Introduction
Ischemic stroke (IS) is the leading cause of
long-term disability and a major cause of morbidity and mortality
worldwide (1,2). Acute ischemic stroke (AIS) accounts
for ~81.9% of the total number of strokes (3). With high morbidity, disability,
recurrence, and mortality rates. AIS may pose a serious threat to
human life and health due to impaired blood flow to the brain,
ischemia, and hypoxia (4,5). This mechanism may be involved in
atherosclerosis, dyslipidemia, and hemodynamic changes in blood
composition. Previous studies have found that the ischemia and
hypoxia-induced injury cascade is the key cause of tissue damage
and long-term neurological dysfunction following cerebral ischemia.
Neuronal injury caused by AIS directly leads to brain parenchymal
injury, and neurons are important target cells for stroke
treatment. Therefore, studying nerve injury and the neuroprotective
mechanism of stroke is of particular importance.
Circular RNAs (circRNAs) are a relatively newfound
class of endogenous non-coding RNAs, and are covalently bonded
closed loops of RNA through a special splicing mechanism. CircRNAs
are widely expressed in eukaryotes, and this expression is highly
conserved, and tissue and spatiotemporal-specific. Due to the
special closed ring structure, circRNAs are less susceptible to
degradation by exonucleases and are more stable than linear RNAs
(6,7). At present, the evaluation of the
effect of circRNAs in ischemic brain injury is still in the initial
stages. It has been reported that a few distinctly expressed
circRNAs were identified in an AIS model (8–10).
However, the mechanisms remain incompletely understood.
Numerous studies have shown that miRNAs can be used
for a variety of diseases as therapeutic targets and biomarkers
(11–13). In fact, miRNAs can also be detected
in several central nervous system diseases, including Parkinson's
disease, Down's syndrome, schizophrenia, and stroke (14–18).
According to previous reports (19), miRNAs are endogenously expressed
RNA molecules that have an important effect in regulating the
pathophysiological process of cerebral infarction, and the
expression levels of miRNAs in the blood can reflect brain damage
and recovery in patients with cerebral infarcts.
In the present study, the functions of circ_0000018
in neuronal cell apoptosis in AIS progression were explored in
vivo and in vitro. It was found that circ_0000018
knockdown alleviated neuronal cell apoptosis by targeting the
miR-871/Bcl-2-like protein 11 (BCL2L11) axis, indicating that
circ_0000018 may serve as a potential strategy for inducing
neuroprotective AIS.
Materials and methods
Analysis of microarrays
A Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) dataset
(GSE115697) that assessed circRNA expression patterns in NSCLC and
was analyzed by using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) was obtained
and used in the present study (20). Raw data were normalized by Quantile
algorithm, limma package the R program (21). R software (version 4.0.3) was used
for the data download and processing (22). The ‘GEOquery’ package was used to
download the dataset expression matrix and platform file. The
‘limma’ package was used for differential analysis of microarray
data. After confirming that the quality of the samples, the
subsequent difference analysis was performed. Significant
differentially expressed transcripts were screened using P<0.05
and fold-change ≥0.5 or ≤-1 The ‘ggplot2’ package was used to
visualize the heatmap and volcano plot.
Cell culture and establishment of the
oxygen-glucose deprivation/reperfusion (OGD/R) model
The mouse neuroblast (Neuro-2a cells; http://www.atcc.org/products/ccl-131)
were obtained from the American Type Culture Collection. Mouse
neuronal cells were cultured in DMEM (Invitrogen; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (Invitrogen; Thermo
Fisher Scientific, Inc.) and antibiotics (100 U/ml penicillin, 100
µg/ml streptomycin, PAN Biotech), and maintained in a humidified
incubator at 37°C and supplied with 5% CO2 air.
For the establishment of the OGD/R cell model, the
original culture medium was discarded, the neuronal cells were
washed with PBS, placed in sugar-free DMEM medium without FBS, and
incubated as above for 2 h. Next, the media was removed, and
supplemented DMEM was added, and the cells were transferred to an
incubator supplied with air consisting of 95% N2, 5%
CO2, and 1% O2 for 6 h (23). Subsequently, the media was replaced
with supplemented DMEM and transferred back to the previous
conditions for 12 h. In addition, cells cultured in normal DMEM at
37°C with 5% CO2 in a humidified incubator were used as
the control.
Cell transfection
To induce circ_0000018 knockdown, OGD/R-treated
cells and control cells were transfected with 50 nM circ_0000018
small interference RNA (si-circ_0000018; Guangzhou RiboBio Co.,
Ltd.) or non-targeting siRNA (si-NC, Guangzhou RiboBio Co., Ltd.).
Similarly, 50 nM miR-871 mimic (agomiR-871), miR-871 inhibitor
(antagomiR-871) and their blank controls (miR-NC and anti-miR-NC;
all from Guangzhou RiboBio Co., Ltd.) were respectively transfected
into neuronal cells. The overexpression vector (pcDNA-BCL2L11) was
obtained by inserting the BCL2-like 11 overexpressed CDS sequence
(oe-BCL2L11) into a pcDNA vector (Invitrogen; Thermo Fisher
Scientific, Inc.), and the empty vector was used as the negative
control. A total of 200 ng vector was transfected into neuronal
cells using Lipofectamine® 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.).
Establishment of the transient middle
cerebral artery occlusion (tMCAO) model
Male C57BL/6J mice (7–8 weeks, 18–25 g) were
obtained from Beijing Vital River Laboratory Animal Technology Co.,
Ltd. The animals were randomly separated into groups and kept in a
temperature-controlled room at 25±2°C with a 40–50% relative
humidity and a 12 h light/dark cycle. All experimental procedures
were approved by the Southeast University Animal Care and Use
Committee (approval no. 20220110026).
The mice were randomly allocated into a sham group,
tMCAO group, tMCAO+sh-NC group, or tMCAO+sh-circ_0000018 group
(n=18 per group). After 7 days of acclimatization, a model of
cerebral AIS was created in mice by tMCAO with modifications based
on earlier reports (24,25). Briefly, 1% sodium pentobarbital (50
mg/kg) was injected intraperitoneally to anesthetize the mice.
Following a midline skin incision, the right common carotid artery,
external carotid artery, and internal carotid artery (ICA) were
explanted, and the L1800 silicone wire (Jialing Biotechnology Co.,
Ltd.) was then inserted 10 mm into the ICA to obscure the origin of
the MCA. Brain reperfusion was performed by withdrawing silicone
filaments after 90 min. In the sham group, the mice underwent the
same tMCAO procedure but without ICA occlusion. The sh-NC and
sh-circ_0000018 (Shanghai GenePharma Co., Ltd.) were injected into
the lateral ventricle 1 day prior to tMCAO. Throughout the
procedure, the temperature of the rectum of the mice was kept at
37.0±0.5°C using a heat lamp (Jiaxing Nomoy Pet Products Co.,
Ltd.). Following the neurobehavioral scoring of all mice, the mice
were euthanized by inhalation of carbon dioxide (35% volume
displacement rate/min) inhalation following isoflurane anesthesia
(induction percentage, 3%; maintenance percentage, 1%), and brain
tissue was obtained for subsequent experiments.
Evaluation of neurological
impairment
The neurological deficits of mice were evaluated
using the Longa biologic score 24 h after 1 h of brain ischemia and
reperfusion (26). The scoring
system used was: 0, both forelimbs are strong and symmetrically
extended to the ground, both shoulders have the same resistance,
and walking is normal; 1, internal rotation on the contralateral
side of the surgery, forelimb tucked in, both shoulders resist in
unison, and walking is normal; 2, internal rotation on the
contralateral side of the surgery, forelimb inversion, decreased
resistance on the contralateral side of the surgery when pushing
both shoulders, and walking is normal; 3, when the surgical
contralateral side is internally rotated, the forelimb is tucked
in, and both shoulders are pushed; when pushing both shoulders, the
resistance of the contralateral side of the surgery decreases, mice
can walk around; and 4, no spontaneous movement on the opposite
side of the surgery. The observers were blinded to the
treatment.
