Introduction
With the development of high-throughput methods, a
number of studies have revealed the roles of non-coding RNAs,
including microRNAs (miRNA) (1),
long non-coding RNAs (lncRNAs) (2)
and small nucleolar RNAs (snoRNAs) (3). lncRNAs are a subclass of RNA
transcripts >200 bps in length (4). Emerging studies have indicated that
lncRNAs are abnormally expressed in various types of human cancer,
including prostate cancer (5),
breast cancer (6), hepatocellular
carcinoma (7) and lung cancer
(8). These lncRNAs were involved in
multiple biological processes, including the cell cycle, invasion,
proliferation and drug resistance, by influencing the targets of
transcriptional (cis/trans) and post-transcriptional regulation
(9). Specific lncRNAs, including
GHET1 (10), LINP1 (11), NEAT1 (12) and MIR100HG (13) were reported to be oncogenes. However,
various other lncRNAs, including GAS5 (14) and LINC00961 (15), were implicated in cancer suppression.
Therefore, exploring the functions of lncRNAs may provide novel
insights to identify diagnostic and prognostic markers of
cancer.
Lung cancer has become the leading cause of
cancer-associated mortality worldwide (16), of which non-small cell lung cancer
(NSCLC) is the primary subtype (16). Despite the development of various
treatment methods, including radiotherapy, chemotherapy and
targeted therapy, the five-year survival rate of NSCLC remains low,
at ~15% (17). In previous reports,
lncRNAs were identified as novel regulators of lung cancer
progression; the transforming growth factor β-induced lncRNA-TBILA
promoted NSCLC progression via cis-regulation of human germinal
center-associated lymphoma HGAL, and activation of S100A7/JAB1
(18). However, in NSCLC, the
potential roles of a great number of lncRNAs remain unknown.
The aim of the present study was to identify novel
therapeutic and prognostic biomarkers for NSCLC. Previously
published datasets were analyzed to identify differentially
expressed lncRNAs (DElnc). To determine the functions of these
DElncs, a series of bioinformatics analyses were performed,
including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and
Genomes analysis.
Materials and methods
lncRNA classification pipeline
A pipeline described by Zhang et al (19) was employed to re-annotate the
microarray data. Entries labeled as ‘NR’ or annotated with
‘lncRNA’, ‘processed transcripts’, ‘non-coding’ or ‘misc_RNA’ in
Ensembl annotations, were retained.
Microarray data and
pre-processing
The following datasets were downloaded from the Gene
Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) to identify DElncs:
GSE19804, GSE27262, GSE18842 and GSE19188. GSE19804 was reported by
Lu et al (20) and contained
65 normal samples and 91 female lung cancer samples; GSE27262
included 25 normal and 25 stage I lung adenocarcinoma patients
(21); GSE18842 included 45 normal
and 46 NSCLC samples (22). A
further dataset, GSE19188, which includes 65 normal and 91 lung
cancer samples, was used for validation of the findings (23). In addition, The Cancer Genome Atlas
Lung Adenocarcinoma dataset (TCGA-LUAD), based on RNA-sequence
methodology was also downloaded to analyze the expression patterns
of lncRNAs in LUAD samples. In the present study, the lncRNAs whose
expression in NSCLC samples were higher than that in normal samples
(with a fold change (FC)≥2 and P<0.05), were considered to be
upregulated. The lncRNAs whose expression in NSCLC samples was
lower than that in normal samples (with an FC≤0.5 and P<0.05),
were considered to be downregulated.
Functional group analysis
Bioinformatics analysis was conducted using the
Database for Annotation, Visualization and Integrated Discovery
system (DAVID version 6.8; http://david.ncifcrf.gov/), to identify the relevant
biological functions of any high-throughput gene functional
analysis (24). P<0.05 was
considered to indicate a statistically significant difference.
Protein-protein interaction (PPI)
network and module analysis
Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING) online software was used to construct PPI
networks (https://string-db.org/cgi/input.pl?sessionId=AUH42ZEZwajP&input_page_show_search=on).
PPIs with a combined score >0.4 were considered to be
significant. Cytoscape software (https://cytoscape.org/) was used to visualize the PPI
networks.
Survival analysis
The Kaplan-Meier plotter (http://www.kmplot.com/analysis) is a public dataset
including 54,675 genes on survival using 2,437 lung cancer samples,
with a mean follow-up period of 49 months. The median expression of
lncRNAs was selected as the cut-off point to divide patients with
NSCLC into high- and low-expression groups.
Statistical analysis
Data are presented as the mean ± standard deviation.
All statistical analyses were performed using SPSS 17.0 software
(SPSS, Inc.). Statistical comparisons between groups were performed
using the Mann-Whitney U test. P<0.05 was considered to indicate
a statistically significant difference.
Results
Identification of DElncs in NSCLC
In order to identify key lncRNAs involved in the
progression of NSCLC, comprehensive analysis of four public
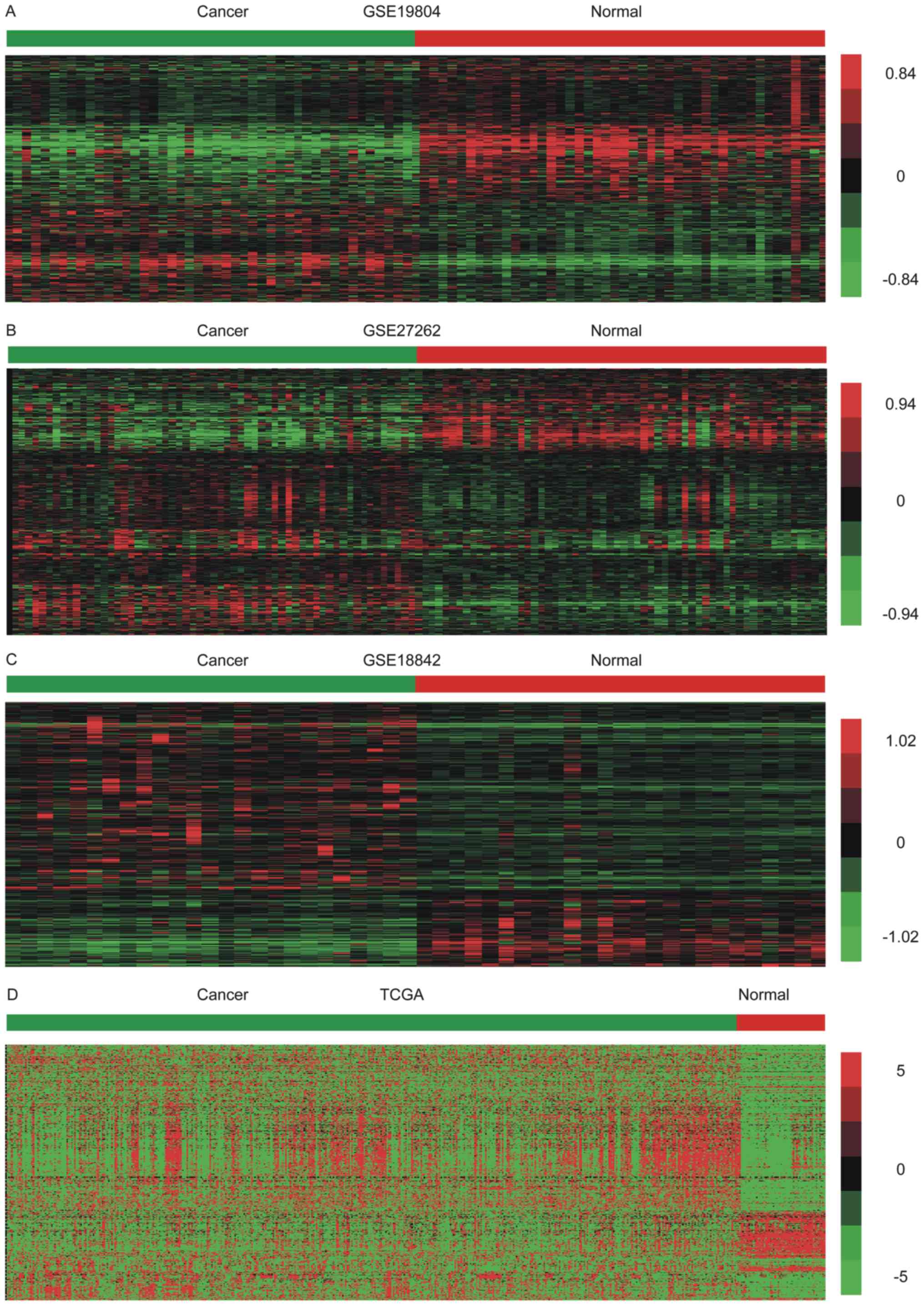
datasets were performed. A total of 638 upregulated and 294
downregulated lncRNAs were identified in the GSE19804 dataset
(Fig. 1A); 525 upregulated and 216
downregulated lncRNAs were identified in the GSE27262 dataset
(Fig. 1B); 379 upregulated and 508
downregulated lncRNAs were identified in the GSE18842 dataset
(Fig. 1C); and 489 upregulated and
223 downregulated lncRNAs were identified in TCGA dataset (Fig. 1D). Hierarchical clustering revealed
systematic variations in the expression of lncRNAs in NSCLC
samples.
