Introduction
Colon cancer is a type of malignant epithelial cell
tumor, and presents a major health concern worldwide. The
inhibition of cancer cell proliferation is an essential strategy in
the treatment of colon cancer (1).
However, the molecular mechanisms of colon cancer cell
proliferation remain unresolved.
Autophagy is an evolutionarily conserved process in
eukaryotes. During autophagy, a nascent double membrane-bound
vesicle called an autophagosome encloses a portion of the cytoplasm
and the outer membrane of autophagosomes then fuses with the
vacuolar or lysosomal membrane to release the inner-membrane
structures called autophagic bodies, into the vacuolar or lysosomal
lumen for digestion (2). Autophagy
serves an important role in the proliferation of colorectal cancer
cells (3), and a number of studies
have suggested that autophagy prevents metabolites from damaging
cells and genomes (4,5). Conversely, other studies have suggested
that autophagy contributes to the supply of nutrients and reused
metabolites to tumor cells, therefore promoting their survival and
proliferation (6,7). Although autophagy has been demonstrated
to affect the proliferation of tumor cells, the regulatory
mechanism underlying autophagy in colon cancer cells has not been
fully investigated.
Sphingosine kinase-1 (SphK1), is an important enzyme
that maintains the intracellular sphingolipid balance and has a
role in the development of multiple malignancies, plays an
important role in resistance to therapies, tumor growth, tumor
neovascularization and metastatic spread (8,9). Recently,
a study reported that SphK1 regulates LC3 expression and autophagy
in neuroblastoma cells (10). A
previous study reported that SphK1 protected the breast cancer cell
line MCF-7, induced autophagy and increased cell death from
mortality through nutrient starvation (11). Despite the involvement of SphK1 in
autophagy, its specific role and associated regulatory mechanism in
colon cancer cells remain unclear.
A number of studies have suggested that increased
extracellular signal-regulated kinase (ERK) phosphorylation levels
induce autophagy in cells (12,13), and
that SphK1 promotes the proliferation of colon cancer cells through
activation of the ERK/phosphorylated (p-)ERK cascade (14). In the present study, the hypothesis
that the activation of the SphK1/ERK/p-ERK pathway promotes
autophagy in HT-29 cells was examined. In order to investigate
this, the protein expression levels of SphK1, ERK1/2 and p-ERK1/2,
and those of the autophagy-associated markers LC3A, ATG5, and ULK1,
were analyzed following the upregulation of SphK1 in HT-29 cells.
Additionally, the protein localization and expression patterns of
intracellular LC3A, a key marker of autophagy, were assessed.
Materials and methods
Cell lines and culture
The human colorectal cancer cell line HT-29, Caco-2,
RKO and HCT116 cells were purchased from the American Type Culture
Collection (Manassas, VA, USA). Cells were cultured in Dulbecco's
modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS;
Excell Bio, Inc., Shanghai, China) at 37°C with 5%
CO2.
Cell transfection
The Lentiviral vector PLenti-SPHK1-IRES-EGFP and the
blank vector (NC; R&S Biotechnology Co., Ltd., Shanghai, China)
were used for infection of cancer cells, the cells inoculated with
lentivirus at a multiplicity of infection (MOI) of 20 for 48 h, and
the percentage of infected cells was approximately 90% at this MOI.
Blasticidins (2 µg/ml) (Merck KGaA, Darmstadt, Germany) was added
for 2 weeks. The SphK1-overexpressing HT-29 cells [SphK1(+)-HT-29]
and the corresponding negative control HT-29 cells (NC-HT-29) were
detected by fluorescence-activated cell sorting. The stabilized
transfected SphK1(+)-HT-29 and NC-HT-29 cells were stored in liquid
nitrogen (Jinfeng liquid Nitrogen Container Co., Ltd., Chengdu,
China) and were used within 3 months between transfection and
subsequent experimentation. Cells were cultured in Dulbecco's
modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% FBS at 37°C with 5%
CO2.
