Introduction
Our bodies are exposed to radiation in numerous
ways, in both high and low doses. The systemic effect of radiation
is directly related to dosage quantity and rate (1). Normal cells are also damaged by
high-dose radiation resulting from accidental radiation leaks, with
detrimental effects on the quality of life in survivors (2). Several research teams are attempting
to identify compounds that can protect radiosensitive organs, such
as the testis (3), intestines
(4) and hippocampus (5). However, there is controversy about
how low-dose radiation exposure (≤0.2 Gy) (6), such as that from imaging diagnosis
(X-ray or computed tomography scan), affects the health of patients
(7,8). Previous studies showed that low-dose
rate (LDR) irradiation (IR) induces testicular damage (9) and aggravates nanoparticle-induced
lung injury (10). However,
several findings also suggest that low-dose radiation ameliorates
asthma symptoms (11) and delays
the progression of Alzheimer's disease (12). In particular, the relationship
between cumulative radiation dose and unfavorable effects,
predicated on ongoing, LDR IR exposure, has not been fully
explained.
The gastrointestinal system is extremely sensitive
to radiation (13). Most radiation
enteritis, or radiation-induced intestinal damage, occurs during
abdominopelvic radiotherapy (14).
Patients with radiation enteritis initially experience diarrhea,
stomach pain and weight loss; however, as the condition progresses,
cancer eventually develops (15).
Studies on low-dose radiation resulting in radiation enteritis are
still inconclusive, with uncertainty on induced alterations.
Finding genes sensitive to low-dose radiation can
provide insights into how LDR radiation affects the human body. RNA
sequencing (RNA-seq) provides an unbiased screening strategy for
potential novel biomarkers associated with radiation response when
searching for a predictor of response. The present study examined
the crucial genes that could be linked to the radiation-induced
intestinal injury response to LDR IR exposure.
Materials and methods
Animals
A total of 90 6-week-old female (weight, 18–21 g)
BALB/c mice were obtained from Dooyeol Biotech and housed at 23±2°C
and relative humidity of 50±5%, with artificial illumination from
08:00-20:00 and 13–18 air changes per h. The mice were classified
into three main categories: control, LDR IR 1 h before, and LDR IR
24 h before. The mice were housed in groups (n=3 mice per group)
and received standard laboratory feed and water at all times. The
experiment involved continuous monitoring of animal health and
behavior, conducted twice daily. All experimental procedures were
conducted according to the National Institutes of Health Guide for
the Care and Use of Laboratory Animals (NIH Publications No. 8023,
8th edition, revised 2011) (16)
and a protocol approved by the Institutional Animal Care and Use
Committee of the Dongnam Institute of Radiological and Medical
Sciences (DIRAMS; approval nos. DI-2015-002 and DI-2021-002).
Efforts were made to reduce suffering and distress
among the animals by providing analgesics (xylazine;
Rompun®; 10 mg/kg; Bayer Koreaa; via intraperitoneal
injection) or anesthetics (alfaxalone; 85 mg/kg (17); Jurox Pty Ltd.; via intraperitoneal
injection) as needed as well as establishing certain housing
conditions. If experimental animals exhibited visible alterations
as well as a weight loss >20%, they were sacrificed in
accordance with criteria for humane endpoints. Following the
administration of respiratory anesthetic induction using 5%
isoflurane for 60 sec, the mouse appeared to be in an unconscious
state. The dose rate of isoflurane was reduced by 2% for
maintenance anesthesia and euthanasia was carried out by applying a
cervical dislocation. Death was confirmed through a physical
examination, which revealed the absence of the cardiac and
respiratory activity. A total of 90 mice were used in the
experiments. No mice were euthanized due to humane endpoints and no
mice were found dead throughout the experiment.
Radiation exposure
The LDR IR exposure was similar to that previously
described (10,11). The DIRAMS LDR IR facility used
radiation exposure with a 137Cs source (370 GBq). The
mouse cages were positioned on shelves with dose rates of 6.0
mGy/h; the total accumulative radiation dose of the set-up places
was 10, 100 and 500 mGy. The group exposed to LDR IR at 10 mGy
undergo three exposures, each occurring at intervals of 3 days. In
addition, mice were high-dose rate (HDR) irradiated with a single
500 mGy dose using 6 MV high energy photon rays (Elekta Instrument
AB) at 3.8 Gy/min. Sham-irradiated mice were treated similarly but
without radiation. After 1 and 24 h of irradiation, the mice were
sacrificed and intestinal tissue samples were taken.
Histological analysis
The small intestine was fixed in 10% neutral
buffered formaldehyde for 24 h at room temperature and four
distinct jejunal segments were sliced into 4 µm thick pieces and
placed on glass slides. To examine morphology, jejunal slices were
stained with hematoxylin and eosin for 2 and 1 min at room
temperature. All samples were sectioned and reoriented in
successive slices to examine the morphological alterations and
ascertain which ones had the longest villi. This method was chosen
because it produced more reliable results than standard techniques,
which only measured the 10 longest villi in a single slice per
sample (4,18). A total of 10 jejunal sections,
including the height of the basal lamina and the length of the 10
longest villi, were measured from each animal. The stained sections
were analyzed using a Motic Easyscan Digital Slide Scanner (Motic
Incorporation, Ltd.).
Dextran sulfate sodium (DSS)-induced
colitis
The induction of colitis in murine models by DSS
(cat. no. 160110; 36–50 kDa; MP Biomedicals) was similar to that
previously described (19); 5% DSS
dissolved in drinking water was used as the treatment method for
mice for 7 days. In the control groups, mice were given normal
drinking water.
Total RNA and mRNA Isolation
Mice (n=1 mouse per group) were sacrificed and their
jejunum tissue were removed. TRIzol® reagent (cat. no.
15596026; Thermo Fisher Scientific, Inc.) was used for total RNA
extraction, according to the manufacturer's instructions.
RNase-Free DNase I (cat. no. EN0521; Thermo Fisher Scientific,
Inc.) was used to degrade genomic DNA. Total RNA levels in the
samples were measured using a Nanodrop 2000 spectrophotometer
(Thermo Fisher Scientific, Inc.). BioAnalyzer2100 (Agilent
Technologies, Inc.) analyzed sample size, quantification and
integrity, eliminating those with a RNA integrity number <7.
Following the manufacturer's instructions, NEXTflex Poly (A) Beads
(Bioo Scientific Corp.) was used to isolate mRNA from total RNA
samples.
mRNA sequencing library preparation
and sequencing
The NEXTflex Rapid Directional RNA-Seq Library Prep
Kit 2.0 (Bioo Scientific Corp.) was used to create an RNA
sequencing library in accordance with the manufacturer's protocols.
In brief, mRNA was chemically fragmented after isolation from 1 µg
of total RNA using NEXTflext Poly (A) Beads. Following the creation
of the double-stranded cDNA from the fragmented mRNA, the library
was purified using magnetic beads made by Agencourt AMPure XP
(Beckman Coulter, Inc.), 3′-end adenylation and sequencing adapter
ligation. The experiment used the Agilent 2100 Bioanalyzer and
reverse transcription-quantitative (RT-q) PCR to verify each
library's successful preparation before moving on to massively
parallel deep sequencing. HiSeq 2500 sequencing platform (Illumina,
Inc.) was used for paired-end sequencing with a typical read size
of 100 nucleotides. The Illumina base-calling pipeline processed
the raw sequencing data and two technical duplicates were created
and sequenced for every sample.
Transcriptome mapping and data
analysis
Adaptor and low-quality bases were trimmed with
Cutadapt ver. 1.3 (20) and Trim
Galore ver. 0.44 (https://www.bioinformatics.babraham.ac.uk) and the
clean reads were aligned to the reference rat genome (rn5)
downloaded from Ensemble (www.ensembl.org) with STAR ver. 2.5 (21). Mapped data from the bam file was
imported to StrandNGS ver. 2.9 (StrandNGS.com) for further
expression analysis. Gene quantitation was normalized using DESEQ
(22) and baseline correction to
the median of gene expression and log2 transform were applied.
Differentially expression genes (DEGs) were determined by
log2-transformed fold changes (≥1 or ≤-1) with one-way analysis of
variance (ANOVA; false discovery rate <0.1). To investigate Gene
ontology (GO) enrichment of DEGs, we utilized Homer ver. 4.5
(23) and Metascape webserver
(http://metascape.org) (24), with Mus musculus used as the
input species and analyzed GO biological process (GO:BP) dataset to
identify upregulated genes. Only terms meeting the criteria of
P<0.05, a minimum count of 3 and an enrichment factor >2 was
considered significant. The most statistically significant term in
a cluster was selected to represent that cluster.