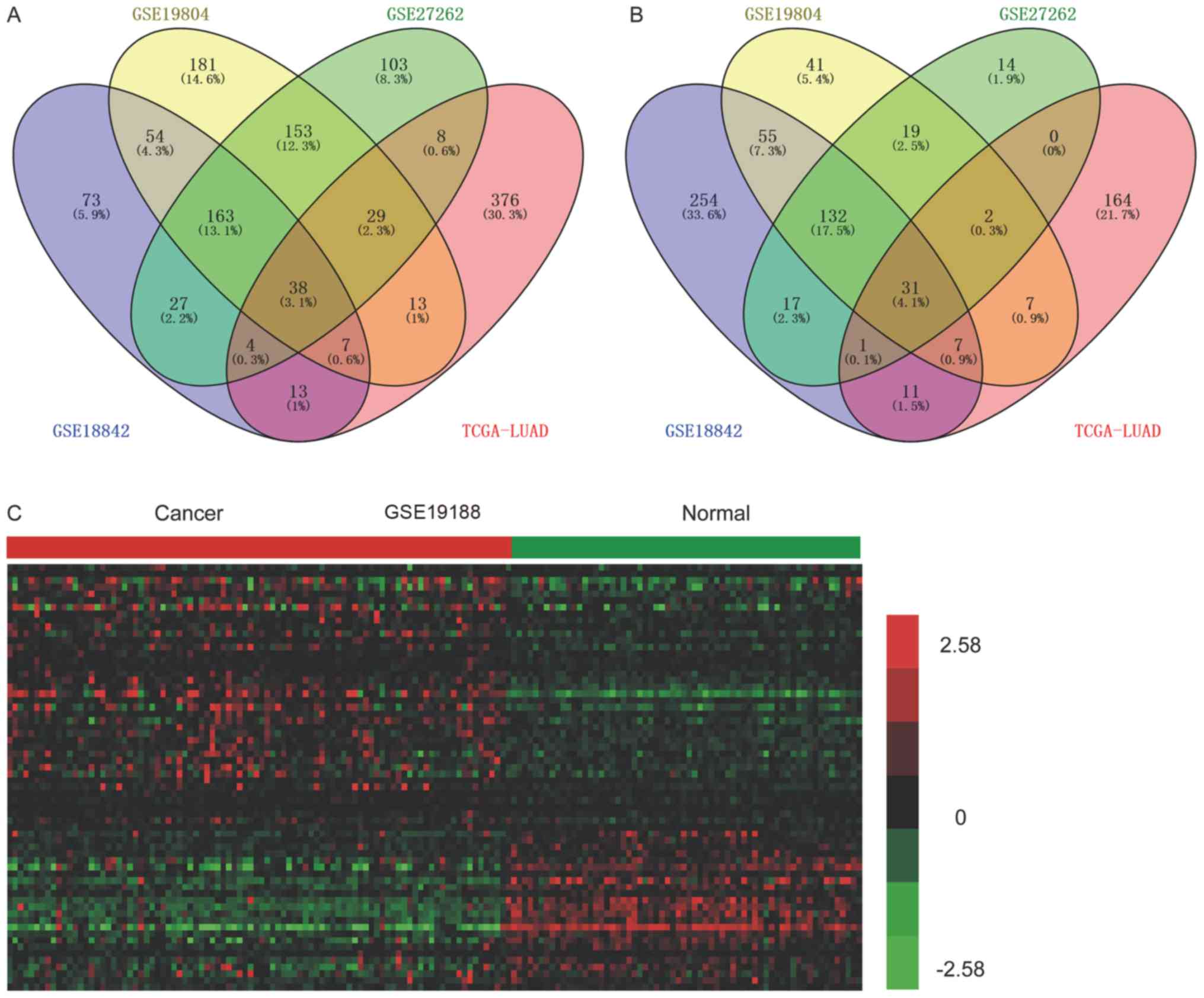
Integrated analysis of DElncs in NSCLC was also
performed. The upregulation of 38 lncRNAs (Fig. 2A), and the downregulation of 31
lncRNAs (Fig. 2B) was observed in
lung cancer compared with normal, healthy samples. Of note, a
number of lncRNAs identified as prominent regulators in lung cancer
progression were upregulated, including TUG1, DLEU2 and DANCR.
However, the functions of the majority of lncRNAs in human disease
require further investigation.
Verification of DElncs in NSCLC
In order to verify the expression pattern of the
aforementioned DElncs in NSCLC, the validation dataset GSE19188 was
analyzed. Upregulation of 38, and downregulation of 31 lncRNAs in
the training datasets were identified as dysregulated in GSE19188.
Hierarchical clustering revealed systematic variations in the
expression of lncRNAs in the NSCLC, by analysis of GSE19188
(Fig. 2C).
Identification of lncRNAs associated
with the progression of NSCLC
Associations between the expression of 69 DElncs and
the clinicopathological features of NSCLC were determined. The
expression levels of lncRNA-GVINP1, RPL32P3, EPB41L4A-AS2,
ILF3-AS1, LINC00938, LINC00667, A2M-AS1, GUSBP11, GGTA1P, BCRP3 and
SIGLEC17P were suppressed, while lncRNA-TMPO-AS1 expression was
upregulated in high grade (stage III and IV) in comparison to low
grade (stage II) NSCLC samples (Fig.
3).