Inverted fluorescence microscopy
analysis
SphK1(+)-HT-29 and NC-HT-29 cells were seeded onto a
6-well plate and cells covered ~95% of each well. The cells were
observed under an inverted fluorescence microscope (TS100-F; Nikon
Corporation, Tokyo, Japan) at ×100 magnification. The NIS-Elements
software (version 4.0; Nikon Corporation, Tokyo, Japan) was used
for cell imaging, according to the manufacturer's protocol. The
transfection efficiency of cells was calculated as follows: The
number of cells in 3 randomly selected fields that expressed green
fluorescent protein (GFP)/the total number of cells.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
RNA isolation was performed using the Total RNA
Extraction Kit (Tiangen Biotech Co., Ltd., Beijing, China),
according to the manufacturer's protocol. cDNA synthesis was
performed using the Reverse Transcription Kit (Takara Bio, Inc.,
Otsu, Japan). A fluorescence-based qPCR method was performed using
2 µl cDNA, 10 µl SYBR Green (Takara Bio, Inc.), 0.6 µl PCR forward
primer (10 µM), 0.6 µl PCR reverse primer (10 µM) and 6.8 µl
dH2O, in a 20 µl PCR reaction volume. The RT-qPCR
reaction was run on a StepOne Real-Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The cycling parameters
were as follows: Denaturing at 95°C for 30 sec, 40 cycles at
denaturing at 95°C for 5 sec, primer annealing at 60°C for 34 sec,
and extension temperature at 95°C for 15 sec; final extension at
60°C for 1 min and final denaturing at 95°C for 15 sec. Gene
expression levels were determined via the 2−ΔΔCq method
(15), using GAPDH as a reference
gene, with the GAPDH gene expression level in NC-HT-29 cells set to
1. The primers of GAPDH and SphK1 were obtained from Takara Bio,
Inc., (Otsu Japan). GAPDH, forward: 5′-GCACCGCAAGGCTGAGAAC-3′, and
reverse: 5′-TGGTGAAGACGCCAGTGGA-3′; SphK1, forward:
5′-GGCTTCATTGCTGATGTGGA-3′, and reverse:
5′-AGGAAGGTGCCCAGAGTGAA-3′.
Western blotting analysis
Total proteins were extracted using
radioimmunoprecipitation buffer (Beyotime Institute of
Biotechnology, Haimen, China). Protein concentrations were measured
by bicinchoninic acid assay (Solarbio Biotech Co., Ltd., Beijing,
China) according to the manufacturer's protocol. A total of 30 µg
of protein from each sample was separated via 12% SDS-PAGE
(Beyotime Institute of Biotechnology, Haimen, China) for 1 h at 100
V, and then transferred onto nitrocellulose membranes. Samples were
blocked with 5% nonfat-milk in Tris-buffered saline with Tween-20
(Solarbio Biotech Co., Ltd., Beijing, China) for 1 h at room
temperature. The membranes were incubated overnight at 4°C with
antibodies diluted in WB Antibody Diluent (Beyotime Institute of
Biotechnology, Haimen, China). Subsequently, the membranes and
secondary antibodies were incubated for 1 h at room temperature.
Bands were quantified by Odyssey infrared imaging (LICOR
Biosciences, Lincoln, NE, USA) and GAPDH acted as an internal
reference. Rabbit polyclonal anti-SphK1 (dilution 1:1,000; catalog
no. A0139), mouse monoclonal anti-ERK1/2 (dilution 1:1,500; catalog
no. A10613), rabbit polyclonal anti-p-ERK1/2 (dilution 1:1,500;
catalog no. AP0472), mouse monoclonal anti-ATG5 (dilution 1:1,000;
catalog no. A2468) and rabbit polyclonal anti-ULK1 (dilution
1:2,000; catalog no. A8529) were purchased from ABclonal, Inc.
(Woburn, MA, USA). Rabbit monoclonal anti-LC3A (dilution 1:1,000,
4599) was purchased from Cell Signaling Technology, Inc. (Danvers,
MA, USA). Rabbit polyclonal anti-GAPDH (dilution 1:2,000; catalog
no. 10494-1-AP) was purchased from ProteinTech Group (Rosemont, IL,
USA). The secondary antibodies horseradish-peroxidase
(HRP)-conjugated Goat Anti-Rabbit IgG (dilution 1:10,000; catalog
no. AS014) and HRP-conjugated Goat Anti-Mouse IgG (dilution
1:10,000; catalog no. AS003) were purchased from ABclonal Inc.
Immunofluorescence
The Cell slide (Solarbio Biotech Co., Ltd., Beijing,
China) was placed in 24-well plates and then cells were seeded at a
density of 1×105. Routinely cultured overnight at 37°C
with 5% CO2, the cells were washed with PBS, fixed in a
4% paraformaldehyde solution (Solarbio Biotech Co., Ltd., Beijing,
China) for 20 min, permeabilized with 0.5% Triton X-100 for 10 min,
sealed with 10% FBS diluted with 10% PBS (Excell Bio, Inc.,
Shanghai, China) for 20 min, and then incubated overnight at 4°C
with rabbit polyclonal anti-LC3A (dilution 1:500; catalog no. 4599,
CST, USA). Subsequently, the cells were incubated with Anti-Rabbit
IgG Fab2 Alexa Flour®594 (dilution 1:500; catalog no.
8889S; Cell Signaling Technology, Inc.) for 1 h at 37°C. Cells were
stained with DAPI (Beyotime Institute of Biotechnology) for 1 min,
and then covered with anti-fluorescent quenching fluid (Beyotime
Institute of Biotechnology). An Olympus BX53 (Olympus Corporation,
Tokyo, Japan) polarizing microscope was used to observe the cells
under ×600 magnification and obtain images for further
analysis.
Statistical analysis
Each immunofluorescence assay was performed a
minimum of three times. Statistical analysis was based on the
unpaired Student's t test or the one-way analysis of variance test
using SPSS v.16.0 software (SPSS, Inc., Chicago, IL, USA). Data are
presented as the mean ± standard deviation. P<0.05 was
considered to indicate a statistically significant difference.
