Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the most
common type of pancreatic cancer in the world (1); these tumors are highly aggressive,
with PDAC projected to be the second leading cause of
cancer-related death by 2030 (2).
Worldwide, the incidence of PDAC is estimated to be fifteenth among
all malignant tumors, with a 5-year survival rate of <5%
(3).
Gemcitabine (GEM) is the first-line chemotherapeutic
drug for patients with pancreatic cancer, as it can improve their
quality of life. However, numerous PDACs are highly resistant to
chemotherapeutic agents, resulting in the failure of pancreatic
cancer chemotherapy. Thus, GEM does not significantly improve the
overall prognosis and survival rate of patients with pancreatic
cancer (4). Identification of the
mechanism underlying the resistance of PDAC to treatment with GEM
may therefore provide clues to more effective therapeutic
methods.
Immunotherapy is a treatment method for the
management and elimination of cancer, which works by restarting and
maintaining the tumor immune cycle and re-establishing the normal
antitumor immune response in the body (5). Present research on pancreatic cancer
has primarily concentrated on early diagnosis, and the exploration
of drug combinations or new treatment targets (6). GEM has been a first-line treatment
for pancreatic cancer for a number of years and holds a significant
position in clinical therapy; however, its associated drug
resistance notably limits its long-term efficacy (7). There is a relative lack of research
on the treatment of pancreatic cancer or the improvement of the
drug-resistant immune microenvironment after drug resistance.
Interferon (IFN)-γ, a member of the type II IFN family, is a
pleiotropic molecule with anti-proliferative, pro-apoptotic and
antitumor activities (8).
Currently, to the best of our knowledge, no studies have assessed
the impact of IFN-γ on GEM-resistant pancreatic cancer. To
investigate the impact of IFN-γ on the cell viability, apoptosis
and migration of GEM-resistant pancreatic cancer cells, the present
study constructed GEM-resistant cells from the PANC-1 pancreatic
cancer cell line, named PANC-1/GEM, and assessed the effects of
IFN-γ on drug resistance.
Materials and methods
Reagents and materials
The human pancreatic cancer cell line PANC-1 (lot
no. SNL-100) was purchased from the American Type Culture
Collection. GEM was obtained from Qilu Pharmaceutical Co., Ltd. The
MTT Cell Proliferation and Cytotoxicity Assay Kit (cat. no. C0009S)
and the BCA protein quantification kit (cat. no. P0009) were
obtained from Beyotime Institute of Biotechnology; DMSO (cat. no.
D8370) was from Beijing Solarbio Science & Technology Co.,
Ltd.; the Apoptosis Detection Kit (cat. no. CA1120) was from
Solarbio Co., Ltd.; IFN-γ (cat. no. 106-06) was from Shanghai Puxin
Biotechnology Co., Ltd.; TRIzol® reagent, DMEM and fetal
bovine serum (FBS) were purchased from Thermo Fisher Scientific,
Inc.; HiScript III RT SuperMix for qPCR (+gDNA wiper) reverse
transcription (RT) kit (cat. no. R323-01) and ChamQ SYBR Color qPCR
Master Mix (cat. no. Q411-02) were from Vazyme Biotech Co., Ltd.;
hematoxylin and eosin (H&E) staining reagent (cat. no. HEH-020)
was from BaSO Biotech; RIPA lysis buffer (cat. no. G2002-100ML) was
from Wuhan Servicebio Technology Co., Ltd.; ColorMixed Protein
Marker (cat. no. RM19001) and anti-β-actin (cat. no. AC026) were
from ABclonal Biotech Co., Ltd.; anti-multidrug
resistance-associated protein (MRP; cat. no. DF8801) and
anti-breast cancer resistance protein (BCRP; cat. no. AF5177) were
from Affinity Biosciences. The BSA (cat. no. GC310001) was from
Servicebio Technology Co., Ltd.; the Goat Anti-Rabbit IgG (H+L) HRP
(cat. no. S0001) was from Affinity Biosciences Co., Ltd.
Study design
The number of repetitions and the duplicate samples
for all cell experiments of the present study were determined
according to the methodologies of other academic studies (9,10).
The sample size calculation for the experiment was validated using
GPower 3.1 software (Düsseldorf University); the cellular
experimental samples were triplicated, achieving a test efficiency
of 0.9. The software was also employed for power analysis, which
was employed to ascertain the required number of experimental
replicates.
Cell viability assay
PANC-1 cells were cultured in DMEM containing 10%
FBS at 37°C in an atmosphere containing 5% CO2, with the
medium replaced every 72 h. Cells were subcultured when their
density reached 80–90%. The cells were trypsinized, their
concentrations were adjusted by counting, and 6,000 cells in 100 µl
medium were added to each well of a 96-well plate. After 48 h, the
medium was removed and fresh DMEM containing 0, 0.1, 0.2, 0.4, 0.8,
1.6 and 3.2 µg/ml GEM was added to the wells, incubate at a
constant temperature of 37°C in a cell incubator. The culture
medium was replaced after 48 h, maintaining the same GEM
concentration as before (11–13).
Each concentration of GEM was evaluated in triplicate. After
culturing in a GEM-containing medium for 96 h, 10 µl of MTT reagent
was added to each well, and DMSO was added to dissolve the formazan
after 4 h. After an additional 4 h, the absorbance of each well was
measured at 570 nm using a microplate reader, and the half-maximal
inhibitory concentration (IC50) of GEM was determined
using the Kärber method (14–16):
IogIC50 = Xm-I [P-(3-Pm-Pn)/4]; where ‘Xm’ represents
the Ig maximum dose, ‘I’ indicates the Ig or maximum dose/near
dose, ‘P’ represents the sum of the positive reaction rate, ‘Pm’
indicates the maximum positive reaction rate, and ‘Pn’ refers to
the minimum positive reaction rate.
The effects of GEM on PANC-1 cell viability were
determined by measuring cell counts at 24, 48 and 72 using the MTT
method. According to the results of the IC50
calculation, the cell viability of PANC-1 cells with and without
GEM (0.8 µg/ml) was assessed. Briefly, 2,000 cells in 100 µl medium
were added to each well of a 96-well plate. The GEM-resistant
strain was designated as PANC-1/GEM cells. The MTT analysis was
also performed to assess the viability of PANC-1/GEM cells and to
evaluate the impact of IFN-γ on viability, using the aforementioned
procedure. The concentrations of IFN-γ used were 0, 0.15625,
0.3125, 0.625, 1.25, 2.5 or 5.0 µg/ml, respectively. MTT detection
was performed after 6 days of constant-temperature culture at 37°C.
% Inhibition = (1-OD value at each concentration/OD value at
concentration 0) ×100.
Induced drug-resistant strains
Drug resistance was induced in PANC-1 cells using a
GEM concentration gradient, as described previously (17,18).