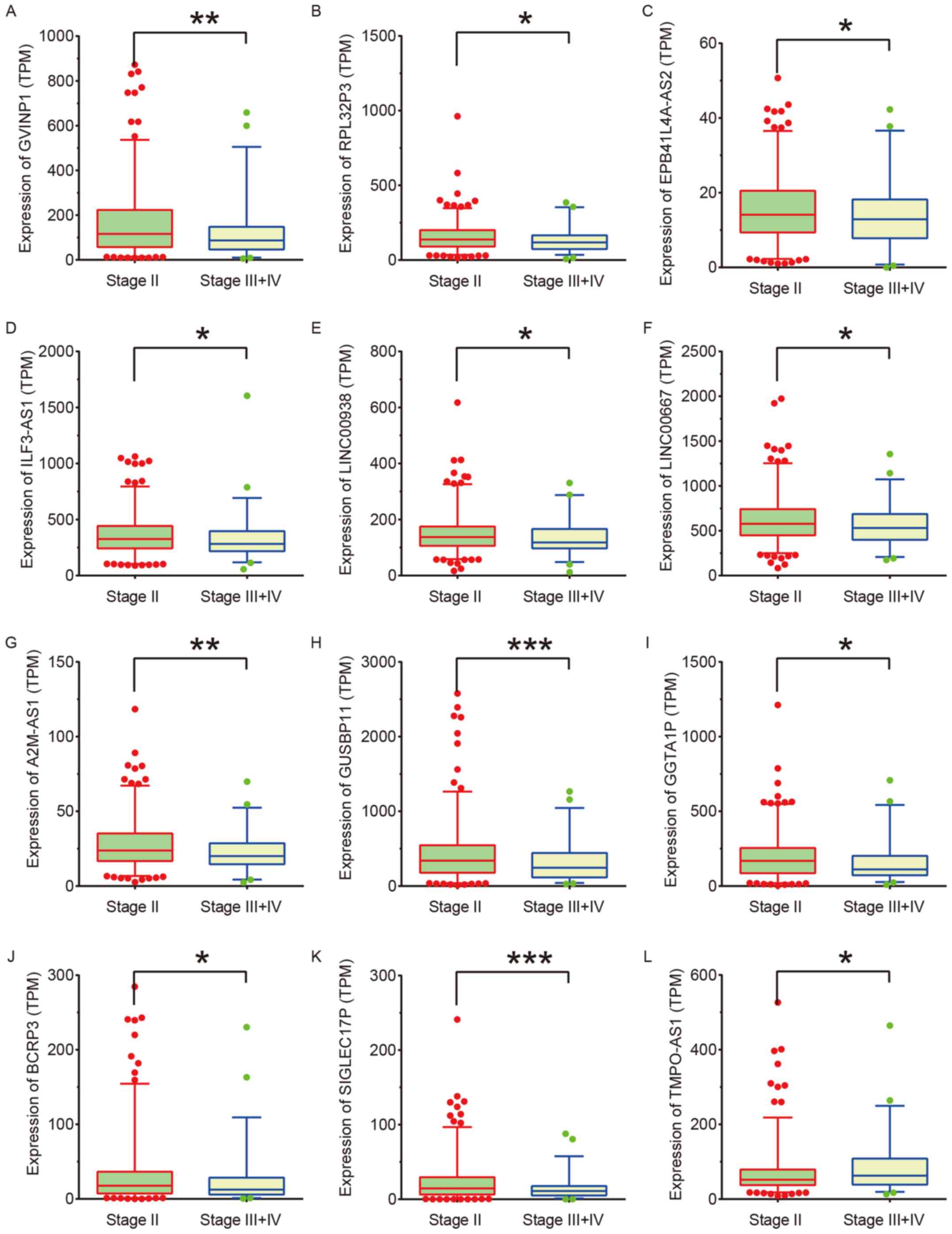 | Figure 3.Identification of lncRNAs associated
with NSCLC progression. (A) lncRNA-GVINP1, (B) RPL32P3, (C)
EPB41L4A-AS2, (D) ILF3-AS1, (E) LINC00938, (F) LINC00667, (G)
A2M-AS1, (H) GUSBP11, (I) GGTA1P, (J) BCRP3, and (K) SIGLEC17P
expression were suppressed, and (L) TMPO-AS1 expression was
upregulated in high grade (stage III and stage IV) compared with
low grade (stage II) NSCLC samples. *P<0.05; **P<0.01;
***P<0.001. DElncs, differentially expressed lncRNAs; NSCLC,
non-small cell lung cancer; TPM, transcripts per kilobase (of exon
model per million mapped reads). |
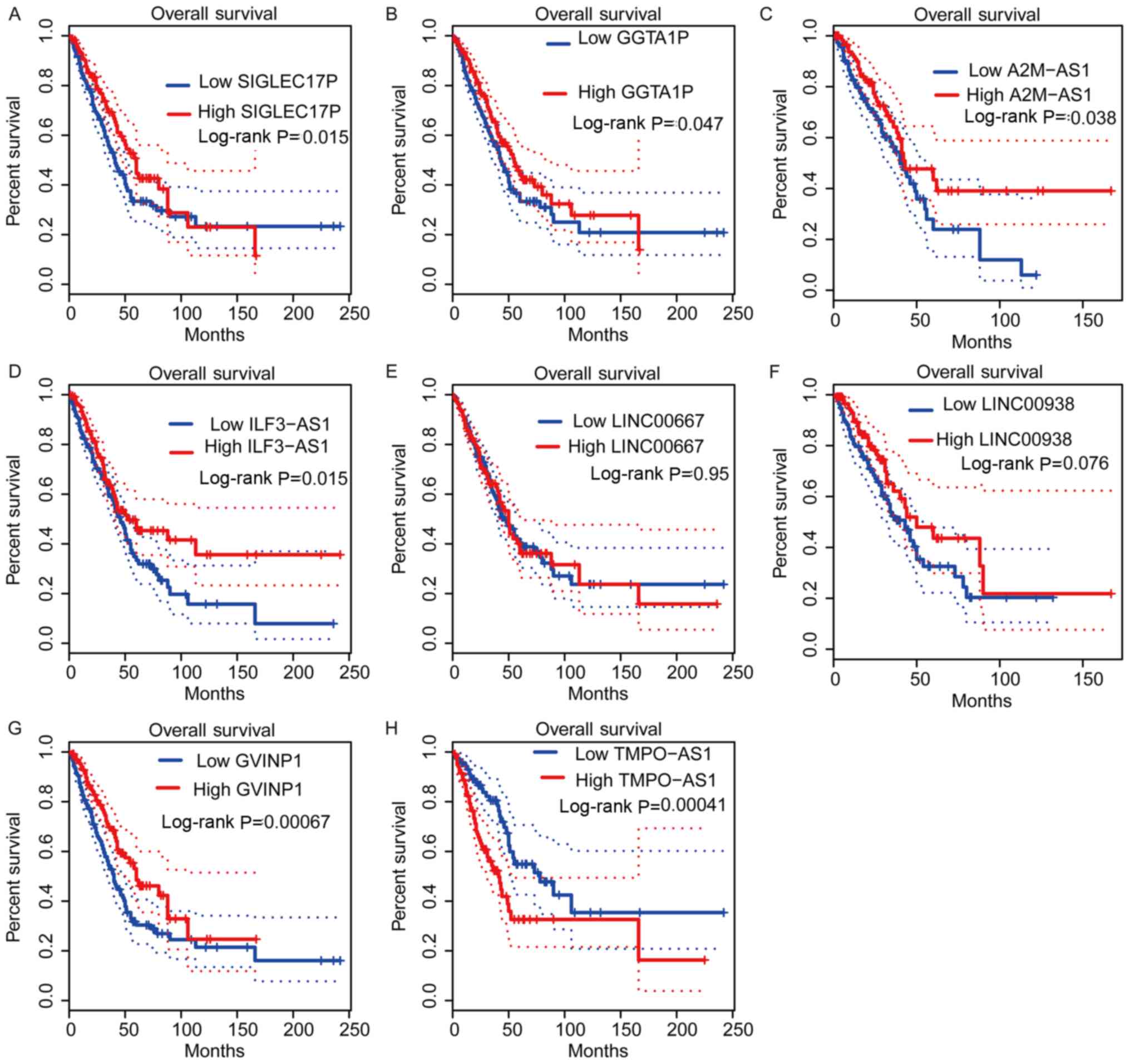
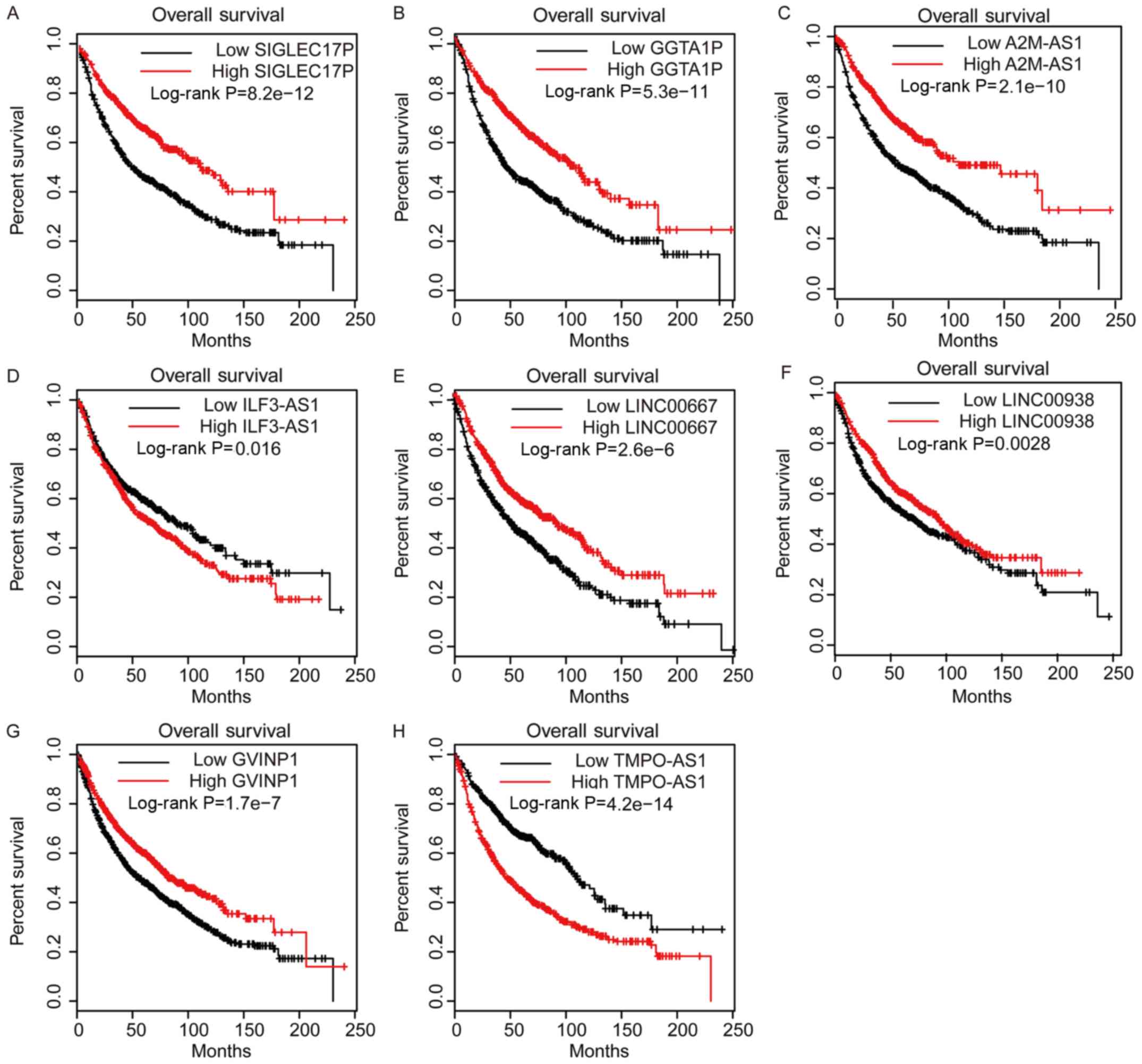
Dysregulation of DElncs is associated
with altered survival time of patients with NSCLC
To evaluate the prognostic value of DElncs in NSCLC,
Kaplan-Meier analysis was performed using TCGA LUAD dataset.
Patients with NSCLC were divided into low- and high-lncRNA
expression groups, with the median expression level selected as the
cut-off. Among these lncRNAs, higher expression of
lncRNA-SIGLEC17P, GGTA1P, A2M-AS1, ILF3-AS1, LINC00938 and GVINP1,
in addition to lower expression of lncRNA-TMPO-AS1, was associated
with longer survival time in patients with LUAD (Fig. 4). Furthermore, the
Kaplan-Meier-plotter database was used to ascertain the potential
prognostic values of the aforementioned lncRNAs. It was revealed
that higher expression levels of lncRNA-SIGLEC17P, GGTA1P, A2M-AS1,
LINC00667 and GVINP1, and lower expression levels of
lncRNA-ILF3-AS1, LINC00938 and TMPO-AS1 were associated with higher
survival rates in patients with NSCLC (Fig. 5). Collectively, these results
suggested that DElncs may serve as novel biomarkers for the
prognosis of patients with NSCLC.
Construction of key lncRNA-associated
PPI networks in NSCLC
Co-expression analysis was performed for the
construction of PPI networks to reveal the potential roles and
molecular mechanisms of key lncRNAs in NSCLC. The Pearson's
correlation coefficient was calculated for the lncRNA-mRNAs, using
their expression levels in TCGA dataset. lncRNA-mRNAs pairs with
|R|>0.7 were considered to be reliable. Subsequently, the STRING
database was used to investigate PPIs among key lncRNA genes that
were co-expressed (combined score >0.4).
