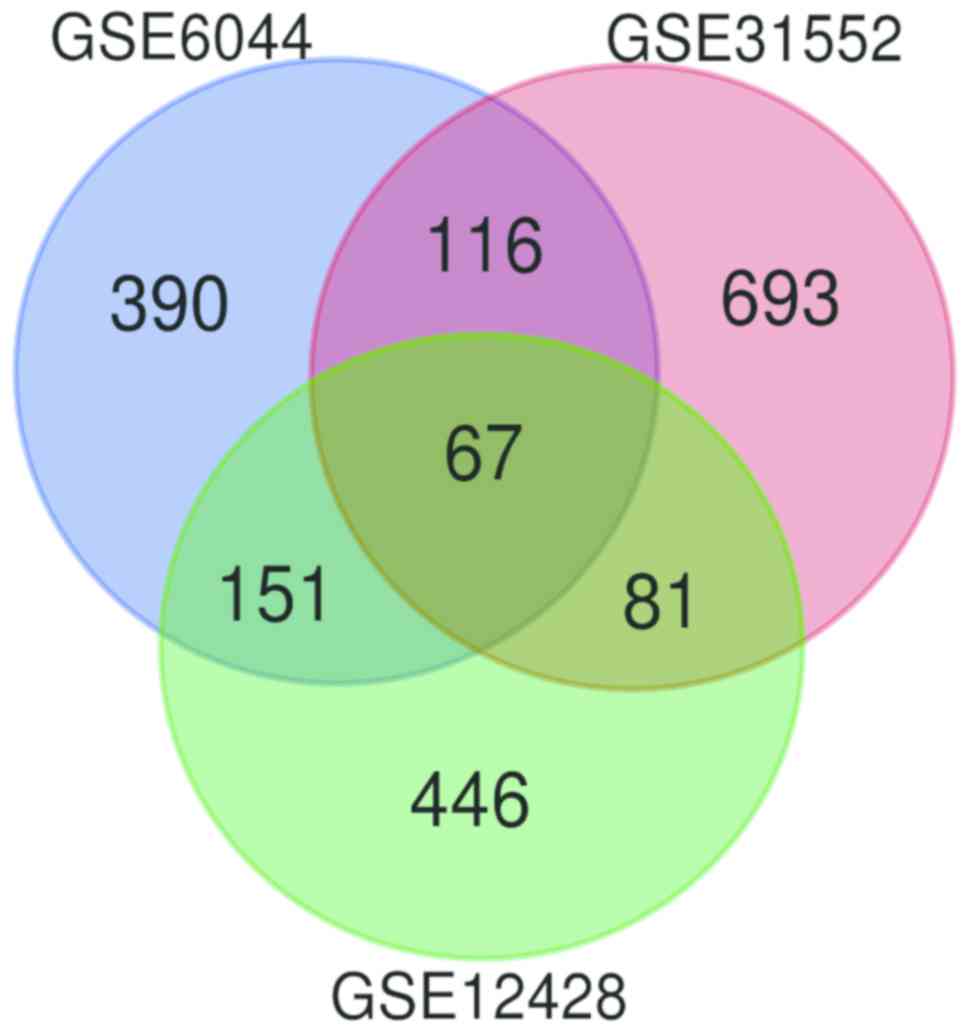
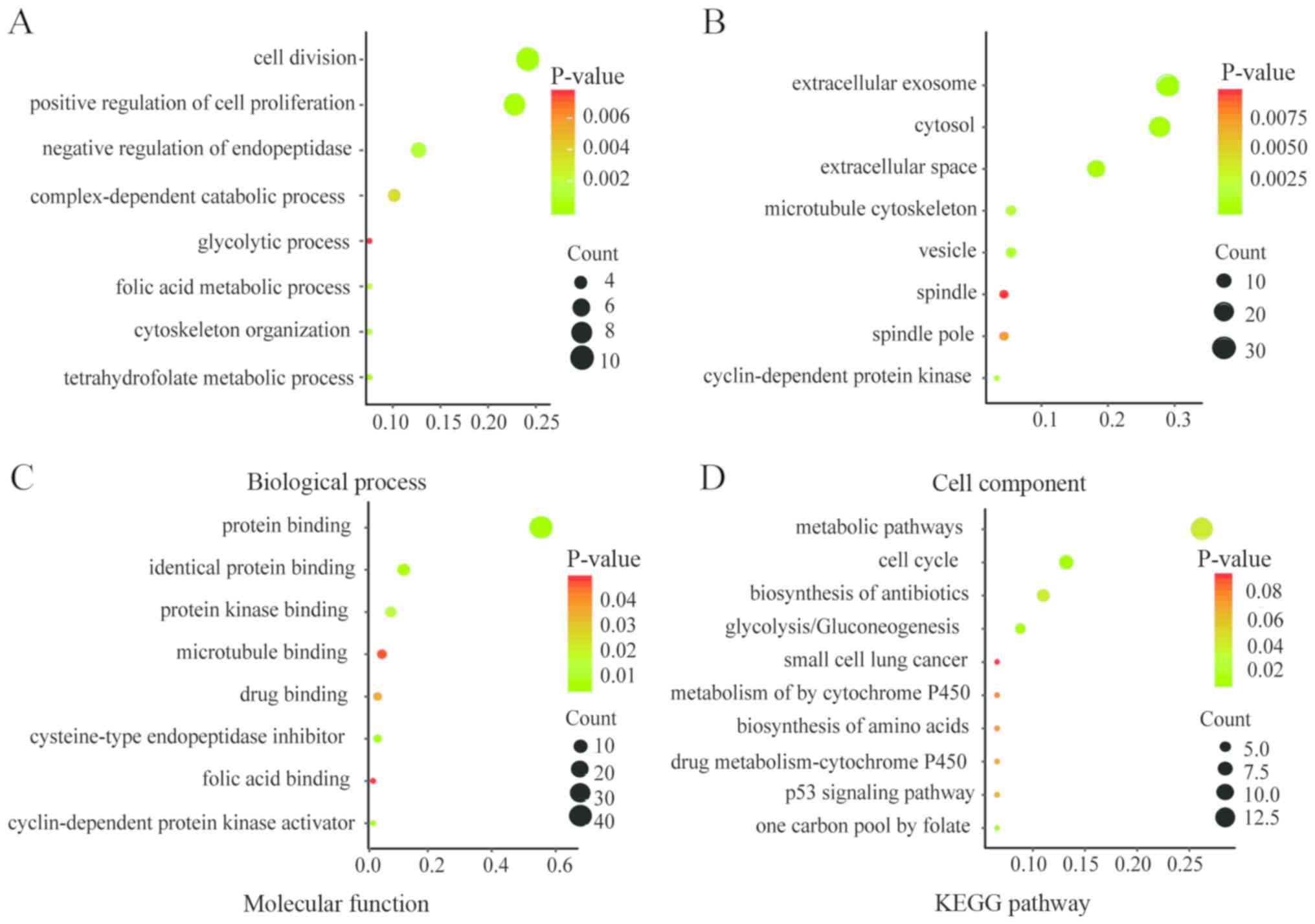
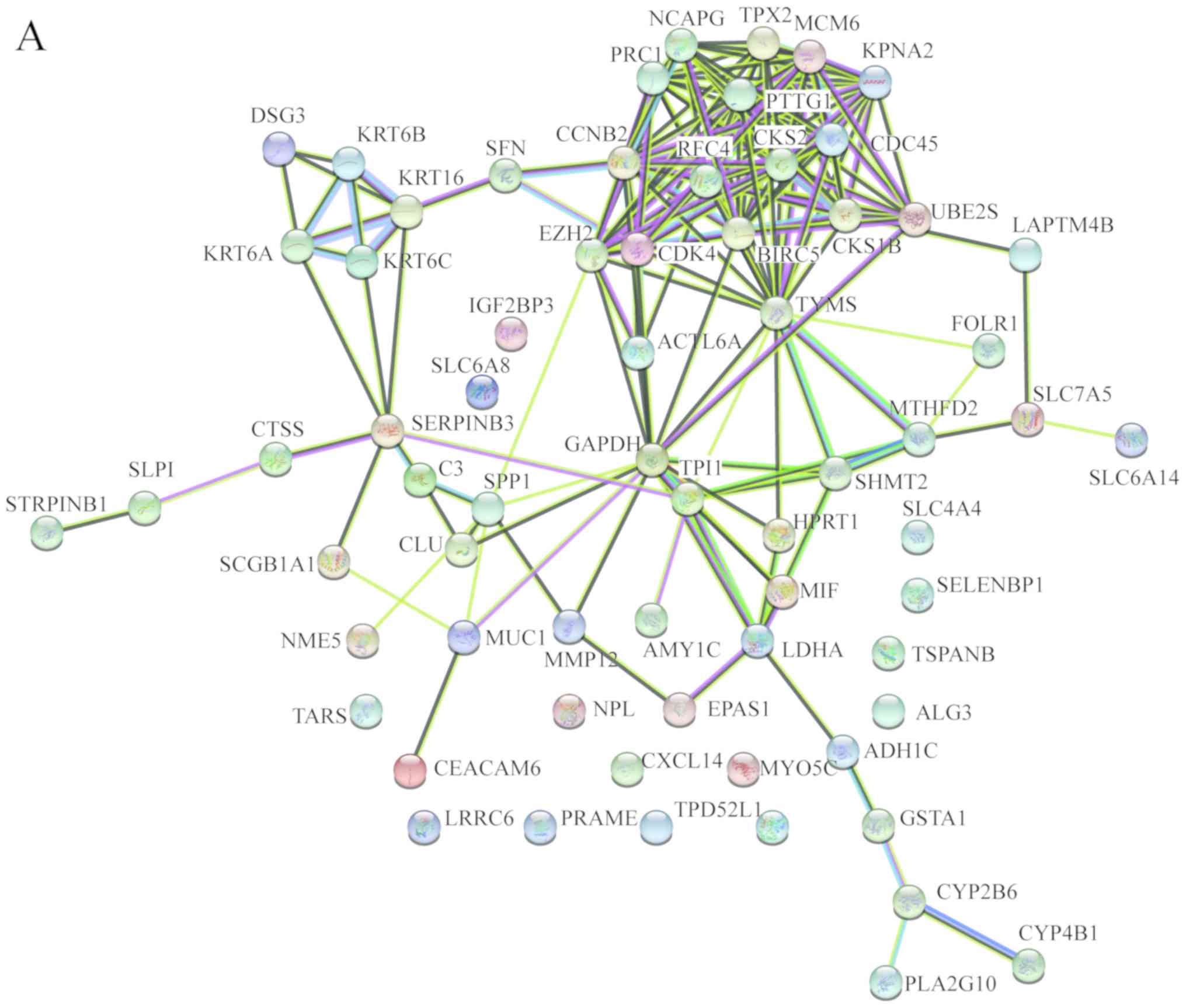
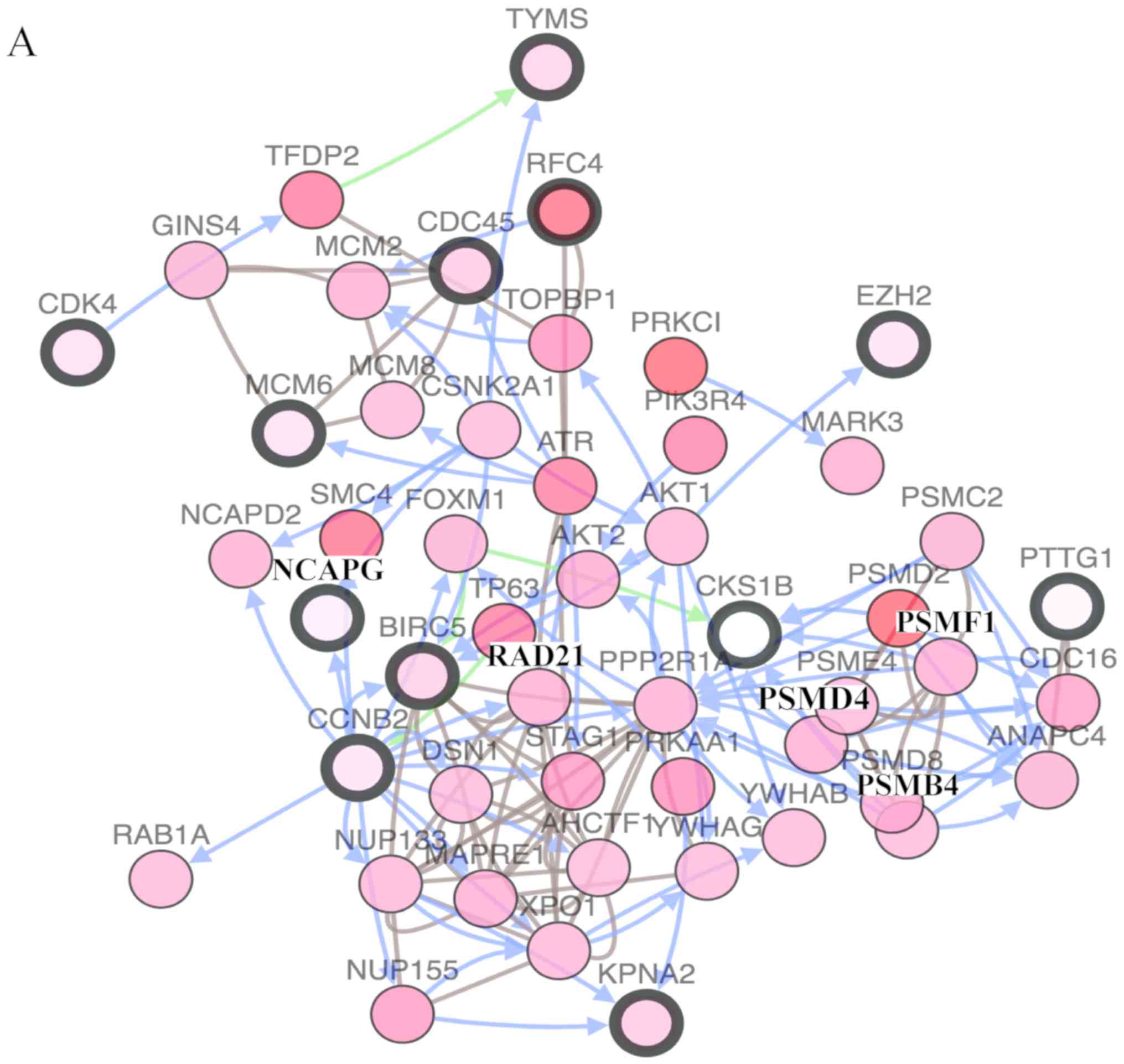
|
1
|
Youlden DR, Cramb SM and Baade PD: The
International Epidemiology of Lung Cancer: Geographical
distribution and secular trends. J Thorac Oncol. 3:819–831. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lazarus KA, Hadi F, Zambon E, Bach K,
Santolla MF, Watson JK, Correia LL, Das M, Ugur R, Pensa S, et al:
BCL11A interacts with SOX2 to control the expression of epigenetic
regulators in lung squamous carcinoma. Nat Commun. 9:33272018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kris MG, Johnson BE, Kwiatkowski DJ,
Iafrate AJ, Wistuba II, Aronson SL, Engelman JA, Shyr Y, Khuri FR,
Rudin CM, et al: Identification of driver mutations in tumor
specimens from 1,000 patients with lung adenocarcinoma: The NCI's
Lung Cancer Mutation Consortium (LCMC). J Clin Oncol.
29:CRA75062011. View Article : Google Scholar
|
|
4
|
Chan BA and Hughes BG: Targeted therapy
for non-small cell lung cancer: Current standards and the promise
of the future. Transl Lung Cancer Res. 4:36–54. 2015.PubMed/NCBI
|
|
5
|
Vayshlya NA, Zinovyeva MV, Sass AV,
Kopantzev EP, Vinogradova TV and Sverdlov ED: Increased expression
of BIRC5 in non-small cell lung cancer and esophageal
squamous cell carcinoma does not correlate with the expression of
its inhibitors SMAC/DIABLO and PML. Mol Biol. 42:579–587. 2008.