Briefly, 2–6×106 PANC-1 cells were cultured in each of
two culture plates containing 5 ml DMEM supplemented with 10% FBS
and 0.8 µg/ml GEM. The concentrations added to both culture dishes
were identical. If the cell density in a single dish was too low,
the cells from both dishes were combined to increase the cell
density and facilitate normal cell proliferation. Cell death was
assessed daily under a light microscope, and the medium was removed
after 48 h. To each plate, DMEM plus 10% FBS without GEM was added
and the cells were cultured until the bottom of the plates was
completely covered. The medium was then removed and the cells were
cultured in medium containing 0.8 µg/ml GEM. After 48 h, the medium
was replaced with medium containing 0.8 µg/ml GEM. After the cells
repopulated the dish, the procedure was repeated. After the cells
became adapted to 0.8 µg/ml GEM, the concentration was gradually
increased to a maximum concentration of 15 µg/ml.
H&E staining
Each well of a 6-well plate was seeded with
1×106 PANC-1 or PANC-1/GEM cells. After 24 h, the medium
was removed and the cells were washed with phosphate-buffered
saline (PBS). Cells were then fixed in absolute ethanol for 20 min
at 37°C and were washed twice with PBS. Subsequently, the cells
were stained with an H&E staining kit at room temperature; with
hematoxylin for 5 min and eosin for 1 min. The cells were then
detected by light microscopy and images were captured using the
NIS-Elements software (v.5.21.00; Nikon Corporation).
RT-quantitative PCR (RT-qPCR)
Total RNA was extracted from 1×106 PANC-1
and PANC-1/GEM cells in the logarithmic growth phase using TRIzol
reagent. After discarding the culture medium from PANC-1/GEM cells,
the cells were rinsed once with pre-cooled sterile PBS, the
supernatant was discarded and then 1 ml single-phase lysis solution
was added for cell lysis. A pipette tip was utilized to ensure
thorough mixing for complete cell lysis. Preparations with an
OD260/280 nm ratio of 1.9–1.95 and with no obvious degradation were
regarded as being suitably pure for further experiments. The RNA
was reverse transcribed to obtain 20 µl aliquots of cDNA, following
the manufacturer's instructions. The mRNA expression levels of MRP,
BCRP and β-actin were determined using the ChamQ SYBR Color qPCR
Master Mix using specific primer sequences (Table I), with each assay performed in
triplicate. The qPCR thermal cycling conditions were as follows:
Initial denaturation at 95°C for 30 sec; cycling reaction at 95°C
for 10 sec, then 60°C for 30 sec; melting curve at 95°C for 15 sec,
then 60°C for 60 sec and 95°C for 15 sec (40 cycles). qPCR was
performed according to the manufacturer's protocol and each group
was tested three times. Analytical data were acquired using CFX
Manager software version 2.0 (Bio-Rad Laboratories, Inc.), with the
mRNA expression levels of MRP and BCRP determined relative to
β-actin mRNA using the 2−ΔΔCq method (19).
 | Table I.Sequences of primers used for reverse
transcription-quantitative PCR. |
Table I.
Sequences of primers used for reverse
transcription-quantitative PCR.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) | Product length,
bp | NCBI reference
no. | Corresponding cDNA
sequence, bp |
|---|
| MRP |
GCGAGTGTCTCCCTCAAACG |
TCCTCACGGTGATGCTGTTC | 118 | NM_004996 | 2,000-2,117 |
| BCRP |
GATATGGATTTACGGCTTTGC |
CGATGCCCTGCTTTACCAA | 135 | NM_004827 | 2,217-2,351 |
| β-actin |
AGTGTGACGTGGACATCCGCAAAG |
ATCCACATCTGCTGGAAGGTGGAC | 220 | NM_001101 | 935-1,154 |
PANC-1/GEM cells in the logarithmic growth phase
were cultured in DMEM containing 0.3 µg/ml IFN-γ for 30 days, with
the medium changed every 2 days; the control group consisted of
PANC-1 cells. Total cell RNA was extracted from the cells with
TRIzol reagent, cDNA was synthesized, and the levels of MRP, BCRP
and β-actin mRNAs were analyzed by fluorescence quantitative PCR
using specific primers for each gene (Table I), with each assay performed in
triplicate. Levels of MRP and BCRP mRNA relative to those of
β-actin mRNA were calculated using the 2−ΔΔCq method, as
aforementioned.
Cell count analysis
PANC-1/GEM cells in the logarithmic growth phase
were digested with trypsin, plated at 5,000 cells/well in 96-well
plates, and cultured in DMEM for 24 h. The medium was then removed
and replaced with medium containing 0, 0.15625, 0.3125, 0.625,
1.25, 2.5 or 5.0 µg/ml IFN-γ, with triplicate wells used for each
concentration. The cells were cultured in a 37°C cell culture
incubator for 6 days, with the medium containing IFN-γ being
renewed every 48 h. Subsequently, the medium was removed, 100 µl
DMEM without IFN-γ was added to each well, images were captured
using the NIS-Elements program.
Hoechst staining
Aliquots containing 2.5×105 PANC-1/GEM
cells in the logarithmic growth phase were added to each well of a
24-well plate, and the cells were cultured in DMEM containing 10%
FBS for 24 h. After the cells adhered to the plate, the medium was
aspirated, and the cells were cultured in medium containing 0.3
µg/ml IFN-γ for 72 h at 37°C. The supernatant was then removed, and
1 ml cell staining buffer, 5 µl Hoechst staining solution (cy5 dye)
and 5 µl PI staining solution (DAPI dye) were added at 4°C (these
are all from the kit). The plates were incubated at 4°C for 4 h and
images were captured under a fluorescence microscope (Olympus
Corporation). Weak red plus weak green staining indicated normal
cells; weak red plus strong blue staining indicated apoptotic
cells; and strong red plus strong blue staining indicated necrotic
cells.
Wound-healing assay
Aliquots containing 1×106 PANC-1/GEM
cells were added to each well of a 6-well plate, and the cells were
cultured for 24 h. Subsequently, upon reaching 90–100% confluence
in the culture plate wells, a monolayer of cells in each well was
mechanically injured using a pipette tip (1,000-µl). The cells in
three wells were then cultured in medium containing 0.3 µg/ml
IFN-γ, whereas the cells in the other three wells were cultured in
medium alone for 72 h. All cells in the culture were maintained in
serum-free media. The images were then captured using an optical
microscope (Nikon Corporation) at 0, 24, 48 and 72 h, and
subsequent analysis was performed using ImageJ software (version
1.8.0; National Institutes of Health). The ImageJ software was
employed to detect the wound area and analyze the percentage of the
healed wound area. Wound healing (%)=(1-unhealed area/initial wound
area) ×100. The ‘unhealed area’ refers to the exposed region within
the wound area measured at 24, 48 and 72 h, whereas the ‘initial
wound area’ represents the area at 0 h (19). In addition, the scratch width was
quantified using Adobe Photoshop 2023 (v24.7.1.741; Adobe Systems,
Inc.). Wound healing % relative to 0 h is shown is presented.