Results
SphK1 expression is upregulated in
transfected HT-29 cells
In a previous study, the relative mRNA expression of
SphK1, when compared with that of the reference gene GAPDH, was
0.96±0.02 in Caco2 (colon adenocarcinoma) cells, 0.61±0.07 in HT-29
cells, 0.92±0.05 in RKO (colon carcinoma) cells and 0.97±0.02 in
HCT116 cells (16). Therefore, the
lowest expression of SphK1 occurred in the HT-29 cell line. To
avoid cell autophagy caused by chemical stress, a
pLenti-SPHK1-IRES-EGFP vector and a blank vector (NC) were used to
transform HT-29 cells in order to obtain an increase in SphK1
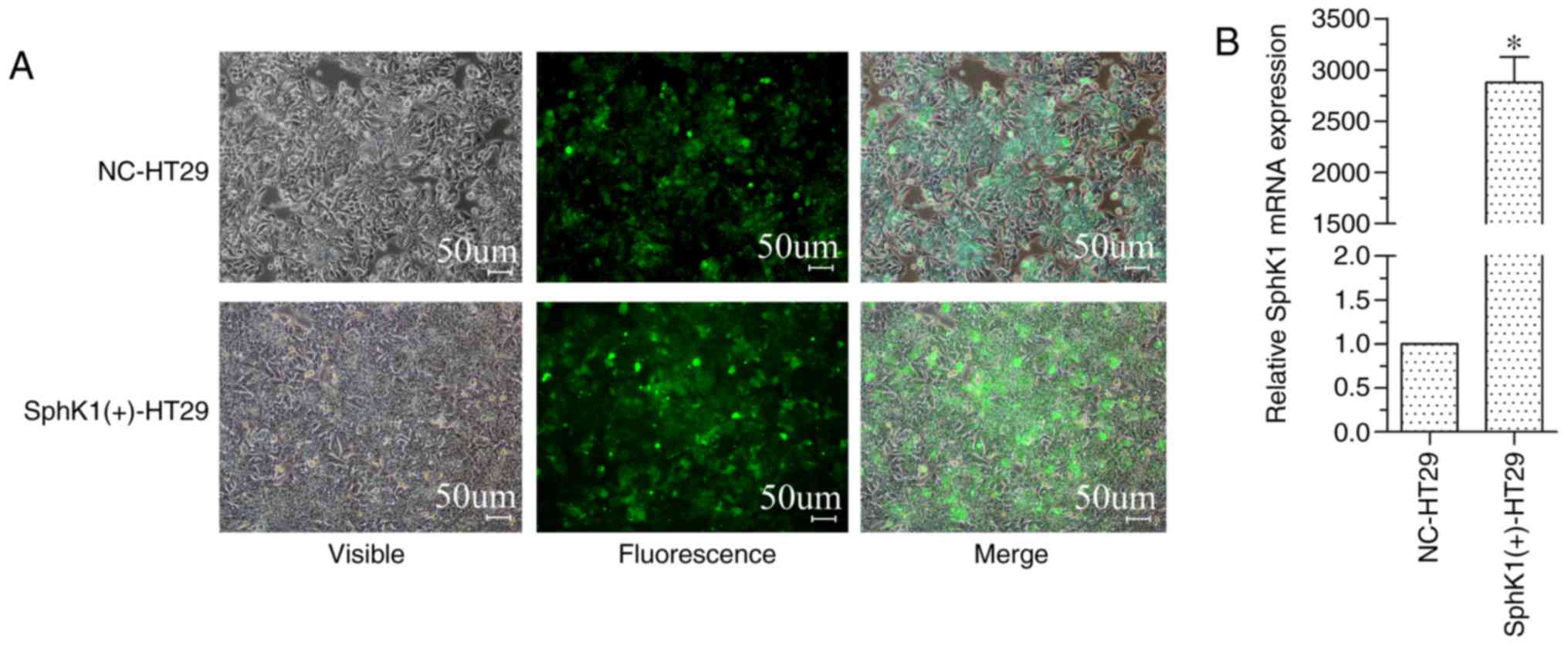
expression. The SphK1(+)-HT-29 and NC-HT-29 cells expressed GFP
(Fig. 1A) with a transfection
efficiency of 92% in NC-HT-29 cells and 95% in SphK1(+)-HT-29
cells. RT-qPCR results, using GAPDH as a reference gene and the
SphK1 expression level of NC-HT-29 cells set to 1, demonstrated
that the relative expression of SphK1 in SphK1(+)-HT-29 cells was
significantly increased (Fig. 1B).
These results also indicated that SphK1(+)-HT-29 and NC-HT-29 cells
were suitable for the subsequent experiments.
