Introduction
Lung cancer is the leading cause of cancer deaths
worldwide due to its high mortality rate (1), with lung adenocarcinoma (LUAD) as the
most frequently occurring subtype, accounting for 38.5% of lung
cancer cases (2). Efforts have
been made to improve the survival outcome for patients with lung
cancer, with effective measures including the implementation of
lung cancer screening by low-dose computed tomography (3), the detection of potential druggable
genetic alterations by modern sequencing methods, and the invention
and application of new targeted therapies and immune checkpoint
inhibitors (4,5). Although these measures have modified
and improved the clinical outcomes, such as overall survival, for
patients with lung cancer, ~70% of patients with lung cancer are
still diagnosed at an advanced stage of disease (6) and with the prognosis falling below
expectations, despite these modern strategies (7). To improve the clinical outcomes,
clinicians should further evaluate experimental treatments and
outcomes to develop more effective therapeutic options for
LUAD.
The enhancer of the rudimentary homolog (ERH)
is a protein-coding gene analog that is highly conserved across
species. For example, human ERH is ~80% identical to its
protein analog in Drosophila melanogaster, DROER (8), located at chromosome 14 and with a
size of ~18,000 base pairs (9).
The translated protein is comprised of 104 amino acids, and it is
mostly localized to the nucleus (10). It is involved in numerous
fundamental biological processes, such as the pyrimidine metabolic
pathway, transcription control, cell cycle progression, DNA damage
response and repair, and mRNA splicing (11), whilst additionally interacting with
RNA Pol II-associated factors that are involved in microRNA
processing (12). The family of
small nuclear ribonucleoprotein polypeptides (SNRPs) has a crucial
role in tumorigenesis and its progression (13). Notably, ERH has been shown to
interact with the spliceosome protein, SNRPD3, and is required for
the mRNA splicing of centrosome-associated protein E (CENP-E)
(14).
ERH has been studied for its role in
malignancy since 2007, and to date, its upregulation has been
reported in numerous cancers, such as breast, ovarian and bladder
cancer (15). ERH
upregulation is associated with poor clinical outcomes in ovarian
cancer (16) and colorectal cancer
(9); however, a conflicting report
illustrated a favorable outcome in patients with gastric carcinoma
with upregulated ERH (17).
Cell behaviors influenced by ERH in cancer have not been
widely studied (18), although
existing evidence has revealed the association between ERH
and cell proliferation, inhibition of apoptosis, migration,
invasion and epithelial-mesenchymal transition (EMT) in different
cancer types (19). The roles of
ERH in LUAD remain unknown or only partially explored
(15), with further elucidation of
the expression pattern, prognostic implications and its functional
roles in pathogenesis required.
Advanced lung cancer, compared with early-stage lung
cancer, carries the worst clinical outcomes despite efforts
invested (20) and further efforts
are required to improve clinical outcomes, such as survival time.
The present study suggests that high-level expression of ERH
confers poor survival time in patients with LUAD with the most
plausible mechanisms dependent on the recruitment of
immunosuppressive cells such as myeloid-derived suppressor cells
(MDSCs) and regulatory T lymphocytes (Tregs), and enhancement of
invasive cellular functions such as migration and EMT. The results
of the present study elucidate the role of ERH in LUAD and
modifying its expression might be of potential benefit as a target
in lung cancer treatment.
Materials and methods
Patient sample collection
Eighteen participants diagnosed with LUAD were
recruited from January 2018 to December 2022 at the Division of
Thoracic Surgery and Division of Pulmonary and Critical Care
Medicine, Kaohsiung Medical University Hospital [approval nos.
KMUH-IRB-20180023, KMU-IRB-20200038, KMU-IRB-E(II)-20220175;
Kaohsiung, Taiwan R.O.C.]. All patients provided written informed
consent to participate. Eight out of eighteen paired adjacent
non-tumor lung and tumor tissues (Case nos. 1-8, Table I) obtained underwent deep RNA
sequencing (RNA-Seq) at a biotechnology company (Welgene, Inc.)
using the Solexa platform (Illumina, Inc.). RNA and small RNA
library construction was performed using a sample preparation kit
(Illumina, Inc.), following the protocol of the TruSeq RNA or Small
RNA Sample Preparation Guide. Data were submitted to the National
Center for Biotechnology Information Gene Expression Omnibus (GEO)
under accession number GSE236816.
 | Table I.Patients' clinicopathological
data. |
Table I.
Patients' clinicopathological
data.
| Case number | Age | Sex | Pathologic
findings | Stage (Tumor, Node
and Metastasis) |
|---|
| 1 | 46 | F | Adenocarcinoma | 1A (T1N0M0) |
| 2 | 50 | F | Adenocarcinoma | 1A (T1aN0M0) |
| 3 | 58 | F | Adenocarcinoma | 1A (T1aN0M0) |
| 4 | 63 | M | Adenocarcinoma | 1B (T2aN0M0) |
| 5 | 64 | F | Adenocarcinoma | 1A (T1aN0M0) |
| 6 | 48 | M | Adenocarcinoma | 1A (T1aN0M0) |
| 7 | 82 | M | Adenocarcinoma | 4 (T1bN2M1b) |
| 8 | 62 | F | Adenocarcinoma | 4 (T1bN0M1b) |
| 9 | 75 | M | Adenocarcinoma | 2B (T2bN1M0) |
| 10 | 65 | F | Adenocarcinoma | 3A (T2aN2M0) |
| 11 | 66 | M | Adenocarcinoma | 1A (T1aN0M0) |
| 12 | 70 | F | Adenocarcinoma | 1B (T2aN0M0) |
| 13 | 75 | F | Adenocarcinoma | 2B (T2bN1M0) |
| 14 | 50 | M | Adenocarcinoma | 1B (T1bN0M0) |
| 15 | 63 | F | Adenocarcinoma | 1A (T1aN0M0) |
| 16 | 57 | F | Adenocarcinoma | 2B (T1N1M0) |
| 17 | 76 | M | Adenocarcinoma | 2B (T2bN1M0) |
| 18 | 65 | M | Adenocarcinoma | 1A (T1aN0M0) |
Immunohistochemical staining
(IHC)
Ten pairs of adjacent non-tumor lung tissues and
lung cancer tissues (Case nos. 9-18, Table I) were utilized for IHC. The
designated tissue samples were fixed with 10% formalin at room
temperature for 1 h and the tissue block was embedded in paraffin.
The formalin-fixed paraffin-embedded tissue block was then split
into sections (8 µm). Xylene was used to dewax the sections and a
descending ethanol gradient was used for rehydration. Antigen
retrieval was heat-mediated using a pressure cooker for 90 sec.
Endogenous peroxidase activity was inactivated by incubation with
3% hydrogen peroxide for 10 min at room temperature, and
non-specific antibody binding was prevented by incubation with 3%
bovine serum albumin (MilliporeSigma) for 20 min at room
temperature. The sections were incubated with primary antibodies
against ERH (1:200; cat. no. NBP1-84976; Novus Biologicals, LLC)
and small nuclear ribonucleoprotein polypeptide G (SNRPG; 1:500;
cat. no. PA5-64155; Invitrogen; Thermo Fisher Scientific, Inc.) to
assess ERH and SNRPG protein expression at 4°C overnight followed
by incubation with horseradish peroxidase conjugated anti-rabbit
secondary antibodies (1:1,000; cat. no. ab6721, Abcam) for 20 min
at room temperature, followed by washing with PBS and 3,3′
diaminobenzidine staining for 2 min at room temperature. The
sections were counterstained with hematoxylin for 1 min at room
temperature. The results of IHC were imaged using an ICC50 HD light
microscope (Leica Microsystems, Inc.) at ×100 magnification.
Data validation using in-house samples
by reverse transcription-quantitative PCR (RT-qPCR) and IHC
To validate the findings from the public database,
the present study collected 8 human LUAD tumor tissues and adjacent
non-tumor tissues. Total RNA was extracted from tissues or cell
lines using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), and 50 ng mRNA was reverse transcribed into cDNA
using an oligo (dT) primer and reverse transcriptase (PrimeScript
RT Reagent Kit; Takara Bio, Inc.) according to the manufacturer's
protocols. RT-qPCR was performed on the human samples to quantify
the ERH mRNA expression levels. The primers used in the
experiment were as follows: ERH forward (F), ERH_ F1,
5′-CCTACCAAGAGGCCAGAAGG-3′ and reverse (R), ERH_ R1,
5′-TAAACCAGGCAGCTGAGGTC-3′; GAPDH F, 5′-TTCACCACCATGGAGAAGGC-3′ and
R, 5′-GGCATGGACTGTGGTCATGA-3′). qPCR was performed at 95°C for 20
sec, followed by 40 cycles at 95°C for 3 sec and 60°C for 30 sec.
The expression levels of specific genes were determined using a
StepOne-Plus PCR instrument (Applied Biosystems; Thermo Fisher
Scientific, Inc.) and SYBR-Green (Thermo Fisher Scientific, Inc.).
The relative expression levels of the specific mRNAs were
normalized to those of GAPDH. The relative standard method
(2−∆∆Cq) was used to calculate relative RNA expression
(21).
