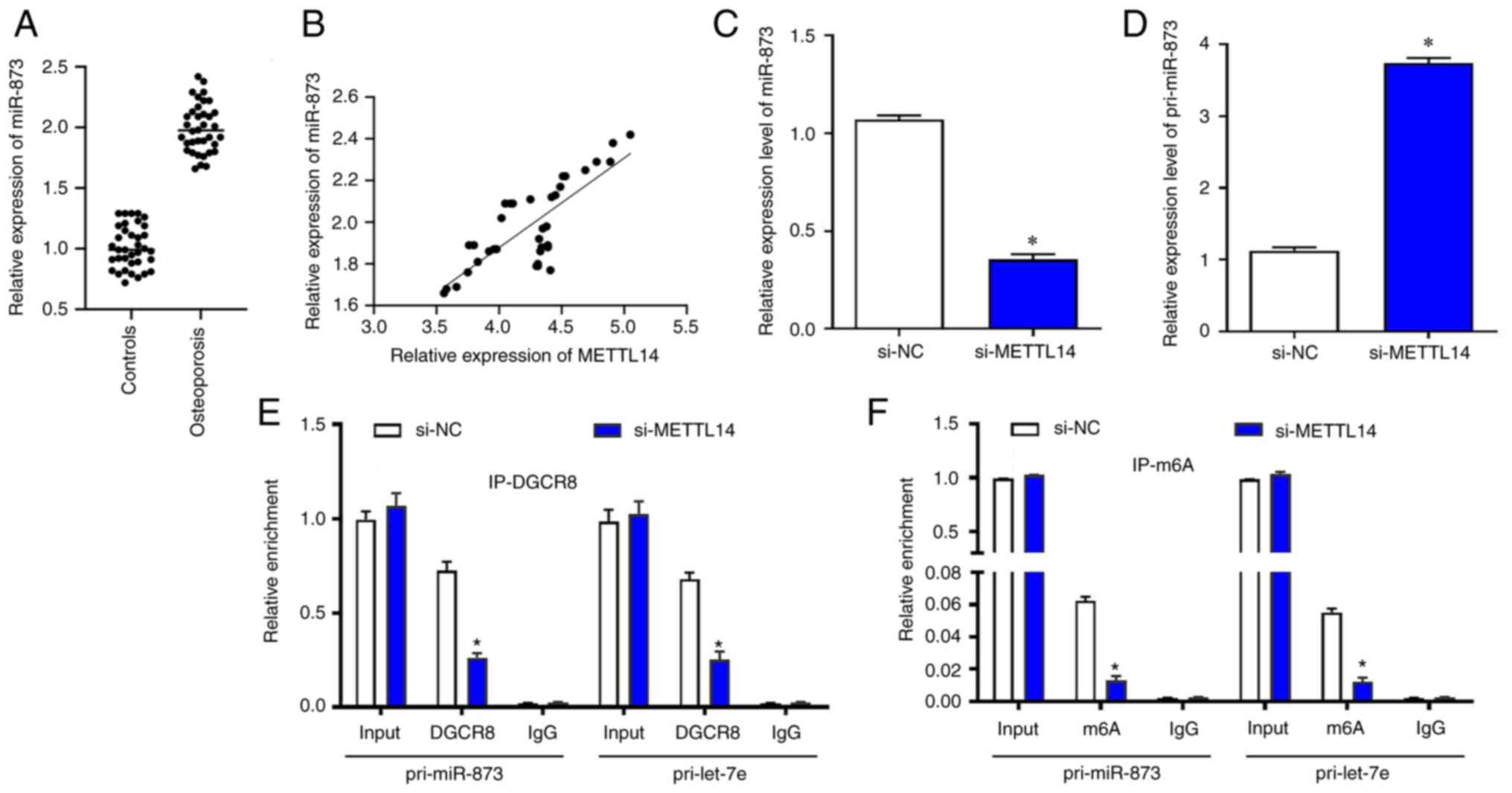
showed that the levels of miR-873 were significantly upregulated in
osteoporosis samples (r=0.589; Fig.