The SphK1/ERK/p-ERK pathway is
activated in HT-29 cells
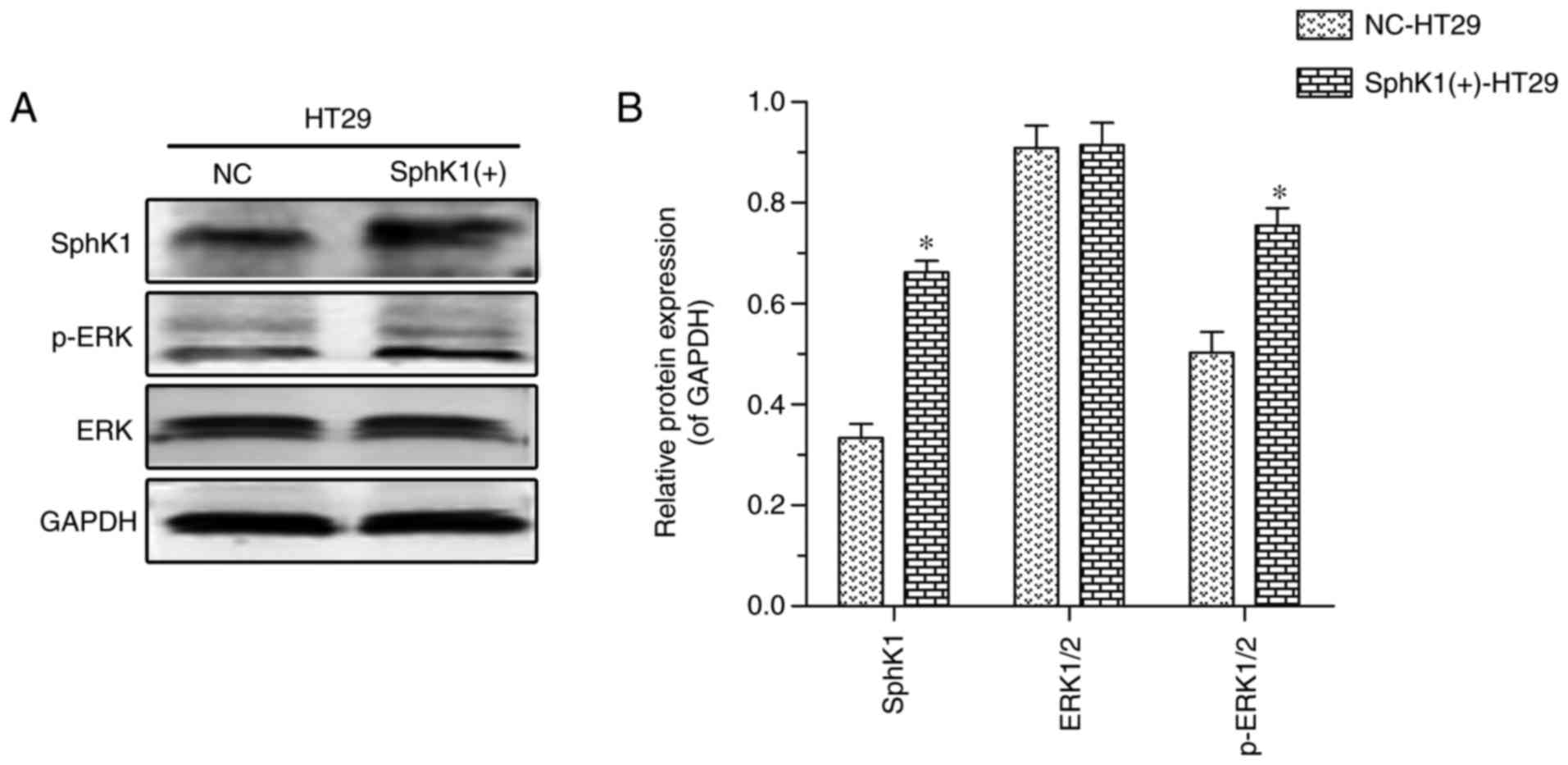
The protein expression levels of SphK1, total ERK1/2
and p-ERK, as detected by western blotting, are illustrated in
Fig. 2. SphK1 and p-ERK protein
expression was increased in SphK1(+)-HT-29 cells, compared with in
NC-HT-29 cells, while there were no significant differences in
total ERK1/2 expression. These results suggest that SphK1 activates
ERK by phosphorylation.