Data collection from public
databases
The gene expression profile of LUAD was obtained
from The Cancer Genome Atlas (TCGA) via the University of Santa
Cruz Xena database (https://xenabrowser.net/datapages/?host=https%3A%2F%2Fpancanatlas.xenahubs.net&removeHub=https%3A%2F%2Fxena.treehouse.gi.ucsc.edu%3A443;
accessed on June 15, 2022). For the protein expression pattern in
LUAD, data were extracted from The Human Protein Atlas website
(https://www.proteinatlas.org/ENSG00000100632-ERH/pathology/lung+cancer#img;
accessed on June 22, 2022). In the TCGA cohort, there were 2
tissues from healthy patients, 57 pairs of adjacent non-tumor and
tumor tissues, and 454 tumor tissues (total normal tissue, n=59;
total tumor tissues, n=511). Further analysis of the gene and
protein expression pattern across tissue types, cancer stages and
the extent of lymph node metastasis were extracted from the public
platform, the University of Alabama at Birmingham CANcer data
analysis Portal [the Clinical Proteomic Tumor Analysis Consortium
(CPTAC) and the International Cancer Proteogenome Consortium
datasets, UALCAN; http://ualcan.path.uab.edu]. The gene sets which
correlated, either positively or negatively, with ERH were
also identified using the UALCAN platform using the cutoff
correlation co-efficiency r>0.3 or r <-0.3, respectively. To
validate the gene expression results from TCGA, the Asian cohort
GSE31210 dataset (control=20, with 15 from adjacent non-tumor
tissues and 5 from normal tissues from healthy individuals;
tumor=226) from the GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE31210)
was used with MAS5 normalization (22). STRING is a database of known and
predicted protein-protein interactions for numerous genes,
including for ERH (https://string-db.org/; version 11.5, accessed on June
15, 2022).
Cell cultures
The Human A549 LUAD cell line (American Type Culture
Collection; CCL185) was cultured in F-12K Medium (Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.), 0.1 mg/ml streptomycin and
100 U/ml penicillin (Thermo Fisher Scientific, Inc.). Human CL1-0
LUAD cell line was donated by Professor Pan-Chyr Yang (National
Taiwan University, College of Medicine, Taiwan). The CL1-0 cell
line was cultured in RPMI 1640 medium supplemented with 10% FBS, 2
mM glutamine and 1% penicillin-streptomycin (Capsugel; Lonza Group,
Ltd.). Both cell lines were confirmed negative for mycoplasma
contamination using mycoplasma test kits (Mycoalert Mycoplasma
Detection Kit; Capsugel; Lonza Group, Ltd.) every 3 months.
Survival analysis of selected genes
using the Kaplan-Meier plotter (KM plotter)
The effect on survival of the genes of interest was
evaluated using the KM plotter (http://kmplot.com/analysis/; accessed on June 19,
2022) using data sourced from the TCGA and GEO databases. In the
survival analysis of LUAD in the KM plotter, data from gene chip
microarray or RNA-Seq were used. Subjects were divided into two
groups based on their high or low RNA expression levels, with the
best cutoff value automatically computed. The hazard ratios for
survival, including overall survival, time to first progression and
post-progression survival, were calculated using the Cox
proportional-hazards model during a pre-defined 60-month period. In
addition, cross-analysis was utilized to compare the survival rates
between patient cohorts with high and low expression of a specific
gene (bisection) (http://kmplot.com/analysis/) (23). P<0.05 was considered to indicate
a statistically significant difference.
Functional analysis
The biological functions of ERH were
evaluated using CancerSEA (http://biocc.hrbmu.edu.cn/CancerSEA/home.jsp; accessed
on June 8, 2023). Gene set enrichment analysis (GSEA) is a
computational tool that computes correlations between biological
functions or pathological states with a pre-defined gene set. In
the present study, the subjects in the TCGA LUAD cohort were
divided into ERH high- and low-expression groups that were
defined as the 1st and 4th quartiles (highest 25% and lowest 25%)
to enhance the significance. GSEA was then used to analyze the
enrichment of biological functions in ERH high- and
low-expression groups. A false discovery rate of <0.05 and
P<0.05 were set as the cutoff criteria. The gene set
‘c2.cp.kegg.v6.2.symbols.gmt’ was used as the reference.
Gene set analysis in Gene Set
Variation Analysis (GSVA) and Metascape
The gene sets either positively or negatively
correlated with ERH were extracted using UALCAN. The
criteria set for the analysis were Pearson correlation coefficient
>0.3/<-0.3 and P<0.05, which were calculated using UALCAN.
The GSVA score of the gene sets with regards to gene expression,
survival rate and immune infiltration was calculated using Gene Set
Cancer Analysis (GSCA) (24)
(http://bioinfo.life.hust.edu.cn/GSCA/#/; accessed on
July 19, 2022). Metascape (https://metascape.org/gp/index.html#/main/step1;
version 3.5, accessed on July 23, 2022) was also used to provide a
comprehensive gene list annotation and analysis for the
ERH-positively correlated gene set. The correlated pathways
enriched with the target gene list were presented as a heatmap and
a clustergram (25).
Analysis of the ERH-associated immune
microenvironment
Tumor Immune Estimation Resource 2.0 (TIMER2.0;
http://timer.cistrome.org/; accessed on
July 15, 2022) was used for the analysis of ERH-associated
infiltrating immune cells (26).
The ‘gene module’ was used to select and visualize ERH
protein expression with its correlated immune cell infiltration
level in LUAD. GSCA was also utilized for immune cell abundance
analysis of the ERH-positively or negatively correlated gene
set (22). Using the ‘Immune cell
abundance’ module, the correlation between immune infiltration and
the calculated GSVA score was calculated.
ERH knockdown by siRNA
transfection
A549 and CL1-0 cells were transfected with
ERH-siRNA or control-siRNA (10 nM) using ON-TARGET plus
SMARTpool siRNA and Dharmafect reagents No1 (GE Healthcare
Dharmacon, Inc.) at 37°C for 48 h. The knockdown efficacy of
ERH siRNA was determined by RT-qPCR at 48 h
post-transfection according to aforementioned methods,
respectively. The sequences were as follows: control siRNA:
UGGUUUACAUGUCGACUAA, UGGUUUACAUGUUGUGUGA, UGGUUUACAUGUUUUCUGA and
UGGUUUACAUGUUUUCCUA and ERH-siRNA: AGACAUACCAGCCUUAUAA,
GGGAAAUAAUUGUGUUGGA, AAGAGAAGAUCUACGUGCU and UAGCCAAGAUUGACUGUAU,
respectively. The subsequent experiments were performed 48 h
post-transfection.
Cell proliferation assay
The cell proliferation of control siRNA or
ERH siRNA transfected A549 cells and CL1-0 cells were
assessed using the WST-1 assay (MilliporeSigma) for a 72 h
incubation, according to the manufacturer's protocol.
Wound healing assay
A total of 2×105 ERH siRNA or
control-siRNA transfected A549 and CL1-0 cells were seeded and
allowed to grow to ~90% confluence in 24-well plates cultured in
media which contained 1% FBS (27). The following day, a uniform scratch
was made down the center of the well using a micropipette tip,
followed by washing once with phosphate-buffered saline (PBS).
Imaging of wound healing at 0 and 24 h was performed using an
Olympus inverted light microscope.
Cell migration assay
A cell migration assay was performed using a
Transwell system, as previously described (28). Briefly, 3×104 ERH
siRNA and control siRNA transfected A549 and CL1-0 cells were
seeded into the top inserts with serum-free medium and incubated
for 10 min and complete medium containing 10% FBS was placed in the
bottom well and cultured at 37°C in an incubator. After 24 h, the
migratory cells on the bottom of inserts then were fixed in 4%
paraformaldehyde for 20 min at room temperature and stained with
0.1% crystal violet overnight at room temperature. The migratory
cells were counted in 10 random microscope fields for each sample
using a light microscope (Nikon Corporation).
Western blotting
The total protein of ERH-knockdown and
control A549 and CL1-0 cells was extracted using RIPA buffer (cat.
no. 20-188; MilliporeSigma), supplemented with a protease inhibitor
mixture (S8830-20TAB, MilliporeSigma). An equal volume of total
protein, 25 µg, was denatured by heating and then separated by
electrophoresis using 12% sodium dodecyl-sulfate polyacrylamide
gels. Proteins in the gel were transferred onto polyvinylidene
difluoride membranes (MilliporeSigma) by electroblotting, which was
probed with primary antibodies overnight after blocking in 5%
non-fat dry milk/0.1% TBST at room temperature (25°C). Primary
antibodies against N-cadherin (1:1,000; cat. no. 610921),
E-cadherin (1:1,000; cat. no. 610182), and Vimentin (1:1,000; cat.
no. 550513) were purchased from Becton, Dickinson and Company.
Anti-ERH antibodies (1:1,000; cat. no. NBP1-84976) were
purchased from Novus Biologicals, LLC and anti-GAPDH (1:5,000; cat.
no. MAB374) antibodies were from MilliporeSigma. After incubation
with HRP-conjugated secondary antibodies (1:5,000; anti-mouse, cat.
no. 7076; anti-rabbit, cat. no. 7074; Cell Signaling Technology,
Inc.). The signal of the specific protein was detected using a
chemiluminescence kit (MilliporeSigma). The western blot was
semi-quantified using ImageJ (version 1.53; National Institutes of
Health) and each experiment was repeated independently at least
three times.