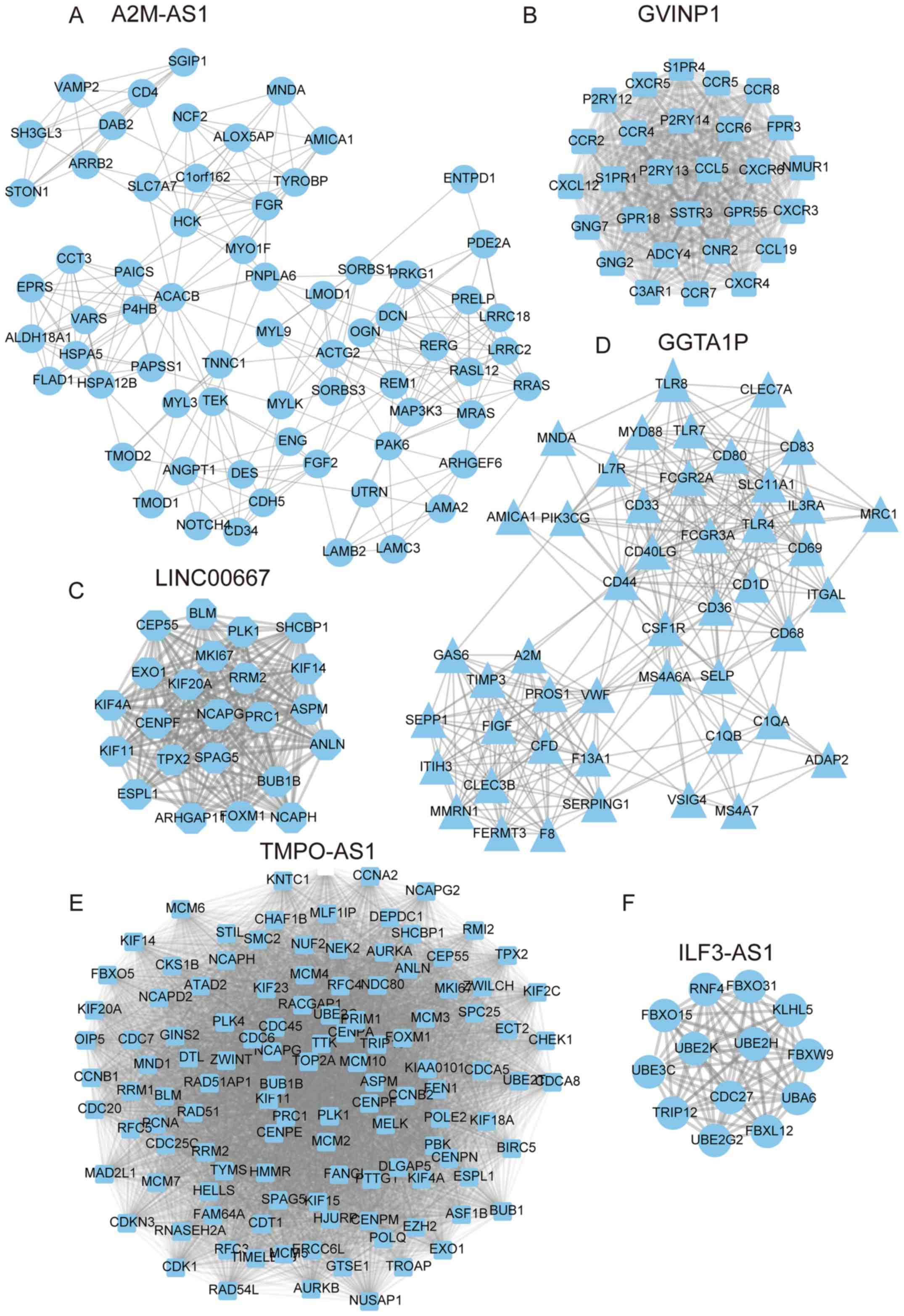
The A2M-AS1 mediated PPI network included 67
proteins and 282 edges (Fig. 6A).
The GVINP1 mediated PPI network included 28 proteins and 378 edges
(Fig. 6B). The LINC00667 mediated
PPI network included 23 proteins and 244 edges (Fig. 6C). The GGTA1P mediated PPI network
included 47 proteins and 329 edges (Fig.
6D). TMPO-AS1mediated PPI network included 118 proteins and
6075 edges (Fig. 6E). The ILF3-AS1
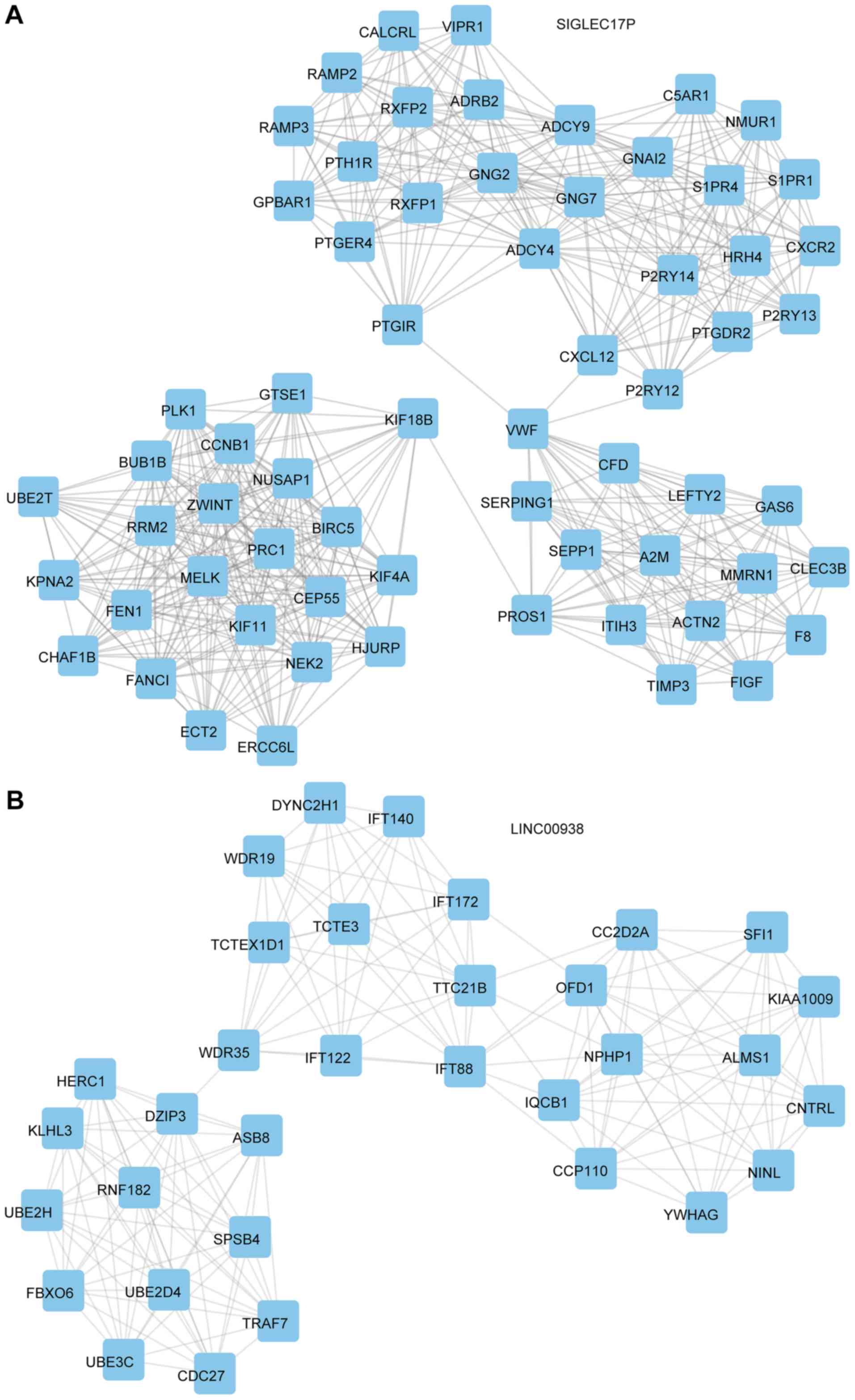
mediated PPI network included 13 proteins and 78 edges (Fig. 6F). The SIGLEC17P mediated PPI network
included 65 proteins and 560 edges (Fig.
7A). The LINC00938 mediated PPI network included 33 proteins
and 176 edges (Fig. 7B).