Western blotting
The PANC-1 cell and PANC-1/GEM cell were lysed in
cold RIPA lysis buffer with 1% protease inhibitor cocktail and
proteasome inhibitors. The supernatant was collected
post-centrifugation (13,800 × g at 4°C for 10 min), and protein
concentration was determined using the BCA protein quantification
kit. The protein samples were then loaded with a marker (20 µg
protein/lane) and separated on 8% gels using SDS-PAGE, followed by
transfer to a PVDF membrane. The membrane was blocked using 5% BSA
in TBS with 0.1% Tween (TBST) for 1 h at room temperature, then
incubated overnight with the following primary antibodies: MRP
(1:1,000), BCRP (1:500) and β-actin (1:50,000). After washing three
times with TBST (5-min intervals), the membrane was incubated with
the secondary antibody for 1 h at room temperature.
Chemiluminescence detection was performed using a western blot
detection system (Shanghai Tianneng Life Sciences Co., Ltd.), and
the chemiluminescence signal was semi-quantified with Image-Pro
Plus 6.0 software (Media Cybernetics, Inc.).
Statistical analysis
Data are presented as the mean ± SD. Experiments
were performed in triplicate and repeated three times with similar
results. Statistical analyses were conducted using SPSS version
26.0 (IBM Corp.). Parametric data were analyzed using unpaired
t-test or one-way analysis of variance, Subsequently, upon
detecting significant differences between groups, Tukey's honestly
significant difference test was employed for post hoc analysis.
(two-tailed P≤0.05 was considered to indicate a statistically
significant difference.
Results
Effect of GEM on the proliferation of
PANC-1 and PANC-1/GEM cells as determined by MTT assay
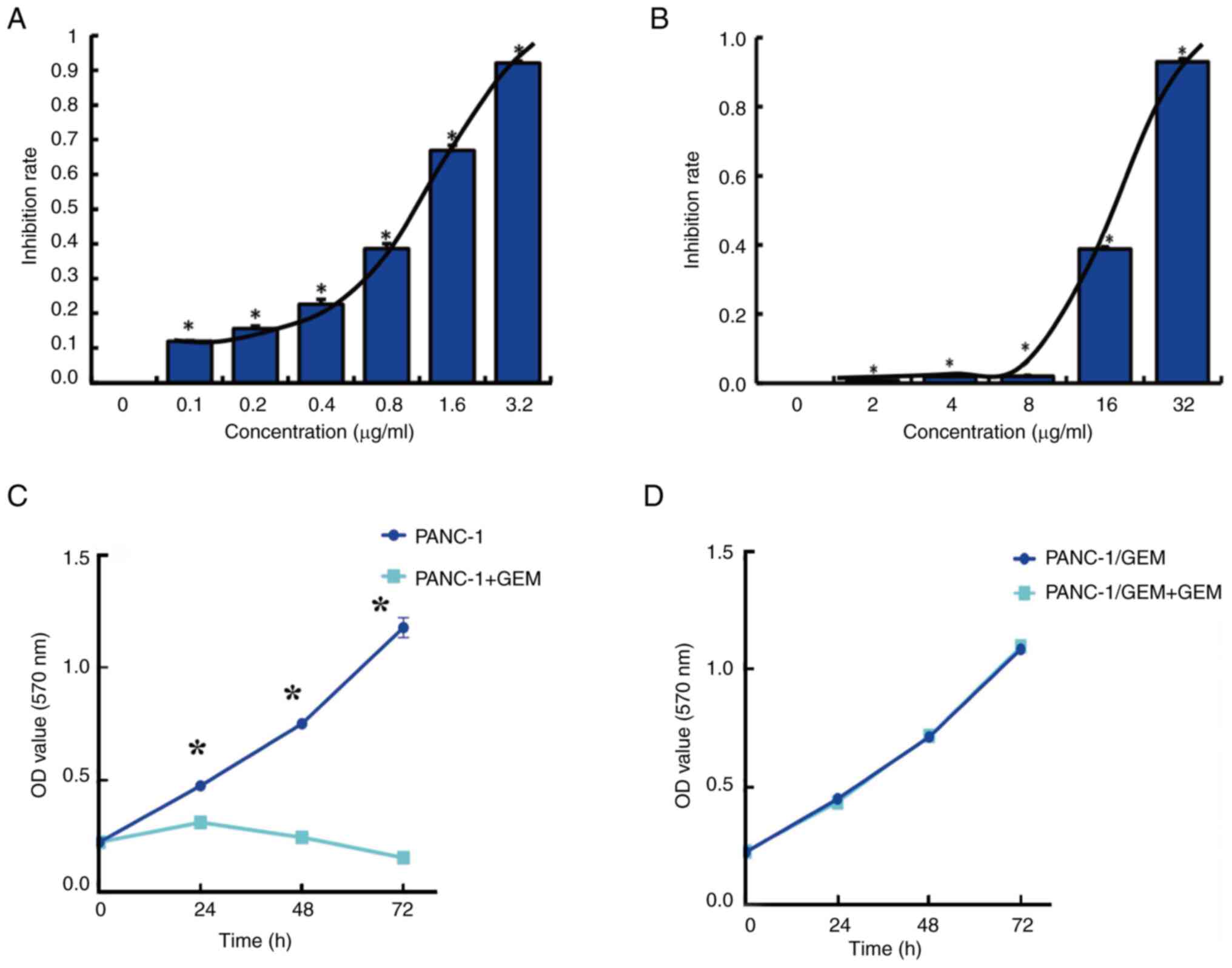
The MTT assay showed that GEM had a median
IC50 of 0.8 µg/ml in PANC-1 cells (Fig. 1A), as calculated using the Kärber
formula. In addition, the MTT assay showed that GEM had a median
IC50 of 18 µg/ml in PANC-1/GEM cells (Fig. 1B) and that the resistance index,
defined as the IC50 of GEM in PANC-1/GEM cells divided
by the IC50 in PANC-1 cells, was 22.4. The proliferation
of PANC-1 cells was subsequently determined with or without
treatment with the IC50 concentration of GEM (0.8
µg/ml). Proliferation of untreated PANC-1 cells exhibited a
time-dependent increase, whereas PANC-1 cells treated with GEM
exhibited inhibited cell proliferation. This suggests that PANC-1
cells were in optimal condition, ensuring the reliability of the
MTT assay results (Fig. 1C).
Notably, the viability of PANC-1/GEM cells remained unchanged in
response to GEM (18 µg/ml), thus indicating the resistance of these
cells to GEM (Fig. 1D).