Statistical analysis
The raw data extracted from GSE31210 and the results
of the cell functional assays were re-plotted using GraphPad Prism
5 software (GraphPad Software; Dotmatics). One-way ANOVA was used
for analysis when comparing more than two groups with Tukey's post
hoc test. Unpaired Student's t-test was used to compare mean values
between different subjects and cell groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
Higher expression of ERH in LUAD
confers a poor prognosis
Using the public transcriptomic database TCGA, the
mRNA-Seq profile of LUAD was obtained and the levels of ERH
mRNA expression were demonstrated to be significantly higher in
tumor tissues (n=511) than in the adjacent non-tumor tissues (n=59)
(Fig. 1A). Significantly elevated
ERH expression was demonstrated across different tumor
stages and at different extents of lymph node involvement, although
the increase did not demonstrate stage- or lymph node
stage-dependence (Fig. 1B and C).
To validate these findings in the in-house cohort, RT-qPCR for
ERH was performed. Higher ERH expression in tumors
was demonstrated in 6 out of 8 samples, ranging from a
log2 fold change of 1.26 to 1.71 compared with the
adjacent non-tumor baseline; by contrast, ERH expression was
lower in 2 of the 8 samples (Fig.
1D).
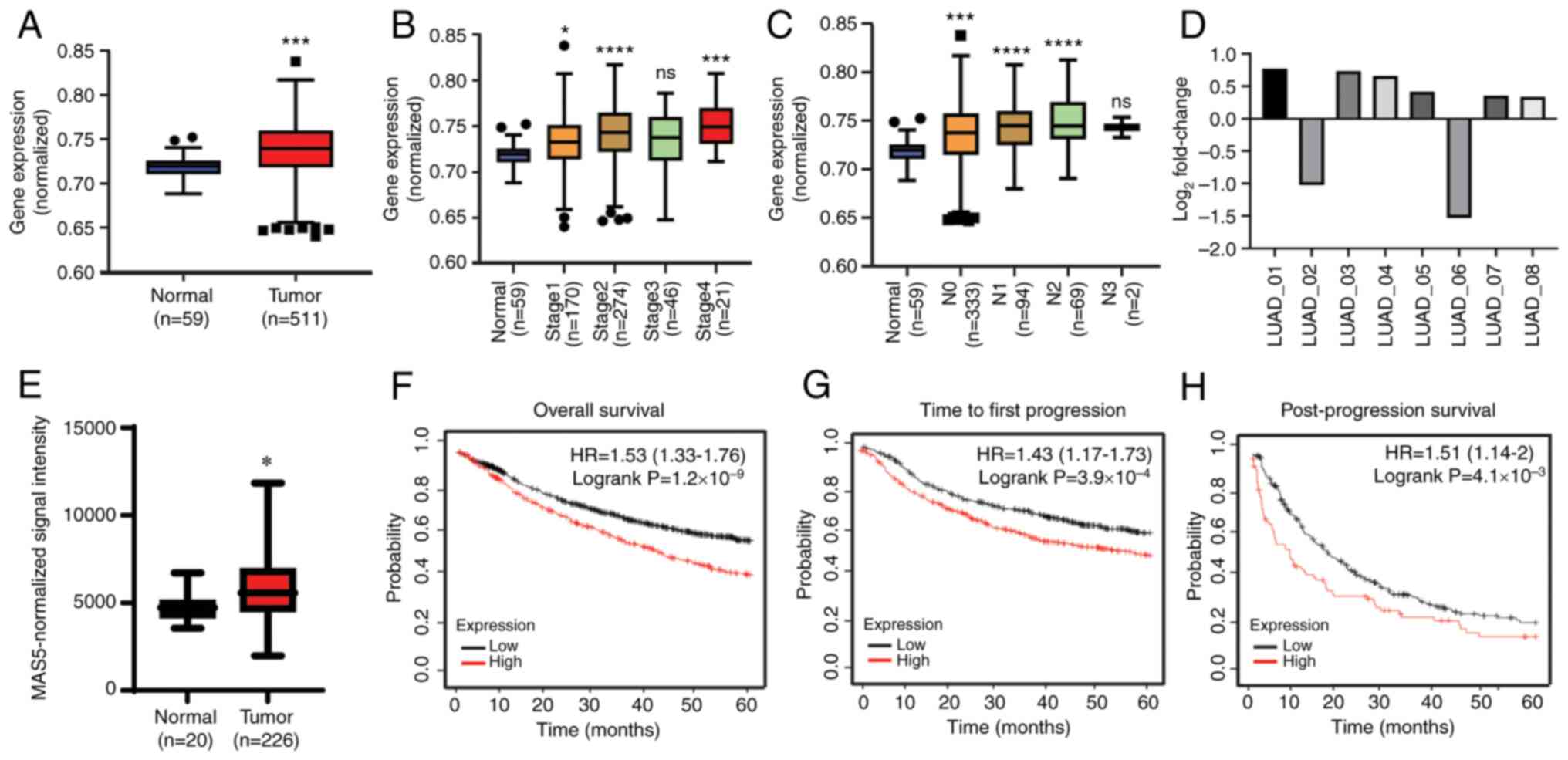 | Figure 1.High expression of ERH in LUAD
confers poor prognosis. The levels of ERH mRNA expression in
non-tumor tissues vs. tumor tissues using the (A) TCGA LUAD
transcriptomic dataset, (B) tumor stages, and (C) extents of lymph
node involvement. N1, ipsilateral hilum; N2, ipsilateral
mediastinal or subcarinal lymphadenopathy; N3, contralateral
mediastinal or contralateral hilar lymphadenopathy or scalene or
supraclavicular node. (D) ERH expression in eight pairs of
adjacent non-tumor vs. tumor in-house LUAD samples and (E)
non-tumor tissues vs. tumor tissues in a public transcriptomic
profile of LUAD, GSE31210, after MAS5 normalization. Survival
analysis based on ERH expression in LUAD in terms of (F)
overall survival, (G) time to first progression and (H)
post-progression survival. *P<0.05; ***P<0.005;
****P<0.001 vs. normal. ERH, enhancer of rudimentary homolog;
LUAD, lung adenocarcinoma; TCGA, The Cancer Genome Atlas; HR,
hazard ratio. |
The results were also validated in another public
transcriptomic dataset, GSE31210, to assess the association between
ERH expression and ethnicity. This dataset exclusively
includes an Asian population and similarly, the expression of
ERH was significantly increased in the tumor tissues
(n=226) compared with the mixed healthy and adjacent normal
tissues [paired adjacent non-tumor tissues (n=15) and normal
tissues from healthy individuals (n=5)] (P<0.05; Fig. 1E). Moreover, the level of
ERH expression in LUAD was significantly linked with worse
outcomes (Fig. 1F-H). The
prognostic profiles regarding overall survival, time to first
progression and time of post-progression survival were obtained
from the KM plotter and as illustrated, for the higher levels of
ERH expressed in tumors, a significantly lower probability
of each survival parameter was notably demonstrated in the
pre-defined 60-month period. To summarize, higher levels of
ERH were expressed in tumor tissues compared with those in
normal tissues across different ethnicities and higher expression
levels of ERH were significantly associated with reduced
survival times. This indicated that ERH may serve a critical
role in LUAD.