Functional analysis of key lncRNAs in
NSCLC
Bioinformatics analysis was performed using the
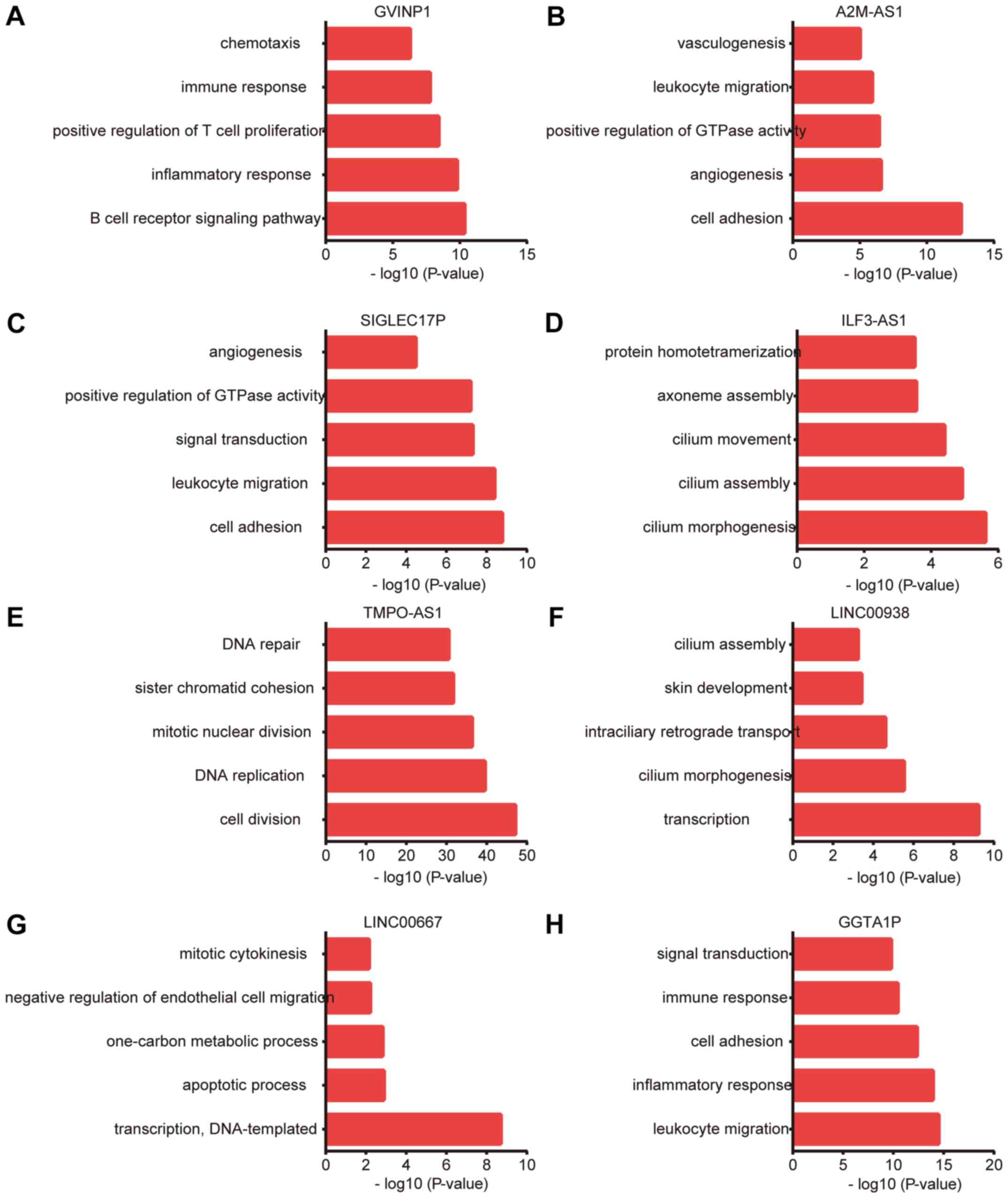
DAVID system. GO analysis revealed that lncRNA-GVINP1 was
associated with the regulation of B cell receptor signaling
pathway, inflammatory response, positive regulation of T cell
proliferation, immune response and chemotaxis (Fig. 8A); A2M-AS1 was associated with cell
adhesion, angiogenesis, positive regulation of GTPase activity,
leukocyte migration and vasculogenesis (Fig. 8B); SIGLEC17P was associated with cell
adhesion, leukocyte migration, signal transduction, positive
regulation of GTPase activity and angiogenesis (Fig. 8C); ILF3-AS1 was involved in
regulating cilium morphogenesis, cilium assembly, cilium movement,
axoneme assembly and protein homotetramerization (Fig. 8D); TMPO-AS1 was associated with cell
division, DNA replication, mitotic nuclear division, sister
chromatid cohesion and DNA repair (Fig.
8E); LINC00938 was involved in regulating transcription, cilium
morphogenesis, intraciliary retrograde transport, skin development
and cilium assembly (Fig. 8F);
LINC00667 was involved in regulating transcription, apoptotic
process, one-carbon metabolic process, negative regulation of
endothelial cell migration and mitotic cytokinesis (Fig. 8G); and GGTA1P was associated with
leukocyte migration, inflammatory response, cell adhesion, immune
response and signal transduction (Fig.
8H).
Discussion
NSCLC is one of the leading causes of
cancer-associated mortality worldwide. Previous studies have
demonstrated that lncRNAs serve a prominent role in the progression
of NSCLC, as either oncogenes or tumor suppressors; lncRNA-MALAT-1
enhanced NSCLC cell motility (25)
and lncRNA-CCAT2 promoted the invasion of NSCLC cells (26). However, lncRNA-SIK1-LNC suppressed
the proliferative and invasive abilities of NSCLC cells (27). lncRNA-PICART1 suppressed the
proliferation of NSCLC cells by inhibiting JAK2/signal transducer
and activator of transcription 3 signaling (28). Of note, other reports have
demonstrated that the dysregulation of specific lncRNAs may serve
as an indicator of NSCLC. For instance, lncRNA-HOTAIR (29), MALAT1 (25), CCAT2 (26) and H19 (30) were upregulated, and lncRNA-MEG3
(31) and TUG1 (32) were downregulated in lung cancer.
However in NSCLC, the expression patterns and molecular functions
of the majority of lncRNAs remained to be investigated.
In the present study, using various public datasets,
DElncs were identified as diagnostic biomarkers for NSCLC. The
upregulation of 38 lncRNAs was observed in NSCLC compared with
normal tissues, in addition to the suppression of a further 31
lncRNAs. Among these lncRNAs, HCG11 (33), CASC15 (34), TUG1 (32), DLEU2 (35) and DANCR (36) were dysregulated during cancer
progression. lncRNA-HCG11 was reported to be downregulated, and to
predict poor prognosis in prostate cancer. Additionally,
lncRNA-CASC15 promoted the progression of various types of cancer,
including melanoma, colon cancer, gastric cancer and hepatocellular
carcinoma. TUG1 is widely reported to be an oncogene, regulating
cancer cell proliferation, apoptosis, invasion and glutamine
metabolism. In NSCLC, lncRNA-TUG1 was reported to be involved in
regulating the growth and chemoresistance of NSCLC cells, in
association with LIMK2b, BAX and HOXB7 (37). Of note, the majority of these
lncRNAs, including EPB41L4A-AS2, ILF3-AS1, LINC00938, LINC00667 and
A2M-AS1, have not been reported in human cancers.