View Article : Google Scholar
|
|
6
|
Li S, Sun X, Miao S, Liu J and Jiao W:
Differential protein-coding gene and long noncoding RNA expression
in smoking-related lung squamous cell carcinoma. Thorac Cancer.
8:672–681. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ganapathy-Kanniappan S, Geschwind JF,
Kunjithapatham R, Buijs M, Vossen JA, Tchernyshyov I, Cole RN, Syed
LH, Rao PP, Ota S and Vali M: Glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) is pyruvylated during 3-bromopyruvate
mediated cancer cell death. Anticancer Res. 29:4909–4918.
2009.PubMed/NCBI
|
|
8
|
Xu RH, Pelicano H, Zhou Y, Carew JS, Feng
L, Bhalla KN, Keating MJ and Huang P: Inhibition of glycolysis in
cancer cells: A novel strategy to overcome drug resistance
associated with mitochondrial respiratory defect and hypoxia.
Cancer Res. 65:613–621. 2005.PubMed/NCBI
|
|
9
|
Dawany NB and Tozeren A: Asymmetric
microarray data produces gene lists highly predictive of research
literature on multiple cancer types. BMC Bioinformatics.
11:4832010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dawany NB, Dampier WN and Tozeren A:
Large-scale integration of microarray data reveals genes and
pathways common to multiple cancer types. Int J Cancer.
128:2881–2891. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lin J, Marquardt G, Mullapudi N, Wang T,
Han W, Shi M, Keller S, Zhu C, Locker J and Spivack SD: Lung cancer
transcriptomes refined with laser capture microdissection. Am J
Pathol. 184:2868–2884. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rohrbeck A, Neukirchen J, Rosskopf M,
Pardillos GG, Geddert H, Schwalen A, Gabbert HE, von Haeseler A,
Pitschke G, Schott M, et al: Gene expression profiling for
molecular distinction and characterization of laser captured
primary lung cancers. J Transl Med. 6:692008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Boelens MC, van den Berg A, Fehrmann RS,
Geerlings M, de Jong WK, te Meerman GJ, Sietsma H, Timens W, Postma
DS and Groen HJ: Current smoking-specific gene expression signature
in normal bronchial epithelium is enhanced in squamous cell lung
cancer. J Pathol. 218:182–191. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang DW, Sherman BT, Tan Q, Kir J, Liu D,
Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC and Lempicki RA:
DAVID Bioinformatics Resources: Expanded annotation database and
novel algorithms to better extract biology from large gene lists.
Nucleic Acids Res. 35:W169–W175. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Scardoni G, Tosadori G, Faizan M, Spoto F,
Fabbri F and Laudanna C: Biological network analysis with
CentiScaPe: Centralities and experimental dataset integration.
F1000Res. 3:1392014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mangan ME, Williams JM, Lathe SM,
Karolchik D and Lathe WC III: UCSC genome browser: Deep support for
molecular biomedical research. Biotechnol Annu Rev. 14:63–108.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hou J, Aerts J, den Hamer B, van Ijcken W,
den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens
JA, Hoogsteden HC, et al: Gene expression-based classification of
non-small cell lung carcinomas and survival prediction. PLoS One.
5:e103122010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Garber ME, Troyanskaya OG, Schluens K,
Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen
GD, Perou CM, Whyte RI, et al: Diversity of gene expression in
adenocarcinoma of the lung. Proc Natl Acad Sci USA. 98:13784–13789.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wachi S, Yoneda K and Wu R:
Interactome-transcriptome analysis reveals the high centrality of
genes differentially expressed in lung cancer tissues.
Bioinformatics. 21:4205–4208. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hou J, Aerts J, den Hamer B, van Ijcken W,
den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens
JA, Hoogsteden HC, et al: Gene expression-based classification of
non-small cell lung carcinomas and survival prediction. PLoS One.