ERH protein expression is upregulated
in LUAD
The ERH protein expression pattern in LUAD was
further evaluated. The proteomic data obtained from the CPTAC
demonstrated a significantly enhanced expression pattern of ERH
protein in tumor tissues compared with adjacent non-tumor tissues
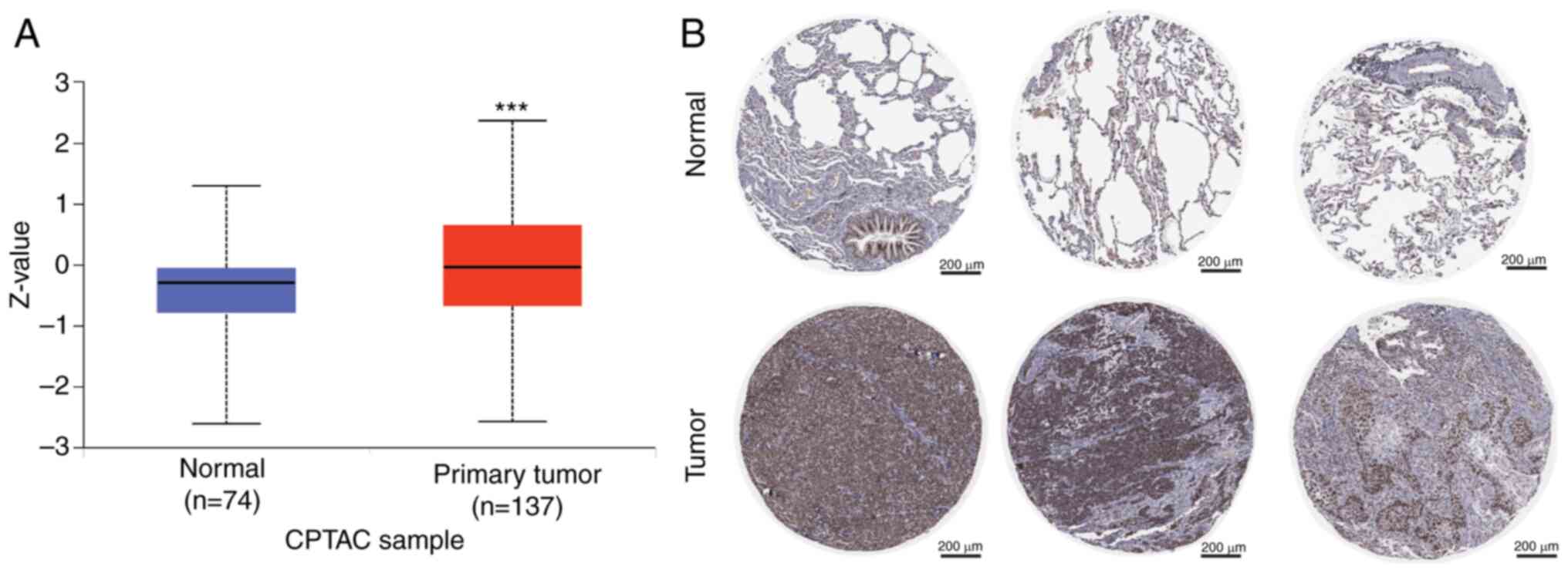
(median Z-score, 0.027 vs. −0.613; Fig. 2A). IHC staining images in the Human
Protein Atlas demonstrated that there was a higher intensity of ERH
expression in tumors compared with in normal tissues (healthy
control) (Fig. 2B). These results
indicated that ERH might be critical in LUAD at the protein
level observed in the public cohorts and the in-house cohort.
ERH expression correlates with
aggressive functional states in LUAD
The CancerSEA website was utilized to elucidate the
corresponding biological function of ERH. It was
demonstrated that in LUAD, ERH expression was significantly
positively correlated with a broad range of functional behaviors
towards cancer progression, such as angiogenesis, apoptosis, cell
cycle, differentiation, DNA damage, DNA repair, EMT, hypoxia,
invasion, metastasis, proliferation, and stemness. Among these, DNA
damage/repair, cell cycle and invasion were the most correlated
aggressive functions with ERH expression after setting a
cutoff correlation strength of 0.3 (Fig. 3A). Furthermore, when linked to
biological processes such as cell cycle, proliferation, invasion,
EMT and metastasis using the GSEA method in lung cancer, the
ERH-correlated gene set was strongly associated with poor
survival and chemo-resistance (Fig.
3B). This bioinformatics evidence demonstrated the roles of ERH
in DNA damage/repair, cell cycle and invasion of LUAD
progression.
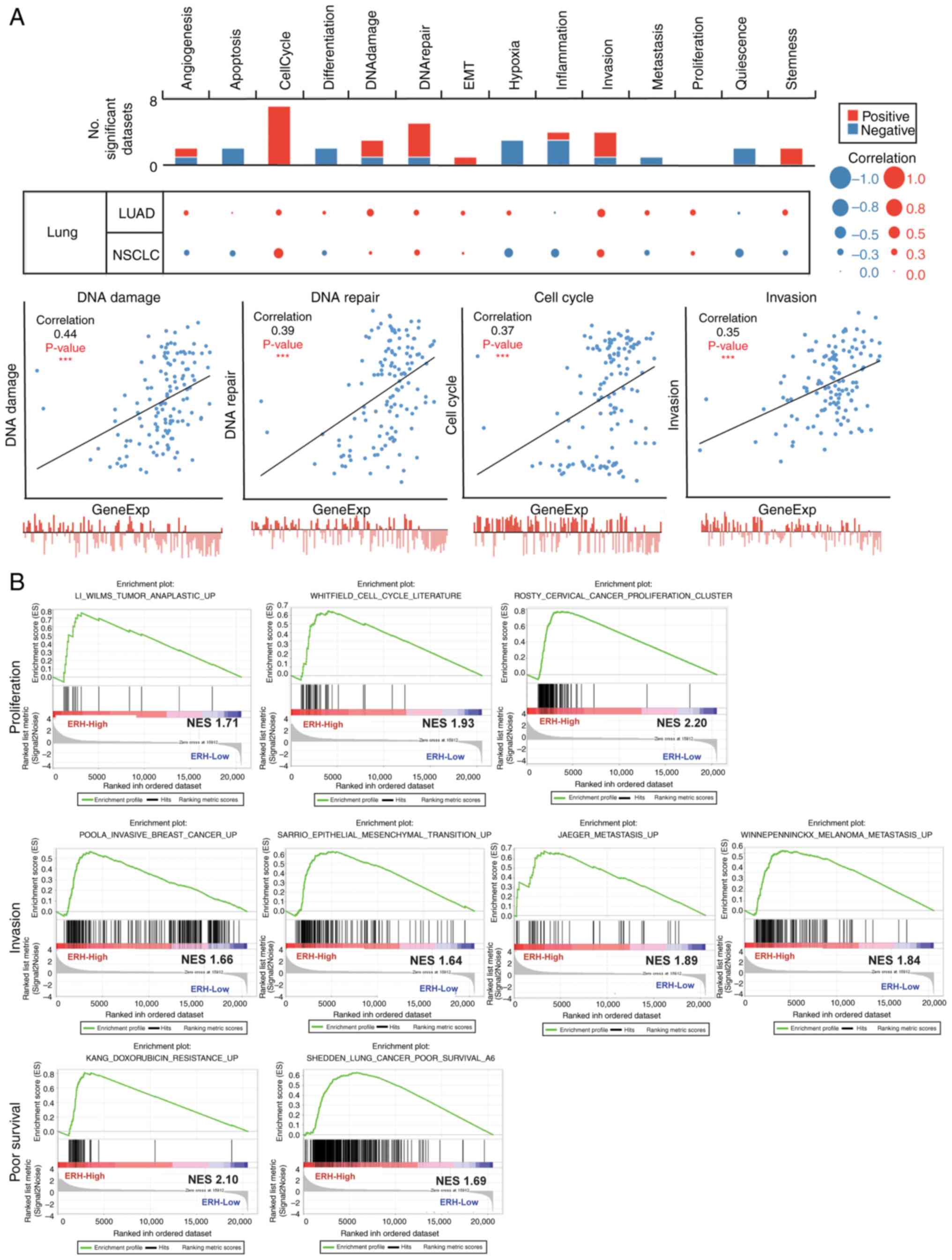 | Figure 3.ERH expression correlates with
aggressive functional states in LUAD. (A) The correlation between
ERH expression and 14 functional states in LUAD, with data obtained
from the CancerSEA database (upper panel). The correlation strength
>0.3 (lower panel). (B) ERH-correlated gene sets and their
functional behaviors toward cancer progression, such as cell cycle
and proliferation (upper panel), invasiveness,
epithelial-mesenchymal transition and metastasis (middle panel),
and chemotherapy resistance and poor survival (lower panel),
obtained using Gene Set Enrichment Analysis. ***P<0.001. ERH,
enhancer of rudimentary homolog; LUAD, lung adenocarcinoma; EMT,
epithelial-mesenchymal transition; NSCLC, non-small cell lung
cancer; NES, normalized enrichment score. |
SNRPG potentially interacts with
ERH
Ten molecules that potentially interact with
ERH were identified using the STRING database data on
protein-protein interactions. These molecules included DDX39B,
CHTOP, HNRNPH1, MATR3, SNRPD1, SNRPD2), SNRPD3, SNRPE, SNRPF, and
SNRPG (Fig. 4A). Moreover, all