TTC staining
After assessment of neurological damage, mice were
euthanized by CO2 inhalation following halothane
anesthesia, and brains were rapidly collected, cut into 2-mm
sections, and incubated with 2% TTC solution (MilliporeSigma) at
37°C for 10 min. Subsequently, 4% paraformaldehyde was used to fix
the brain tissue for 1–2 days at room temperature, and then an
image was taken. The infarct volume was calculated using ImageJ
1.8.0.345 (National Institutes of Health). Normal brain tissue
appeared red when TTC reacted with dehydrogenase, while in the
ischemic brain tissue, it appeared white due to reduced
dehydrogenase activity. The infarct volume ratio as a percentage
was calculated as follows: Infarct volume (%)=[(Sum of infarct area
×2 mm3)/(sum of total brain area ×2 mm3)
×100.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from cultured cells, serum,
and brain tissue using TRIzol® reagent according to the
manufacturer's protocol (Invitrogen; Thermo Fisher Scientific,
Inc.). The PrimeScript RT kit was used to reverse transcribe total
RNA to cDNA according to the manufacturer's protocol (Takara Bio,
Inc.). The levels of circ_0000018, miR-871, and BCL2L11 were
measured using qPCR with a SYBR green kit (Takara Bio, Inc.) in a
Bio-Rad CFX96 system (Bio-Rad, Laboratories, Inc.). The procedure
was as follows: 95°C for 3 min; followed by 39 cycles of 95°C for
15 sec, 60°C for 60 sec and 72°C-30 sec for mRNA, or 95°C for 15
sec and 60°C for 60 sec for miRNA; 95°C for 10 sec, followed by a
melt curve analysis (60–95°C, 0.5°C increments for 20 sec) to
confirm specificity of the PCR primers. The expression levels were
normalized to the expressions of β-actin or U6. The
2−∆∆Cq method was used to determine gene expression in
the neuronal cells and brain tissues (27).
The sequences of the primers used were: circ_0000018
forward, 5′-CAAGATCACCTCCGATTGGT-3′ and reverse,
5′-TGTCTTCTGCTCCAGGATCTTT-3′; miR-871 forward,
5′-TGCGGTCTGACCGTGGTAAGACC-3′ and reverse,
5′-CCAGTGCAGGGTCCGAGGT-3′; BCL2L11 forward,
5′-CCCGGAGATACGGATTGCAC-3′ and reverse, 5′-GCCTCGCGGTAATCATTTGC-3;
and U6 forward, 5′-CTCGCTTCGGCAGCAC-3′ and reverse,
5′-ACGCTTCACGAATTTGC-3′; cicr_0001646 forward,
5′-GGCAGATGGAAGCTTCTTGA-3′ and reverse,
5′-AGCAAGTTGACCCATTTTCC-3′.
Western blot analysis
Total protein was extracted from cells using RIPA
lysis buffer (Cell Signaling Technology, Inc.) and quantified using
a BCA Protein Quantification Kit (Abbkine Scientific Co., Ltd.).
Equivalent quantities of protein (30 µg) was loaded per lane on a
10% SDS-gel, resolved using SDS-PAGE, and transferred onto PVDF
membranes (MilliporeSigma), followed by blocking with 5% milk in
TBST at room temperature for 2 h, and subsequent incubation with
one of the primary antibodies at 4°C overnight. The corresponding
secondary antibodies were used to incubate the membranes for 1 h at
room temperature, followed by visualization using a
chemiluminescence detection kit (Beyotime Institute of
Biotechnology). The primary antibodies used in the present study
were: Toll-like receptor 4 (TLR4; ab22048; 1:300), β-tubulin III
(ab18207; 1:3,000), Bax (ab32503; 1;300), Bcl-2 (ab182858;
1:3,000), Caspase-3 (ab32351; 1:1,000), H2A.X Variant Histone
(H2AX) (ab124781; 1:1,000), γH2AX (ab81299; 1:5,000) and β-actin
(ab8226; 1:1,000) (all Abcam). The secondary antibody used in the
present study were as follows: HRP-Conjugated AffiniPure Goat
Anti-Rabbit/Mouse IgG H&L (cat. no. BA1056; 1:5,000) and
HRP-Conjugated AffiniPure Donkey Anti-Rabbit IgG H&L (cat. no.
BA1061; 1:2,000) (both Wuhan Boster Biological Technology, Co.,
Ltd.).
RNase R treatment
A total of 2 µg RNA was analyzed by incubation at
37°C for 30 min with or without 5 U/µg RNase R (Epicenter;
Illumina, Inc), following purification with an RNeasy MinElute
Cleaning Kit (Qiagen GmbH), and then assessed using RT-qPCR.
Actinomycin D assay
The nerve cells in logarithmic growth stage were
used, and when the cell density reached 80–90%, the adherent cells
were enzymatically dissociated into a single-cell suspension. The
neuronal cells were exposed to 2 µg/ml actinomycin D
(MilliporeSigma). The cells were then collected and total RNA was
withdrawn. circRNA and mRNA stability was analyzed using
RT-qPCR.
Cell counting kit-8 (CCK-8) assay
The cells in each group following treatment were
seeded in 96 well plates with a cell density of 2×104
cells/well. Following 0, 24, and 48 h of culture, 10 µl CCK-8
solution (Beyotime Institute of Biotechnology) was added to each
well. Following culture at 37°C for 1.5 h, the absorbance of the
cells at a wavelength of 450 nm was measured using an Elx800 reader
(Omega Bio-Tek, Inc.) as a measure of the viability of the cells
(28).
Flow cytometry assay
The OGD/R-treated neuronal cells were cultured for
48 h following transfection to assess apoptosis (18). After the culturing, cells were
stained using an Annexin V FITC Apoptosis Detection Kit according
to the manufacturer's protocol (BD Biosciences). The proportion of
apoptotic cells in each group was analyzed using a flow cytometer
(CytoFLEX; Beckman Coulter, Inc.). The flow data were analyzed by
FlowJoTM v10.6.1 (BD Biosciences).
Dual-luciferase assay
The online tool StarBase (https://starbase.sysu.edu.cn/) was used to predict the
putative miR-871 binding sites in circ_0000018 and BCL2L11. A
fragment of circ_0000018 untranslated region (3′-UTR) with
wild-type (circ_0000018-WT) and mutant (circ_0000018-MUT) was
introduced into the pmirGLO luciferase vector (E1330, Promega
Corporation). Similarly, the BLC2L11-3′-UTR fragment with wild type
(BLC2L11-WT) and mutant (BLC2L11-MUT) was introduced into the
pmirGLO luciferase vector. Lipofectamine® 3000 was used
to co-transfect circ_0000018-WT/circ_0000018-MUT with
BLC2L11-WT/BLC2L11-MUT and agomiR-871 or agomiR-NC into neuronal
cells. Following transfection for 48 h, cells were collected, and
the activity of luciferase was determined using a SpectraMax L
fluorometer (Molecular Devices, LLC).
RNA pull-down assay
RNA pull-down assays were performed as previously
reported (29). Briefly, neuronal
cells (1×107 cells) were collected and lysed.
Glycosylated miR-871 probes were synthesized by Shanghai GenePharma
Co., Ltd. and cultured with streptavidin agarose beads (Thermo
Fisher Scientific, Inc.). The cell lysate of the miR-871 probe or
oligo probe (control) was incubated overnight at 4°C. The
bead-bound RNA complexes were purged with wash buffer, and the
degree of enrichment of circ_0000018 extracted by the miR-871 probe
was examined using RT-qPCR.
Statistical analysis
Data were analyzed using GraphPad Prism version 7.0
(GraphPad Software, Inc.). Comparisons between two groups were
performed using a Student's t-test, and comparisons between
multiple groups were performed using a one-way ANOVA with a
post-hoc Tukey's test. Data are presented as the mean ± standard
deviation of three repeats.