Autophagy in HT-29 cells is induced by
activation of the SphK1/ERK/p-ERK pathway
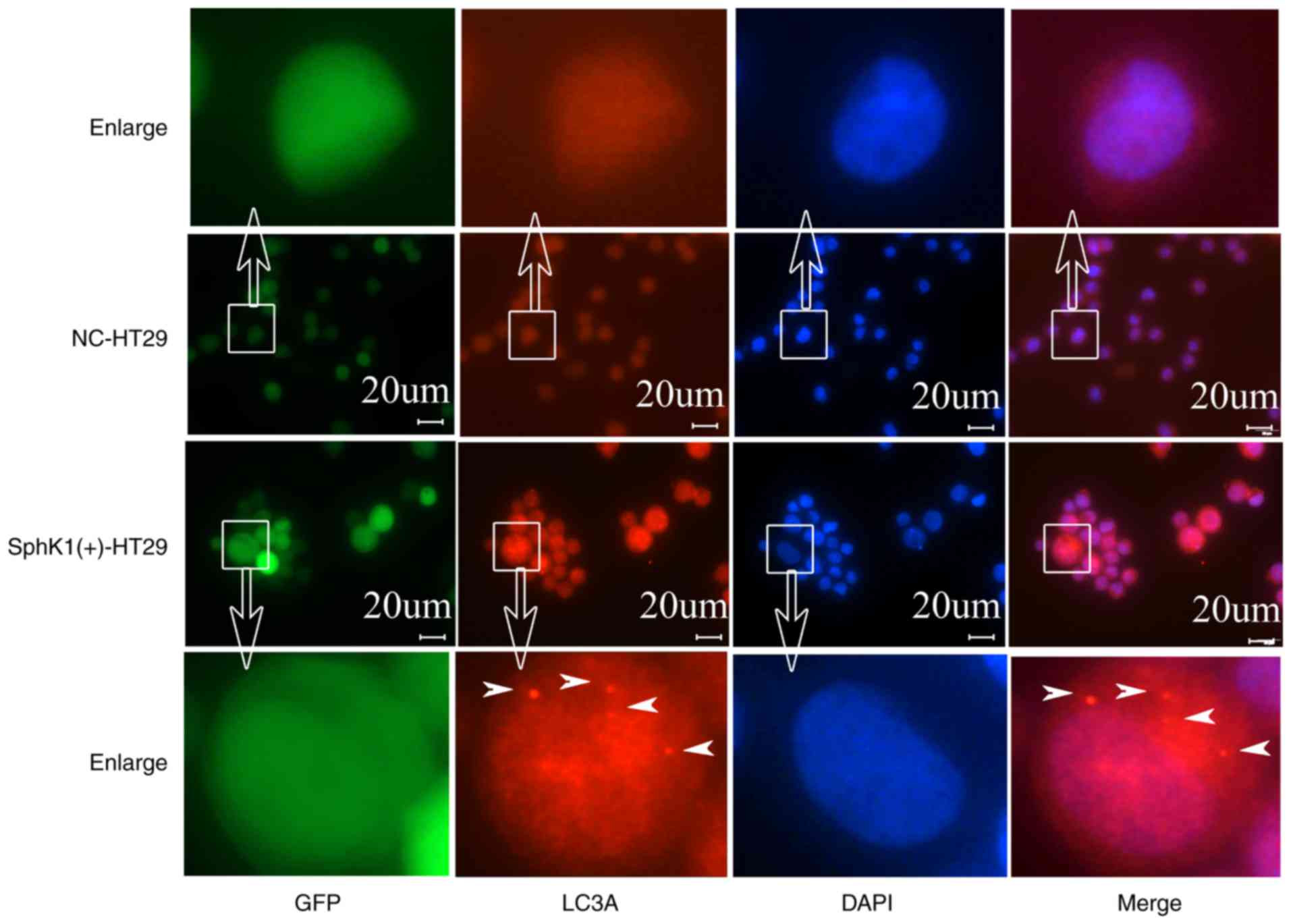
LC3A protein is a recognized marker for autophagy
(17–19), which, upon staining, presents a
spotted aggregation pattern under fluorescence microscopy (20,21). As
shown in Fig. 3, the protein
expression of LC3A appeared as a spotted aggregation in the
cytoplasm of SphK1(+)-HT-29 cells, but not in NC-HT-29 cells,
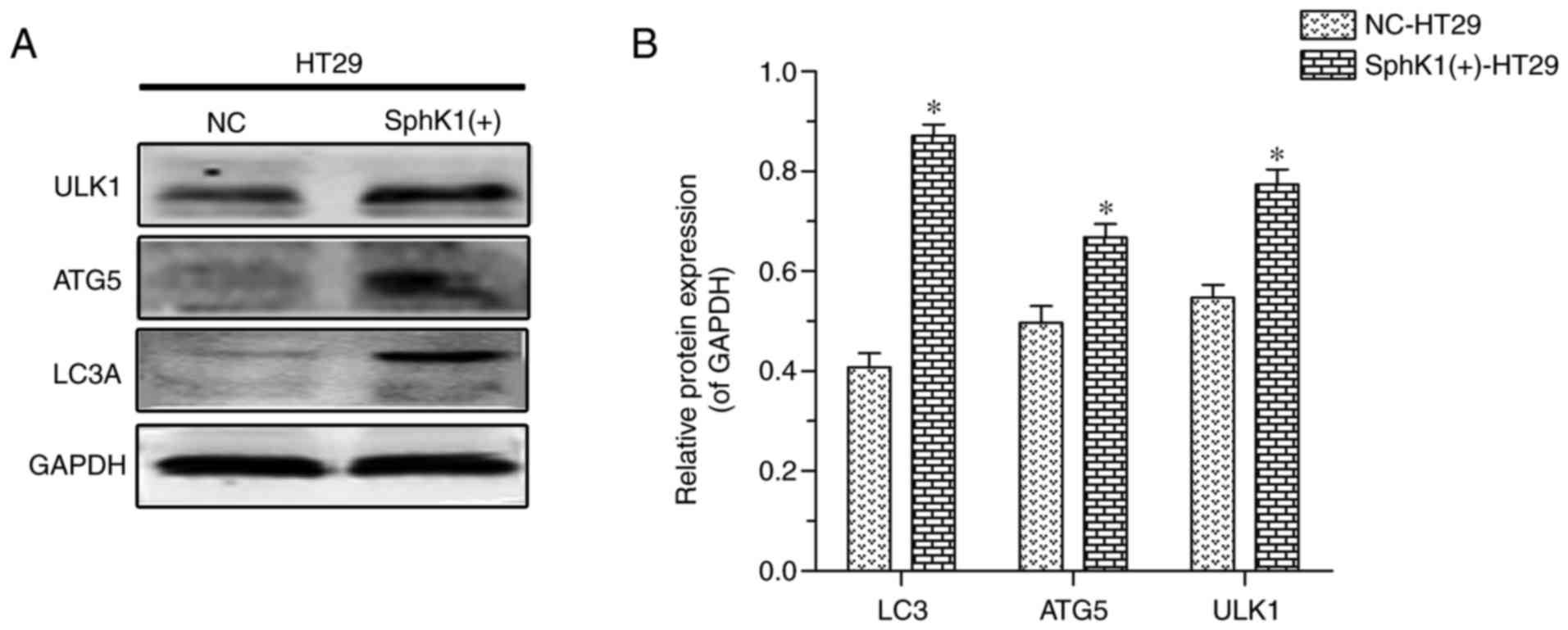
suggesting that SphK1 promotes autophagy in HT-29 cells. ATG5 and
ULK1 proteins are other important autophagy-associated markers
(21–23). In the present study, the protein
expression of LC3A, ATG5 and ULK1 was increased in SphK1(+)-HT-29
cells when compared with in NC-HT-29 cells (Fig. 4). Previous studies have reported that
increased expression of p-ERK decreases the levels of mTOR/p-mTOR,
which then results in increased expression of ULK1 (24,25). When
analyzed together, these results suggested that autophagy in HT-29
cells is induced via activation of the SphK1/ERK/p-ERK pathway.
Discussion
Increasing evidence suggests that the sphingosine
kinase-1 (SphK1) serves an important role in the development of
cancer, including cell proliferation, apoptosis, metastasis,
angiogenesis and chemotherapeutic resistance (9). It has also been reported that SphK1
promotes the proliferation and metastasis of colon cancer (14). SphK1 is involved in the regulation of
sphingolipid metabolism via the production of sphingosine-1 (S1P)
(26,27). SphK1/S1P regulates cell proliferation
through multiple pathways including the ERK, P38 mitogen-activated
protein kinase (MAPK) and Akt pathways (28). In the present study, SphK1 expression
upregulated ERK phosphorylation, as previously hypothesized. This
is consistent with the results of a previous study, which reported
that the activation ERK is elevated by the upregulation of SphK1
and attenuated by the suppression of SphK1, while the blocking of
the ERK pathway suppressed the effects that are mediated by the