were strongly correlated with ERH expression in the TCGA
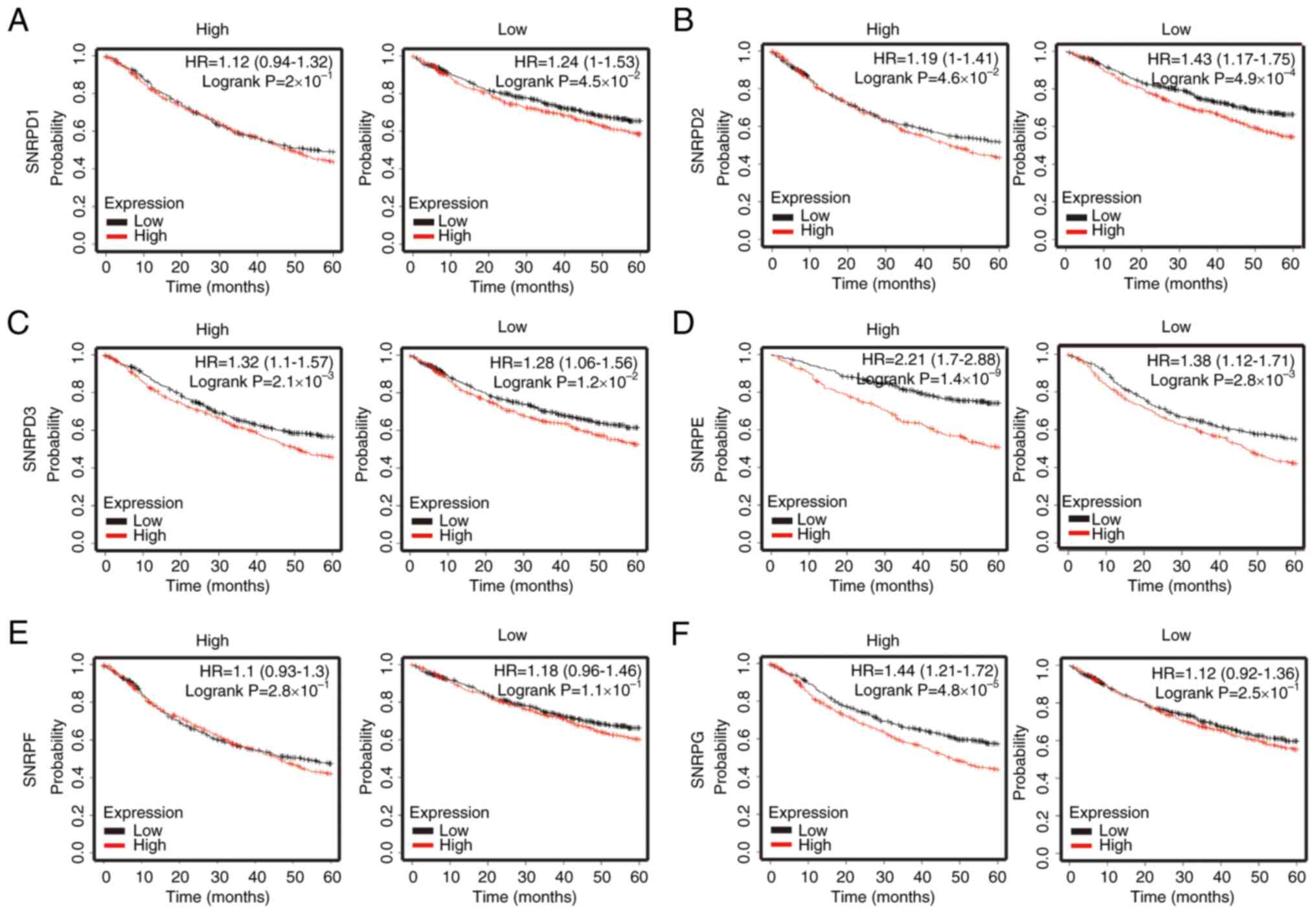
LUAD dataset with a correlation strength r>0.4 (Fig. 4B). Among the potentially
interactive factors, the upregulation of SNRPD1, SNRPD2, SNRPD3,
SNRPE, SNRPF and SNRPG were linked to significantly
shorter survival times, determined by the best cutoff method,
whereas upregulation of MATR3 was associated with a
significantly better prognosis (Fig.
4C). Survival-associated molecules were validated using the
CPTAC database, from which the available protein expressions of
SNRPD3, SNRPE and SNRPG were demonstrated to be significantly
different between tumor and non-tumor samples, whereas SNRPD1 and
SNRPD2 were expressed in similar amounts between tumor and
non-tumor parts. However, only SNRPG protein demonstrated greater
expression in primary tumor samples (Fig. 4D), which was in accordance with the
presumed expression pattern of mRNA in the TCGA. In addition, 7 out
of 10 of the tumor samples (with the exception of patient Nos 1, 8
and 9) in the in-house cohort were strongly stained by ERH or SNRPG
antibodies compared with adjacent non-tumor samples. In these 7
patients, higher expression of ERH was accompanied by elevated
SNRPG expression (Fig. 4E). SNRPG
expression was associated with ERH expression and shorter survival
duration.
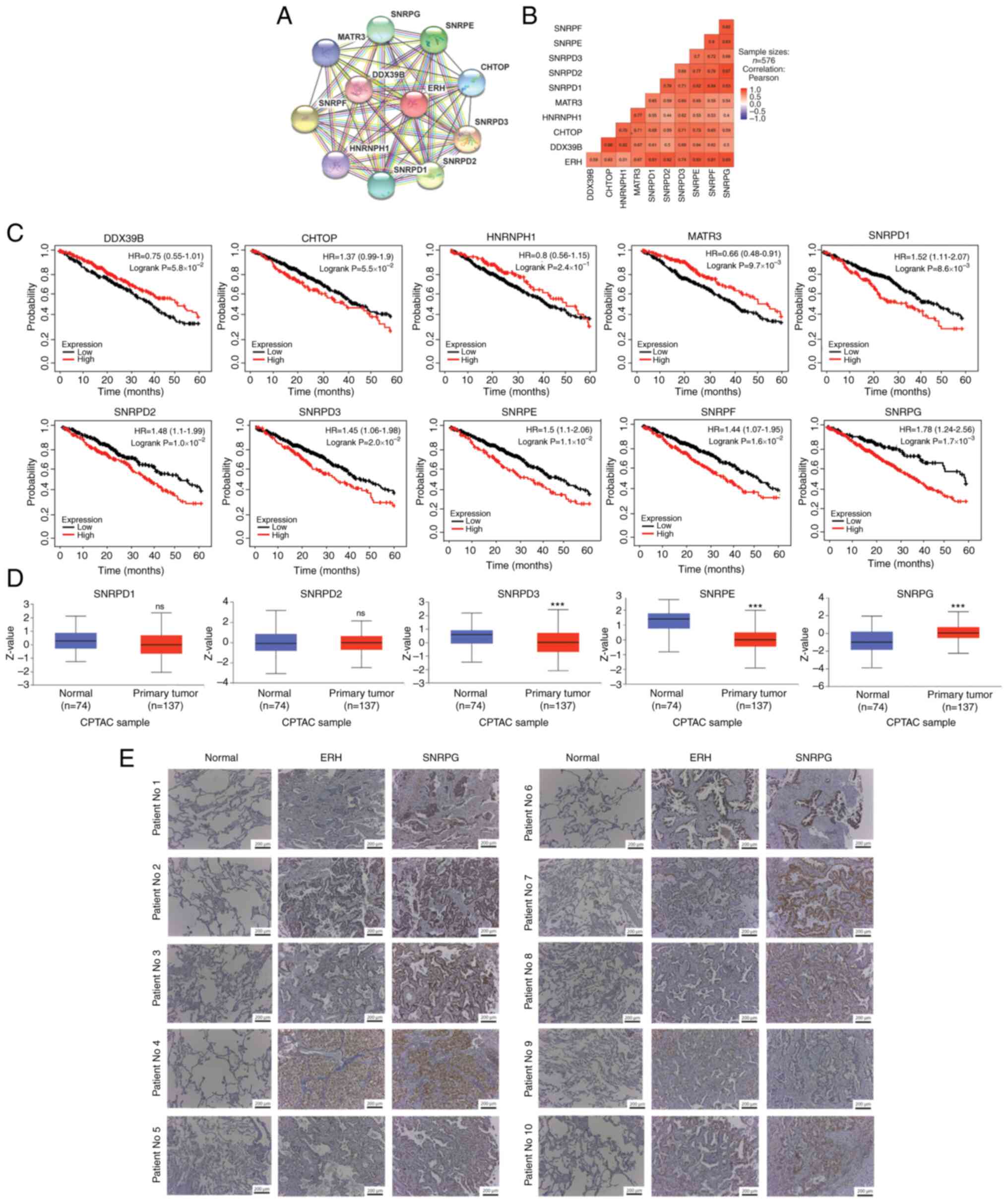 | Figure 4.SNRPG and its potential interacting
molecules with ERH. (A) Ten molecules potentially interacted with
ERH using data from the STRING database. (B) The correlations
between ERH and each candidate gene at the mRNA level in the LUAD
cohort of TCGA. (C) Survival analysis of DOX39B, CHTOP, HNRNPH1,
MATR3, SNRPD1, SNRPD2, SNRPD3, SNRPE, SNRPF and SNRPG. (D)
Validation of the survival-associated molecules in tumor and
non-tumor parts using data from the CPTAC database. (E)
Immunohistochemistry staining of ERH and SNRPG collected from the
in-house paired LUAD samples, magnification ×10. ***P<0.001; NS,
non-significant; SNRPG, small nuclear ribonucleoprotein polypeptide
G; ERH, enhancer of rudimentary homolog; CPTAC, Clinical Proteomic
Tumor Analysis Consortium; HR, hazard ratio. |
The survival effect of ERH is altered
by SNRPG mRNA expression levels
To assess the potential interacting effect of small
nuclear ribonucleoproteins (SNRs) with ERH, the effect of
ERH expression on the survival time of patients with LUAD
based on high or low levels of various SNRs was evaluated using
cross-analysis. Analysis using the KM plotter demonstrated that the
overall survival time of patients with high ERH expression
was significantly shorter than that of patients with low ERH
expression in the high expression subgroups of SNRPD2, SNRPD3,
SNRPE and SNRPG (Fig. 5B-D
and F). These data indicated a potential SNRPG-dependent
interaction that governed the survival impact of ERH.