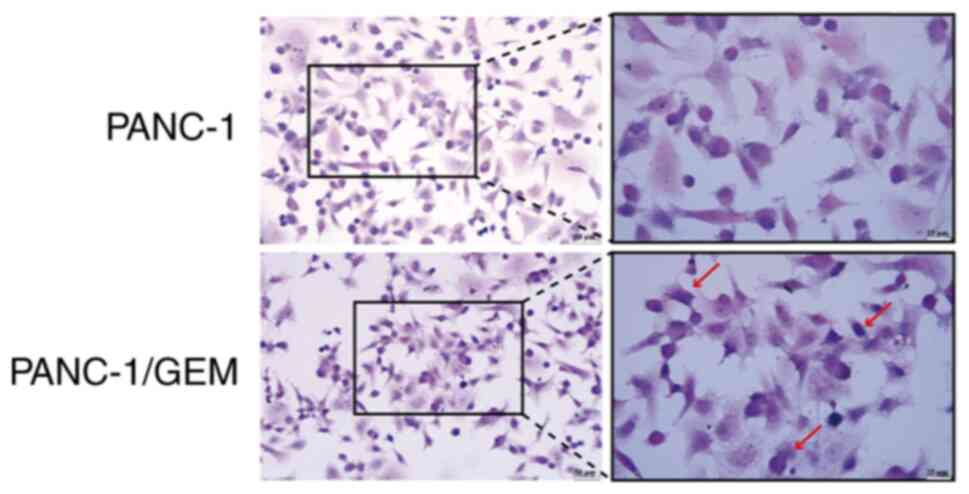
Cell morphology
Hematoxylin stained the nuclei of PANC-1 cells a
vivid blue, whereas eosin stained the eosinophilic granules in the
cytoplasm brilliant red with significant light reflection, and the
cytoplasm was stained various colors ranging from pink to peach.
H&E staining of PANC-1/GEM cells showed an altered morphology,
with the appearance of irregular forms, such as circles; however,
there were no marked differences in their nucleocytoplasmic ratios
(Fig. 2).
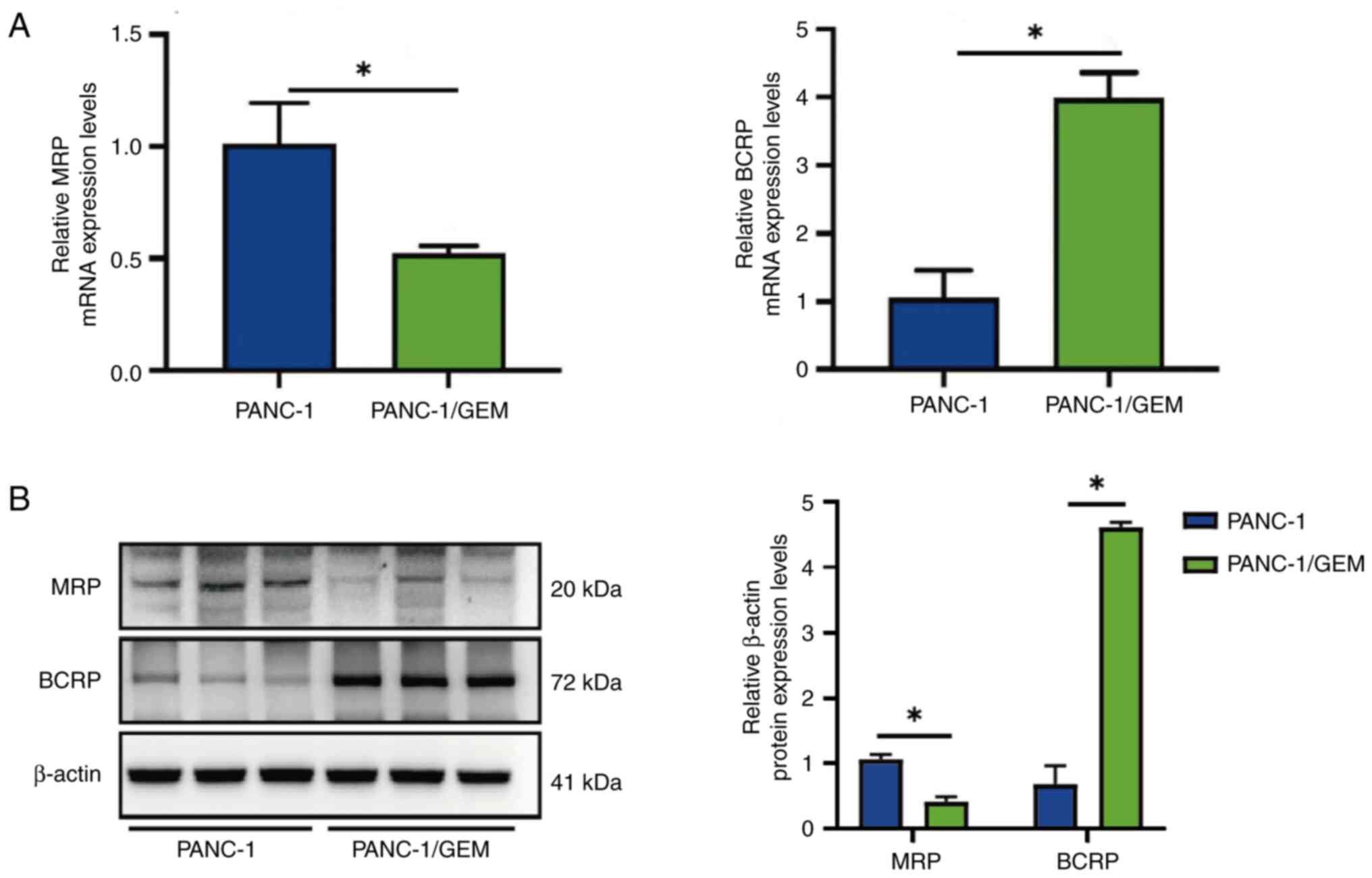
Effects of GEM on the mRNA expression
levels of MRP and BCRP
RT-qPCR showed that the mRNA expression levels of
MRP were 2-fold lower, whereas the mRNA expression levels of BCRP
were 4-fold higher in PANC-1/GEM cells compared with those in
PANC-1 cells; both differences were statistically significant
(Fig. 3A). In addition, western
blotting revealed a decrease in MRP protein expression and an
increase in BCRP protein expression after PANC-1 cells developed
resistance to GEM. The findings imply that PANC-1 cells developed
resistance to GEM due to changes in the expression of
resistance-related genes (Fig.
3B). This result indicated the successful establishment of
GEM-resistant pancreatic cancer cells, denoted as PANC-1/GEM
cells.