overexpression of SphK1 (14). These
results suggest that SphK1 modulates the ERK/p-ERK pathway.
ERK is an important component of the MAPK system,
which is one of the most important signaling cascades, and has been
identified as frequently dysregulated in tumors (29). Additionally, an increase in the
expression of ERK and p-ERK led to a decrease in the levels of mTOR
and p-mTOR (24). mTOR is an
inhibitor of ULK1 (25), which
induces the initiation of autophagy (23). Similarly, in the present study,
increased levels of p-ERK were observed to promote an increase in
ULK1 protein expression. Furthermore, it has been reported that ERK
and its upstream kinase MEK are localized in the extra-luminal face
of the autophagosomes, and that ERK interacts with autophagy
proteins via its substrate-binding domains (20). Overall, these results suggest that
increasing ERK phosphorylation induces cell autophagy (12,13).
Autophagy is a degradation system that supplies
cytoplasmic components into the lysosome or vacuole, where the
degradation of lipid droplets is known to occur (30). Autophagy eliminates incorrectly
translated proteins, metabolic waste and toxic oxygen free-radicals
in cancer cells in order to achieve self-renewal and to promote the
growth and development of cells (31). Cell autophagy begins with the
formation of a bilayer structure termed an autophagosome.
Subsequently, two ubiquitination systems are activated, including
the ATG8/LC3 phosphatidylethanolamine conjugate system and the
ATG12-ATG5 conjugate system (22).
LC3 and ATG5 proteins are, consequently, regarded as the principal
autophagy-associated proteins. Ubiquitination systems are widely
known to be involved in various physiological processes, including
cell proliferation, apoptosis and autophagy (32). In the present study, the LC3 protein
formed a spotted aggregation pattern in SphK1(+)-HT-29 cells under
fluorescent microscopy, which suggested autophagy; this was in
accordance with the results of previous studies (20,21).