Gene set positively correlated with
ERH is associated with poor survival and an immunosuppressive tumor
microenvironment (TME)
The top 200 genes either positively or negatively
correlated with ERH were extracted from the LUAD cohort of
TCGA. The positive and negative gene sets were calculated via the
GSCA website to acquire the GSVA scores. The scores provide the
significance of the gene set in terms of survival (29). It was demonstrated that the GSVA
scores for the ERH positively correlated gene set were
significantly higher in LUAD tumor samples than normal tissue
samples (Fig. 6A). Inversely, the
GSVA scores for the ERH negatively correlated gene set were
significantly lower in LUAD tumor samples than normal tissue
samples (Fig. 6B). Markedly higher
GSVA scores for the ERH positively correlated gene set were
linked to advanced cancer stages (Fig.
6C). However, higher GSVA scores for the ERH negatively
correlated gene set demonstrated a declining trend as the cancer
stage advanced (Fig. 6D). Higher
GSVA scores for the ERH positively-correlated gene set were
associated with significantly worse patient outcomes in terms of
overall survival (OS; HR=1.45), progression-free survival (PFS;
HR=1.37) and disease-specific survival (DSS; HR=1.79), but not with
disease-free survival (DFI; HR=1.28, Cox P=0.25) (Fig. 6E). In a reverse pattern, higher
GSVA scores for the ERH negatively correlated gene set were
linked to significantly better patient outcomes in terms of OS
(HR=0.66) and DSS (HR=0.57), but not PFS (HR=0.78, Cox P=0.05) and
DFI (HR=0.90, Cox P=0.62) (Fig.
6F). Analysis using the Metascape website demonstrated that the
ERH positively correlated gene set was associated with
certain biological processes, such as the ‘cell cycle, DNA
metabolic process, S phase and retinoblastoma gene in cancer’
(Fig. 6G). Associated gene
clusters were illustrated as an enriched ontology cluster network
(Fig. 6H). Further immune
microenvironment analysis was undertaken using the TIMER2.0
website, which demonstrated that ERH gene expression was
significantly positively correlated with the infiltration of MDSCs
in tumors with a ρ-value of 0.435 but not purity (ρ=0.097)
(Fig. 6I). Using the GSCA website,
the infiltrating immune cells corresponding to the GSVA scores for
the ERH positively correlated gene set were demonstrated to
include natural regulatory T cells (nTregs), effector memory T
cells, T helper type 1 cells and exhausted T cells. Among those,
nTregs demonstrated the highest correlation (Fig. 6J). The ERH negatively
correlated gene set correlated with the infiltration of cells such
as CD4 T cells, follicular helper T cells, natural killer T (NKT)
cells and natural killer (NK) cells (Fig. 6K). Based on the aforementioned
findings, ERH and its correlated genes expressed in LUAD
were associated with the infiltration of immune-suppressive cells,
such as MDSCs and nTregs, which led to a microenvironment against
immune surveillance.
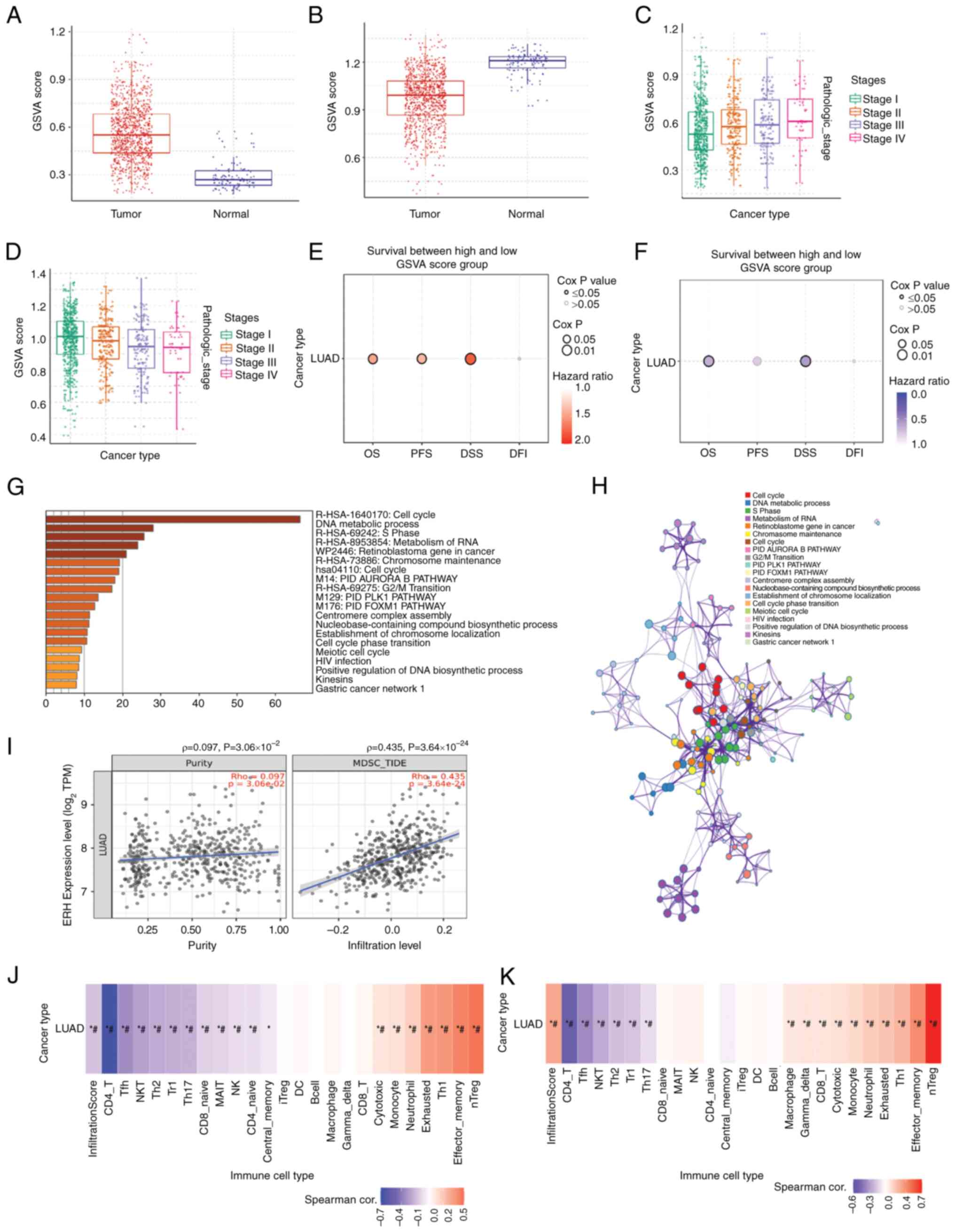 | Figure 6.ERH positively correlated gene set is
associated with worse survival and an immunosuppressive tumor
microenvironment. (A) The GSVA score of the ERH positively
correlated gene and (B) ERH negatively correlated gene set between
LUAD tumor samples and normal samples. The association between the
advanced stage and GSVA score of (C) the ERH positively correlated
gene or (D) the ERH negatively correlated gene set. The association
between survival analysis and GSVA score of (E) the ERH positively
correlated gene set or (F) the ERH negatively correlated gene set.