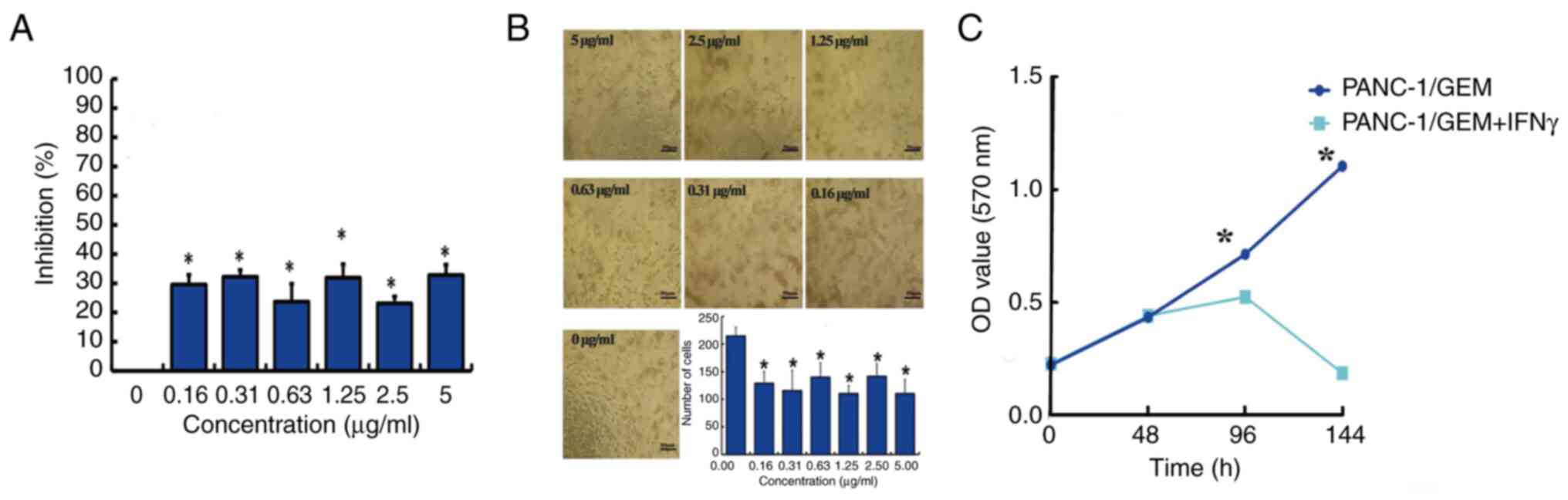
Effect of IFN-γ on PANC-1/GEM cell
viability
Treatment of PANC-1/GEM cells with 0.16–5.0 µg/ml
IFN-γ for 6 days inhibited cell viability, as shown by MTT assays,
with the lowest effective concentration being 0.31 µg/ml (Fig. 4A). To exclude the influence of the
MTT method on cell viability, the numbers of cells were counted
directly. The findings indicated that a concentration of 0.31 µg/ml
effectively inhibited PANC-1/GEM cell viability (Fig. 4B). In order to exclude poor
cellular activity or inactivation of the IFN-γ protein from
interfering with the accuracy of the MTT results, the viability of
PANC-1/GEM cells was detected with and without the addition of
IFN-γ. The results revealed that 0.31 µg/ml IFN-γ significantly
reduced PANC-1/GEM cell viability, in contrast to the viability of
cells without IFN-γ treatment (Fig.
4C).
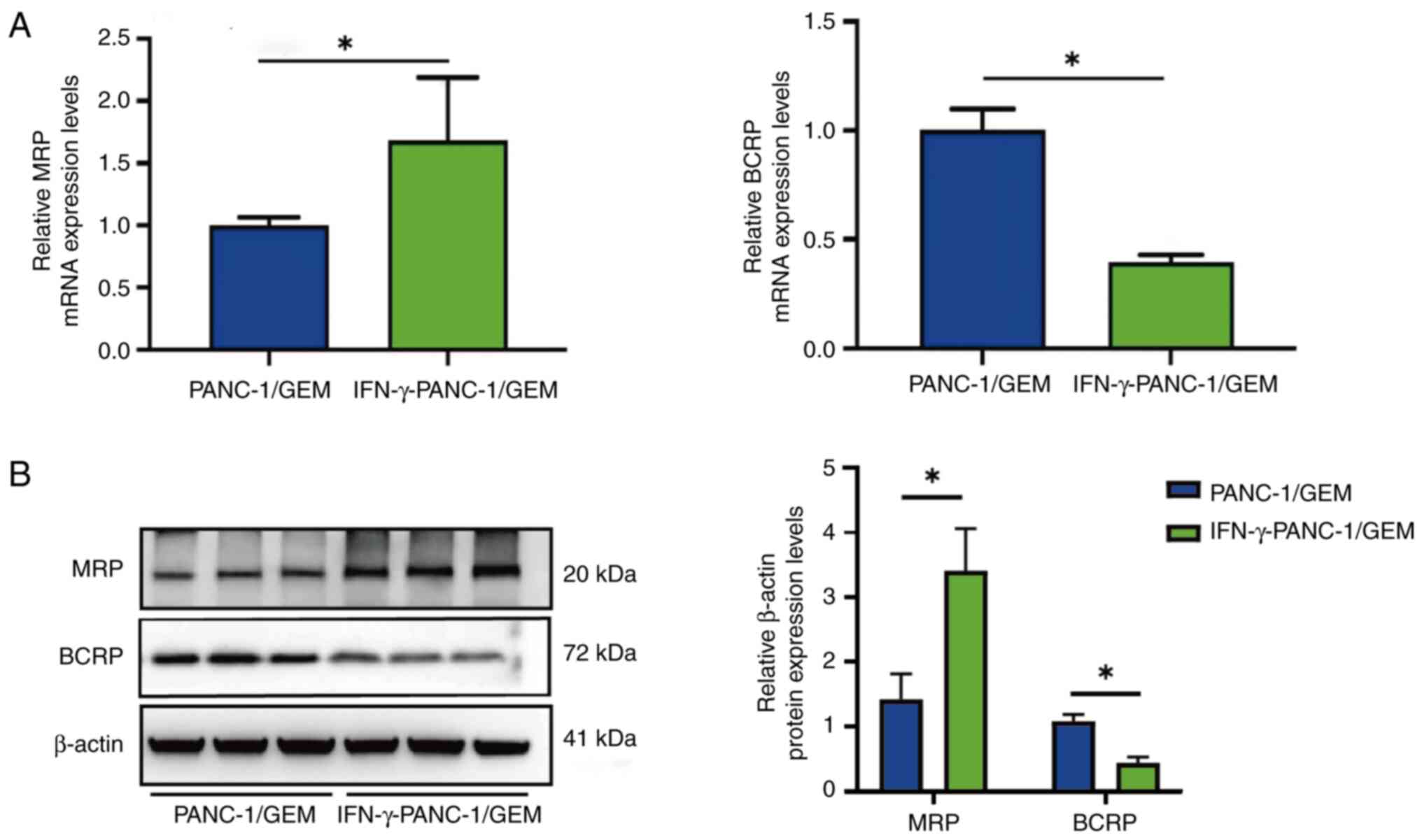
Effects of IFN-γ on MRP and BCRP mRNA
expression levels in PANC-1/GEM cells
RT-qPCR assays showed that treatment of PANC-1/GEM
cells with IFN-γ resulted in 1.61-fold higher expression levels of
MRP mRNA and 2.5-fold lower expression levels of BRCP mRNA than in
the control PANC-1/GEM cells (Fig.