RT-qPCR analysis
Using an ISOGEN kit (311-02501; Nippon Gene, Tokyo,
Japan), total RNA was isolated from jejunal samples. RT-qPCR
analyses were performed as described in a previous study (4). RT-qPCR was performed duplicate using
CFX Opus 96 instrument (Bio-Rad Laboratories) and
TOPreal™ SYBR Green qPCR 2× PreMIX (Cat. No. RT500M;
Enzynomics) and the according the manufacturer's guidelines. The
RT-qPCR reaction was carried out in a 25 µl reaction, including 5
µl of cDNA (40 ng), 1 µl of each primer (6 µM), 12.5 µl of 2× SYBR
green PreMix, and 5.5 µl of double distilled water. The thermal
cycling profile comprised a preincubation step at 94°C for 10 min,
followed by 45 cycles of denaturation (94°C, 15 sec), annealing
(55–60°C, 30 sec), and elongation (72°C, 20 sec). The software
generated amplification curves and determined threshold cycle
values. Primer candidates were acquired utilizing an application
available on the PRIMER3 ver. 0.4.0 website (https://bioinfo.ut.ee/primer3-0.4.0/). The primers
were validated using the NCBI BLAST (basic local alignment search
tool) ver. +2.15.0 database (https://blast.ncbi.nlm.nih.gov/). The expression
levels of mRNAs for ATPase NA+/K+
transporting subunit alpha 4 (ATP1A4), glucose-6-phosphatase
catalytic subunit 2 (G6PC2), mucin 6 (MUC6), mucin 20 (MUC20) and
transient receptor potential cation channel subfamily V member 6
(TRPV6) were measured, normalized to the expression level of
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA expression
and expressed relative to the corresponding mean value for the
jejunum tissue of control mice that was unexposed to radiation
using the 2−ΔΔCq method (25). Mouse primers used in analysis were
as follows: ATP1A4-(NM_013734) forward 5′-CTGGTGAGCTGAATCAGAAACC-3′
and reverse 5′-AAGACCCTTGGTCAAGTCCAC-3′; G6PC2-(NM_001289857)
forward 5′-CAGGAGGACTACCGGACTTAC-3′ and reverse
5′-TCAACTGAAACCAAAGTGGGAA-3′; MUC6-(NM_001368953) forward
5′-AGCCCACATTCCCTATCAGC-3′ and reverse 5′-CACAGTGGAAGATTGCGAGAG-3′;
TRPV6-(NM_022413) forward 5′-AGGGGTTAATACTCTGCCTATGG-3′ and
reverse, 5′-GCACCTCACATCCTTCAAACTT-3′; GAPDH-(NM_008084.2) forward
5′-TCCATGACAACTTTGGCATT-3′ and reverse
5′-GTTGCTGTTGAAGTCGCAGG-3′.
Western blot analysis
The jejunum was homogenized in buffer H (50 mM
β-glycerophosphate, 1.5 mM ethylene glycol tetra acetic acid, 0.1
mM Na3VO4, 1 mM dithiothreitol, 10 µg/ml
aprotinin, 2 µg/ml pepstatin, 10 µg/ml leupeptin, and 1 mM
phenylmethanesulfonyl fluoride, pH 7.4) for western blotting
examination. The protein content of each sample was determined
using a Bio-Rad protein assay reagent (cat. no. 500-0006; Bio-Rad
Laboratories, Hercules, CA, USA). Western blotting was performed as
described previously (10).
Briefly, each sample's total protein equivalent was separated on
10% SDS-polyacrylamide gels (40 µg of protein per lane) and
transferred to an Immobilon-PSQ transfer membrane (cat. no.
ISEQ00010; MilliporeSigma). The membrane was soaked for 1 h at room
temperature in a blocking solution containing 5% non-fat milk.
Primary antibodies against ATP1A4 (diluted 1:1,000; cat. no.
STJ117451; St John's Laboratory Ltd.), G6PC2 (diluted 1:1,000; cat.
no. bs-13386R; BIOSS), MUC6 (diluted 1:1,000; cat. no. 224329;
United States Biological) and TRPV6 (diluted 1:1,000; cat. no.
ACC-036; Alomone Labs) were incubated at 4°C with consistent
rocking overnight. After four 10-min Tris-buffered saline (TBS) and
0.1% Tween-20 (TBS-T) washes, the membrane was incubated with
secondary polyclonal anti-rabbit antibodies (diluted 1:10,000; cat.
no. 12-348; MilliporeSigma) in TBS-T buffer at ambient temperature
for 1 h. Using a chemiluminescence reagent (cat. no. 34580;
SuperSignal West Pico PLUS; Thermo Fisher Scientific) and the
ChemiDoc MP Imaging system (Bio-Rad Laboratories, Inc.), the
labeling of the secondary antibody conjugated with horseradish was
detected. The membrane was probed with β-actin (diluted 1:5,000;
cat. no. A5441; MilliporeSigma), a housekeeping gene, to measure
protein expression levels. Using Image Lab ver. 6.0.1 (Bio-Rad
Laboratories, Inc.) software, protein bands were quantified.
Statistical analysis
Results are presented as mean ± standard error of
mean (SEM). Radiation-induced response screening was analyzed using
one-way analysis of variance (ANOVA) with additional Tukey's post
hoc testing. Dose-dependent response validation,
frequency-dependent response validation and evaluation of
correlation between LDR IR and inflammatory bowel disease (IBD)
were analyzed using two-way ANOVA followed by Tukey's post hoc
testing. P<0.05 was considered to indicate a statistically
significant difference.
Results
Gene ontology biological process
analysis of RNA-seq screening data
To screen for genes changed by LDR IR exposure,
RNA-seq was performed 1 and 24 h after irradiation at 10 mGy
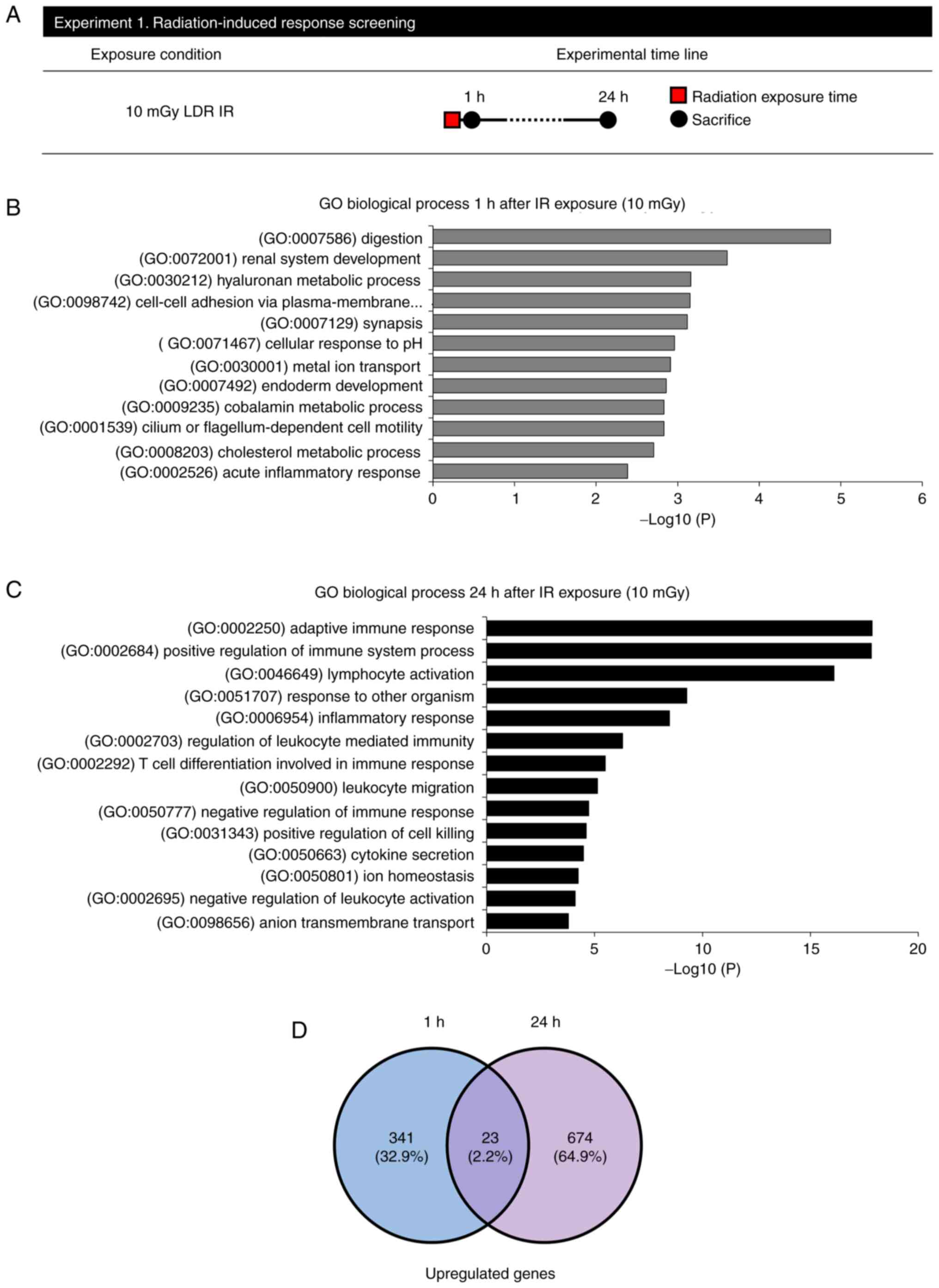
(Fig. 1A). At 1 h following
radiation exposure, 12 enriched GO:BP terms were obtained when 0 Gy
was compared with 1 h after exposure to 10 mGy (Fig. 1B and Table I). Furthermore, there was an
increase in the BP associated with the digestion system. After 24 h
of radiation exposure, 14 enriched GO:BP terms were obtained when 0
Gy was compared with 24 h after 10 mGy (Fig. 1C and Table II). Moreover, there was an
observed increase in the BP associated with the immune system.
However, no meaningful biological process demonstrated an expected
increase across a period of 1 and 24 h following exposure. RNA-seq
was used to find genes responsive to LDR IR exposure to identify
DEGs affected in the LDR IR jejunum (364 DEGs upregulated in the 1
h after LDR irradiated jejunum; 697 DEGs upregulated in the 24 h
after LDR of the jejunum, with 23 genes in common with the
comparison of 1 and 24-h IR groups; Fig. 1D and Table III).
 | Table I.GO term enrichment analysis of DEGs 1
h after 10 mGy radiation exposure. |
Table I.