Results
circ_0000018 levels in the in vivo AIS
model
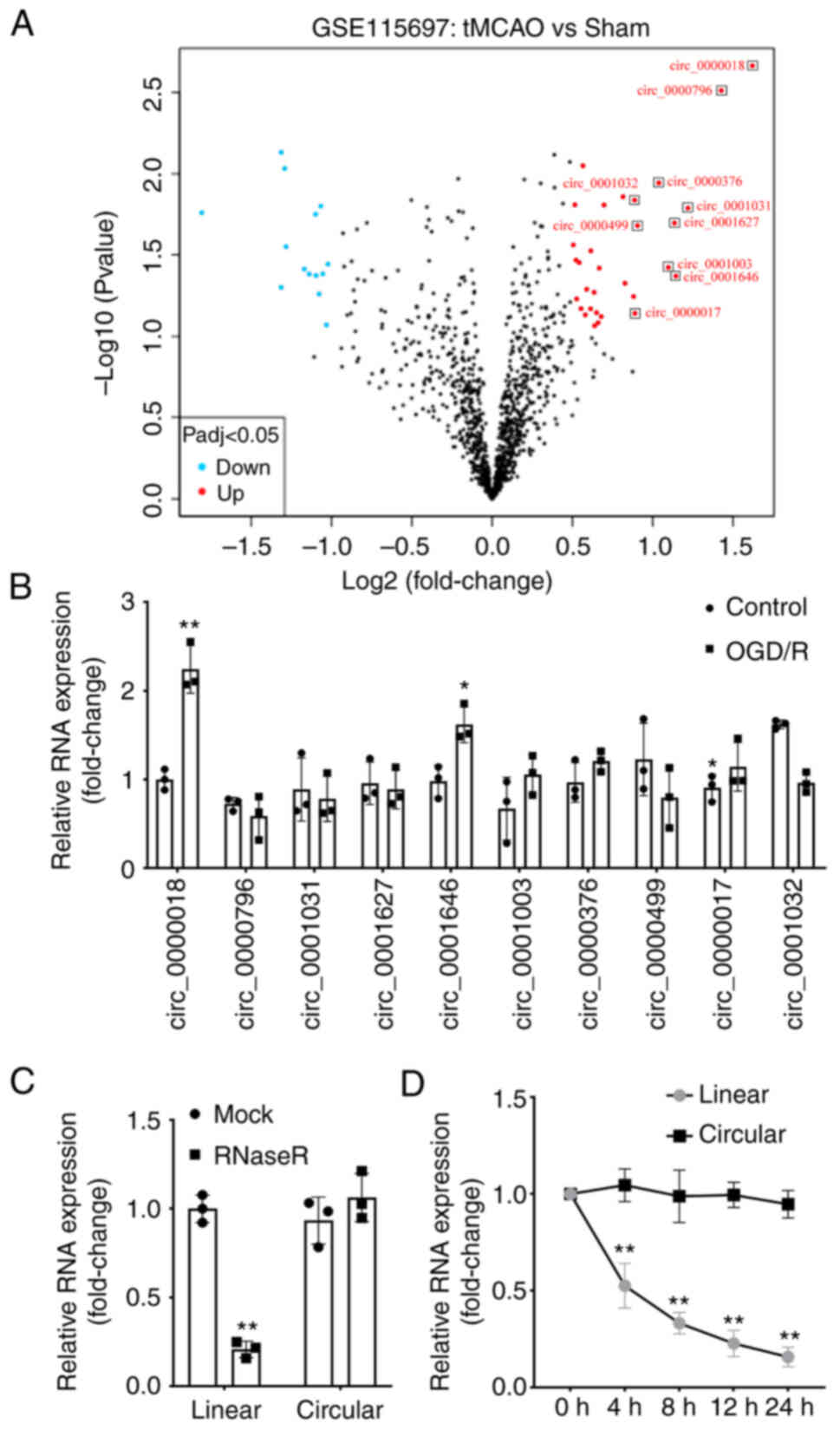
First, the GSE115697 dataset was downloaded from GEO
and analyzed in R. The differentially expressed circRNAs in the
tMCAO mice were obtained and expressed as a Volcano plot (Fig. 1A). Among these differentially
expressed circRNAs, 10 circRNAs with the most notable differences
in expression were selected and their levels in the OGD/R-treated
cells were determined using RT-qPCR. The findings demonstrated that
circ_0000018 and circ_0001646 levels were significantly increased
in OGD/R treated cells compared with those in normal neuronal
cells, while circ_0000018 levels were decreased. Of these two
circRNAs, the difference in circ_0000018 levels expression was
greater (Fig. 1B). Therefore,
circ_0000018 was selected for subsequent experiments. In addition,
following ribonuclease R (Fig. 1C)
or actinomycin D (Fig. 1D)
treatment, the linear RNA levels were significantly reduced, while
no significant change was observed in circular RNA levels.
circ_0000018 downregulation relieves
OGD/R-treated neuronal cell damage in vitro
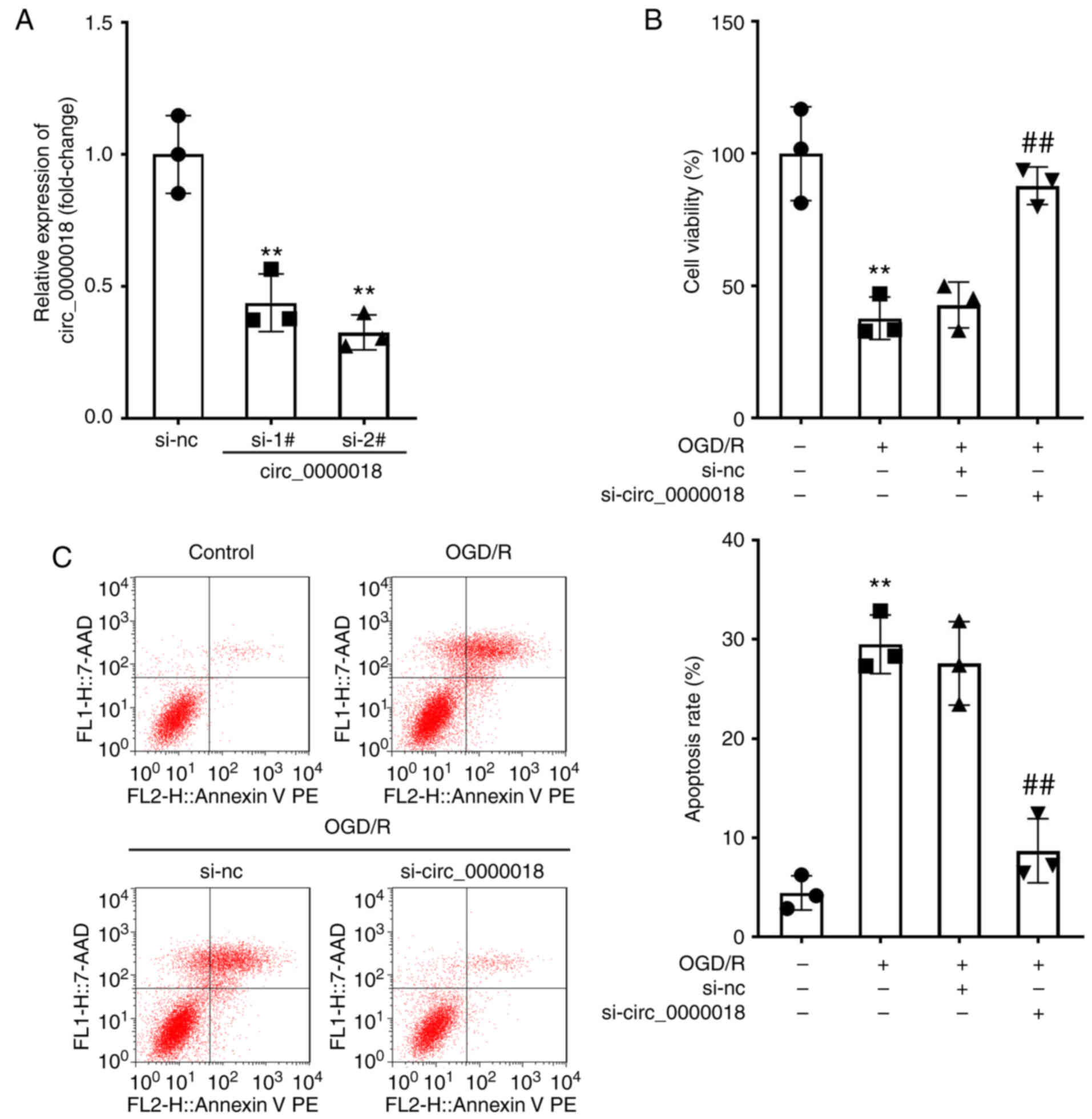
To explore the effect of circ_0000018 in
vitro, neuronal cells were transfected with si-circ_0000018 1#,
si-circ_0000018 2#, or si-NC. The RT-qPCR results confirmed that
transfection with siRNA plasmids targeting circ_0000018 markedly
reduced the levels of circ_0000018 in neuronal cells, and
si-circ_0000018 2# was selected for the subsequent experiments
given the better knockdown efficiency (Fig. 2A). Furthermore, to explore the
effects of circ_0000018 knockdown on AIS in vitro, an ODG/R
cell model was established using transfected neuronal cells. CCK-8
assays showed that the knockdown of circ_0000018 markedly reduced
neuronal cell activity (Fig. 2B).
In addition, flow cytometry analysis indicated that apoptosis of
neuronal cells was induced by OGD/R treatment, whereas circ_0000018
knockdown attenuated the apoptosis of OGD/R-stimulated neuronal
cells (Fig. 2C).