3A). The expression of METTL14 and miR-873 were positively
correlated in osteoporosis samples using Pearson's correlation test
(Fig. 3B). Following transfection
with si-METTL14, miR-873 was significantly reduced in BMSCs
(Fig. 3C), whereas the levels of
pri-miR-873 were significantly increased (Fig. 3D). RIP was used to verify the
relationship between METTL14/m6a modification and
pri-miR-873. The results suggested that in BMSCs transfected with
si-METTL14, the content of pri-miR-873 bound to DGCR8 was
significantly reduced (Fig. 3E).
Similarly, the content of pri-miR-873 modified by m6a
was also markedly decreased after the transfection with si-METTL14
(Fig. 3F).
Overexpression of miR-873
significantly inhibits BMSCs cell proliferation
To investigate the effects of miR-873 on BMSCs,
gain- and loss-of function experiments were performed. The
transfection efficiencies of miR-873 inhibitors and mimics were
confirmed by RT-qPCR (Fig. 4A).
CCK-8 assays were used to examine proliferation. The results showed
that the proliferation of BMSCs after transfection with
miR-873-mimics was significantly lower compared with the miR-NC
group at the 72 h timepoint (Fig.
4B). Following overexpression of miR-873, the mRNA and protein
expression levels of PCNA in BMSCs were markedly reduced compared
with miR-NC group (Fig. 4C). Flow
cytometry analysis indicated that the percentage of cells in the S
phase was markedly decreased in BMSCs transfected with
miR-873-mimics compared with miR-NC, whereas the percentage of
cells in the G0/G1 phase was significantly
increased (Fig. 4D-F).
Furthermore, the results of Alizarin Red S and Oil Red O staining
revealed that overexpression of miR-873 markedly inhibited the
differentiation of BMSCs (Fig. 4G and
I). Consistent with these findings, the mRNA and protein levels
of corresponding differentiation markers osteocalcin, Bglap2, Sp7,
Runx2, SREBP1, adioponectin, C/EBPa and PPARr were also reduced in
cells treated with miR-873 mimics (Fig. 4H and J).