5:e103122010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hou GX, Liu P, Yang J and Wen S: Mining
expression and progQixing M, Gaochao D, Wenjie X, Anpeng W, Bing C,
Weidong M, Lin X and Feng J: Microarray analyses reveal genes
related to progression and prognosis of esophageal squamous cell
carcinoma. Oncotarget. 8:78838–78850. 2017.PubMed/NCBI
|
|
29
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: a cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sholl LM, Aisner DL, Varella-Garcia M,
Berry LD, Dias-Santagata D, Wistuba II, Chen H, Fujimoto J, Kugler
K, Franklin WA, et al: Multi-institutional Oncogenic Driver
Mutation Analysis in Lung Adenocarcinoma: The Lung Cancer Mutation
Consortium ExperienceJ Thorac Oncol; 10. pp. 768–777. 2015,
PubMed/NCBI
|
|
32
|
Mantripragada K and Khurshid H: Targeting
genomic alterations in squamous cell lung cancer. Front Oncol.
3:1952013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Heist RS, Sequist LV and Engelman JA:
Genetic changes in squamous cell lung cancer: A review. J Thorac
Oncol. 7:924–933. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cancer Genome Atlas Research Network, .
Comprehensive genomic characterization of squamous cell lung
cancers. Nature. 489:519–525. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Brambilla C, Laffaire J, Lantuejoul S,
Moro-Sibilot D, Mignotte H, Arbib F, Toffart AC, Petel F, Hainaut
P, Rousseaux S, et al: Lung squamous cell carcinomas with basaloid
histology represent a specific molecular entity. Clin Cancer Res.
20:5777–5786. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yao W, Bai Y, Li Y, Guo L, Zeng P, Wang Y,
Qi B, Liu S, Qin X, Li Y and Zhao B: Upregulation of MALAT-1 and
its association with survival rate and the effect on cell cycle and
migration in patients with esophageal squamous cell carcinoma.
Tumour Biol. 37:4305–4312. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee CH, Lee MK, Kang CD, Kim YD, Park DY,
Kim JY, Sol MY and Suh KS: Differential expression of hypoxia
inducible factor-1 alpha and tumor cell proliferation between
squamous cell carcinomas and adenocarcinomas among operable
non-small cell lung carcinomas. J Korean Med Sci. 18:196–203. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hirano T, Franzén B, Kato H, Ebihara Y and
Auer G: Genesis of squamous cell lung carcinoma. Sequential changes
of proliferation, DNA ploidy, and p53 expression. Am J Pathol.
144:296–302. 1994.PubMed/NCBI
|
|
39
|
Liu GM and Zhang YM: Targeting FBPase is
an emerging novel approach for cancer therapy. Cancer Cell Int.
18:362018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Keku T, Millikan R, Worley K, Winkel S,
Eaton A, Biscocho L, Martin C and Sandler R:
5,10-methylenetetrahydrofolate reductase codon 677 and 1298
polymorphisms and colon cancer in African Americans and whites.
Cancer Epidemiol Biomarkers Prev. 11:1611–1621. 2002.PubMed/NCBI
|
|
42
|
Ferlazzo N, Currò M, Zinellu A, Caccamo D,
Isola G, Ventura V, Carru C, Matarese G and Ientile R: Influence of
MTHFR genetic background on p16 and MGMT methylation in oral
squamous cell cancer. Int J Mol Sci. 18:E7242017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fujii T, Toyooka S, Ichimura K, Fujiwara
Y, Hotta K, Soh J, Suehisa H, Kobayashi N, Aoe M, Yoshino T, et al:
ERCC1 protein expression predicts the response of cisplatin-based
neoadjuvant chemotherapy in non-small-cell lung cancer. Lung
Cancer. 59:377–384. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kasai D, Ozasa H, Oguri T, Miyazaki M,
Uemura T, Takakuwa O, Kunii E, Ohkubo H, Maeno K and Niimi A:
Thymidylate synthase gene copy number as a predictive marker for
response to pemetrexed treatment of lung adenocarcinoma. Anticancer
Res. 33:1935–1940. 2013.PubMed/NCBI
|
|
45
|
Lu Y, Zhuo C, Cui B, Liu Z, Zhou P, Lu Y
and Wang B: TYMS serves as a prognostic indicator to predict the
lymph node metastasis in Chinese patients with colorectal cancer.