Targeting association between
circ_0000018 and miR-871
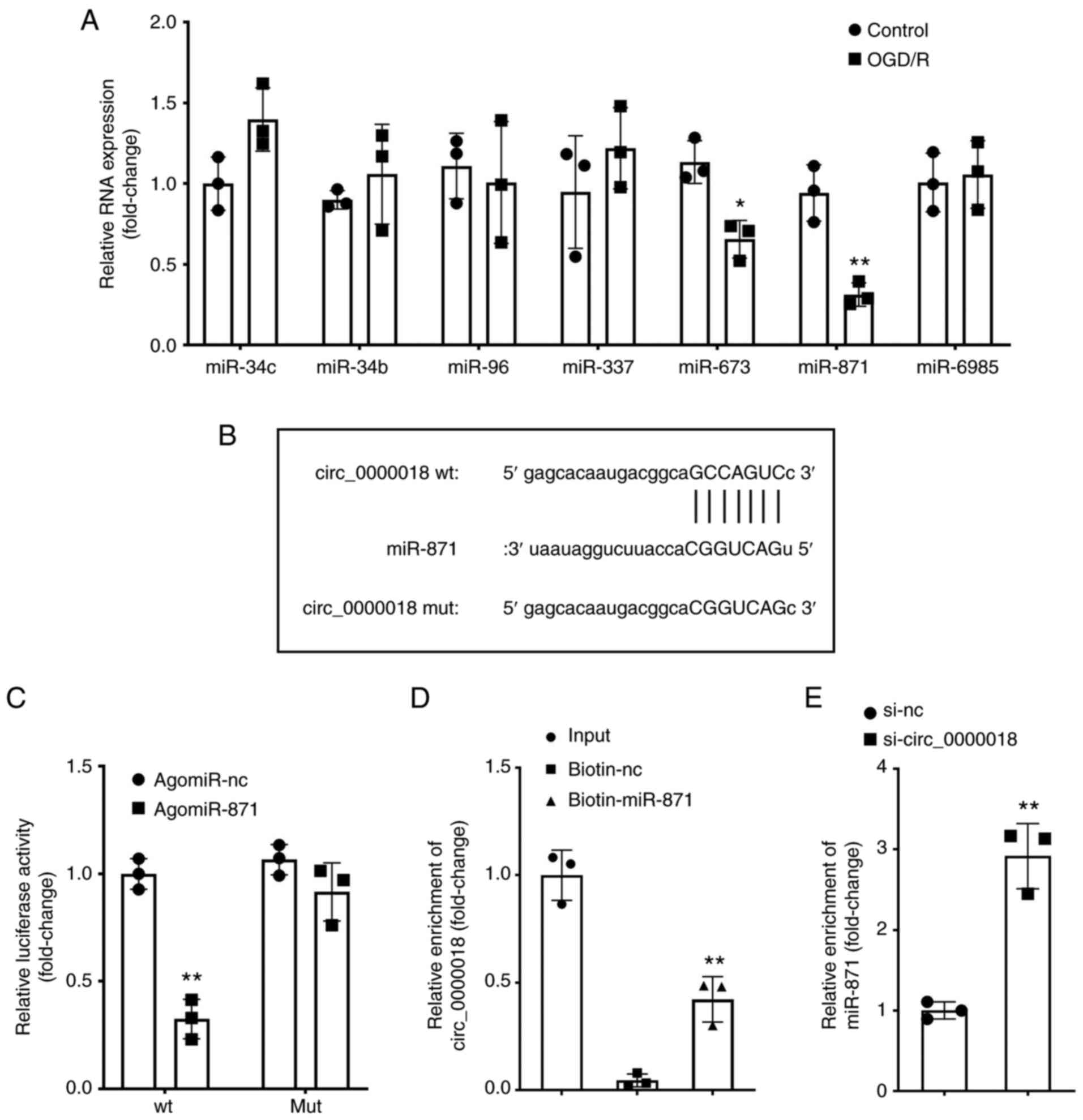
Using bioinformatics analysis, the top-7 miRNAs
targeted by circ_0000018 were obtained. Next, based on PCR, only
miR-871 expression was decreased in the OGD/R-treated neuronal
cells (Fig. 3A). miR-871 was
selected for further study. In Fig.
3B, the binding site between circ_0000018 and miR-871 is shown.
Dual luciferase assays indicated that agomiR-871 markedly decreased
the luciferase activity of the circ_0000018-wt reporter vector,
with no influence on the MUT reporter vector (Fig. 3C). The RNA pull-down assays also
showed that miR-871 and circ_0000018 could bind to each other
(Fig. 3D). Furthermore, it was
discovered that the levels of miR-871 were markedly increased in
the OGD/R-treated neuronal cells transfected with si-circ_0000018
compared with cells transfected with si-NC (Fig. 3E). These findings revealed that
circ_0000018 negatively regulated the expression of miR-871 by
specifically binding to miR-871.
miR-871 downregulation alleviates the
impact of circ_0000018 on the growth and apoptosis of OGD/R-treated
neuronal cells
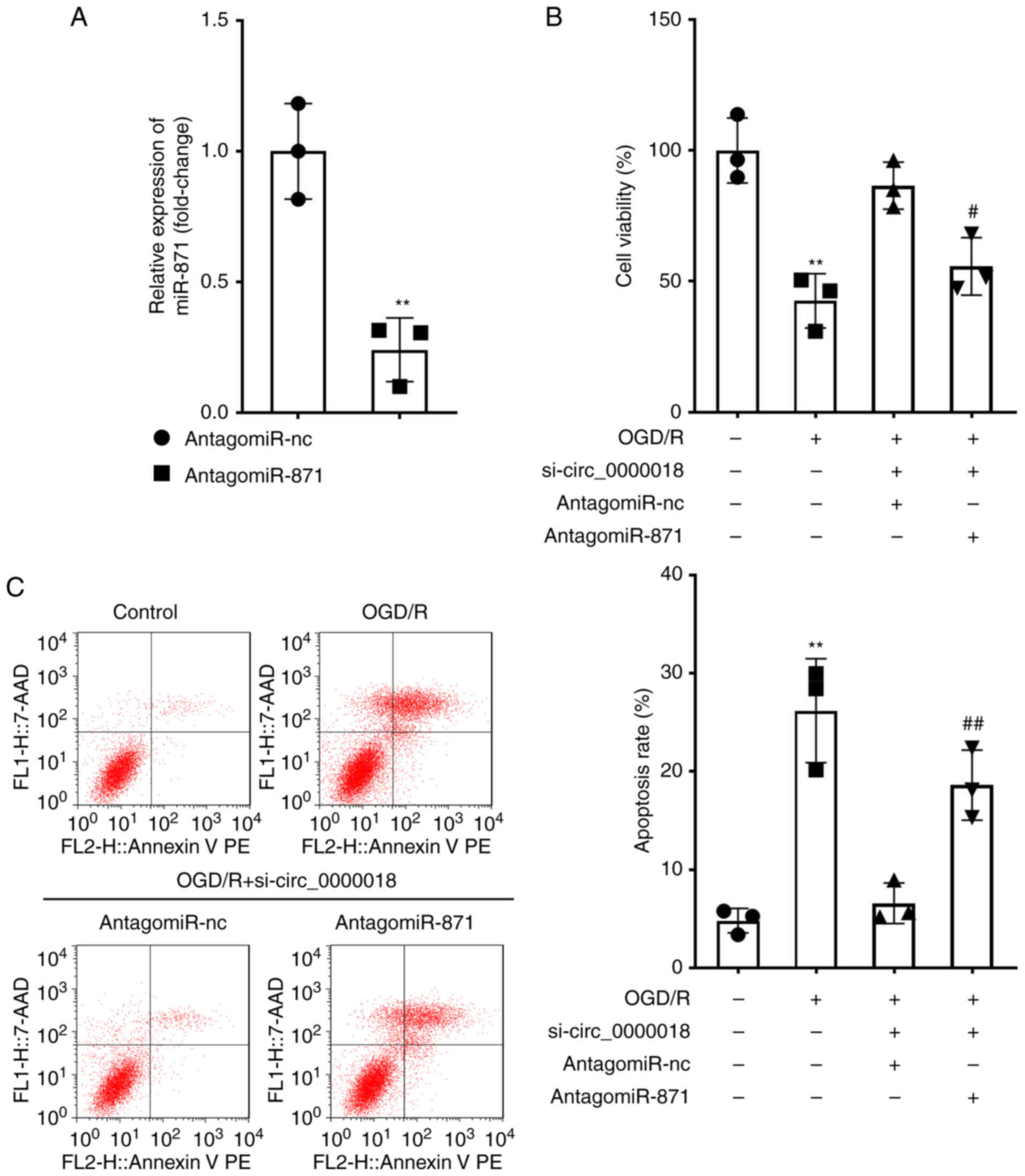
For the purpose of further investigating the roles
of a circ_0000018/miR-871 axis on the injury of the OGD/R-treated
cells, the OGD/R-treated neuronal cells were transfected with
si-circ_0000018 and antogomiR-871. The transfection efficiency of
antogomiR-871 was verified by RT-qPCR, and it was confirmed that
antogomiR-871 transfection significantly decreased the miR-871
levels (Fig. 4A). The results of
the CCK-8 assay indicated that si-circ_0000018 markedly reduced the
survival of OGD/R-treated neuronal cells, while co-transfection
with antagomiR-871 reduced this inhibitory effect (Fig. 4B). Similarly, the inhibitory effect
on cell apoptosis induced by circ_0000018 downregulation was
markedly alleviated by antagomiR-871 in OGD/R-treated neuronal
cells (Fig. 4C). Together, these
results showed that downregulation of miR-871 can partially reverse
the effects of si-circ_0000018 on the apoptosis and proliferation
of neuronal cells following treatment with OGD/R. Proof of
transfection for agomiR-871 and agomiR-NC in neuronal cells was
obtained by RT-qPCR (Fig.