GO term enrichment analysis of DEGs 1
h after 10 mGy radiation exposure.
| Pathways | P-value | DEGs/Total | Genes |
|---|
| (GO:0007586)
digestion |
1.34×10−5 | 10/164 | AMY2A, AMY2B,
CCKAR, CEL, CLPS, CTRL, MUC6, PPY, PRSS3, WNK3 |
| (GO:0072001) renal
system development |
2.47×10−4 | 13/277 | HMGCS2, LHX1,
NPHS1, PKHD1, UMOD, TBX18, CRLF1, KLHL3, KLF15, SIX4, PKD1L3, FZD3,
RNF207 |
| (GO:0030212)
hyaluronan metabolic process |
6.48×10−4 | 4/37 | EGF, ITIH2, ITIH4,
HYAL3 |
| (GO:0098742)
cell-cell adhesion via plasma-membrane adhesion molecules |
7.04×10−4 | 20/219 | FAT2, UMOD,
PCDHGB4, CLDN10, PCDHGC5, PCDHGB5, PCDHGB1, PCDHGA4, CDHR4, CEL,
CTLA4, IGFBP2, INS, ITGA2B, PKHD1, CCL2, EOMES, MMP24, CLEC4E,
ADGRV1 |
| (GO:0007129)
synapsis |
7.63×10−4 | 4/38 | SYCP2, STAG3,
SPO11, RNF212 |
| (GO:0071467)
cellular response to pH |
1.06×10−3 | 3/19 | INSRR, SLC38A3,
PKD1L3 |
| (GO:0030001) metal
ion transport |
1.23×10−3 | 21/796 | ATP1A4, CCKAR,
CNGA1, EGF, KCNJ11, SCN4A, CCL2, SLC12A3, SLC38A3, KLHL3, SLC40A1,
TRPV6, WNK3, CATSPER2, KCNH8, SLC9B2, NKAIN2, PKD1L3, RNF207,
CPT1B, CHRNA2 |
| (GO:0007492)
endoderm development |
1.38×10−3 | 6/75 | COL7A1, ONECUT1,
LHX1, VTN, EOMES, CRB2 |
| (GO:0009235)
cobalamin metabolic process |
1.48×10−3 | 3/21 | PRSS3, CUBN,
CTRC |
| (GO:0001539) cilium
or flagellum-dependent cell motility |
1.48×10−3 | 3/21 | DNAH6, DNAH7,
CFAP54 |
| (GO:0008203)
cholesterol metabolic process |
1.97×10−3 | 10/119 | CEL, HMGCS2, STAR,
CUBN, CELA3B, APOL2, ALDH3A1, CCKAR, IGFBP2, CCL2 |
| (GO:0002526) acute
inflammatory response |
4.12×10−3 | 11/138 | C4B, INS, ITIH4,
VTN, APOL2, NUPR1, COL7A1, ITIH2, SSPO, SERPINA10, CRB2 |
 | Table II.GO term enrichment analysis of DEGs
24 h after 10 mGy radiation exposure. |
Table II.
GO term enrichment analysis of DEGs
24 h after 10 mGy radiation exposure.
| Pathways | P-value | DEGs/Total | Genes |
|---|
| (GO:0002250)
adaptive immune response |
1.40×10−18 | 76/371 | BTK, C4B, CD27,
CD40, CD79B, CLU, CR2, CTLA4, CD55, FCER2, IGHD, IGHG1, IGHM,
IGLC2, IL12A, JAK3, LY9, POU2F2, CCL19, SLAMF1, TNF, LAT2, EOMES,
IL27RA, MASP2, LILRB4, CLCF1, LAT, SIT1, FOXP3, LEF1, IL20RB, LAX1,
LIME1, AICDA, MYO1G, FCAMR, KLHL6, SLAMF6, TNFRSF13C, BTLA, CLEC4D,
THEMIS, KLRD1, NOS2, CCL2, STAP1, IL21, CA7, IFI16, IL2RA, INS,
MX1, MYLK, PPY, MYOM1, CHST3, TSPAN32, CLEC4E, ALPK1, NLRC5,
PGLYRP2, TRIM6, CXCR5, MS4A1, IKZF3, BANK1, TNIP2, CCR6, LTF, PAX5,
SERPINC1, F8, ABCA1, PRTN3, MARCO |
| (GO:0002684)
positive regulation of immune system process |
1.49×10−18 | 74/943 | BTK, C4B, CD2,
CD19, CD27, CD40, CD79B, CLU, CCR6, MAP3K8, CR2, CTLA4, CD55, COCH,
FCER2, CXCL1, CXCL2, IFI16, IGHD, IGHG1, IGHM, IGLC2, IL2RA, IL12A,
ITGA2B, JAK3, LTF, NOS2, CCL2, CCL19, CCL20, CXCL5, SLAMF1, TNF,
LAT2, MARCO, IL27RA, MASP2, CLCF1, STAP1, CLEC4E, LAT, ICOS, FOXP3,
LEF1, UBASH3A, LAX1, LIME1, IL21, TRPV4, MYO1G, RASAL3, TNIP2,
TNIP3, FCRL5, NLRC5, KLHL6, PGLYRP2, SLAMF6, TNFRSF13C, TRIM6,
REG3G, SLC9B2, CARMIL2, BTLA, GCSAM, CLEC4D, ABCB5, TICAM2, THEMIS,
RASAL1, RASGRP3, SPTBN5, IL20RB |
| (GO:0046649)
lymphocyte activation |
8.02×10−17 | 103/618 | CXCR5, BTK, CD2,
MS4A1, CD27, CD40, MAP3K8, CR2, CTLA4, CD55, HLA-DOA, IGHD, IGHG1,
IGHM, IGLC2, IL2RA, IL12A, INS, JAK3, LY9, POU2F2, SATB1, CCL2,
CCL19, SLAMF1, LAT2, EOMES, IL27RA, IKZF3, KIAA0922, CLCF1, CLEC4E,
LAT, SIT1, ICOS, IL21R, FOXP3, LEF1, IL20RB, LAX1, BANK1, AICDA,
IL21, RASAL3, TNIP2, NFKBID, PGLYRP2, SLAMF6, TNFRSF13C, CARMIL2,
BTLA, CLEC4D, THEMIS, CLU, CNR2, TSPAN32, STAP1, F8, ITGA2B, PLEK,
TTN, SLC7A11, C6orf25, CELSR3, REG3A, S100A9, TNF, UMOD, PCDHGB4,
PCDHGC5, PCDHGB5, PCDHGB1, PCDHGA7, PCDHGA4, TRPV4, CDH26, PCDH15,
ADGRV1, CDHR4, IFI16, LTF, PRTN3, LILRB4, SLC9B2, APOD, HLA-DOB,
UBASH3A, FCRL5, NLRC5, GCSAM, TICAM2, ABCA1, ALOX15B, TNFRSF8, LTB,
NOS2, CCL20, WNT11, TRIM6, SLC40A1, SIX4, NUGGC, RELL2 |
| (GO:0051707)
response to other organism |
5.33×10−10 | 71/922 | ABCA1, C4B, CA7,
CD27, TNFRSF8, CD40, CLU, CNR2, CYP27B1, CD55, COCH, CXCL1, CXCL2,
HMGCS2, IFI16, IGHD, IGHG1, IGHM, IGLC2, IL2RA, IL12A, LTF, MX1,
MYLK, NOS2, PPY, S100A9, CCL2, CCL19, CCL20, CXCL5, STAR, TIMP4,
TNF, CXCR4, MYOM1, SOCS3, IL27RA, CHST3, TSPAN32, STAP1, CLEC4E,
FOXP3, AICDA, TNIP2, TNIP3, ALPK1, NLRC5, PGLYRP2, TRIM6, REG3G,
CLEC4D, TICAM2, IFITM10, APOD, SERPINC1, BTK, MAP3K8, INS, MARCO,
IL20RB, IL21, TRPV4, SLAMF6, GIPR, NPY1R, WNT11, NCOA4, GPR83,
LEF1, CCDC62 |
| (GO:0006954)
inflammatory response |
5.37×10−9 | 61/646 | APOD, SERPINC1,
C4B, CD27, TNFRSF8, CD40, CNR2, CD55, F8, FPR2, CXCL1, CXCL2,
IFI16, IL2RA, INS, ITIH4, NOS2, REG3A, S100A9, CCL2, CCL19, CCL20,
CXCL5, TNF, CXCR4, NPFF, SOCS3, CHST4, APOL2, LAT, FOXP3, IL20RB,
IL21, TRPV4, TNIP2, TNIP3, NFKBID, PGLYRP2, REG3G, TICAM2, PLA2G4B,
CA7, CCR6, CNGA1, CYP27B1, IL12A, LTF, MYLK, PDE6G, PLEK, PPY, RHO,