Furthermore, the results of the present study demonstrated that the
protein expression of LC3A, ATG5 and ULK1 were increased in the
SphK1-upregulated HT-29 cells. These results suggested that SphK1
regulates the expression of LC3 and promotes the autophagy process
in colon cancer cells, which is consistent with SphK1 induced
autophagy in neuroblastoma and breast cancer cells (13,14).
In summary, the present study demonstrated that
activation of the SphK1/ERK/p-ERK pathway promotes autophagy in
colon cancer HT-29 cells. An increase in SphK1 lead to an
upregulation of ERK/p-ERK by increasing ERK phosphorylation, which
in turn resulted in an increase in the expression level of the
autophagy-associated markers LC3, ATG5 and ULK1 in SphK1(+)-HT-29
cells. These findings provide a rationale for the development of
SphK1 inhibitors, or other cell autophagy inhibitors, as part of a
therapeutic strategy for patients with colorectal cancer or other
epithelial tumor types. Furthermore, in order to further
investigate the role of autophagy in colorectal cancer cells, gene
regulation of ERK expression or change cells culture conditions is
need in future studies.
Acknowledgements
Not applicable.
Funding
This work was supported by grants from the National
Natural Science Foundation of China (grant nos. 81460380 and
g81260365) and the Innovation Project of Guangxi Graduate Education
(grant nos. YCBZ2017035 and YCSW2017100).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
CYX and WLZ conceived and designed the study and
conducted the experiments. CYX assisted with drafting the
manuscript. WHW and MBQ performed the statistical analysis. JAH and
SQL interpreted the statistical analysis, reviewed and made final
approval of the version to be published. All authors read and
approved the manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nagappan A, Lee WS, Yun JW, Lu JN, Chang
SH, Jeong JH, Kim GS, Jung JM and Hong SC: Tetraarsenic hexoxide
induces G2/M arrest, apoptosis, and autophagy via PI3K/Akt
suppression and p38 MAPK activation in SW620 human colon cancer
cells. PLoS One. 12:e01745912017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Izumi M and Nakamura S: Chloroplast
protein turnover: The influence of extraplastidic processes,
including autophagy. Int J Mol Sci. 19:E8282018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lampada A, O'Prey J, Szabadkai G, Ryan KM,
Hochhauser D and Salomoni P: mTORC1-independent autophagy regulates
receptor tyrosine kinase phosphorylation in colorectal cancer cells
via an mTORC2-mediated mechanism. Cell Death Differ. 24:1045–1062.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
He M, Luo M, Liu Q, Chen J, Li K, Zheng M,
Weng Y, Ouyang L and Liu A: Combination treatment with fasudil and
clioquinol produces synergistic anti-tumor effects in U87
glioblastoma cells by activating apoptosis and autophagy. J
Neurooncol. 127:261–270. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cheng YC, Hueng DY, Huang HY, Chen JY and
Chen Y: Magnolol and honokiol exert a synergistic anti-tumor effect
through autophagy and apoptosis in human glioblastomas. Oncotarget.
7:29116–29130. 2016.PubMed/NCBI
|
|
6
|
Zhang L, Yu Y, Xia X, Ma Y, Chen XW, Ni ZH
and Wang H: Transcription factor E2-2 inhibits the proliferation of
endothelial progenitor cells by suppressing autophagy. Int J Mol
Med. 37:1254–1262. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Samardzija G, Stevovic TK, Djuricic S,
Djokic D, Djurisic M, Ciric D, Martinovic T, Bumbasirevic V and
Vujic D: Aggressive human neuroblastomas show a massive increase in
the numbers of autophagic vacuoles and damaged mitochondria.
Ultrastruct Pathol. 40:240–248. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Long J, Xie Y, Yin J, Lu W and Fang S:
SphK1 promotes tumor cell migration and invasion in colorectal
cancer. Tumour Biol. 37:6831–6836. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Almejún MB, Borge M, Colado A, Elías EE,
Podaza E, Risnik D, De Brasi CD, Stanganelli C, Slavutsky I,
Cabrejo M, et al: Sphingosine kinase 1 participates in the
activation, proliferation and survival of chronic lymphocytic
leukemia cells. Haematologica. 102:e257–e260. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Manchon Moruno JF, Uzor NE, Finkbeiner S
and Tsvetkov AS: SPHK1/sphingosine kinase 1-mediated autophagy
differs between neurons and SH-SY5Y neuroblastoma cells. Autophagy.
12:1418–1424. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lavieu G, Scarlatti F, Sala G, Carpentier
S, Levade T, Ghidoni R, Botti J and Codogno P: Regulation of
autophagy by sphingosine kinase 1 and its role in cell survival
during nutrient starvation. J Biol Chem. 281:8518–8527. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ren H, Guo H, Thakur A, Zhang S, Wang T,
Liang Y, Shi P, Gao L, Liu F, Feng J, et al: Blockade efficacy of
MEK/ERK-dependent autophagy enhances PI3K/Akt inhibitor
NVP-BKM120's therapeutic effectiveness in lung cancer cells.