S1A).
BCL2L11 is a target of miR-871
There is a general consensus that miRNAs exert their
biological effects by targeting mRNAs (30–32).
Therefore, StarBase was used to find potential target genes for
miR-871. Several binding sites were obtained between miR-871 and
BCL2L11 (Fig. 5A). Next, a dual
luciferase assay performed in neuronal cells confirmed this binding
relationship. The findings demonstrated that the upregulation of
miR-871 markedly decreased the luciferase activity of the
BCL2L11-wt reporter vector, whereas the luciferase activity of
BCL2L11-mut was not decreased (Fig.
5B). The RNA pull-down assay demonstrated that miR-871 and
BCL2L11 could specifically bind to each other (Fig. 5C). In addition, RT-qPCR and western
blotting showed that the introduction of agtagomiR-871 decreased
the effects of si-circ_0000018 on the mRNA levels of BCL2L11 in
neuronal cells (Fig. 5D and F),
revealing that circ_0000018 could regulate the levels of BCL2L11 by
sponging miR-871. Consistently, it was found that BCL2L11
expression was increased in OGD/R-treated neuronal cells compared
with the corresponding control group (Fig. 5E and G), which demonstrated that
BCL2L11 may be involved in OGD/R-treated cell trauma in
vitro.
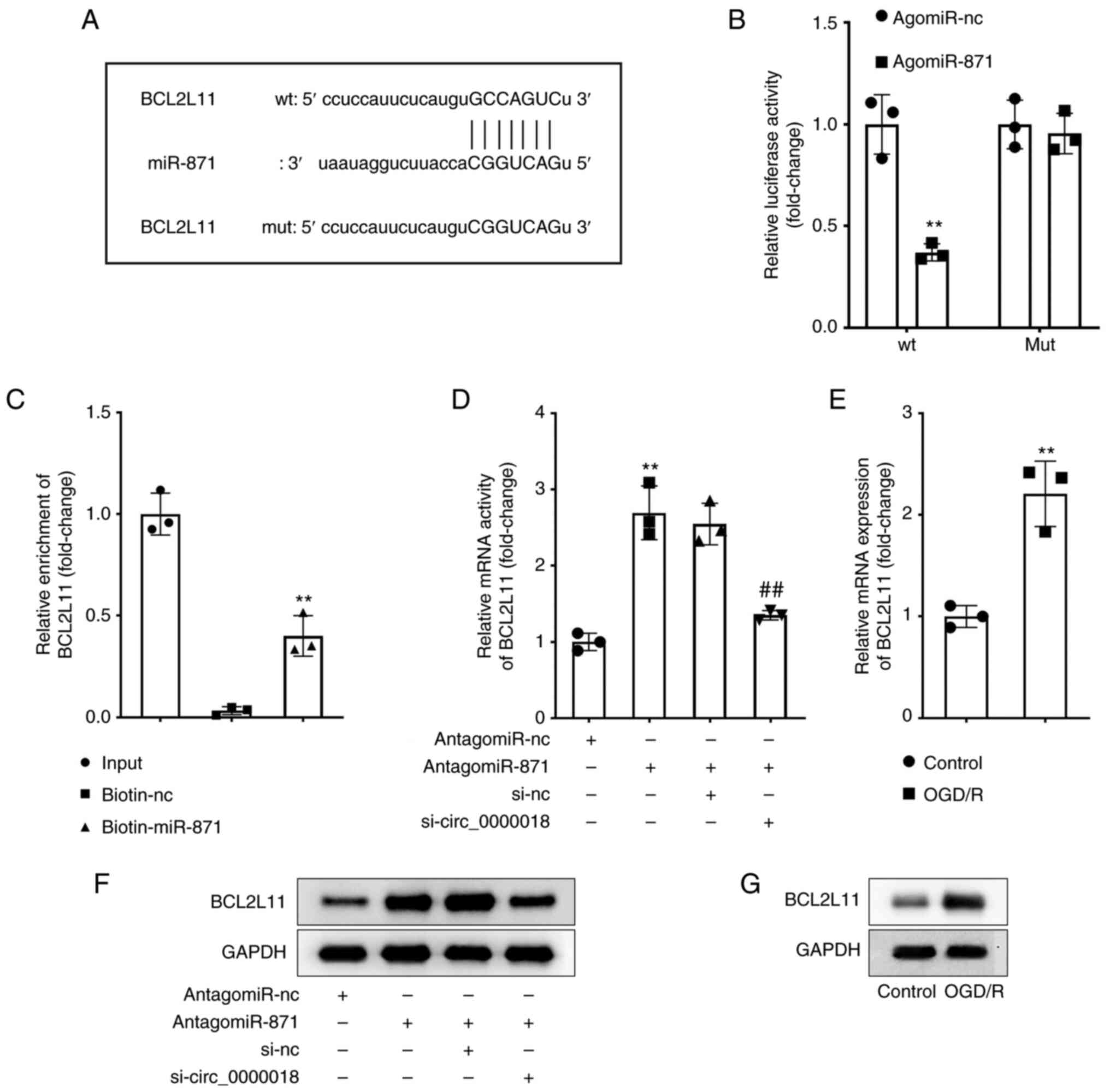 | Figure 5.BCL2L11 acts as a target of miR-871.
(A) The structure of the predicted binding site to miR-871 in the
3′-UTR sequence of BCL2L11 is shown in the schematic diagram. (B)
The binding between miR-871 and BCL2L11 was confirmed using a dual
luciferase reporter assay. **P<0.01 vs. agomiR-NC. (C) RNA
pull-down assays were performed to validate the relationship
between miR-871 and BCL2L11. **P<0.01 vs. biotin-NC. (D) The
levels of BCL2L11 were assessed in neuronal cells transfected with
antagomiR-NC, antagomiR-871, si-NC, and si-circ_0000018 using
RT-qPCR. **P<0.01 vs. antagomiR-NC; ##P<0.01 vs.
antagomiR-871 + si-NC. (E) The levels of BCL2L11 were assessed in
neuronal cells treated with or without OGD/R using RT-qPCR.
**P<0.01 vs. control. (F) Western blotting was performed to
determine the protein expression levels of BCL2L11 in neuronal
cells transfected with antagomiR-NC, antagomiR-871, si-NC, and
si-circ_0000018. (G) Western blotting was performed to determine
the protein expression levels of BCL2L11 in neuronal cells treated
with or without OGD/R. BCL2L11,Bcl-2-like protein 11; miR,
microRNA; RT-qPCR, reverse transcription-quantitative PCR; OGD/R,
oxygen-glucose deprivation; UTR, untranslated region. |
Overexpression of BCL2L11 reverses the
effects of miR-871 on the growth and apoptosis of OGD/R-treated
neuronal cells
To verify the potential influence of the
circ_0000018/miR-871/BCL2L11 axis on the neuronal injury induced by
ODG/R, rescue experiments were performed. Transfection efficiencies
were determined by RT-qPCR (Fig. 6A
and B). In addition, CCK-8 assays revealed that the
proliferation of OGD/R-treated neuronal cells transfected with
agomiR-871 was significantly increased, and this was abrogated by
BCL2L11 overexpression (Fig. 6C).
The flow cytometry results suggested that the treatment with ODG/R
promoted apoptosis, whereas transfection of agomiR-871 in
ODG/R-treated cells decreased apoptosis; further overexpression of
BCL2L11 significantly reversed the effects of agomiR-871 on
stimulating cell apoptosis (Fig.
6D).