MYOM1, CHST3, TSPAN32, STAP1, ALPK1, SPX, TRIM6, ATP6V0D2,
NLRC5 |
| (GO:0002703)
regulation of leukocyte-mediated immunity |
5.03×10−7 | 32/155 | BTK, CD40, FCER2,
IL12A, JAK3, NOS2, CCL2, SLAMF1, TNF, IL27RA, CLCF1, STAP1, FOXP3,
IL20RB, IL21, SLAMF6, CCL19, TNFRSF13C, CD55, POU2F2, VPREB3, LAX1,
AICDA, TRIM6, NUGGC, MS4A1, CR2, CTLA4, IKZF3, LEF1, CRLF1,
TEX15 |
| (GO:0002292) T cell
differentiation involved in immune response |
3.16×10−6 | 24/54 | JAK3, LY9, CCL19,
EOMES, CLEC4E, FOXP3, LEF1, SLAMF6, CLEC4D, CD55, IL12A, INS,
SATB1, RASAL3, CD40, SLAMF1, LAT2, IL27RA, CLCF1, LAT, AICDA,
PGLYRP2, IL21, THEMIS |
| (GO:0050900)
leukocyte migration |
7.22×10−6 | 18/355 | APOD, CD2, CCR6,
CXCL1, CXCL2, IL12A, ITGA2B, MAG, S100A9, CCL2, CCL19, CCL20,
CXCL5, SELL, TNF, UMOD, CXCR4, SLC7A1 |
| (GO:0050777)
negative regulation of immune response | 1.85 ×
10−5 | 20/120 | CD55, IFI16, IL2RA,
INS, JAK3, SLAMF1, TNF, IL27RA, FOXP3, IL20RB, NLRC5, PGLYRP2, BTK,
CD40, CLCF1, TRIM6, SATB1, IL12A, MYO1G, BANK1 |
| (GO:0031343)
positive regulation of cell killing |
2.42×10−5 | 7/39 | FCER2, IL12A, NOS2,
CCL2, STAP1, IL21, SLAMF6 |
| (GO:0050663)
cytokine secretion |
3.27×10−5 | 45/169 | ABCA1, ALOX15B,
CD2, INS, NOS2, CCL19, SLAMF1, TNF, CLEC4E, FOXP3, BANK1, TRPV4,
TRIM6, CARMIL2, CD40, CLU, F8, GIPR, ITGA2B, NPY1R, PLEK, POU2F2,
PPY, SLC1A7, TFR2, TRPM2, TTN, LAT2, SYN3, NPFF, CACNA1I, LAT,
SERPINA10, CKLF, LAX1, TRPV6, G6PC2, MYO1G, SYT8, ILDR1, CYP4A11,
NPHS1, S100A9, WNK3, SPX |
| (GO:0050801) ion
homeostasis |
5.66×10−5 | 60/664 | ABCA1, ATP1A4, CA7,
CD40, CCR6, CYP4A11, CYP27B1, CD55, GIPR, INS, JAK3, LTF, NPY1R,
RYR1, S100A9, CCL2, CCL19, TFR2, TRPM2, UMOD, CXCR4, NPFF, KLHL3,
SLC40A1, TRPV4, WNK3, SPX, FCRL5, SLC4A11, SLC9A7, ATP6V0D2,
SLC26A11, CNGA1, GRM6, KCNJ15, MYLK, SCN4A, SCN8A, SLC13A1,
CACNA1I, CACNG4, TRPV6, KCNT1, DPP10, SLC13A3, MFSD4B, CATSPER2,
SLC9B2, SLC38A8, PKD1L3, CATSPER3, CLCN1, CPT1B, GABRA1, GABRE,
SLC37A2, APOC4, AQP9, STAR, G6PC2 |
| (GO:0002695)
negative regulation of leukocyte activation |
7.94×10−5 | 17/139 | CNR2, CTLA4, IL2RA,
JAK3, TSPAN32, KIAA0922, FOXP3, IL20RB, LAX1, BANK1, NFKBID,
PGLYRP2, LTF, LILRB4, LEF1, TRPV4, CARMIL2 |
| (GO:0098656) anion
transmembrane transport |
1.60×10−4 | 31/245 | ABCA1, AQP9, CLCN1,
CPT1B, GABRA1, GABRE, SLC1A7, SLC13A1, SLC7A11, SLC13A3, LRRC8E,
SLC4A11, SLC38A8, SLC37A2, SLC26A11, SLC6A18, CA7, CYP4A11, NOS2,
PLA2G5, SLCO1C1, ATP8B4, SPX, SLC16A9, APOC4, APOD, CLU, STAR, TNF,
APOL2, PLIN5 |
 | Table III.Common upregulated genes. |
Table III.
Common upregulated genes.
|
|
|
| Fold change |
|---|
|
|
|
|
|
|---|
| Gene ID | Gene symbol | Description | 1 h | 24 h |
|---|
| 239552 | APOL2 | Apolipoprotein
L2 | 8.3 | 4.4 |
| 480 | ATP1A4 | ATPase Na+/K+
transporting subunit alpha 4 | 2.8 | 3.0 |
| 20293 | CCL2 | C-C motif chemokine
ligand 2 | 3.4 | 3.5 |
| 6364 | CCL20 | C-C motif chemokine
ligand 20 | 2.1 | 3.5 |
| 26253 | CLEC4E | C-type lectin
domain family 4 member E | 2.1 | 6.5 |
| 9244 | CRLF1 | Cytokine receptor
like factor 1 | 2.6 | 2.1 |
| 57818 | G6PC2 |
Glucose-6-phosphatase catalytic subunit
2 | 9.3 | 8.7 |
| 16334 | INS | Insulin | 140.0 | 100.0 |
| 3700 | ITIH4 | Inter-alpha-trypsin
inhibitor heavy chain family member 4 | 2.7 | 6.9 |
| 26249 | KLHL3 | Kelch-like family
member 3 | 3.1 | 2.3 |
| 200958 | MUC20 | Mucin 20, cell
surface associated | 4.8 | 4.9 |
| 4588 | MUC6 | Mucin 6, oligomeric
mucus/gel-forming | 4.7 | 6.6 |
| 4868 | NPHS1 | NPHS1 nephrin | 3.9 | 2.5 |
| 5539 | PPY | Pancreatic
polypeptide | 29 | 11 |
| 19693 | REG1B | Regenerating family
member 1 beta | 2.3 | 140 |
| 51156 | SERPINA10 | Serpin family A
member 10 | 8.1 | 6.3 |
| 30061 | SLC40A1 | Solute carrier
family 40 member 1 | 2.2 | 3.0 |
| 133308 | SLC9B2 | Solute carrier
family 9 member B2 | 4.3 | 4.9 |
| 80763 | SPX | Spexin hormone | 5.5 | 7.2 |
| 943 | TNFRSF8 | Tumor necrosis
factor receptor superfamily member 8 | 11.0 | 18.0 |
| 55503 | TRPV6 | Transient receptor
potential cation channel subfamily V member 6 | 11.0 | 11.0 |
| 7369 | UMOD | Uromodulin | 6.5 | 9.1 |
| 65267 | WNK3 | WNK lysine
deficient protein kinase 3 | 3.7 | 3.9 |
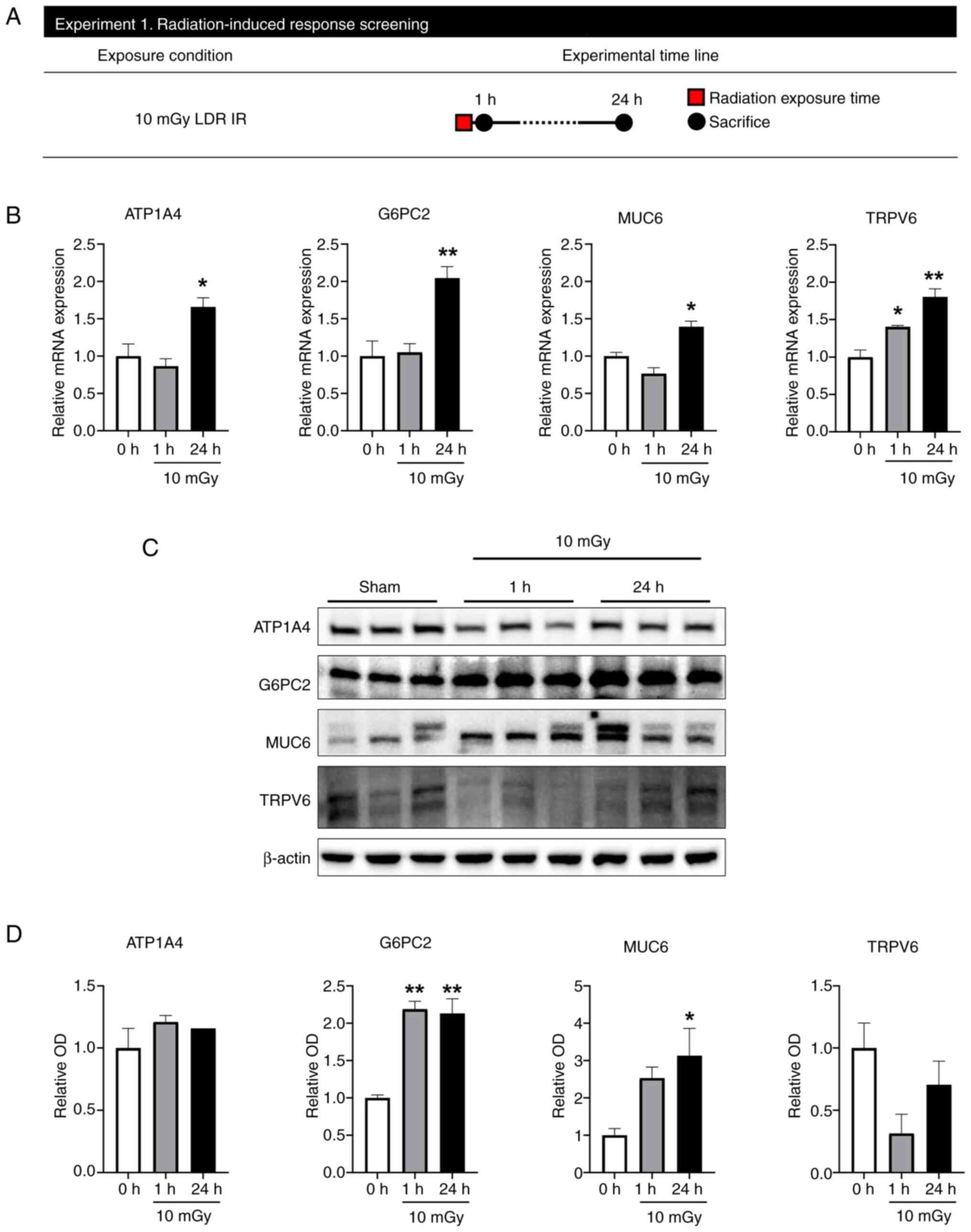
Immediately following LDR IR exposure,
G6PC2 and MUC6 mRNA and protein expression are increased
To verify changes in genes and proteins caused by
LDR IR exposure, RT-qPCR and western blot were performed on mice 1
and 24 h after irradiation at 10 mGy (Fig. 2A). A total of 23 DEGs were selected
for qPCR detection to validate the outcome of RNA-seq analysis:
APOL2, ATP1A4, CCL2, CCL20, CLEC4E, CRLF1, G6PC2, INS, ITIH4,
KLHL3, MUC20, MUC6, NPHS1, PPY, REG1B, SERPINA10, SLC40A1, SLC9B2,
SPX, TNFRSF8, TRPV6, UMOD and WNK3. Among these, qPCR results
showed that only ATP1A4, G6PC2, MUC6 and TRPV6 levels increased
significantly 24 h after LDR IR (Fig.