5A). Furthermore, following IFN-γ treatment of PANC-1/GEM
cells, western blotting demonstrated an increase in MRP protein
expression and a decrease in BCRP protein expression (Fig. 5B). These findings indicated that
IFN-γ treatment reversed the effects of GEM on GEM-resistant
cells.
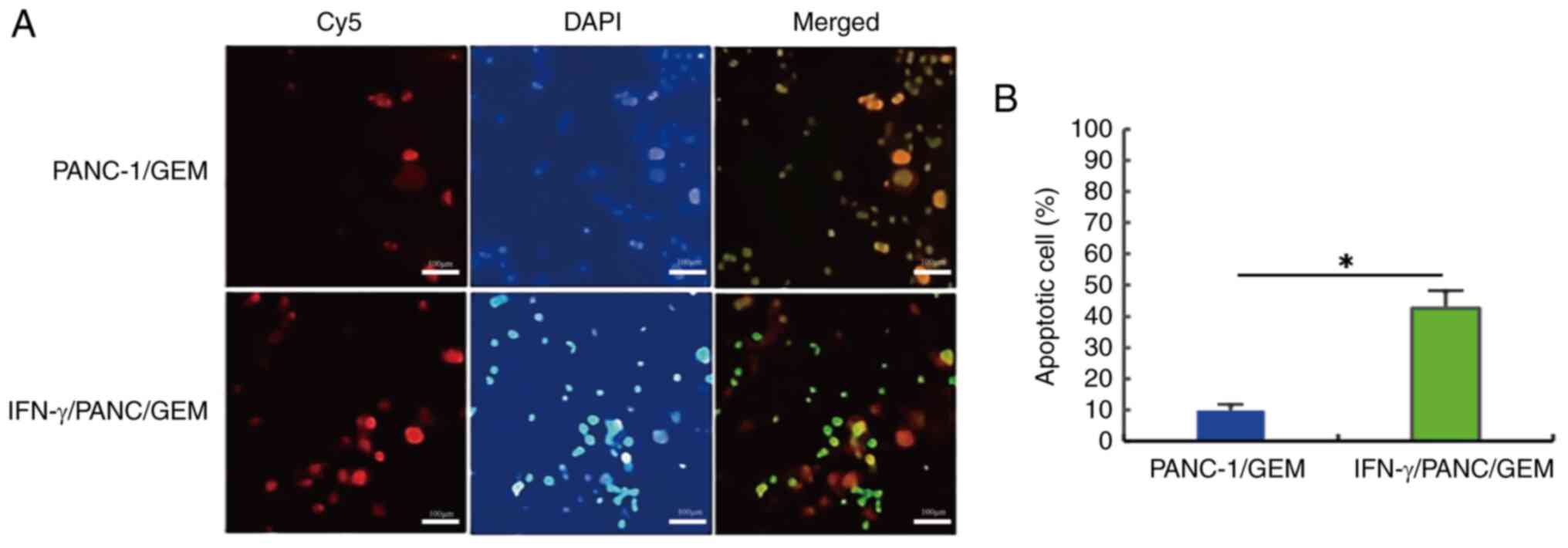
Effects of IFN-γ on cell
apoptosis
PANC-1/GEM cells were treated with 0.3 µg/ml IFN-γ
for 6 h, and cell apoptosis was determined using a cell apoptosis
detection kit (Fig. 6A). The
apoptotic count of PANC-1/GEM cells increased by ~4.3 times
following treatment with IFN-γ compared with that in the untreated
group, and this difference was statistically significant (Fig. 6B). These findings indicated that
drug-resistant PANC-1/GEM cells can undergo IFN-γ-induced
apoptosis.
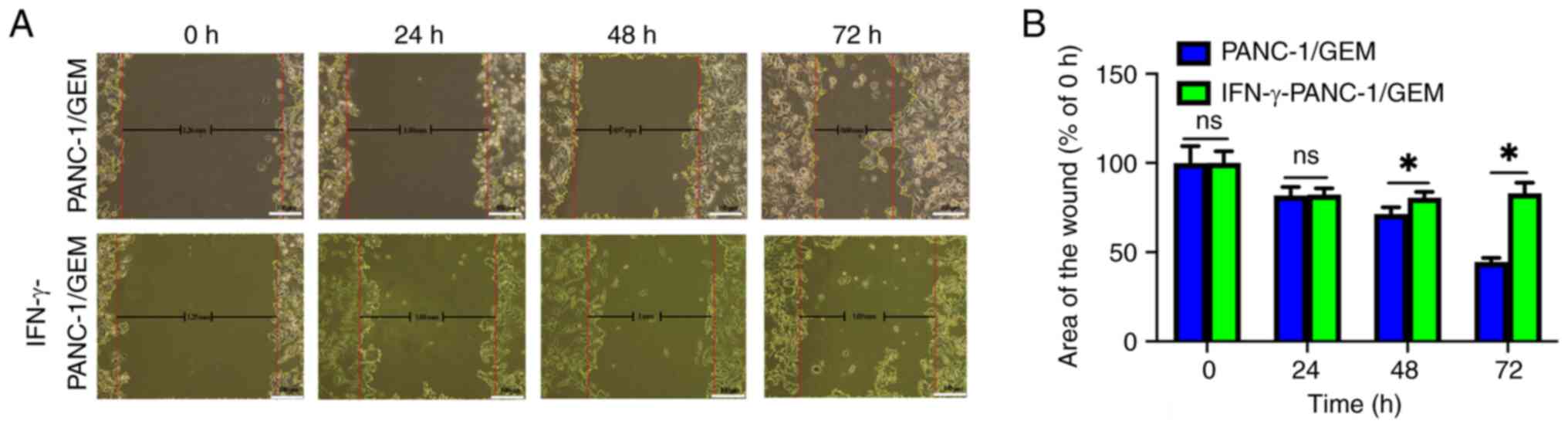
IFN-γ exhibits the ability to inhibit
the migration of PANC-1/GEM cells
In the control group, the initial wound width of
PANC-1/GEM cells was 1.26 mm, reducing to 0.60 mm at 72 h.
Meanwhile, the IFN-γ-treated group exhibited a wound width of 1.25
mm at 0 h and 1.03 mm at 72 h (Fig.