(G) The biological processes associated with the ERH positively
correlated gene set, using data from the Metascape database. (H)
The ERH-associated gene clusters using an enriched ontology cluster
network. (I) Infiltration of myeloid-derived suppressor cells in
tumors based on ERH expression using TIMER2.0. Infiltrating immune
cells corresponding to the GSVA score of the ERH (J) positively-
and (K) negatively-correlated gene sets. *P≦0.05;
#FDR≦0.05. ERH, enhancer of rudimentary homolog; GSVA,
Gene Set Variation Analysis; LUAD, lung adenocarcinoma; Tfh, T
follicular helper; NKT, natural killer T; Th2, T helper 2; Tr1,
Type 1 regulatory; Th17, T helper 17; MAIT, mucosal-associated
invariant; NK, natural killer; iTreg, induced regulatory T; DC,
dendritic; Th1, T helper 2; nTreg, natural regulatory T. |
ERH promotes EMT and cell migration in
vitro
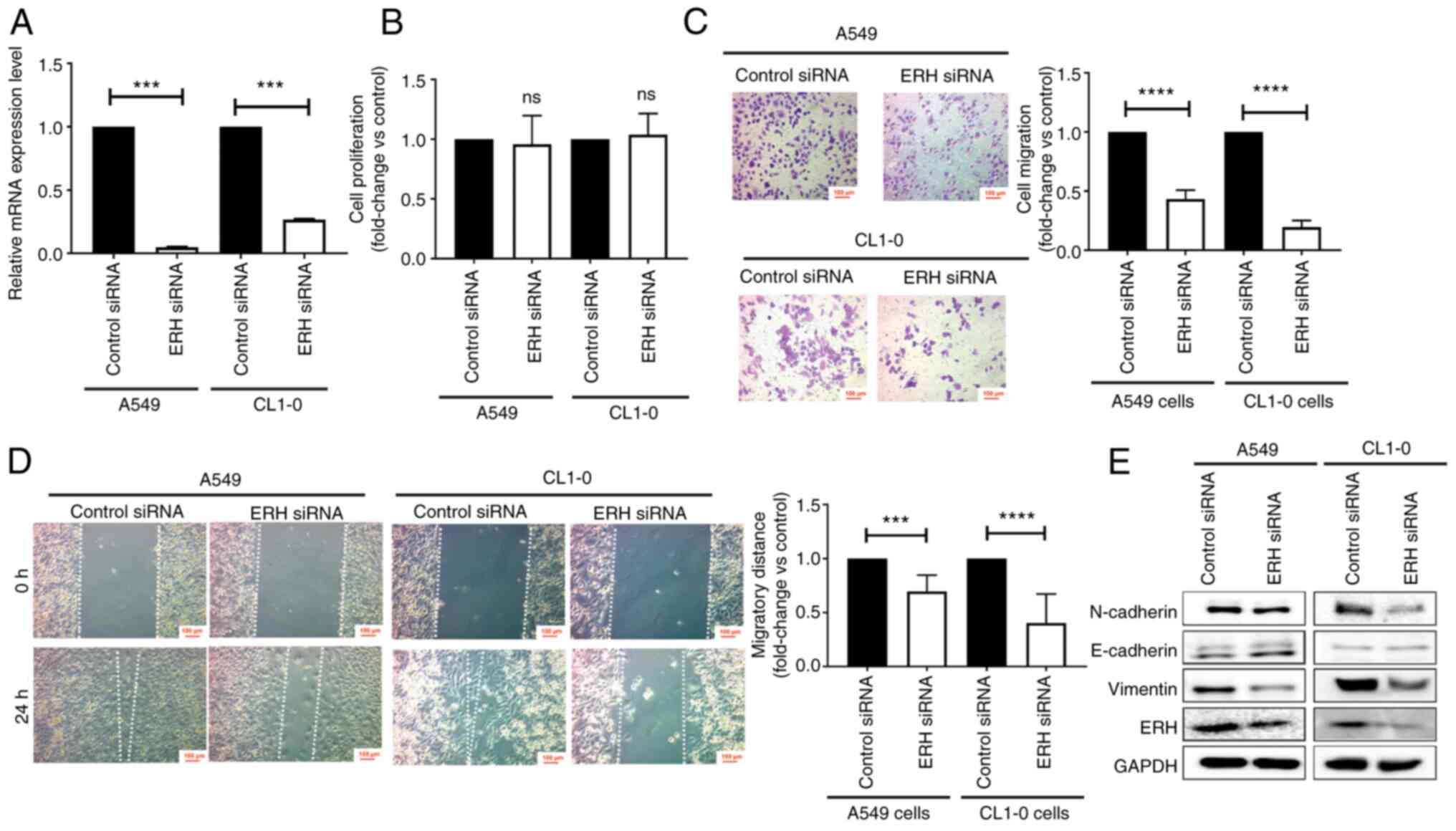
GSEA revealed an association between high ERH
expression and cancer EMT and invasiveness. To validate the
findings in vitro, cellular functional studies were designed
using siRNAs to knock down ERH with high knockdown
efficiency (Fig. 7A). There was no
significant difference in the proliferation of A549 and CL1-0 cells
with ERH knockdown compared with that of the control cells
(Fig. 7B). To evaluate cell
migration, Transwell migration and wound healing assays were
performed, with the percentage of migrated cells and the distance
of migration both significantly reduced in ERH-knockdown
A549 and CL1-0 cells (Fig. 7C and
D). These data supported the potential of ERH to enhance
cell migration. Concerning EMT, the expression of mesenchymal
markers, such as N-cadherin and vimentin, notably declined, whereas
the epithelial marker E-cadherin was slightly elevated in
ERH-knockdown A549 and CL1-0 cells (Fig. 7E). These results suggested that
ERH regulated promotion and progression steps in LUAD, but
not proliferation.
Discussion
To identify potential cures for lung cancer, more
research efforts into the underlying mechanism of lung cancer and
drug discovery are required. In the present study, ERH was
demonstrated to be highly expressed in LUAD and was significantly
associated with a poor prognosis. Meanwhile, upregulated ERH
and its correlated gene set were linked to LUAD progression via
control of the cell cycle, proliferation, invasion, EMT and
metastasis. In addition, ERH was enriched in an immune
suppressive microenvironment with a high fraction of MDSCs and
nTregs. The SNRPG was the most critical interactive molecule
which mediated the effects of ERH as the survival influence
of ERH was likely reliant on an SNRPG-dependent
mechanism. Therefore, ERH could be a potential target for
LUAD treatment and manipulation of ERH and its interactive
partner, SNRPG, could alter LUAD cellular behaviors and the
immune microenvironment, consequently enhancing the sensitivity of
immunotherapy.
Although ERH-related proliferation, invasion
and migration have been previously studied in numerous cancer types
(15,16), previous reports of the role of
ERH in lung cancer are limited. To the best of our
knowledge, only one previous study has reported the retarded growth
of lung cancer cells resulting from the reduction of ERH and
this was mediated by microRNA-574-3p over-expression secondary to
irradiation in vitro (30).
In the present study, it was proposed that functions of ERH
might promote LUAD via cell cycle progression, proliferation,
invasion, EMT and metastasis as determined by bioinformatics
analysis. ERH-associated cellular behaviors were assessed
using A549 and CL1-0 cells, and the knockdown of ERH reduced
cancer cell migratory ability and reversed the expression pattern
of EMT markers. ERH has been linked to cellular
proliferation (30,31), however the present study did not
observe that knockdown of ERH affected cellular
proliferation. Silencing of ERH led to reduced migration in
the present study, which is compatible with the effects of
ERH on tumor cell promotion and progression in bladder
cancer (15) and ovarian cancers
(16). These results add to the
knowledge concerning lung cancer behaviors.
The TME is critical in the formation of immunity,
tumor progression and metastasis, where chronic inflammatory status
might alter immune cell adaptation which imbalances anti-cancer
activity and favors immune evasion (32). MDSCs and Tregs serve major roles in
tumor-associated immunosuppression and are associated with poor
clinical outcomes in patients with lung cancer (33). In the present study, the
observations of ERH overexpression and the
immune-suppressive microenvironment were also unprecedented. First,
ERH gene expression was demonstrated to be positively
correlated with the infiltration of MDSCs in tumors using TIMER2.0.
Second, the infiltrating immune cells corresponding to the GSVA
score of the ERH positively correlated gene set were nTregs.
Finally, the ERH negatively correlated gene set correlated
with the infiltration of cancer killing NKT and NK cells.
ERH was also demonstrated to have influenced the
infiltration of immune cells in the lung cancer microenvironment,
where high ERH expression was associated with a high
percentage of MDSCs and nTregs. In contrast, low ERH
expression was related to a high percentage of NKT and NK cells.
Therefore, downregulation of ERH might increase infiltration
and functions of NKT/NK cells in lung cancer, thereby improving the
disease outcome of LUAD patients.
SNRPG, also termed as Smith Protein G,
belongs to the SNRP family which constructs the main unit of
spliceosomes and manages mRNA splicing (34). SNRPG is a part of the U1, U2, U4
and U5 small nuclear ribonucleoprotein (snRNP) complexes, whilst
SNRPG is reportedly also a component of the U7 snRNP complex that
takes part in the splicing of the 3′ end of histone transcripts
(35). Therefore, both ERH
and SNRPs might be involved in the RNA splicing process.
Previously, ERH was reported to interact with SNRPD3 for
splicing of CENP-E in the context of KRAS-mutant cancer cells.
Interference in CENP-E RNA splicing leads to the defects of
chromosome congression, hampering further cell cycle progress and
proliferation (14). In the
present study, numerous SNRPs interacting with ERH were
identified using the STRING website. Among these, only SNRPG
was positively correlated with ERH expression, at both mRNA
and protein expression levels, as demonstrated by data from the
TCGA and CPTAC databases, respectively. Moreover, the survival
impact of ERH was only seen in cells with high SNRPG
expression, which indicated an SNRPG-dependent modulating
process. The results of the present study suggest the
ERH-SNRPG interaction could serve a role in lung cancer
treatment. There are limitations of the present study and further
validation is required. First, the protein-protein interaction
between ERH and SNRPG should be confirmed by
co-immunoprecipitation. Second, the influence of ERH on MDSC
infiltration in the tumor immune ecosystem should be validated by
IHC staining.
To conclude, the prognosis for lung cancer is poor
and novel therapies are worthy of investigation. The results from
the present study demonstrated that ERH may serve a critical
role in promotion, progression and alteration of the tumor-immune
microenvironment. Therefore, drugs targeting ERH would be
worth investigating and developing.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the Ministry of Science
and Technology (grant nos. MOST 110-2314-B-037-124-MY3, MOST
110-2314-B-037-126-MY2 and MOST 111-2314-B-037-089), the Kaohsiung
Medical University Hospital Research Funding (grant nos.
KMUH-108-8R15, KMUH-110-0R14, and KMUH-110-0R17) and the Kaohsiung
Municipal Ta-Tung Hospital Research Funding (grant no.
KMTTH-110TA-04).