2B). The protein expression levels of ATP1A4, G6PC2, MUC6,
MUC20 and TRPV6 were examined (Fig. 2C
and D). G6PC2 expression showed significant upregulation in the
1 and 24 h following 10 mGy radiation exposure to the jejunum. MUC6
was also significantly upregulated in the 24 h following LDR IR
exposure. However, MUC20 was not detectable by western blotting and
ATP1A4 and TRPV6 did not alter after LDR IR exposure. Therefore, it
was hypothesized that G6PC2 and MUC6 are important genes triggered
by LDR IR exposure.
G6PC2 mRNA increases in low doses of
LDR IR and repeated LDR IR exposure conditions in the jejunum
To further validate the dose-dependent change in
G6PC2 gene expression, qPCR analysis was performed on mice
intestinal tissues exposed to LDR IR (10, 100 and 500 mGy) and HDR
(500 mGy; Fig. 3A). In the
jejunum, G6PC2 gene expression was significantly increased
following 10 and 100 mGy exposure compared with the sham group
(Fig. 3B). However, 500 mGy of LDR
IR and 500 mGy of HDR IR did not alter G6PC2 expression levels.
Furthermore, G6PC2 expression of the colon also did not change
after LDR IR and HDR IR (Fig. 3C).
Therefore, it was concluded that G6PC2 only reacts at LDR IR and
this reaction is limited to the jejunum. In an additional
experiment, the expression level of G6PC2 was validated by repeated
LDR IR treatment (Fig. 3D). A
single 10 mGy exposure significantly increased G6PC2 expression
after 24 h and three 10 mGy exposures increased G6PC2 expression
explosively after 24 h (Fig. 3E),
suggesting that frequent exposure to LDR effectively increases
G6PC2 expression.
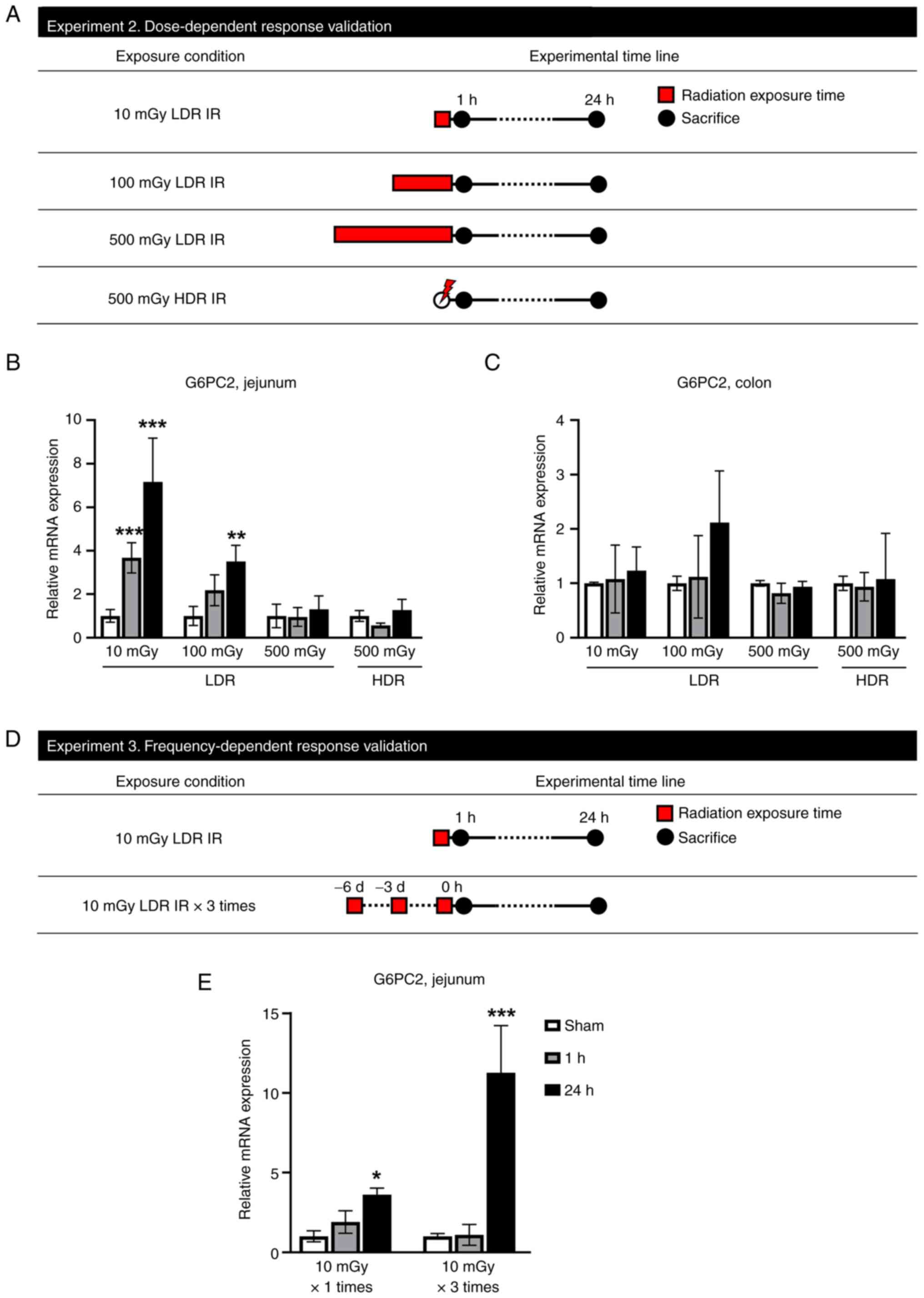 | Figure 3.G6PC2 mRNA increases in low dose of
LDR IR and repeated LDR IR exposure conditions in the jejunum. (A)
Schematic diagram of the experimental procedure. In experiment 2,
mice exposed to LDR IR with 0, 10, 100, or 500 mGy or HDR IR with
500 mGy, then sacrificed at 1 or 24 h following IR. Red square
indicates radiation exposure time; black circles indicate the times
of tissue collection from test animals. Changes in the mRNA
expression of G6PC2 in mice exposed to LDR IR. Dose-related changes
in G6PC2 mRNA expression level in (B) jejunum and (C) colon. (D)
Schematic diagram of the experimental procedure. In experiment 3,
mice irradiated with sham, 10 mGy once, or 10 mGy three times were
sacrificed for tissue sampling at 1 or 24 h following LDR IR. Red
square indicates radiation exposure time; black circles indicate
the times of tissue collection from test animals. (E)
Repetition-related changes in G6PC2 mRNA expression level in
jejunum. Data are expressed as the mean ± SEM (n=3 mice per group).
*P<0.05, **P<0.01 and ***P<0.01 vs. the sham group. G6PC2,
glucose-6-phosphatase catalytic subunit 2; LDR IR, low-dose-rate
irradiation; HDR IR, high-dose-rate irradiation. |
MUC6 mRNA increases in low doses of
LDR IR and repeated LDR IR exposure conditions in the jejunum
To validate the dose-dependent change in MUC6 gene
expression, qPCR analysis was performed on mice intestinal tissue
exposed to LDR IR (10, 100 and 500 mGy) and HDR (500 mGy; Fig. 4A). Identifying MUC6 mRNA expression
for dose-dependent alterations revealed outcomes comparable to
G6PC2. In the jejunum, MUC6 gene expression was significantly
higher following 10 mGy exposure compared with the sham group
(Fig. 4B). However, 100 and 500
mGy of LDR IR and 500 mGy of HDR IR did not alter MUC6 expression
levels. Furthermore, MUC6 expression of the colon also did not
change following LDR IR and HDR IR (Fig. 4C). Therefore, it can be concluded
that MUC6 only reacts at LDR IR and that this reaction is limited
to the jejunum. In an additional experiment, the expression level
of MUC6 was validated by repeated LDR IR treatment (Fig. 4D). A single 10 mGy exposure
significantly increased MUC6 expression after 24 h and three 10 mGy
exposures increased MUC6 expression markedly after 24 h (Fig. 4E), suggesting that frequent
exposure to LDR IR effectively increases MUC6 expression.