In the present study, the association of these
abnormally expressed lncRNAs with patient stage and survival time
was assessed. It was revealed that the expression of lncRNA-GVINP1,
RPL32P3, EPB41L4A-AS2, ILF3-AS1, LINC00938, LINC00667, A2M-AS1,
GUSBP11, GGTA1P, BCRP3 and SIGLEC17P was suppressed, and that
lncRNA-TMPO-AS1 expression was upregulated in high grade (stage III
and IV) compared with low grade (stage II) NSCLC samples.
Furthermore, Kaplan-Meier analysis illustrated that the
overexpression of GVINP1, A2M-AS1, GGTA1P, SIGLEC17P, ILF3-AS1 and
LINC00938, and the downregulation of TMPO-AS1 were associated with
longer survival times in patients with LUAD. This demonstrated that
these lncRNAs may serve as novel biomarkers for NSCLC.
For the most part, the functions of lncRNAs in
cancer remain uninvestigated. Co-expression and bioinformatics
analysis has been widely used to identify the potential functions
and molecular mechanisms of novel lncRNAs in human diseases
(38,39); using bioinformatics analysis, Jiao
et al (40) revealed the
roles of lncRNAs associated with polycystic ovary syndrome. In the
present study, lncRNA-associated PPI modules in NSCLC progression
were constructed. GO bioinformatics analysis indicated that
lncRNA-LINC00667 was involved in the regulation of transcription
and apoptotic process, and that lncRNA-GVINP1 was involved in
regulating B cell receptor signaling and the inflammatory response.
lncRNA-A2M-AS1 and SIGLEC17P were associated with cell adhesion
regulation, and GGTA1P was associated with leukocyte migration,
inflammatory response and immune response. lncRNA-ILF3-AS1 was
involved in regulating cilium morphogenesis and cilium assembly,
whilst LINC00938 was involved in regulating transcription and
cilium morphogenesis, and TMPO-AS1, with cell cycle and DNA repair
progression.
The present study identified potential, novel
biomarkers for the progression of NSCLC. However, one limitation
should be noted; the expression levels of these lncRNAs in NSCLC
tumor and serum samples were not detected using clinical samples.
Further validation of their expression in NSCLC samples is
required, and may strengthen their prognostic value as potential
biomarkers for NSCLC.
In conclusion, 38 upregulated, and 31 downregulated
NSCLC-associated lncRNAs were identified through comprehensive
analysis of public datasets. A total of 12 lncRNAs were associated
with NSCLC progression. Furthermore, Kaplan-Meier-plotter analysis
illustrated for the first time, that higher expression levels of
lncRNA-GVINP1, A2M-AS1, GGTA1P, SIGLEC17P, ILF3-AS1 and LINC00938,
and lower expression levels of lncRNA-TMPO-AS1 were associated with
longer survival time in patients with NSCLC. Key lncRNA-associated
PPI networks were constructed, and bioinformatics analysis
performed to provide insights for the identification of novel
biomarkers for NSCLC.
Acknowledgements
The authors would like to thank Dr Xuechao Wan
(School of Life Science, Fudan University, China) for excellent
technical and graphic design expertise.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81700092), the
National Natural Science Foundation of China (grant no. 81700049),
the China Postdoctoral Science Foundation (grant no. 2016M601603),
the Shanghai Science and Technology Innovation Fund (grant no.
18140903802), the Shanghai Municipal Health and Family Planning
Commission Research Fund (grant no. 20174Y0234), the Shanghai
Municipal Health and Family Planning Commission Research Fund
(grant no. 20174Y0094) and the Huashan Hospital affiliated to Fudan
University Research Start-up Fund (grant no. 2016QD087).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WF and HZ conceived and designed the study. GS
acquired the data, and TL analyzed the data. WF and WZ drafted the
manuscript and were involved in the conception of the study. LL
performed the statistical analysis. All authors read and approved
the manuscript and agree to be accountable for all aspects of the
research, ensuring that the accuracy and integrity of any part of
the work was appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lu Y, Zhao X, Liu Q, Li C, Graves-Deal R,
Cao Z, Singh B, Franklin JL, Wang J, Hu H, et al: lncRNA
MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance
via Wnt/β-catenin signaling. Nat Med. 23:1331–1341. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang M, Weng W, Zhang Q, Wu Y, Ni S, Tan
C, Xu M, Sun H, Liu C, Wei P and Du X: The lncRNA NEAT1 activates
Wnt/β-catenin signaling and promotes colorectal cancer progression
via interacting with DDX5. J Hematol Oncol. 11:1132018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Giraldez MD, Spengler RM, Etheridge A,
Godoy PM, Barczak AJ, Srinivasan S, De Hoff PL, Tanriverdi K,
Courtright A, Lu S, et al: Comprehensive multi-center assessment of
small RNA-seq methods for quantitative miRNA profiling. Nat
Biotechnol. 36:746–757. 2018. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Beermann J, Piccoli MT, Viereck J and Thum
T: Non-coding RNAs in development and disease: Background,
mechanisms, and therapeutic approaches. Physiol Rev. 96:1297–1325.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang Y, Su X, Kong Z, Fu F, Zhang P, Wang
D, Wu H, Wan X and Li Y: An androgen reduced transcript of lncRNA
GAS5 promoted prostate cancer proliferation. PLoS One.
12:e01823052017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Luo L, Tang H, Ling L, Li N, Jia X, Zhang
Z, Wang X, Shi L, Yin J, Qiu N, et al: LINC01638 lncRNA activates
MTDH-Twist1 signaling by preventing SPOP-mediated c-Myc degradation
in triple-negative breast cancer. Oncogene. 37:6166–6167. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yuan JH, Liu XN, Wang TT, Pan W, Tao QF,
Zhou WP, Wang F and Sun SH: The MBNL3 splicing factor promotes
hepatocellular carcinoma by increasing PXN expression through the
alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol. 19:820–832.
2017. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou W, Chen X, Hu Q, Chen X, Chen Y and
Huang L: Galectin-3 activates TLR4/NF-κB signaling to promote lung
adenocarcinoma cell proliferation through activating lncRNA-NEAT1
expression. BMC Cancer. 18:5802018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xia Y, Yan Z, Wan Y, Wei S, Bi Y, Zhao J,
Liu J, Liao DJ and Huang H: Knockdown of long noncoding RNA GHET1
inhibits cellcycle progression and invasion of gastric cancer
cells. Mol Med Rep. 18:3375–3381. 2018.PubMed/NCBI
|
|
11
|
Wu HF, Ren LG, Xiao JQ, Zhang Y, Mao XW
and Zhou LF: Long non-coding RNA LINP1 promotes the malignant
progression of prostate cancer by regulating p53. Eur Rev Med
Pharmacol Sci. 22:4467–4476. 2018.PubMed/NCBI
|
|
12
|
Wang P, Wu T, Zhou H, Jin Q, He G, Yu H,
Xuan L, Wang X, Tian L, Sun Y, et al: Long noncoding RNA NEAT1
promotes laryngeal squamous cell cancer through regulating
miR-107/CDK6 pathway. J Exp Clin Cancer Res. 35:222016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zou SM, Li WH, Wang WM, Li WB, Shi SS,
Ying JM and Lyu N: The gene mutational discrepancies between
primary and paired metastatic colorectal carcinoma detected by
next-generation sequencing. J Cancer Res Clin Oncol. 144:2149–2159.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cheng K, Zhao Z, Wang G, Wang J and Zhu W:
lncRNA GAS5 inhibits colorectal cancer cell proliferation via the
miR1825p/FOXO3a axis. Oncol Rep. 41:2371–2380. 2018.
|
|
15
|
Huang Z, Lei W, Tan J and Hu HB: Long
noncoding RNA LINC00961 inhibits cell proliferation and induces
cell apoptosis in human non-small cell lung cancer. J Cell Biochem.