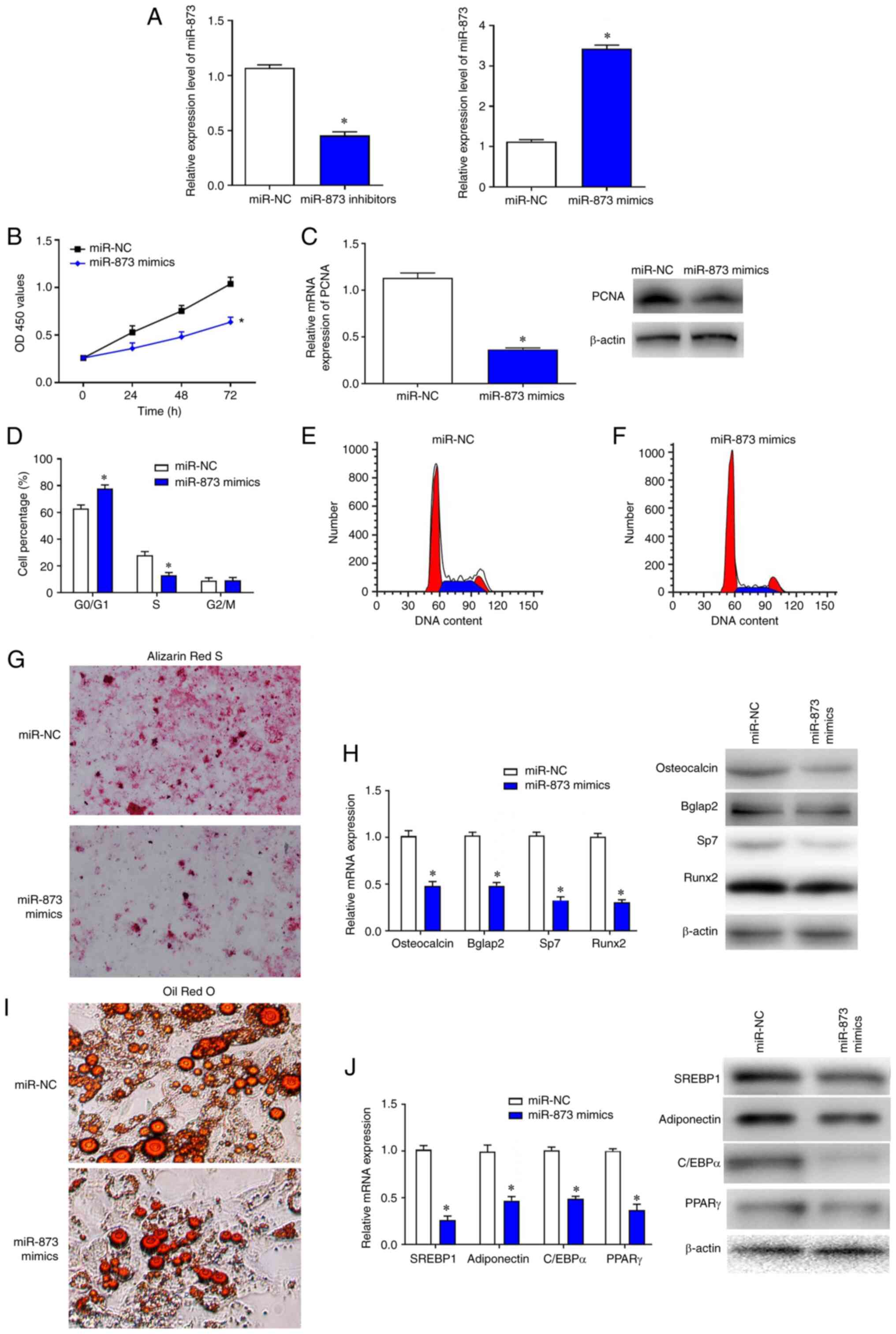 | Figure 4.Overexpression of miR-873
significantly inhibits BMSC proliferation. (A) The transfection
efficiencies of miR-873 inhibitors and mimics were confirmed by
RT-qPCR. (B) The proliferation of BMSCs was detected by Cell
Counting Kit-8 assay. (C) RT-qPCR and western blotting were used to
measure the relative expression levels of PCNA mRNA and protein,
respectively. (D) Flow cytometry was used to evaluate the
proportion of BMSCs at different phases. (E and F) The DNA contents
in miR-NC and miR-873 mimics transfected cells were evaluated. (G
and I) The differentiation of cells was inhibited by the
transfection with miR-873 mimics (magnification, ×200). (H and J)
Western blots revealed the expression of differentiation-related
markers were reduced in miR-873 mimic transfected cells. The data
are presented as the mean ± SD; *P<0.05 vs. miR-NC. Bglap2, bone
γ-carboxyglutamate protein 2; BMSCs, bone marrow mesenchymal stem
cells; C/EBPα, CCATT/enhancer binding protein α; miR, microRNA; NC,
negative control; OD, optical density; PCNA, proliferating cell
nuclear antigen; PPARγ, peroxisome proliferator-activated receptor
γ; RT-qPCR, reverse transcription-quantitative PCR; RunX,
runt-related transcription factor; SREBP1, sterol regulatory
element binding protein 1. |
METTL14/miR-873 signaling is involved
in the regulation of BMSC proliferation
To further explore the effects of the
METTL14/miR-873 signaling on BMSCs, cells were transfected with
si-NC, si-METTL14 or si-METTL14 + miR-873 mimics and cultured for 3
days. The results revealed that the proliferative ability and PCNA
mRNA expression levels, (Fig.
5A-C) in cells co-transfected with si-METTL14 + miR-873 mimics
were significantly reduced compared with those transfected with
si-METTL14 alone. In addition, there were fewer cells at the
G0/G1 stage and more cells at S stage after
the transfection with si-METTL14, which was reversed by the
treatment with miR-873 mimics (Fig.