7A). Additionally, statistical analysis revealed that, over
time, the area of the wound in the IFN-γ-treated group was
significantly higher than that in the control group at 48 and 72 h,
suggesting that cell migration was inhibited in the IFN-γ-treated
group (Fig. 7B). These findings
suggested that IFN-γ has the potential to inhibit the migration of
PANC-1/GEM cells.
Discussion
Most patients with PDAC exhibit local progression
and/or metastasis at diagnosis, preventing surgical resection of
the primary tumor. Early diagnosis is hampered by the highly
aggressive nature of these tumors and the lack of screening
techniques for early identification (20,21).
Although patients with both resectable and non-resectable PDAC can
benefit from chemotherapy, these benefits are minor due to the high
prevalence of drug resistance (22). Therefore, it is crucial to identify
agents that can successfully treat PDAC following the development
of chemotherapeutic drug resistance. Immunotherapy, which targets
mechanisms of carcinogenesis, may be successful in treating these
patients. Recent studies have demonstrated a significant
correlation between immunotherapy-related biomarkers, including
NLRP3, PARP1, NOX1, NOX2, eNOS and iNOS, and the treatment
outcomes, prognoses and survival rates of patients with pancreatic
cancer (23,24). Since the cytokine IFN-γ has been
shown to promote cell apoptosis and limit cell proliferation, lung,
breast and several other types of cancer are treated with this
protein, which serves a key role in activating cellular immunity
and stimulating antitumor immune responses (25). Consequently, IFN-γ may improve the
treatment of patients with pancreatic cancer resistant to GEM.
The present study investigated the effects of IFN-γ
on a PANC-1 pancreatic cancer cell line that had developed
resistance to the chemotherapeutic agent GEM, and quantified the
impact of IFN-γ on PANC-1/GEM cell resistance. These findings may
establish a basis for the treatment of patients with cancer who
develop resistance to chemotherapeutic agents. Relative to PANC-1
cells, PANC-1/GEM cells had a resistance index of 22.4.
H&E staining was performed to compare the
morphological characteristics of PANC-1 and PANC-1/GEM cells. The
findings showed that cells became round and irregular in shape
after becoming resistant to GEM, which is consistent with the
hallmarks of cellular malignancy (26). GEM may rapidly induce resistance by
altering the expression levels or inducing mutations in
resistance-related genes in cancer stem cells (27,28).
RT-qPCR showed that tumor cell resistance to GEM was associated
with a significant reduction in MRP mRNA expression levels
(29) and a significant increase
in BCRP mRNA expression levels (30). This further supports the
credibility of the constructed PANC-1/GEM cells in the present
study.
IFN-γ is a key immune response controller, which has
a major effect on tumors by regulating and activating the cellular
immune responses, thereby activating tumor-killing activity
(8). IFN-γ may have a greater
impact on GEM-resistant tumor cells as its therapeutic mechanism
differs from that of GEM. Screening of IFN-γ concentrations to
treat PANC-1/GEM cells showed that 0.3 µg/ml IFN-γ exhibited
optimal activity against drug-resistant cells. Because genes
associated with tumor cell resistance have been frequently
associated with tumor resistance to chemotherapeutic agents,
PANC-1/GEM cells were cultured for 1 month in the presence of 0.3
µg/ml IFN-γ, and it was revealed that IFN-γ treatment reversed the
effects of GEM on the mRNA expression levels of MRP and BCRP. IFN-γ
has been reported to enhance the antigenicity of tumor cells by
upregulating the expression of major histocompatibility complex
class Ia (31), suggesting that
the antitumor activity of IFN-γ against PANC-1/GEM cells may be due
to its enhancement of the antigenicity of drug-resistant tumor
cells and its immunomodulatory action. Thus, the drug resistance of
these tumor cells was partially reversed, and their susceptibility
to chemotherapeutic agents was enhanced. This could explain the
observed reversal in the expression of resistance-related genes in
PANC-1/GEM cells after IFN-γ treatment, as compared to the
post-resistance period in the present study.
Both direct cell counting and MTT assays showed that
IFN-γ greatly decreased the viability of PANC-1/GEM cells,
suggesting that IFN-γ has a considerable impact on the viability of
pancreatic cancer cells, even after those cells have developed
resistance to GEM. Thus, IFN-γ may be an additional and/or
alternative option for the treatment of patients with pancreatic
cancer for whom chemotherapy has failed. Moreover, IFN-γ was able
to promote the apoptosis of drug-resistant pancreatic cancer cells
and to markedly decrease the migration of PANC-1/GEM cells,
suggesting that IFN-γ may reduce the invasiveness of GEM-resistant
pancreatic cancer cells. Based on previous studies, we propose that
IFN-γ, acting directly on GEM-resistant pancreatic cancer cells,
may influence their proliferation, migration and apoptosis by
impacting the cell cycle and chemotaxis.
In conclusion, the present study showed that IFN-γ
could reduce the migration and viability, and enhance the apoptosis
of GEM-resistant pancreatic cancer cells. The reversal of
resistance-related gene expression in PANC-1/GEM cells following
IFN-γ treatment suggests that IFN-γ may have reversed the
resistance of pancreatic cancer cells to chemotherapy. These
findings suggested that IFN-γ may improve the condition of patients
with GEM-resistant pancreatic cancer.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Chinese University Student
Innovation and Entrepreneurship Fund (grant no. 201810634026).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
XK and DC were responsible for the study design,
development and molecular biology experiments. XX, YZ and XL
conducted molecular biology experiments and participated in data
analysis. WP participated in the experimental design and
supervision of the experiment. All authors read and approved the
final manuscript. WP and XK confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Luchini C, Grillo F, Fassan M, Vanoli A,
Capelli P, Paolino G, Ingravallo G, Renzulli G, Doglioni C, D'Amuri
A, et al: Malignant epithelial/exocrine tumors of the pancreas.
Pathologica. 112:210–226. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Haeberle L and Esposito I: Pathology of
pancreatic cancer. Transl Gastroenterol Hepatol. 4:502019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lu H, Niu F, Liu F, Gao J, Sun Y and Zhao
X: Elevated glypican-1 expression is associated with an unfavorable
prognosis in pancreatic ductal adenocarcinoma. Cancer Med.