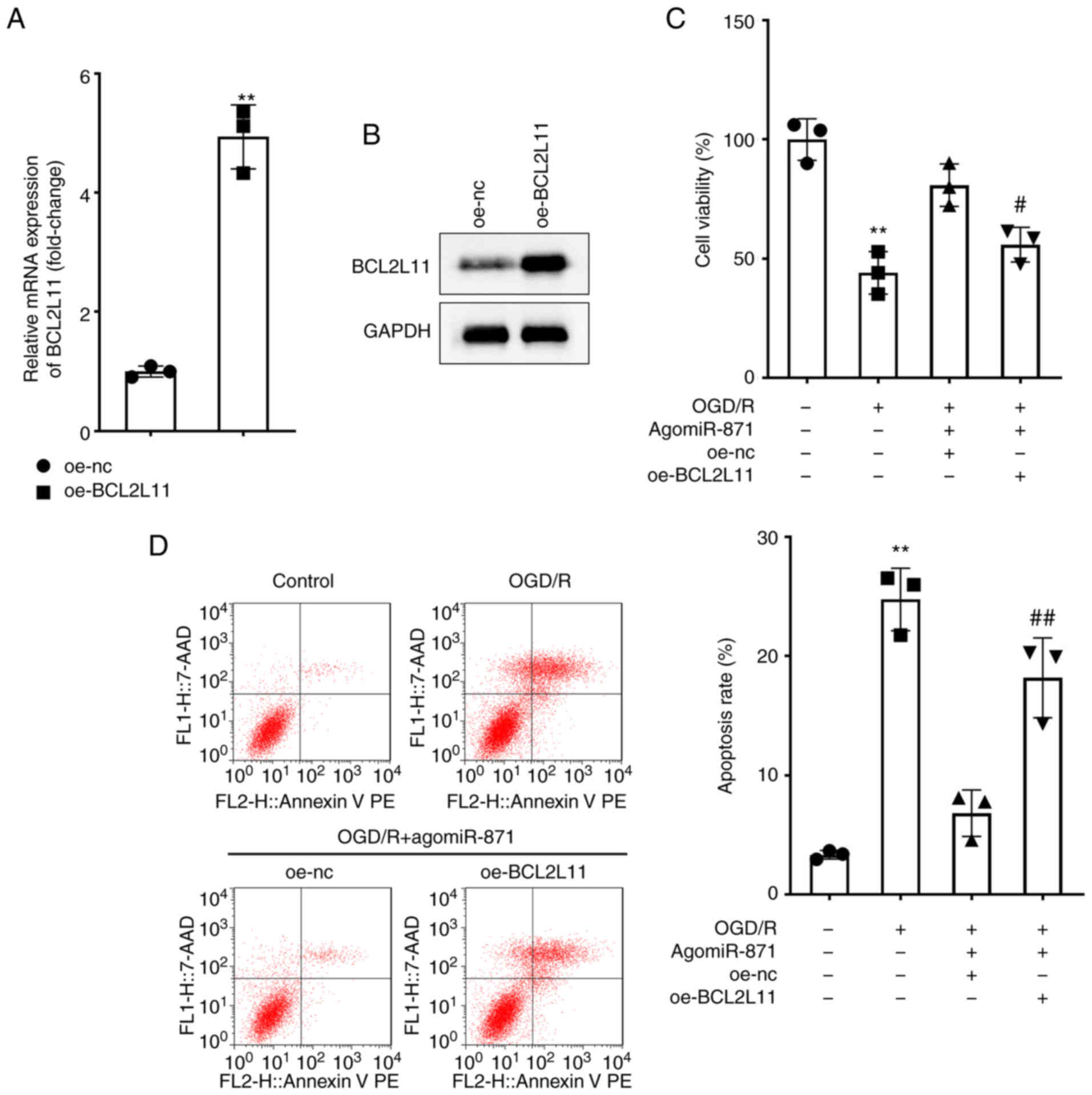 | Figure 6.Overexpression of BCL2L11 reverses
the effects of miR-871 on the growth and death of OGD/R-treated
neuronal cells. (A) Expression of BCL2L11 following transfection
with oe-NC and oe-BCL2L11 in neuronal cells was assessed by
RT-qPCR. **P<0.01 vs. oe-NC. (B) Expression of BCL2L11 following
transfection with oe-NC and oe-BCL2L11 in neuronal cells was
detected by western blotting. (C and D) Neuronal cells were treated
with control, OGD/R, OGD/R+agomiR-871+oe-NC, or
OGD/R+agomiR-871+oe-BCL2L11. (C) Proliferation of neuronal cells
was evaluated using a CCK-8 assay. (D) Apoptosis of neuronal cells
following treatment was examined using flow cytometry. **P<0.01
vs. control; #P<0.05, ##P<0.01 vs.
OGD/R + agomiR-871 +oe-NC. BCL2L11, Bcl-2-like protein 11; miR,
microRNA; OGD/R, oxygen-glucose deprivation; RT-qPCR, reverse
transcription-quantitative PCR; CCK-8, cell counting kit-8; oe,
overexpression; NC, negative control. |
circ_0000018 knockdown reduces
cerebral ischemia/reperfusion injury in tMCAO mice
Finally, the contribution of circ_0000018 in
vivo in the tMCAO mouse models was evaluated. RT-qPCR showed
that circ_0000018 (Fig. 7A) and
BCL2L11 (Fig. 7C) expression were
significantly increased while miR-871 expression (Fig. 7B) was decreased in mouse brain
tissues in the tMCAO group. Mice in the tMCAO group presented with
increased neurological damage and brain infarction when compared
with the sham group (Fig. 7D-F),
and knockdown of circ_0000018 significantly ameliorated these
changes. RT-qPCR results showed that the expression of circ_0000018
in tissues decreased after transfection with the plasmid with
knockdown of circ_0000018 (Fig.
S1B).
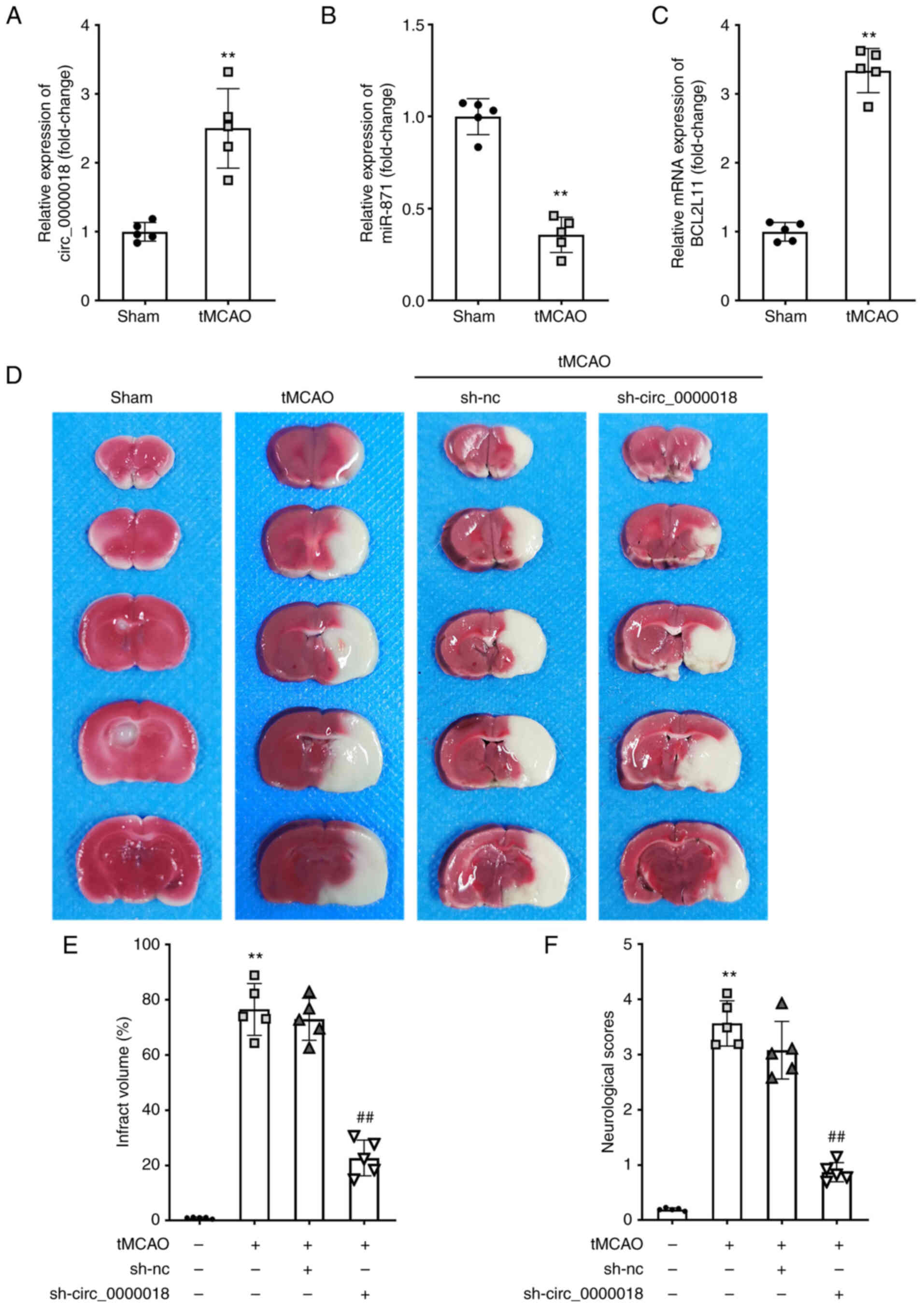 | Figure 7.circ_0000018 knockdown relieves
cerebral ischemia-reperfusion injury in tMCAO mice. (A-C) The
levels of circ_0000018, miR-871, and BCL2L11 in cerebral tissue
from tMCAO mice were examined using RT-qPCR. The mice were randomly
allocated to a sham, tMCAO, tMCAO+sh-NC, or tMCAO+sh-circ_0000018
group (n=18, per group). **P<0.01 vs. sham. (D) TTC staining
revealed focal ischemia induced by tMCAO in a typical brain
section. (E) Following ischemia/reperfusion injury, the brain
infarct volume in mice was analyzed using TTC staining. (F)
Neurological impairment assessment. **P<0.01 vs. control;
##P<0.01 vs. tMCAO + sh-NC. circRNA, circular RNA;
tMCAO, transient middle cerebral artery occlusion; BCL2L11,
Bcl-2-like protein 11; RT-qPCR, reverse transcription-quantitative
PCR. |
Discussion
Herein, OGD/R-treated neuronal cells and tMCAO mice
were employed to investigate the function of circ_0000018. This
study demonstrated that circ_0000018 levels were markedly increased
both in vitro and in vivo in the AIS model and that
the knockdown of circ_0000018 reduced the apoptosis of neuronal
cells induced by OGD/R treatment. This indicated that circ_0000018
was involved in the pathogenesis of AIS and may have potential as a
novel therapeutic target.