5C). However, proliferation and PCNA expression were markedly
increased in cells co-transfected with si-METTL14 + miR-873
inhibitors compared with those transfected with si-METTL14 alone
(Fig. 5D-F). In addition, there
were fewer cells at G0/G1 stage and more
cells at S stage after the transfection with si-METTL14, which was
strengthened after the treatment with miR-873 inhibitors (Fig. 5F). The expression of corresponding
differentiation markers osteocalcin, Bglap2, Sp7, Runx2, SREBP1,
adioponectin, C/EBPa and PPARr were enhanced in cells treated the
si-METTL14, and these effects were reversed by miR-873 mimics
co-transfection and strengthened by miR-873 inhibitors (Fig. 5G).
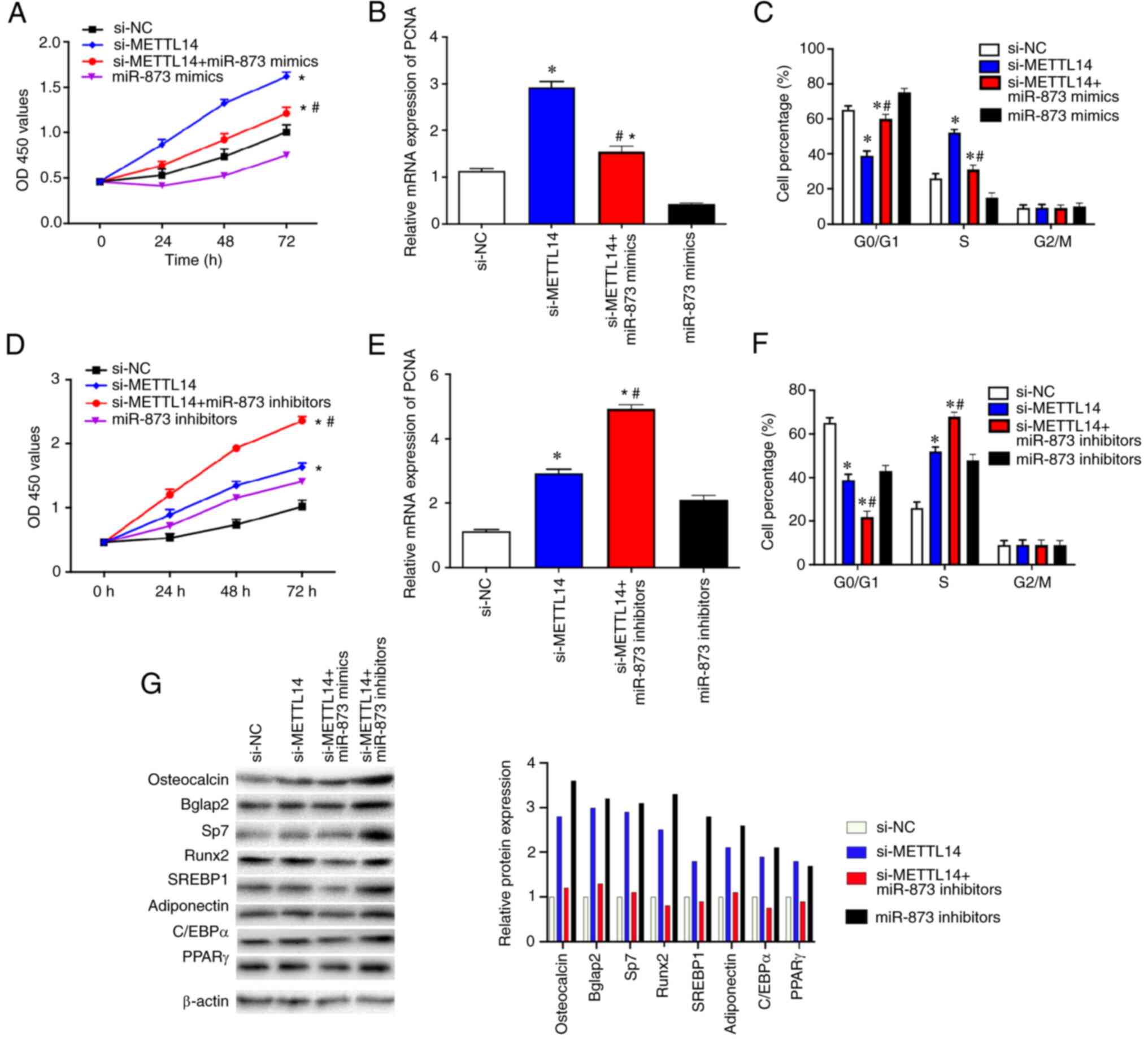 | Figure 5.METTL14/miR-873 axis serves an
important role in the proliferation of BMSCs. (A) CCK-8 assay was
used to examine the proliferation of BMSCs transfected with si-NC,
si-METTL14 or si-METTL14 + miR-873 mimics. (B) RT-qPCR was used to
determine the mRNA expression levels of PCNA in transfected BMSCs.