119:9072–9080. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gandhi L, Rodriguez-Abreu D, Gadgeel S,
Esteban E, Felip E, De Angelis F, Domine M, Clingan P, Hochmair MJ,
Powell SF, et al: Pembrolizumab plus chemotherapy in metastatic
non-small-cell lung cancer. N Engl J Med. 378:2078–2092. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Miller VA, Hirsh V, Cadranel J, Chen YM,
Park K, Kim SW, Zhou C, Su WC, Wang M, Sun Y, et al: Afatinib
versus placebo for patients with advanced, metastatic
non-small-cell lung cancer after failure of erlotinib, gefitinib,
or both, and one or two lines of chemotherapy (LUX-Lung 1): A phase
2b/3 randomised trial. Lancet Oncol. 13:528–538. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lu Z, Li Y, Che Y, Huang J, Sun S, Mao S,
Lei Y, Li N, Sun N and He J: The The TGFβ-induced lncRNA TBILA
promotes non-small cell lung cancer progression in vitro and in
vivo via cis-regulating HGAL and activating S100A7/JAB1 signaling.
Cancer Lett. 432:156–168. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang X, Sun S, Pu JK, Tsang AC, Lee D,
Man VO, Lui WM, Wong ST and Leung GK: Long non-coding RNA
expression profiles predict clinical phenotypes in glioma.
Neurobiol Dis. 48:1–8. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lu TP, Tsai MH, Lee JM, Hsu CP, Chen PC,
Lin CW, Shih JY, Yang PC, Hsiao CK, Lai LC and Chuang EY:
Identification of a novel biomarker, SEMA5A, for non-small cell
lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers
Prev. 19:2590–2597. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wei TY, Hsia JY, Chiu SC, Su LJ, Juan CC,
Lee YC, Chen JM, Chou HY, Huang JY, Huang HM and Yu CT: Methylosome
protein 50 promotes androgen- and estrogen-independent
tumorigenesis. Cell Signal. 26:2940–2950. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sanchez-Palencia A, Gomez-Morales M,
Gomez-Capilla JA, Pedraza V, Boyero L, Rosell R and Fárez-Vidal ME:
Gene expression profiling reveals novel biomarkers in nonsmall cell
lung cancer. Int J Cancer. 129:355–364. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hou J, Aerts J, den Hamer B, van Ijcken W,
den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens
JA, Hoogsteden HC, et al: Gene expression-based classification of
non-small cell lung carcinomas and survival prediction. PLoS One.
5:e103122010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu J, Weng Y, He F, Liang D and Cai L:
lncRNA MALAT-1 competitively regulates miR-124 to promote EMT and
development of non-small-cell lung cancer. Anticancer Drugs.
29:628–636. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qiu M, Xu Y, Yang X, Wang J, Hu J, Xu L
and Yin R: CCAT2 is a lung adenocarcinoma-specific long non-coding
RNA and promotes invasion of non-small cell lung cancer. Tumour
Biol. 35:5375–5380. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang L, Xie N, Huang J, Huang H, Xu S,
Wang Z and Cai J: SIK1-LNC represses the proliferative, migrative,
and invasive abilities of lung cancer cells. Onco Targets Ther.
11:4197–4206. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao JM, Cheng W, He XG, Liu YL, Wang FF
and Gao YF: Long non-coding RNA PICART1 suppresses proliferation
and promotes apoptosis in lung cancer cells by inhibiting
JAK2/STAT3 signaling. Neoplasma. 65:779–789. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu XH, Liu ZL, Sun M, Liu J, Wang ZX and
De W: The long non-coding RNA HOTAIR indicates a poor prognosis and
promotes metastasis in non-small cell lung cancer. BMC Cancer.
13:4642013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang Q, Li X, Li X, Li X and Chen Z:
lncRNA H19 promotes epithelial-mesenchymal transition (EMT) by
targeting miR-484 in human lung cancer cells. J Cell Biochem.
119:4447–4457. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang Z, Liu T, Wang K, Qu X, Pang Z, Liu
S, Liu Q and Du J: Down-regulation of long non-coding RNA MEG3
indicates an unfavorable prognosis in non-small cell lung cancer:
Evidence from the GEO database. Gene. 630:49–58. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lin PC, Huang HD, Chang CC, Chang YS, Yen
JC, Lee CC, Chang WH, Liu TC and Chang JG: Long noncoding RNA TUG1
is downregulated in non-small cell lung cancer and can regulate
CELF1 on binding to PRC2. BMC Cancer. 16:5832016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang Y, Zhang P, Wan X, Su X, Kong Z,
Zhai Q, Xiang X, Li L and Li Y: Downregulation of long non-coding
RNA HCG11 predicts a poor prognosis in prostate cancer. Biomed
Pharmacother. 83:936–941. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yao XM, Tang JH, Zhu H and Jing Y: High
expression of lncRNA CASC15 is a risk factor for gastric cancer
prognosis and promote the proliferation of gastric cancer. Eur Rev
Med Pharmacol Sci. 21:5661–5667. 2017.PubMed/NCBI
|
|
35
|
Chen CQ, Chen CS, Chen JJ, Zhou LP, Xu HL,
Jin WW, Wu JB and Gao SM: Histone deacetylases inhibitor
trichostatin A increases the expression of Dleu2/miR-15a/16-1 via
HDAC3 in non-small cell lung cancer. Mol Cell Biochem. 383:137–148.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shi H, Shi J, Zhang Y, Guan C, Zhu J, Wang
F, Xu M, Ju Q, Fang S and Jiang M: Long non-coding RNA DANCR
promotes cell proliferation, migration, invasion and resistance to
apoptosis in esophageal cancer. J Thorac Dis. 10:2573–2582. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Niu Y, Ma F, Huang W, Fang S, Li M, Wei T
and Guo L: Long non-coding RNA TUG1 is involved in cell growth and
chemoresistance of small cell lung cancer by regulating LIMK2b via
EZH2. Mol Cancer. 16:52017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen W, Zhang X, Li J, Huang S, Xiang S,
Hu X and Liu C: Comprehensive analysis of coding-lncRNA gene
co-expression network uncovers conserved functional lncRNAs in
zebrafish. BMC Genomics. 19 (Suppl 2):S1122018. View Article : Google Scholar
|
|
39
|
Fang L, Wang H and Li P: Systematic
analysis reveals a lncRNA-mRNA co-expression network associated
with platinum resistance in high-grade serous ovarian cancer.
Invest New Drugs. 36:187–194. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jiao J, Shi B, Wang T, Fang Y, Cao T, Zhou
Y, Wang X and Li D: Characterization of long non-coding RNA and
messenger RNA profiles in follicular fluid from mature and immature
ovarian follicles of healthy women and women with polycystic ovary
syndrome. Hum Reprod. 33:1735–1748. 2018. View Article : Google Scholar : PubMed/NCBI
|