According to recent studies, circRNAs not only exert
significant effects in the regulation of gene expression but also
take part in the pathogenesis of AIS. For instance, Mehta et
al (33) studied the
expressions profile of circ RNAs in the pneumonic cortex of MCAO
mice at 6, 12, and 24 h after reperfusion by circRNA chip, and
found that 283 circRNAs were significantly differentially expressed
(based on a 2-fold change in expression). Bioinformatics analysis
determined that 16/283 circRNAs were associated with a multitude of
miRNA binding sites, as well as with stroke pathophysiology in a
functional manner (33). In
addition, Liu et al (34)
also investigated the characteristics of circRNA expression in IS
mouse tissue, and demonstrated that circRNA, such as
mmu_circRNA_40001, mmu_circRNA_013120 and mmu_circRNA_40806 has the
potential of becoming a target for the diagnosis and treatment of
IS. Duan et al (35) more
recently reported the characteristics of circRNA expression in rats
following brain ischemia and examined the relationship between
circRNA caused by MCAO and IS in rats. Peng et al (10) reported the findings of a clinical
investigation, which found that in peripheral blood mononuclear
cells separated from blood samples in patients with AIS and healthy
controls to detect the levels of circ_HECT domain E3
ubiquitin-protein ligase (HECTD), the levels of circ_HECTD were
associated with a higher risk of disease, disease severity,
inflammation, and recurrence of AIS. It is worth noting that the
OGD/R-treated neuronal injury model has been extensively used to
study circRNA in IS. circ_HECTD1 expression was decreased in
OGD/R-treated mouse brain neuronal cells (HT-22), and the
overexpression of circ_HECTD1 alleviated the death of OGD/R-treated
HT-22 cells. Similarly, the present study demonstrated that
circ_0000018 downregulation relieved OGD/R-treated neuronal cell
damage in vitro.
There is substantial evidence to show that circRNAs
exert various biological roles through their interactions with
miRNAs (36,37). For example, circ_HECTD1 affected
cellular injury following cerebral infarction by acting on the
miR-27a-3p/FSTL1 axis (38). In
addition, circ_0101874 overexpression increased the levels of
phosphodiesterase 4D (PDE4D) by targeting miR-335-5p, which
promoted neuronal damage in IS (39). To further validate the functional
mechanism of circ_0000018 in the pathological process of AIS, the
target mRNA, miR-871, was determined and validated. Luciferase
reporter assays also showed the targeting association between
circ_0000018 and miR-871. Moreover, the downregulation of miR-871
reduced the suppressive functions of circ_0000018 knockdown on
OGD/R-treated cell injury, which confirmed that the protective
effect of circ_0000018 deficiency on AIS may be attributed, in
part, to the association with miR-871. As a result, the inhibition
of miR-871 can reverse the effect of knockdown circ_0000018 on
neuronal cells, suggesting that miR-871 may inhibit the development
of AIS.
As a member of the BCL-2 protein family, BCL2L11 is
located in the outer mitochondrial membrane, and plays an important
regulatory role in mediating excitatory apoptosis, induction of
gene sequence translocation, and mitochondrial depolarization
(40,41). It was found that miR-29b inhibited
the pro-apoptotic protein BCL2L11, Bcl-2 modifier, Bcl-2
interacting protein Harakir, and Bcl-2 binding component 3
(p53-upregulated modulator of apoptosis) during neuronal
development, and played an essential role in neuronal maturation
and inhibition of neuronal apoptosis (42). In the apoptotic pathway, Bax and
Bcl-2 are two important regulatory genes; Bcl-2 inhibits cell death
while Bax promotes it, and the ratio of Bcl-2/Bax is closely
associated with the sensitivity of a cell to undergoing apoptosis
(43). BCL2L11 has been validated
as a key regulator of the apoptosis of B-lymphocytes,
T-lymphocytes, macrophages, and granulocytes (44). Cheng et al (45) revealed the role of lncRNA-TUG1 in
promoting neuronal apoptosis through the regulation of the
miR-9/BCL2L11 axis in the context of cerebral ischemia, thus
potentially providing a novel therapeutic target for stroke. Of
note, in the present study, bioinformatics software predictions
revealed that miR-871 has a conserved binding site in the 3′-UTR
region of BCL2L11. The binding was further verified by luciferase
reporter gene assays. Furthermore, the present study confirmed that
BCL2L11 overexpression partially eliminated the inhibitory effect
of miR-871 on OGD/R-treated neuronal cell injury. Furthermore, in
neuronal cells, circ_00000018/miR-871 could regulate the expression
of BCL2L11, which supports the regulatory functions of the
circ_00000018/miR-871/BCL2L11 axis in AIS.
In conclusion, the findings of the present study
indicated that circ_0000018, the expression of which was
upregulated in the AIS model, may serve as a ceRNA for miR-871 to
influence the levels of BCL2L11 and participate in the progression
of AIS. Mechanistically, circ_0000018 knockdown relieved AIS in
vivo and in vitro by regulating the miR-871/BCL2L11
axis. Thus, circ_0000018 may serve as a novel target for AIS
treatment.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MJ and XBW conceived the study. MJ, XBW and SJ
performed the experiments. MJ analyzed the data. MJ wrote the
manuscript. All authors have read and approved the final
manuscript. SJ and XBW confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
This study was approved by the Southeast University
Animal Care and Use Committee (approval no. 20220110026).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Feigin VL, Lawes CM, Bennett DA,
Barker-Collo SL and Parag V: Worldwide stroke incidence and early
case fatality reported in 56 population-based studies: A systematic
review. Lancet Neurol. 8:355–369. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Inohara T, Liang L, Kosinski AS, Smith EE,
Schwamm LH, Hernandez AF, Bhatt DL, Fonarow GC, Peterson ED and
Xian Y: Recent myocardial infarction is associated with increased
risk in older adults with acute ischemic stroke receiving
thrombolytic therapy. J Am Heart Assoc. 8:e0124502019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang YJ, Li ZX, Gu HQ, Zhai Y, Jiang Y,
Zhao XQ, Wang YL, Yang X, Wang CJ, Meng X, et al: China stroke
statistics 2019: A report from the national center for healthcare
quality management in neurological diseases, China national
clinical research center for neurological diseases, the Chinese
stroke association, national center for chronic and
non-communicable disease control and prevention, Chinese center for
disease control and prevention and institute for global
neuroscience and stroke collaborations. Stroke Vasc Neurol.
5:211–239. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
GBD 2019 Stroke Collaborators, . Global,
regional, and national burden of stroke and its risk factors,
1990–2019: A systematic analysis for the global burden of disease
study 2019. Lancet Neurol. 20:795–820. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen Z, Jiang B, Ru X, Sun H, Sun D, Liu
X, Li Y, Li D, Guo X and Wang W: Mortality of stroke and its
subtypes in China: Results from a nationwide population-based
survey. Neuroepidemiology. 48:95–102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ebbesen KK, Kjems J and Hansen TB:
Circular RNAs: Identification, biogenesis and function. Biochim
Biophys Acta. 1859:163–168. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ostolaza A, Blanco-Luquin I,
Urdánoz-Casado A, Rubio I, Labarga A, Zandio B, Roldán M,
Martínez-Cascales J, Mayor S, Herrera M, et al: Circular RNA
expression profile in blood according to ischemic stroke etiology.