(C) Flow cytometry was used to investigate the cell cycle in
transfected BMSCs. (D) CCK-8 assay was used to detect the
proliferation of BMSCs after the transfection with si-NC,
si-METTL14, or si-METTL14 + miR-873 inhibitors. (E) RT-qPCR was
used to measure the PCNA mRNA expression levels in transfected
BMSCs. (F) Flow cytometry was used to investigate the cell cycle of
transfected BMSCs. (G) The expression of corresponding
differentiation markers osteocalcin, Bglap2, Sp7, Runx2, SREBP1,
adioponectin, C/EBPa and PPARr were examined by western blotting.
The data are presented as the mean ± SD; *P<0.05 vs. si-NC;
#P<0.05 vs. si-METLL14. BMSCs, bone marrow
mesenchymal stem cells; C/EBPα, CCATT/enhancer binding protein α;
METTL14, methyltransferase-like 14; miR, microRNA; NC, negative
control; OD, optical density; PCNA, proliferating cell nuclear
antigen; PPARγ, peroxisome proliferator-activated receptor γ; RNA;
RT-qPCR, reverse transcription-quantitative PCR; RunX, runt-related
transcription factor; si, small interfering SREBP1, sterol
regulatory element binding protein 1. |
Discussion
Osteoporosis is a multifactorial bone disease and is
characterized by loss of bone mass and reduced bone strength
(1). The consequences of
osteoporosis-related fracture can be life threatening (2). Impaired proliferation and
differentiation in endogenous BMSCs is considered as one of the
causes of osteoporosis (3).
RNA m6a modification was first reported
in 1974 (13). Subsequent research
found that RNA m6a modification widely exists in mouse
bovine and rabbit zygotes and exhibits dynamic and reversible
changes in different developmental stages and different tissues
(7). RNA m6a
modification is jointly regulated by methyltransferases including
METTL3, METTL14, WT1-associated protein and KIAA1429, and by
demethylases such as FTO and ALKBH5 (17–21).
The DGCR8 gene encodes a subunit of the microprocessor complex that
mediates the biogenesis of miRNAs from the pri-miRNA transcript
(9,10). It has also been reported that
METTL14/m6a modification can regulate the processing of
pri-miR-126 by binding to DGCR8 protein and serve an important role
in tumor metastasis (18).
Previous studies have shown that m6a modification can
mark pri-miRNA molecules and recognize DGCR8 molecules in a
METTL3/m6a-dependent manner, thereby participating in
the maturation of pri-miRNAs and leading to differential expression
of miRNAs in various biological processes (19,20).
The present study found that the level of
m6a modification was significantly higher in
osteoporosis samples compared with controls, and that METTL14
expression levels in the osteoporosis group were markedly
upregulated compared with other methyltransferases and
demethylases. The results of Co-IP indicated that METTL14 could
bind to DGCR8 in BMSCs. Silencing of METTL14 not only reduced the
METTL14 expression but also decreased the level of m6a
modification. These findings suggested that METTL14 regulated the
m6a methylation modification by binding to DGCR8.