6:1181–1191. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou C, Yi C, Yi Y, Qin W, Yan Y, Dong X,
Zhang X, Huang Y, Zhang R, Wei J, et al: LncRNA PVT1 promotes
gemcitabine resistance of pancreatic cancer via activating
Wnt/β-catenin and autophagy pathway through modulating the
miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes. Mol Cancer. 19:1182020.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu Z, Li S and Zhu X: The mechanism of
stimulating and mobilizing the immune system enhancing the
anti-tumor immunity. Front Immunol. 12:6824352021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
McDonald OG: The biology of pancreatic
cancer morphology. Pathology. 54:236–247. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Amrutkar M and Gladhaug IP: Pancreatic
cancer chemoresistance to gemcitabine. Cancers. 9:1572017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Castro F, Cardoso AP, Gonçalves RM, Serre
K and Oliveira MJ: Interferon-gamma at the crossroads of tumor
immune surveillance or evasion. Front Immunol. 9:8472018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Narayanan P, Farghadani R, Nyamathulla S,
Rajarajeswaran J, Thirugnanasampandan R and Bhuwaneswari G: Natural
quinones induce ROS-mediated apoptosis and inhibit cell migration
in PANC-1 human pancreatic cancer cell line. J Biochem Mol Toxicol.
36:e230082022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
He J, Bugde P, Li J, Biswas R, Li S, Yang
X, Tian F, Wu Z and Li Y: Multidrug resistance protein 5 affects
cell proliferation, migration and gemcitabine sensitivity in
pancreatic cancer MIA Paca-2 and PANC-1 cells. Oncol Rep. 51:72024.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen M, He M, Song Y, Chen L, Xiao P, Wan
X, Dai F and Shen P: The cytoprotective role of gemcitabine-induced
autophagy associated with apoptosis inhibition in triple-negative
MDA-MB-231 breast cancer cells. Int J Mol Med. 34:276–282. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nowak AK, Robinson BW and Lake RA:
Gemcitabine exerts a selective effect on the humoral immune
response: Implications for combination chemo-immunotherapy. Cancer
Res. 62:2353–2358. 2002.PubMed/NCBI
|
|
13
|
Zhang QB, Ye RF, Ye LY, Zhang QY and Dai
NG: Isocorydine decrease gemcitabine-resistance by inhibiting
epithelial-mesenchymal transition via STAT3 in pancreatic cancer
cells. Am J Transl Res. 12:3702–3714. 2020.PubMed/NCBI
|
|
14
|
Liu L, Yang L, Chang H, Chen YN, Zhang F,
Feng S, Peng J, Ren CC and Zhang XA: CP-31398 attenuates
endometrial cancer cell invasion, metastasis and resistance to
apoptosis by downregulating MDM2 expression. Int J Oncol.
54:942–954. 2019.PubMed/NCBI
|
|
15
|
Zhu F, Wu Q, Ni Z, Lei C, Li T and Shi Y:
miR-19a/b and MeCP2 repress reciprocally to regulate multidrug
resistance in gastric cancer cells. Int J Mol Med. 42:228–236.
2018.PubMed/NCBI
|
|
16
|
Zhu K, Fang W, Chen Y, Lin S and Chen X:
TNF-related apoptosis-inducing ligand enhances vinorelbine-induced
apoptosis and antitumor activity in a preclinical model of
non-small cell lung cancer. Oncol Rep. 32:1234–1242. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang B, Shen C, Li Y, Zhang T, Huang H,
Ren J, Hu Z, Xu J and Xu B: Oridonin overcomes the gemcitabine
resistant PANC-1/Gem cells by regulating GST pi and LRP/1 ERK/JNK
signalling. Onco Targets Ther. 12:5751–5765. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou Y, Zhou Y, Yang M, Wang K, Liu Y,
Zhang M, Yang Y, Jin C, Wang R and Hu R: Digoxin sensitizes
gemcitabine-resistant pancreatic cancer cells to gemcitabine via
inhibiting Nrf2 signaling pathway. Redox Biol. 22:1011312019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pekarek L, Fraile-Martinez O,
Garcia-Montero C, Saez MA, Barquero-Pozanco I, Del Hierro-Marlasca
L, de Castro Martinez P, Romero-Bazán A, Alvarez-Mon MA, Monserrat
J, et al: Clinical applications of classical and novel biological
markers of pancreatic cancer. Cancers. 14:18662022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Desai NV, Sliesoraitis S, Hughes SJ,
Trevino JG, Zlotecki RA, Ivey AM and George TJ Jr:
Multidisciplinary neoadjuvant management for potentially curable
pancreatic cancer. Cancer Med. 4:1224–1239. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Principe DR, Underwood PW, Korc M, Trevino
JG, Munshi HG and Rana A: The current treatment paradigm for
pancreatic ductal adenocarcinoma and barriers to therapeutic
efficacy. Front Oncol. 11:6883772021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fraile-Martinez O, García-Montero C,
Pekarek L, Saz JV, Álvarez-Mon MÁ, Barrena-Blázquez S,
García-Honduvilla N, Buján J, Asúnsolo Á, Coca S, et al: Decreased
survival in patients with pancreatic cancer may be associated with
an increase in histopathological expression of inflammasome marker
NLRP3. Histol Histopathol. 39:35–40. 2024.PubMed/NCBI
|
|
24
|
Ortega MA, Fraile-Martinez O, Pekarek L,
García-Montero C, Alvarez-Mon MA, Castellanos AJ, García-Honduvilla
N, Buján J, Alvarez-Mon M, Sáez MA, et al: Oxidative stress markers
are associated with a poor prognosis in patients with pancreatic
cancer. Antioxidants (Basel). 11:7592022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jorgovanovic D, Song M, Wang L and Zhang
Y: Roles of IFN-γ in tumor progression and regression: A review.
Biomark Res. 8:492020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xin L, Xiao W, Che L, Liu J, Miccio L,
Bianco V, Memmolo P, Ferraro P, Li X and Pan F: Label-free
assessment of the drug resistance of epithelial ovarian cancer
cells in a microfluidic holographic flow cytometer boosted through
machine learning. ACS Omega. 6:31046–31057. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Patel H, Wu ZX, Chen Y, Bo L and Chen ZS:
Drug resistance: From bacteria to cancer. Mol Biomed. 2:272021.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Qin T, Chen K, Li J, Qian W, Xiao Y, Wu E,
Ma J, Chen Z, Wang Z, Ma Q and Wu Z: Heat shock factor 1 inhibition
sensitizes pancreatic cancer to gemcitabine via the suppression of
cancer stem cell-like properties. Biomed Pharmacother.
148:1127132022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Miller DW, Fontain M, Kolar C and Lawson
T: The expression of multidrug resistance-associated protein (MRP)
in pancreatic adenocarcinoma cell lines. Cancer Lett. 107:301–306.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Washio I, Nakanishi T, Ishiguro N,
Yamamura N and Tamai I: Impact of breast cancer resistance protein
expression on the in vitro efficacy of anticancer drugs in
pancreatic cancer cell lines. Drug Metab Dispos. 46:214–222. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mojic M, Takeda K and Hayakawa Y: The dark
side of IFN-γ: Its role in promoting cancer immunoevasion. Int J
Mol Sci. 19:892017. View Article : Google Scholar : PubMed/NCBI
|