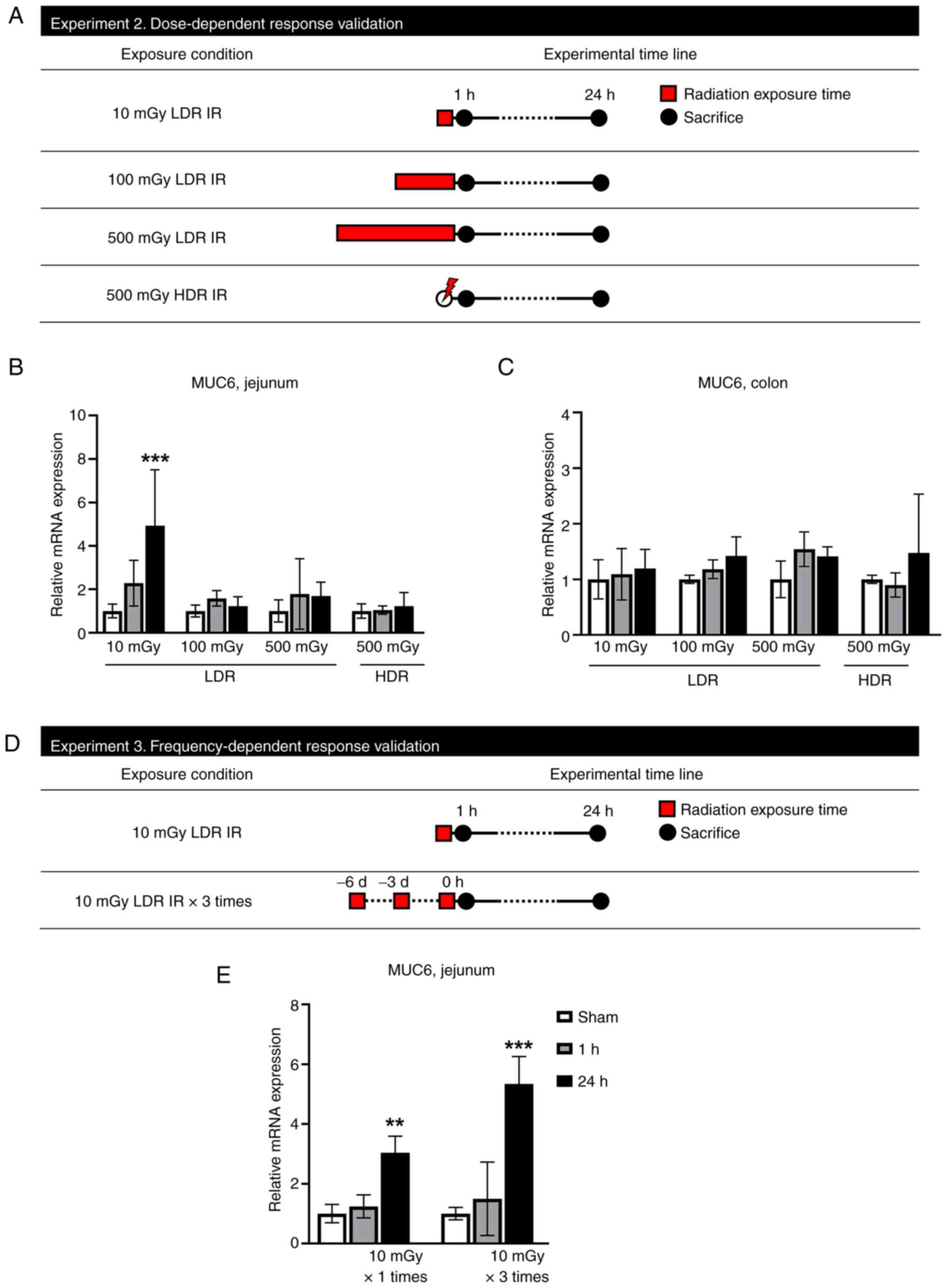 | Figure 4.MUC6 mRNA increases in low dose of
LDR IR and repeated LDR IR exposure conditions in the jejunum. (A)
Schematic diagram of the experimental procedure. In experiment 2,
mice exposed to LDR IR with 0, 10, 100, or 500 mGy or HDR IR with
500 mGy, then sacrificed at 1 or 24 h following IR. Red square
indicates radiation exposure time; black circles indicate the times
of tissue collection from test animals. Changes in the mRNA
expression of MUC6 in the intestine of mice exposed to LDR IR.
Dose-related changes in MUC6 mRNA expression level in (B) jejunum
and (C) colon. (D) Schematic diagram of the experimental procedure.
In experiment 3, mice irradiated with sham, 10 mGy once, or 10 mGy
three times were sacrificed for tissue sampling at 1 or 24 h
following LDR IR (n=3 mice per group). Red square indicates
radiation exposure time; black circles indicate the times of tissue
collection from test animals. (E) Repetition-related changes in
MUC6 mRNA expression level in jejunum. Data are expressed as the
mean ± SEM (n=3 mice per group). **P<0.01 and ***P<0.001 vs.
the sham group. MUC6, mucin 6; LDR IR, low-dose-rate irradiation;
HDR IR, high-dose-rate irradiation. |
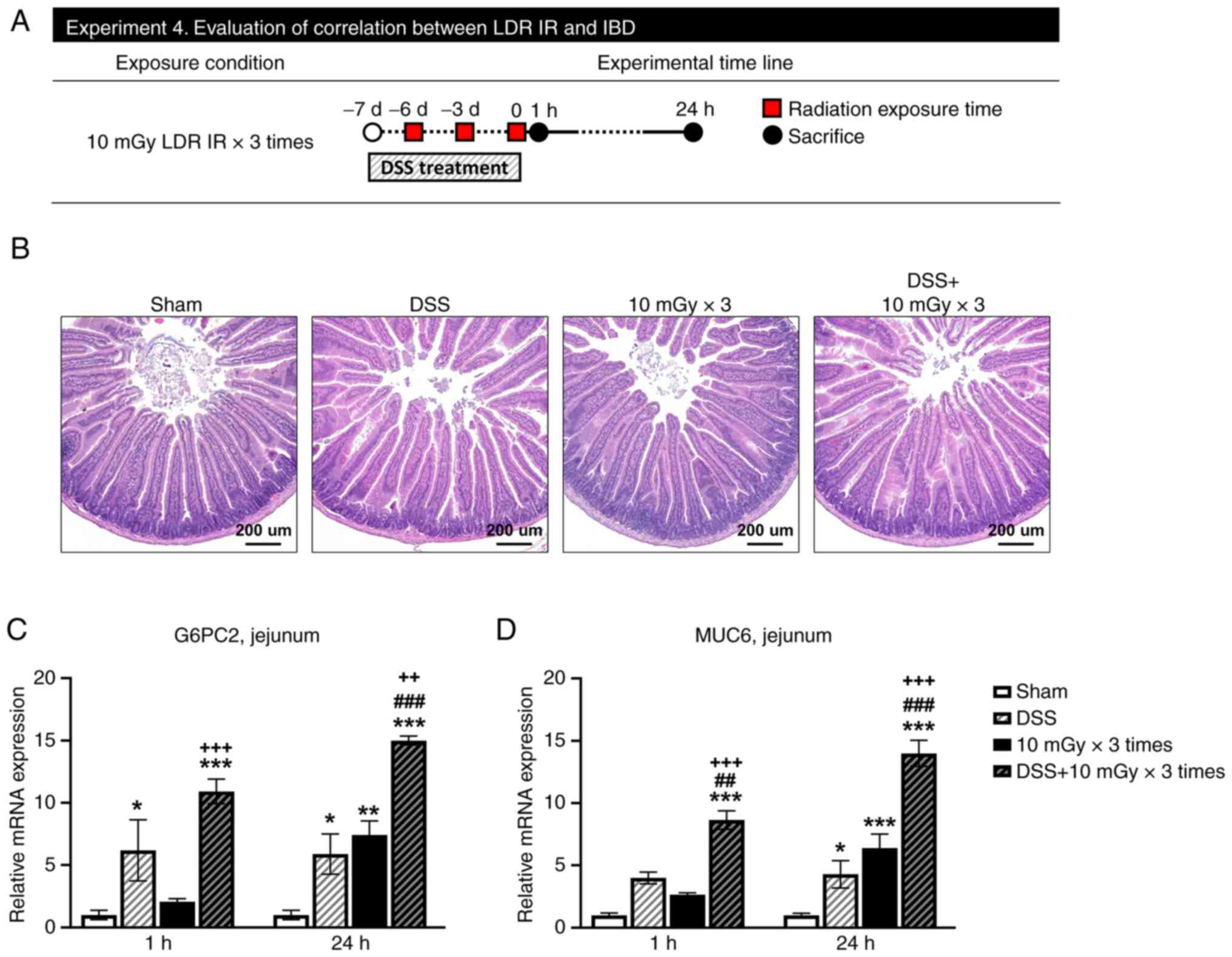
DSS and LDR IR did not alter jejunal
morphology but increased G6PC2 and MUC6 mRNA expression
In order to establish the effect of LDR IR on the
intestines with inflammatory conditions, infectious bowel disease
was induced by DSS, the jejunal area received radiation and the
expression levels of G6PC2 and MUC6 were verified (Fig. 5A). DSS-induced IBD and LDR IR did
not cause morphological alterations in the small intestine when
processed independently or even after combined treatments;
subsequently, neither the length of the villus nor the number of
crypts changed, observed 24 h after irradiation (Fig. 5B). Thus, DSS and LDR IR in the
jejunum did not act as a powerful stimulus to alter morphological
modifications. However, after 24 h, repeated irradiation of LDR IR
and DSS significantly boosted G6PC2 expression. Furthermore, when
the two stimuli were combined, G6PC2 expression increased with
greater efficacy, observed 1 and 24 h after irradiation (Fig. 5C). The expression of MUC6 was
significantly increased by exposure to LDR IR and stimulation with
DSS. In addition, the combination of these two stimuli led to a
substantial increase in MUC6 expression (Fig. 5D).