Furthermore, knockdown of METTL14 significantly promoted the
proliferation of BMSCs, increased the proportion of cells at S
phase and decreased the percentage of cells at
G0/G1 phase. These results indicated that
METTL14 may affect the proliferation of BMSCs. However, the
detailed mechanisms remain unclear and require further
investigation. Consistent with the present findings, METTL14 could
regulate the proliferation of tumor cells (11). Furthermore, METTL14 suppressed the
metastatic potential of hepatocellular carcinoma by modulating
N6-methyladenosine-dependent primary microRNA processing (19).
miR-873 is an endogenous, non-coding single-stranded
RNA molecule that is involved in the occurrence and development of
various diseases. Gao et al (14) found that miR-873 was highly
expressed in lung adenocarcinoma cell lines and tissues, and the
overexpression of miR-873 could significantly facilitate the
proliferation and migration of these cells. miR-873 is also found
to inhibit colorectal cancer cell by directly targeting TAB1 and
TRAF5 (15). Recently, it has been
reported that miR-873 is significantly upregulated in congenital
heart disease tissues and can significantly inhibit the
proliferation of H9C2 cardiomyocytes (16).
The present study found that miR-873 was
significantly overexpressed in patients with osteoporosis.
Silencing of METTL14 significantly reduced the levels of
pri-miR-873 bound to DGCR8. Knockdown of METTL14 also decreased the
expression of pri-miR-873 modified by m6a in BMSCs.
Furthermore, overexpression of miR-873 inhibited the proliferation
of BMSCs, reduced the proportion of cells at S phase and increased
the percentage of cells at G0/G1 phase.
Further experiments revealed that miR-873 mimics significantly
inhibited the upregulated proliferation of BMSCs caused by
si-METTL14. In addition, miR-873 inhibitors further promoted the
proliferation of BMSCs transfected with si-METTL14. These data
suggested that METTL14 may regulate the proliferation of BMSCs
through m6a/miR-873. Consistent with the present
findings, previous studies indicate that miR-873 is involved in the
regulation of different types of cells (14–16).
However, there are some limitations in the present study. For
example, other YTH protein family members could also serve a role
in this process to recognize the m6a modification.
In conclusion, the results of the present study
showed that METTL14 and m6a modification levels were
significantly higher in bone samples from patients with
osteoporosis compared with normal individuals. METTL14 promoted the
processing of pri-miR-873 into mature miR-873 by mediating
m6a modification, thereby inhibiting the proliferation
of BMSCs. Thus, METTL14/m6a/miR-873 axis may be a novel
candidate for the treatment of osteoporosis. However, the treatment
of osteoporosis may need to be developed by the promotion of
osteoblasts by BMSCs and the inhibition of osteoclast
differentiation; these issues should be addressed in a future
study.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BL initiated this study. XD, BL, JZ, XL, KY, KR, XZ,
XB and WG conducted the experiments and data analyses. XD and BL
confirm the authenticity of all the raw data. All authors drafted
the manuscript and all authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Informed consents were signed by the patients and
healthy donors, and all research protocols were reviewed and
approved by the Medical Ethics Committee of Tangdu Hospital
(approval no. 202109616).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
BMSCs
|
bone marrow mesenchymal stem cells
|
|
DGCR8
|
DiGeorge syndrome critical region
8
|
|
m6a
|
N6-methyl-adenosine
|
|
METTL14
|
methyltransferase-like 14
|
|
miR/miRNA
|
microRNA
|
References
|
1
|
Xiao PL, Cui AY, Hsu CJ, Peng R, Jiang N,
Xu XH, Ma YG, Liu D and Lu HD: Global, regional prevalence, and
risk factors of osteoporosis according to the World Health
Organization diagnostic criteria: A systematic review and
meta-analysis. Osteoporos Int. 33:2137–2153. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Johnell O and Kanis J: An estimate of the
worldwide prevalence and disability associated with osteoporotic
fractures. Osteoporos Int. 17:1726–1733. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Consensus Development Conference, .
Diagnosis, prophylaxis, and treatment of osteoporosis. Am J Med.
94:646–650. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cooper C, Campion G and Melton L III: Hip
fractures in the elderly: A world-wide projection. Osteoporos Int.