Oncotarget. 7:67277–67287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao Y, Fan D, Zheng ZP, Li ET, Chen F,
Cheng KW and Wang M: 8-C-(E-phenylethenyl)quercetin from onion/beef
soup induces autophagic cell death in colon cancer cells through
ERK activation. Mol Nutr Food Res. 61:2017. View Article : Google Scholar
|
|
14
|
Liu SQ, Huang JA, Qin MB, Su YJ, Lai MY,
Jiang HX and Tang GD: Sphingosine kinase 1 enhances colon cancer
cell proliferation and invasion by upregulating the production of
MMP-2/9 and uPA via MAPK pathways. Int J Colorectal Dis.
27:1569–1578. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu CY, Liu SQ, Qin MB, Zhuge CF, Qin L,
Qin N, Lai MY and Huang JA: SphK1 modulates cell migration and
EMT-related marker expression by regulating the expression of p-FAK
in colorectal cancer cells. Int J Mol Med. 39:1277–1284. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nihira K, Miki Y, Iida S, Narumi S, Ono K,
Iwabuchi E, Ise K, Mori K, Saito M, Ebina M, et al: An activation
of LC3A-mediated autophagy contributes to de novo and acquired
resistance to EGFR tyrosine kinase inhibitors in lung
adenocarcinoma. J Pathol. 234:277–288. 2014.PubMed/NCBI
|
|
18
|
Miyamoto M, Takano M, Aoyama T, Soyama H,
Yoshikawa T, Tsuda H and Furuya K: Inhibition of autophagy protein
LC3A as a therapeutic target in ovarian clear cell carcinomas. J
Gynecol Oncol. 28:e332017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Giatromanolaki A, Koukourakis MI,
Pouliliou S, Gatter KC, Pezzella F, Harris AL and Sivridis E:
Overexpression of LC3A autophagy protein in follicular and diffuse
large B-cell lymphomas. Hematol Oncol Stem Cell Ther. 6:20–25.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Martinez-Lopez N, Athonvarangkul D,
Mishall P, Sahu S and Singh R: Autophagy proteins regulate ERK
phosphorylation. Nat Commun. 4:27992013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tanida I, Ueno T and Kominami E: LC3 and
autophagy. Methods Mol Biol. 445:77–88. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mowers EE, Sharifi MN and Macleod KF:
Autophagy in cancer metastasis. Oncogene. 36:1619–1630. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yu Y, Hou L, Song H, Xu P, Sun Y and Wu K:
Akt/AMPK/mTOR pathway was involved in the autophagy induced by
vitamin E succinate in human gastric cancer SGC-7901 cells. Mol
Cell Biochem. 424:173–183. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang R, Shen Z, Yang L, Yin M, Zheng W, Wu
B, Liu T and Song H: Protective effects of heme
oxygenase-1-transduced bone marrow-derived mesenchymal stem cells
on reducedsize liver transplantation: Role of autophagy regulated
by the ERK/mTOR signaling pathway. Int J Mol Med. 40:1537–1548.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nazio F and Cecconi F: Autophagy up and
down by outsmarting the incredible ULK. Autophagy. 13:967–968.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bao Y, Li K, Guo Y, Wang Q, Li Z, Yang Y,
Chen Z, Wang J, Zhao W, Zhang H, et al: Tumor suppressor PRSS8
targets Sphk1/S1P/Stat3/Akt signaling in colorectal cancer.
Oncotarget. 7:26780–26792. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
González-Fernández B, Sánchez DI, Crespo
I, San-Miguel B, Álvarez M, Tuñón MJ and González-Gallego J:
Inhibition of the SphK1/S1P signaling pathway by melatonin in mice
with liver fibrosis and human hepatic stellate cells. Biofactors.
43:272–282. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gao Z, Wang H, Xiao FJ, Shi XF, Zhang YK,
Xu QQ, Zhang XY, Ha XQ and Wang LS: SIRT1 mediates
Sphk1/S1P-induced proliferation and migration of endothelial cells.
Int J Biochem Cell Biol. 74:152–160. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Buchegger K, Silva R, López J, Ili C,
Araya JC, Leal P, Brebi P, Riquelme I and Roa JC: The ERK/MAPK
pathway is overexpressed and activated in gallbladder cancer.
Pathol Res Pract. 213:476–482. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Maeda Y, Oku M and Sakai Y:
Autophagy-independent function of Atg8 in lipid droplet dynamics in
yeast. J Biochem. 161:339–348. 2017.PubMed/NCBI
|
|
31
|
Li L, Chen H, Gao Y, Wang YW, Zhang GQ,
Pan SH, Ji L, Kong R, Wang G, Jia YH, et al: Long noncoding RNA
MALAT1 promotes aggressive pancreatic cancer proliferation and
metastasis via the stimulation of autophagy. Mol Cancer Ther.
15:2232–2243. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kwon YT and Ciechanover A: The ubiquitin
code in the ubiquitin-proteasome system and autophagy. Trends
Biochem Sci. 42:873–886. 2017. View Article : Google Scholar : PubMed/NCBI
|