Availability of data and materials
The data generated in the present study can be
accessed from the National Center for Biotechnology Information
Gene Expression Omnibus (accession number, GSE236816) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE236816).
All other datasets used and/or analyzed during the current study
are available from the corresponding author on reasonable
request.
Authors' contributions
YMT and JYH conceptualized the present study. YYW,
YCH, CYC, YYC and LXL provided technical support, performed the
experiments and acquired the data. YMT, KLW and JYH confirmed the
authenticity of all of the raw data. CYC and HHC provided the
software management and analyzed the data. YMT and KLW performed
the formal analysis. JYH pursued the investigation and provided the
resources. YMT, KLW and JYH performed data curation and interpreted
the data. YMT and KLW wrote the original draft. JYH wrote, reviewed
and edited the final manuscript. JYH supervised the study, was the
project administrator and acquired the funding. All authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
The protocol of the present study was approved
[approval nos. KMUH-IRB-20180023, KMU-IRB-20200038 and KMU-IRB-E
(II)-20220175] by the Institutional Review Board of Kaohsiung
Medical University Hospital (Kaohsiung, Taiwan) and written
informed consent was acquired from all enrolled patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Skřičková J, Kadlec B, Venclíček O and
Merta Z: Lung cancer. Cas Lek Cesk. 157:226–236. 2018.PubMed/NCBI
|
|
3
|
Bonney A, Malouf R, Marchal C, Manners D,
Fong KM, Marshall HM, Irving LB and Manser R: Impact of low-dose
computed tomography (LDCT) screening on lung cancer-related
mortality. Cochrane Database Syst Rev. 8:CD0138292022.PubMed/NCBI
|
|
4
|
Deb D, Moore AC and Roy UB: The 2021
global lung cancer therapy landscape. J Thorac Oncol. 17:931–936.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Singh N, Temin S, Baker S Jr, Blanchard E,
Brahmer JR, Celano P, Duma N, Ellis PM, Elkins IB, Haddad RY, et
al: Therapy for stage IV non-small-cell lung cancer without driver
alterations: ASCO living guideline. J Clin Oncol. 40:3323–3343.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nicholson AG, Tsao MS, Beasley MB, Borczuk
AC, Brambilla E, Cooper WA, Dacic S, Jain D, Kerr KM, Lantuejoul S,
et al: The 2021 WHO classification of lung tumors: Impact of
advances since 2015. J Thorac Oncol. 17:362–387. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Luo YH, Liang KH, Huang HC, Shen CI,
Chiang CL, Wang ML, Chiou SH and Chen YM: State-of-the-art
molecular oncology of lung cancer in Taiwan. Int J Mol Sci.
23:70372022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Isomura M, Okui K, Fujiwara T, Shin S and
Nakamura Y: Cloning and mapping of a novel human cDNA homologous to
DROER, the enhancer of the Drosophila melanogaster
rudimentary gene. Genomics. 32:125–127. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Weng MT and Luo J: The enigmatic ERH
protein: Its role in cell cycle, RNA splicing and cancer. Protein
Cell. 4:807–812. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Krzyzanowski MK, Kozlowska E and Kozlowski
P: Identification and functional analysis of the erh1(+) gene
encoding enhancer of rudimentary homolog from the fission yeast
Schizosaccharomyces pombe. PLoS One. 7:e490592012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang X, Xie H, Zhu Z, Zhang J and Xu C:
Molecular basis for the recognition of CIZ1 by ERH. FEBS J.
290:712–723. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fang W and Bartel DP: MicroRNA clustering
assists processing of suboptimal MicroRNA hairpins through the
action of the ERH protein. Mol Cell. 78:289–302.e6. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Y, Wang X, Wang H, Jiang Y, Xu Z and
Luo L: Elevated small nuclear ribonucleoprotein polypeptide an
expression correlated with poor prognosis and immune infiltrates in
patients with hepatocellular carcinoma. Front Oncol. 12:8931072022.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Weng MT, Lee JH, Wei SC, Li Q, Shahamatdar
S, Hsu D, Schetter AJ, Swatkoski S, Mannan P, Garfield S, et al:
Evolutionarily conserved protein ERH controls CENP-E mRNA splicing
and is required for the survival of KRAS mutant cancer cells. Proc
Natl Acad Sci USA. 109:E3659–E3667. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pang K, Zhang Z, Hao L, Shi Z, Chen B,
Zang G, Dong Y, Li R, Liu Y, Wang J, et al: The ERH gene regulates
migration and invasion in 5637 and T24 bladder cancer cells. BMC
Cancer. 19:2252019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang D, Chu YJ, Song KJ, Chen YL, Liu W,
Lv T, Wang J, Zhao H, Ren YZ, Xu JX, et al: Knockdown of enhancer
of rudimentary homolog inhibits proliferation and metastasis in
ovarian cancer by regulating epithelial-mesenchymal transition.
Biomed Pharmacother. 125:1099742020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Park JH, Park M, Park SY, Lee YJ, Hong SC,
Jung EJ, Ju YT, Jeong CY, Kim JY, Ko GH, et al: ERH overexpression
is associated with decreased cell migration and invasion and a good
prognosis in gastric cancer. Transl Cancer Res. 9:5281–5291. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Park C, Lee WS, Go SI, Jeong SH, Yoo J,
Cha HJ, Lee YJ, Kim HS, Leem SH, Kim HJ, et al: Apoptotic effects
of anthocyanins from vitis coignetiae pulliat are enhanced by
augmented enhancer of the rudimentary homolog (ERH) in human
gastric carcinoma MKN28 cells. Int J Mol Sci. 22:30302021.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pang K, Li ML, Hao L, Shi ZD, Feng H, Chen
B, Ma YY, Xu H, Pan D, Chen ZS and Han CH: ERH gene and its role in
cancer cells. Front Oncol. 12:9004962022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Blandin Knight S, Crosbie PA, Balata H,
Chudziak J, Hussell T and Dive C: Progress and prospects of early
detection in lung cancer. Open Biol. 7:1700702017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lim WK, Wang K, Lefebvre C and Califano A:
Comparative analysis of microarray normalization procedures:
Effects on reverse engineering gene networks. Bioinformatics.
23:i282–i288. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ghufran SM, Sharma P, Roy B, Jaiswal S,
Aftab M, Sengupta S, Ghose S and Biswas S: Transcriptome wide
functional analysis of HBx expressing human hepatocytes stimulated
with endothelial cell cross-talk. Genomics. 115:1106422023.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu CJ, Hu FF, Xia MX, Han L, Zhang Q and
Guo AY: GSCALite: A web server for gene set cancer analysis.
Bioinformatics. 34:3771–3772. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhou Y, Zhou B, Pache L, Chang M,
Khodabakhshi AH, Tanaseichuk O, Benner C and Chanda SK: Metascape
provides a biologist-oriented resource for the analysis of
systems-level datasets. Nat Commun. 10:15232019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48((W1)): W509–W514. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Saadoun S, Papadopoulos MC, Watanabe H,
Yan D, Manley GT and Verkman AS: Involvement of aquaporin-4 in
astroglial cell migration and glial scar formation. J Cell Sci.
118:5691–5698. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chang WA, Yen MC, Hung JY, Yang CJ, Jian
SF, Yeh IJ, Liu KT, Hsu YL and Kuo PL: Investigation of the role of
tumor necrosis factor-like weak inducer of apoptosis in non-small
cell lung cancer. Oncol Rep. 39:573–581. 2018.PubMed/NCBI
|
|
29
|
Liu CJ, Hu FF, Xie GY, Miao YR, Li XW,
Zeng Y and Guo AY: GSCA: An integrated platform for gene set cancer
analysis at genomic, pharmacogenomic and immunogenomic levels.
Brief Bioinform. 24:bbac5582023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ishikawa K, Ishikawa A, Shoji Y and Imai
T: A genotoxic stress-responsive miRNA, miR-574-3p, delays cell
growth by suppressing the enhancer of rudimentary homolog gene in
vitro. Int J Mol Sci. 15:2971–2990. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fujimura A, Kishimoto H, Yanagisawa J and
Kimura K: Enhancer of rudimentary homolog (ERH) plays an essential
role in the progression of mitosis by promoting mitotic chromosome
alignment. Biochem Biophys Res Commun. 423:588–592. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Quail DF and Joyce JA: Microenvironmental
regulation of tumor progression and metastasis. Nat Med.
19:1423–1437. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Binnewies M, Roberts EW, Kersten K, Chan
V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI,
Ostrand-Rosenberg S, Hedrick CC, et al: Understanding the tumor
immune microenvironment (TIME) for effective therapy. Nat Med.
24:541–550. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mabonga L and Kappo AP: The oncogenic
potential of small nuclear ribonucleoprotein polypeptide G: A
comprehensive and perspective view. Am J Transl Res. 11:6702–6716.
2019.PubMed/NCBI
|
|
35
|
Bucholc K, Aik WS, Yang XC, Wang K, Zhou
ZH, Dadlez M, Marzluff WF, Tong L and Dominski Z: Composition and
processing activity of a semi-recombinant holo U7 snRNP. Nucleic
Acids Res. 48:1508–1530. 2020. View Article : Google Scholar : PubMed/NCBI
|