Cell Biosci. 10:342020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu F, Han B, Wu S, Yang L, Leng S, Li M,
Liao J, Wang G, Ye Q, Zhang Y, et al: Circular RNA TLK1 aggravates
neuronal injury and neurological deficits after ischemic stroke via
miR-335-3p/TIPARP. J Neurosci. 39:7369–7393. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peng X, Jing P, Chen J and Xu L: The role
of circular RNA HECTD1 expression in disease risk, disease
severity, inflammation, and recurrence of acute ischemic stroke. J
Clin Lab Anal. 33:e229542019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Van Rooij E, Purcell AL and Levin AA:
Developing micro RNA therapeutics. Circ Res. 110:496–507. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ghoreishy A, Khosravi A and Ghaemmaghami
A: Exosomal microRNA and stroke: A review. J Cell Biochem.
120:16352–16361. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen Y, Gao C, Sun Q, Pan H, Huang P, Ding
J and Chen S: MicroRNA-4639 is a regulator of DJ-1 expression and a
potential early diagnostic marker for Parkinson's disease. Front
Aging Neurosci. 9:2322017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Arena A, Iyer AM, Milenkovic I, Kovacs GG,
Ferrer I, Perluigi M and Aronica E: Developmental expression and
dysregulation of miR-146a and miR-155 in Down's syndrome and mouse
models of Down's syndrome and Alzheimer's disease. Curr Alzheimer
Res. 14:1305–1317. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Beveridge NJ, Tooney PA, Carroll AP,
Gardiner E, Bowden N, Scott RJ, Tran N, Dedova I and Cairns MJ:
Dysregulation of miRNA 181b in the temporal cortex in
schizophrenia. Hum Mol Genet. 17:1156–1168. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jeyaseelan K, Lim KY and Armugam A:
MicroRNA expression in the blood and brain of rats subjected to
transient focal ischemia by middle cerebral artery occlusion.
Stroke. 39:959–966. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dharap A, Bowen K, Place R, Li LC and
Vemuganti R: Transient focal ischemia induces extensive temporal
changes in rat cerebral microRNAome. J Cereb Blood Flow Metab.
29:675–687. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu DZ, Tian Y, Ander BP, Xu H, Stamova
BS, Zhan X, Turner RJ, Jickling G and Sharp FR: Brain and blood
microRNA expression profiling of ischemic stroke, intracerebral
hemorrhage, and kainate seizures. J Cereb Blood Flow Metab.
30:92–101. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Abe A, Tanaka M, Yasuoka A, Saito Y, Okada
S, Mishina M, Abe K, Kimura K and Asakura T: Changes in whole-blood
microRNA profiles during the onset and treatment process of
cerebral infarction: A human study. Int J Mol Sci. 21:31072020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Han B, Zhang Y, Zhang Y, Bai Y, Chen X,
Huang R, Wu F, Leng S, Chao J, Zhang JH, et al: Novel insight into
circular RNA HECTD1 in astrocyte activation via autophagy by
targeting MIR142-TIPARP: Implications for cerebral ischemic stroke.
Autophagy. 14:1164–1184. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
R Core Team, . R: A language and
environment for statistical computing. R Foundation for Statistical
Computing; Vienna, Austria: ISBN 3-900051-07-0. 2012
|
|
22
|
RStudio Team, . Rstudio: Integrated
development for R. Rstudio Inc.; Boston, MA: 2015
|
|
23
|
Liu W, Miao Y, Zhang L, Xu X and Luan Q:
MiR-211 protects cerebral ischemia/reperfusion injury by inhibiting
cell apoptosis. Bioengineered. 11:189–200. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wibrand K, Pai Bl, Siripornmongcolchai T,
Bittins M, Berentsen B, Ofte ML, Weigel A, Skaftnesmo KO and
Bramham CR: MicroRNA regulation of the synaptic plasticity-related
gene Arc. PLoS One. 7:e416882012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fiore R, Khudayberdiev S, Christensen M,
Siegel G, Flavell SW, Kim TK, Greenberg ME and Schratt G:
Mef2-mediated transcription of the miR379-410 cluster regulates
activity-dependent dendritogenesis by fine-tuning Pumilio2 protein
levels. EMBO J. 28:697–710. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Magill ST, Cambronne XA, Luikart BW, Lioy
DT, Leighton BH, Westbrook GL, Mandel G and Goodman RH:
microRNA-132 regulates dendritic growth and arborization of newborn
neurons in the adult hippocampus. Proc Natl Acad Sci USA.
107:20382–20387. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Olde Loohuis NFM, Kos A, Martens GJM, Van
Bokhoven H, Nadif Kasri N and Aschrafi A: MicroRNA networks direct
neuronal development and plasticity. Cell Mol Life Sci. 69:89–102.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Broderick JA and Zamore PD: MicroRNA
therapeutics. Gene Ther. 18:1104–1110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hu J, Xie C, Xu S, Pu Q, Liu H, Yang L,
Wang W, Mao L, Li Z and Chen W: Liver fibrosis-derived exosomal
miR-106a-5p facilitates the malignancy by targeting SAMD12 and
CADM2 in hepatocellular carcinoma. PLoS One. 18:e02860172023.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tuo X, Zhou Y, Yang X, Ma S, Liu D, Zhang
X, Hou H, Wang R, Li X and Zhao L: miR-532-3p suppresses
proliferation and invasion of ovarian cancer cells via
GPNMB/HIF-1α/HK2 axis. Pathol Res Pract. 237:1540322022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cheng Q, Chen M, Wang H, Chen X, Wu H, Du
Y and Xue J: MicroRNA-27a-3p inhibits lung and skin fibrosis of
systemic sclerosis by negatively regulating SPP1. Genomics.
114:1103912022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mehta SL, Pandi G and Vemuganti R:
Circular RNA expression profiles alter significantly in mouse brain
after transient focal ischemia. Stroke. 48:2541–2548. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu C, Zhang C, Yang J, Geng X, Du H, Ji X
and Zhao H: Screening circular RNA expression patterns following
focal cerebral ischemia in mice. Oncotarget. 8:86535–86547. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Duan X, Li L, Gan J, Peng C, Wang X, Chen
W and Peng D: Identification and functional analysis of circular
RNAs induced in rats by middle cerebral artery occlusion. Gene.
701:139–145. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dai Q, Ma Y, Xu Z, Zhang L, Yang H, Liu Q
and Wang J: Downregulation of circular RNA HECTD1 induces
neuroprotection against ischemic stroke through the
microRNA-133b/TRAF3 pathway. Life Sci. 264:1186262021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pei L, Xu X and Yuan T: Circ_0101874
overexpression strengthens PDE4D expression by targeting miR-335-5p
to promote neuronal injury in ischemic stroke. J Stroke Cerebrovasc
Dis. 31:1068172022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang Z, He J and Wang B: Circular RNA
circ_HECTD1 regulates cell injury after cerebral infarction by
miR-27a-3p/FSTL1 axis. Cell Cycle. 20:914–926. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Concannon CG, Tuffy LP, Weisová P, Bonner
HP, Dávila D, Bonner C, Devocelle MC, Strasser A, Ward MW and Prehn
JH: AMP kinase-mediated activation of the BH3-only protein Bim
couples energy depletion to stress-induced apoptosis. Cell Biol.
189:83–94. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kilbride SM, Farrelly AM, Bonner C, Ward
MW, Nyhan KC, Concannon CG, Wollheim CB, Byrne MM and Prehn JH:
AMP-activated protein kinase mediates apoptosis in response to
bioenergetic stress through activation of the pro-apoptotic Bcl-2
homology domain-3-only protein BMF. J Biol Chem. 285:36199–36206.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ouyang YB, Xu L, Lu Y, Sun X, Yue S, Xiong
XX and Giffard RG: Astrocyte-enriched miR-29a targets PUMA and
reduces neuronal vulnerability to forebrain ischemia. Glia.
61:1784–1794. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu L, Liu Y, Zhang T, Wu H, Lin M, Wang
C, Zhan Y, Zhou Q, Qiao B, Sun X, et al: Synthetic Bax-Anti Bcl2
combination module actuated by super artificial hTERT promoter
selectively inhibits malignant phenotypes of bladder cancer. Exp
Clin Cancer Res. 35:32016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Strasser A: The role of BH3-only proteins
in the immune system. Nat Rev Immunol. 5:189–200. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen S, Wang M, Yang H, Mao L, He Q, Jin
H, Ye ZM, Luo XY, Xia YP and Hu B: LncRNA TUG1 sponges microRNA-9
to promote neurons apoptosis by up-regulated Bcl2l11 under
ischemia. Biochem Biophys Res Commun. 485:167–173. 2017. View Article : Google Scholar : PubMed/NCBI
|