Discussion
The present study aimed to identify genes that were
changed rapidly and consistently in the jejunum of mice exposed to
LDR IR. The following important conclusions were noted: i) A total
of 23 genes consistently showed elevated expression levels at both
1 and 24 h following exposure to an LDR IR dose of 10 mGy. ii) qPCR
and western blot analyses confirmed the expression patterns of the
G6PC2 and MUC6 genes. iii) Significantly, the upregulation of G6PC2
and MUC6 gene expressions was limited to the jejunum following
exposure to LDR IR, whereas it was not observed in the colon. iv)
The increases in the expressions of G6PC2 and MUC6 were more
noticeable when exposed to repeated LDR IR, indicating a link
between the radiation dosage and the response. v) Furthermore, the
present study revealed that in the presence of inflammatory bowel
illness, the levels of G6PC2 and MUC6 expressions were increased
significantly, emphasizing the possible significance of these genes
in developing the disease.
The small intestine is highly vulnerable to
radiation (26). The main symptoms
of radiation-induced intestinal injury include vomiting, diarrhea,
anorexia, systemic infections and, in severe cases, septic shock
and death (27). Several research
teams are investigating the identification and therapy of the
underlying mechanisms responsible for radiation-induced
gastrointestinal problems. The influence of the gastrointestinal
system on radiation has mainly studied in the jejunum, specifically
3 days after exposure to a radiation dose of 10 Gy (4,28).
Radiation causes changes in the structure and function of the
intestines, including the death of intestinal crypt cells,
disruption of the barrier and inflammation of the mucous membrane,
resulting in a decrease in the height of the villi (29). However, the effect of LDR IR on the
jejunum remains uncertain.
RNA-seq was used to determine the molecular
differences between the jejunum of sham- and LDR IR-exposed mice.
Through GO:BP analysis, a notable variation in 12 GO:BP terms was
detected 1 h after exposure to 10 mGy irradiation, with the
digestion (GO:0007586) showing the most significant change. At 24 h
following exposure to 10 mGy irradiation, a notable difference was
detected in 14 GO:BP terms, with the adaptive immune response
(GO:0002250) being the most significant alteration. Although no
common GO:BP term were identified between these two time points, it
was found that 23 genes exhibited an expected increase. PCR and
western blot analyses confirmed that the gene expressions of G6PC2
and MUC6 were upregulated following irradiation. This study
revealed that the gene expressions of G6PC2 and MUC6 increased in
response to LDR IR (10 mGy) and their expression further increased
when the LDR IR was exposed repeatedly.
G6PC2, known as islet-specific
glucose-6-phosphate-catalytic-subunit-related protein (IGRP), is
one of three G6PC isoforms that catalyze the hydrolysis of glucose
6-phosphate to glucose and inorganic phosphate. G6PC2 is expressed
almost exclusively in pancreatic islet beta cells, which are
supposed to act as a negative regulator of the beta cell glucose
sensor glucokinase (30,31). Instead of employing its enzymatic
function, G6PC2 acts as a major autoantigen in type I diabetes
patients and mouse models of human disease. Studies show that G6PC2
(IGRP206-214)-reactive CD8+ T cells may modulate the
interaction between type I diabetes and colitis through molecular
mimicry in non-obese diabetic (NOD) mice. Antigen mimics from gut
microbes could activate or repress G6PC2
(IGRP206-214)-reactive CD8+ T cells, resulting in the
modulation of type I diabetes and colitis (32,33).
MUC6, an isoform of mucin glycoproteins, is found
only in the stomach and duodenum of healthy individuals and
generates gel-forming mucin that covers the digestive epithelium
(34). Several investigations have
found MUC6 in breast mucinous carcinoma, stomach adenocarcinoma,
colon neuroendocrine carcinoma and pulmonary adenocarcinoma
(34–37). In addition to its association with
cancer, MUC6 plays a role in epithelial wound healing after mucosal
injury in IBDs, such as Crohn's disease (38).
In the present study, G6PC2 and MUC6 upregulation
was observed in the jejunum following exposure to LDR IR, whereas
no comparable alteration was observed in the colon. These two genes
have been linked to the pathogenesis of IBD, such as colitis
(33) and Crohn's disease
(38). Therefore, the present
study aimed to induce IBD using DSS, a chemical that induces
colitis, and to validate the expression of G6PC2 and MUC6 in
radiation-exposed jejunum. While the colon is well recognized as
the main location of DSS-induced damage, reports indicate that the
small intestine, specifically the jejunum-ileum, is also
susceptible to slight effects (39). However, in the present study,
histological confirmation did not reveal any damage in the jejunum
induced by DSS. Although no studies are available on the expression
and roles of G6PC2 in the intestine, the results of the present
study showed that G6PC2 was expressed in the jejunum and its
expression could be enhanced by LDR IR or inflammation (DSS
treatment) and more significantly when both were applied
simultaneously. Crohn's disease can affect any part of the
gastrointestinal tract. Although ulcerative colitis (UC) generally
occurs in the colon, it can also affect the part of the intestine
during severe colitis (40). Thus,
G6PC2 can be induced by LDR IR- and/or disease-associated
inflammation and may play some role in modulating inflammation in
the intestine and type I diabetes. At present, it is not known
whether proteins of any microbes in the intestine can act as
antigenic mimics of G6PC2-reactive CD8+ T cells. The possibility
and role of LDR IR-induced G6PC2 in the intestine may need further
investigation. Consistent with these reports, the results of the
present study showed that MUC6 expression in the intestine can be
enhanced by LDR IR- or inflammation (DSS treatment). Furthermore,
simultaneous treatment with both stimuli can further increase its
expression. MUC6 may play protective roles in LDR IR-induced, or
disease- and cancer-associated inflammation. However, it may be
possible that inflammation-induced MUC6 expression in unexpected
regions contributes to cancer development. Thus, further
investigations on the roles of enhanced MUC6 expression in
inflammatory environments are required.
During the present study, it was shown that when
exposed to LDR IR, the genes associated with inflammation in the
intestines, namely G6PC2 and MUC6, were increased in the jejunum.
Furthermore, it was determined that this rise occurred under
repeated exposure to LDR IR. Moreover, these genes exhibited a
further increase in the presence of pre-existing enteritis.
Nevertheless, due to the absence of clinical symptoms or
morphological alterations in the jejunum, establishing a clear
relationship between the G6PC2 and MUC6 genes and intestinal
inflammation could not be achieved. Future research should modify
these irradiation settings and expand the post-irradiation period
to validate the long-term alteration pattern in the jejunum.
The present study revealed particular genes that
were upregulated by LDR IR. It is recognized that in gene
expression analysis, it is essential to take into account both
upregulated and downregulated genes. The present study tried to
validate thoroughly the RNA-seq results with RT-qPCR but
encountered difficulties in confirming the downregulated genes.
Despite attempts to resolve and deal with these problems, achieve
reliable validation for the downregulated genes could not be
achieved in the present study. Therefore, it was decided to focus
on reporting and discussing the confirmed upregulated genes in the
results. The limitation of excluding downregulated genes from the
validation procedure is recognized and the valuable insights that
may have been obtained by investigating them. Although limited, the
present study provided useful insights into the upregulated gene
expression alterations in the small intestine under LDR IR.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Dongnam Institute of
Radiological & Medical Sciences (DIRAMS) grant funded by the
Korean government (MSIT), grant nos. 50496-2017 and 50491-2021. It
was also supported by grants from the National Research Foundation
(grant nos. NRF-2020M2C8A2069337 and NRF-2020R1A2C1004272) funded
by the Ministry of Science and ICT (MSIT), Republic of Korea.
Availability of data and materials
The metagenomic sequencing data may be found in the
sequence read archive (SRA) database under accession number PRJNA
1086250 or at the following URL: ncbi.nlm.nih.gov/sra/PRJNA1086250.
The data generated in the present study may be requested from the
corresponding author.
Authors' contributions
CGL and JSK conceived the present study. SK, MJB and
YS performed the investigations. MJB, WSJ, CM and HJL were involved
in data analysis and interpretation. MKK, HJK, YRK, BJ and JL
curated the data by collecting, arranging and processing the data.
CGL and JSK confirm the authenticity of all the raw data. SK, MJB,
CGL and JSK wrote the original draft. SK and JSK reviewed and
edited the manuscript. All authors have read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
All experimental procedures were approved by the
Institutional Animal Care and Use Committee of Dongnam Institute of
Radiological and Medical Sciences (DIRAMS; approval nos.