2:285–289. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Aghebati-Maleki L, Dolati S, Zandi R,
Fotouhi A, Ahmadi M, Aghebati A, Nouri M, Kazem Shakouri S and
Yousefi M: Prospect of mesenchymal stem cells in therapy of
osteoporosis: A review. J Cell Physiol. 234:8570–8578. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Su P, Tian Y, Yang C, Ma X, Wang X, Pei J
and Qian A: Mesenchymal stem cell migration during bone formation
and bone diseases therapy. Int J Mol Sci. 19:23432018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wossidlo M, Nakamura T, Lepikhov K,
Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W
and Walter J: 5-Hydroxymethylcytosine in the mammalian zygote is
linked with epigenetic reprogramming. Nat Commun. 2:2412011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Baylin SB and Jones PA: A decade of
exploring the cancer epigenome-biological and translational
implications. Nat Rev Cancer. 11:726–734. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Clancy MJ, Shambaugh ME, Timpte CS and
Bokar JA: Induction of sporulation in Saccharomyces cerevisiae
leads to the formation of N6-methyladenosine in mRNA: A potential
mechanism for the activity of the IME4 gene. Nucleic Acids Res.
30:4509–4518. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Krug RM, Morgan MA and Shatkin AJ:
Shatkin, Influenza viral mRNA contains internal N6-methyladensine
and 5′-terminal 7-methylguanosine in cap structures. J Virol.
20:45–53. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shi B, Liu WW, Yang K, Jiang GM and Wang
H: The role, mechanism, and application of RNA methyltransferase
METTL14 in gastrointestinal cancer. Mol Cancer. 21:1632022.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Malavika D, Shreya S, Priya V, Rohini M,
He Z, Partridge NC and Selvamurugan N: miR-873-3p targets HDAC4 to
stimulate matrix metalloproteinase-13 expression upon parathyroid
hormone exposure in rat osteoblasts. J Cell Physiol. 235:7996–8009.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gao Y, Xue Q, Wang D, Du M, Zhang Y and
Gao S: MiR-873 induces lung adenocarcinoma cell proliferation and
migration by targeting SRCIN1. Am J Transl Res. 7:2519–2526.
2016.PubMed/NCBI
|
|
15
|
Gong H, Fang L, Li Y, Du J, Zhou B, Wang
X, Zhou H, Gao L, Wang K and Zhang J: miR-873 inhibits colorectal
cancer cell proliferation by targeting TRAF5 and TAB1. Oncol Rep.
39:1090–1098. 2018.PubMed/NCBI
|
|
16
|
Zhang JS, Zhao Y, Lv Y, Liu PY, Ruan JX,
Sun YL, Gong TX, Wan N and Qiu GR: miR-873 suppresses H9C2
cardiomyocyte proliferation by targeting GLI1. Gene. 626:426–432.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W,
Xie ZG, Shi L, He X, Jin SG, et al: The role of Tet3 DNA
dioxygenase in epigenetic reprogramming by oocytes. Nature.
477:606–610. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao X, Yang Y, Sun BF, Shi Y, Yang X,
Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, et al: FTO-dependent
demethylation of N6-methyladenosine regulates mRNA splicing and is
required for adipogenesis. Cell Res. 24:1403–1419. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH,
Wang F, Wang TT, Xu QG, Zhou WP and Sun SH: METTL14 suppresses the
metastatic potential of hepatocellular carcinoma by modulating
N6-methyladenosine-dependent primary MicroRNA processing.
Hepatology. 65:529–543. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Feng Z, Li Q, Meng R, Yi B and Xu Q:
METTL3 regulates alternative splicing of MyD88 upon the
lipopolysaccharide-induced inflammatory response in human dental
pulp cells. J Cell Mol Med. 22:2558–2568. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lin S, Choe J, Du P, Triboulet R and
Gregory RI: The m(6)A Methyltransferase METTL3 Promotes Translation
in Human Cancer Cells. Molecular Cell. 62:335–345. 2016. View Article : Google Scholar : PubMed/NCBI
|