DI-2015-002 and DI-2021-002) and the animals were cared for in
accordance with the NIH Guide for the Care and Use of Laboratory
Animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hall EJ: Weiss lecture. The dose-rate
factor in radiation biology. Int J Radiat Biol. 59:595–610. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Narendran N, Luzhna L and Kovalchuk O: Sex
difference of radiation response in occupational and accidental
exposure. Front Genet. 10:2602019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nam HH, Kang S, Seo YS, Lee J, Moon BC,
Lee HJ, Lee JH, Kim B, Lee S and Kim JS: Protective effects of an
aqueous extract of Protaetia brevitarsis seulensis larvae against
radiation-induced testicular injury in mice. Food Sci Nutr.
10:3969–3978. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kang S, Lee AY, Nam HH, Lee SI, Kim HY,
Lee JM, Moon C, Shin IS, Chae SW, Lee JH, et al: Protective effect
of bojungikki-tang against radiation-induced intestinal injury in
mice: Experimental verification and compound-target prediction.
Evid Based Complement Alternat Med. 2023:54178132023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee HJ, Son Y, Lee M, Moon C, Kim SH, Shin
IS, Yang M, Bae S and Kim JS: Sodium butyrate prevents
radiation-induced cognitive impairment by restoring pCREB/BDNF
expression. Neural Regen Res. 14:1530–1535. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ren H, Shen J, Tomiyama-Miyaji C, Watanabe
M, Kainuma E, Inoue M, Kuwano Y and Abo T: Augmentation of innate
immunity by low-dose irradiation. Cell Immunol. 244:50–56. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schultz CH, Fairley R, Murphy LS and Doss
M: The risk of cancer from CT scans and other sources of low-dose
radiation: A critical appraisal of methodologic quality. Prehosp
Disaster Med. 35:3–16. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hong JY, Han K, Jung JH and Kim JS:
Association of exposure to diagnostic low-dose ionizing radiation
with risk of cancer among youths in South Korea. JAMA Netw Open.
2:e19105842019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gong EJ, Shin IS, Son TG, Yang K, Heo K
and Kim JS: Low-dose-rate radiation exposure leads to testicular
damage with decreases in DNMT1 and HDAC1 in the murine testis. J
Radiat Res. 55:54–60. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kang S, Lee HJ, Son Y, Bae MJ, Jo WS, Park
JH, Jeong S, Moon C, Shin IS, Lee CG and Kim JS: Low-dose-rate
gamma radiation aggravates titanium dioxide nanoparticle-induced
lung injury in mice. Mol Cell Toxicol. 20:389–398. 2024. View Article : Google Scholar
|
|
11
|
Jo WS, Kang S, Jeong SK, Bae MJ, Lee CG,
Son Y, Lee HJ, Jeong MH, Kim SH, Moon C, et al: Low dose rate
radiation regulates M2-like macrophages in an allergic airway
inflammation mouse model. Dose Response. 20:155932582211173492022.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Son Y, Lee CG, Kim JS and Lee HJ:
Low-dose-rate ionizing radiation affects innate immunity protein
IFITM3 in a mouse model of Alzheimer's disease. Int J Radiat Biol.
99:1649–1659. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bachmann R, Heinzelmann F, Müller AC,
Ladurner R, Schneider CC, Königsrainer A and Zdichavsky M:
Laparoscopic pelvic mesh placement with closure of pelvic floor
entrance to prevent small intestine radiation trauma-a
retrospective cohort analysis. Int J Surg. 23:62–67. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Moussa L, Usunier B, Demarquay C,
Benderitter M, Tamarat R, Sémont A and Mathieu N: Bowel radiation
injury: Complexity of the pathophysiology and promises of cell and
tissue engineering. Cell Transplant. 25:1723–1746. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Loge L, Florescu C, Alves A and Menahem B:
Radiation enteritis: Diagnostic and therapeutic issues. J Visc
Surg. 157:475–485. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
National Research Council (US) Committee
for the Update of the Guide for the Care and Use of Laboratory
Animals, . Guide for the care and use of laboratory animals. 8th
edition. The National Academies Press; Washington, DC, USA:
2011
|
|
17
|
Siriarchavatana P, Ayers JD and Kendall
LV: Anesthetic activity of alfaxalone compared with ketamine in
mice. J Am Assoc Lab Anim Sci. 55:426–430. 2016.PubMed/NCBI
|
|
18
|
Kim JS, Ryoo SB, Heo K, Kim JG, Son TG,
Moon C and Yang K: Attenuating effects of granulocyte-colony
stimulating factor (G-CSF) in radiation induced intestinal injury
in mice. Food Chem Toxicol. 50:3174–3180. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kang S, Son Y, Shin IS, Moon C, Lee MY,
Lim KS, Park SJ, Lee CG, Jo WS, Lee HJ and Kim JS: EFFECT of
abdominal irradiation in mice model of inflammatory bowel disease.
Radiat Prot Dosimetry. 199:564–571. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kechin A, Boyarskikh U, Kel A and
Filipenko M: cutPrimers: A new tool for accurate cutting of primers
from reads of targeted next generation sequencing. J Comput Biol.
24:1138–1143. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dobin A, Davis CA, Schlesinger F, Drenkow
J, Zaleski C, Jha S, Batut P, Chaisson M and Gingeras TR: STAR:
Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Anders S and Huber W: Differential
expression analysis for sequence count data. Genome Biol.
11:R1062010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Heinz S, Benner C, Spann N, Bertolino E,
Lin YC, Laslo P, Cheng JX, Murre C, Singh H and Glass CK: Simple
combinations of lineage-determining transcription factors prime
cis-regulatory elements required for macrophage and B cell
identities. Mol Cell. 38:576–589. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tripathi S, Pohl MO, Zhou Y,
Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che
J, Mulder LCF, et al: Meta- and orthogonal integration of influenza
‘OMICs’ data defines a role for UBR4 in virus budding. Cell Host
Microbe. 18:723–735. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guo J, Liu Z, Zhang D, Chen Y, Qin H, Liu
T, Liu C, Cui J, Li B, Yang Y, et al: TLR4 agonist monophosphoryl
lipid a alleviated radiation-induced intestinal injury. J Immunol
Res. 2019:21210952019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lu L, Jiang M, Zhu C, He J and Fan S:
Amelioration of whole abdominal irradiation-induced intestinal
injury in mice with 3,3′-Diindolylmethane (DIM). Free Radic Biol
Med. 130:244–255. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livanova AA, Fedorova AA, Zavirsky AV,
Bikmurzina AE, Krivoi II and Markov AG: Dose and time dependence of
functional impairments in rat jejunum following ionizing radiation
exposure. Physiol Rep. 9:e149602021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang J, Zheng H, Kulkarni A, Ou X and
Hauer-Jensen M: Regulation of early and delayed radiation responses
in rat small intestine by capsaicin-sensitive nerves. Int J Radiat
Oncol Biol Phys. 64:1528–1536. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hutton JC and O'Brien RM:
Glucose-6-phosphatase catalytic subunit gene family. J Biol Chem.
284:29241–29245. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Boortz KA, Syring KE, Pound LD, Mo H,
Bastarache L, Oeser JK, McGuinness OP, Denny JC and O'Brien RM:
Effects of G6pc2 deletion on body weight and cholesterol in mice. J
Mol Endocrinol. 58:127–139. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tai N, Peng J, Liu F, Gulden E, Hu Y,
Zhang X, Chen L, Wong FS and Wen L: Microbial antigen mimics
activate diabetogenic CD8 T cells in NOD mice. J Exp Med.
213:2129–2146. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hebbandi Nanjundappa R, Ronchi F, Wang J,
Clemente-Casares X, Yamanouchi J, Sokke Umeshappa C, Yang Y, Blanco
J, Bassolas-Molina H, Salas A, et al: A gut microbial mimic that
hijacks diabetogenic autoreactivity to suppress colitis. Cell.
171:655–667.e17. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dwertmann Rico S, Schliesser SJA, Gorbokon
N, Dum D, Menz A, Büscheck F, Hinsch A, Lennartz M, von Bargen C,
Bawahab AA, et al: Pattern of MUC6 expression across 119 different
tumor types: A tissue microarray study on 15 412 tumors. Pathol
Int. 73:281–296. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kufe DW: Mucins in cancer: Function,
prognosis and therapy. Nat Rev Cancer. 9:874–885. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bhatia R, Gautam SK, Cannon A, Thompson C,
Hall BR, Aithal A, Banerjee K, Jain M, Solheim JC, Kumar S and
Batra SK: Cancer-associated mucins: Role in immune modulation and
metastasis. Cancer Metastasis Rev. 38:223–236. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hamamoto A, Abe Y, Nishi M, Fujimori S,
Ohnishi Y, Yamazaki H, Oida Y, Miyazaki N, Inada KI, Ueyama Y, et
al: Aberrant expression of the gastric mucin MUC6 in human
pulmonary adenocarcinoma xenografts. Int J Oncol. 26:891–896.
2005.PubMed/NCBI
|
|
38
|
Buisine MP, Desreumaux P, Leteurtre E,
Copin MC, Colombel JF, Porchet N and Aubert JP: Mucin gene
expression in intestinal epithelial cells in Crohn's disease. Gut.
49:544–551. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Geier MS, Smith CL, Butler RN and Howarth
GS: Small-intestinal manifestations of dextran sulfate sodium
consumption in rats and assessment of the effects of Lactobacillus
fermentum BR11. Dig Dis Sci. 54:1222–1228. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mourad FH, Barada KA and Saade NE:
Impairment of small intestinal function in ulcerative colitis: Role
of enteric innervation. J Crohns Colitis. 11:369–377.
2017.PubMed/NCBI
|