Clin Biochem. 46:1478–1483. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
da Silva Nogueira J Jr, de Lima Marson FA
and Sílvia Bertuzzo C: Thymidylate synthase gene (TYMS)
polymorphisms in sporadic and hereditary breast cancer. BMC Res
Notes. 5:6762012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shichijo S, Azuma K, Komatsu N, Ito M,
Maeda Y, Ishihara Y and Itoh K: Two proliferation-related proteins,
TYMS and PGK1, could be new cytotoxic T lymphocyte-directed
tumor-associated antigens of HLA-A2+ colon cancer. Clin Cancer Res.
10:5828–5836. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chao YL and Anders CK: TYMS gene
polymorphisms in breast cancer patients receiving
5-fluorouracil-based chemotherapy. Clin Breast Cancer.
18:e301–e304. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
He Y, Penney ME, Negandhi AA, Parfrey PS,
Savas S and Yilmaz YE: XRCC3 Thr241Met and TYMS variable number
tandem repeat polymorphisms are associated with time-to-metastasis
in colorectal cancer. PLoS One. 13:e01923162018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Knizhnik AV, Kovaleva OB, Laktionov KK,
Mochal'nikova VV, Komel'kov AV, Chevkina EM and Zborovskaia IB:
Arf6, RalA and BIRC5 protein expression in non small cell lung
cancer. Mol Biol (Mosk). 45:307–315. 2011.(In Russian). View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yu X, Zhang Y, Cavazos D, Ma X, Zhao Z, Du
L and Pertsemlidis A: miR-195 targets cyclin D3 and survivin to
modulate the tumorigenesis of non-small cell lung cancer. Cell
Death Dis. 9:1932018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Phiboonchaiyanan PP, Petpiroon N,
Sritularak B and Chanvorachote P: Phoyunnanin E induces apoptosis
of non-small cell lung cancer cells via p53 activation and
down-regulation of survivin. Anticancer Res. 38:6281–6290. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ganapathy-Kanniappan S: Evolution of GAPDH
as a druggable target of tumor glycolysis? Expert Opin Ther
Targets. 22:295–298. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Hao L, Zhou X, Liu S, Sun M, Song Y, Du S,
Sun B, Guo C, Gong L, Hu J, et al: Elevated GAPDH expression is
associated with the proliferation and invasion of lung and
esophageal squamous cell carcinomas. Proteomics. 15:3087–3100.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Mo ML, Chen Z, Li J, Li HL, Sheng Q, Ma
HY, Zhang FX, Hua YW, Zhang X, Sun DQ, et al: Use of serum
circulating CCNB2 in cancer surveillance. Int J Biol Markers.
25:2010. View Article : Google Scholar
|
|
56
|
Takashima S, Saito H, Takahashi N, Imai K,
Kudo S, Atari M, Saito Y, Motoyama S and Minamiya Y: Strong
expression of cyclin B2 mRNA correlates with a poor prognosis in
patients with non-small cell lung cancer. Tumour Biol.
35:4257–4265. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lei CY, Wang W, Zhu YT, Fang WY and Tan
WL: The decrease of cyclin B2 expression inhibits invasion and
metastasis of bladder cancer. Urol Oncol. 34:237.e1–e10. 2016.
View Article : Google Scholar
|
|
58
|
Wang XC, Yue X, Zhang RX, Liu TY, Pan ZZ,
Yang MJ, Lu ZH, Wang ZY, Peng JH, Le LY, et al: Genome-wide RNAi
screening identifies RFC4 as a factor that mediates radioresistance
in colorectal cancer by facilitating nonhomologous end joining
repair. Clin Cancer Res. 25:4567–4579. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Xiang J, Fang L, Luo Y, Yang Z, Liao Y,
Cui J, Huang M, Yang Z, Huang Y, Fan X, et al: Levels of human
replication factor C4, a clamp loader, correlate with tumor
progression and predict the prognosis for colorectal cancer. J
Transl Med. 12:3202014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Liu YZ, Wang BS, Jiang YY, Cao J, Hao JJ,
Zhang Y, Xu X, Cai Y and Wang MR: MCMs expression in lung cancer:
Implication of prognostic significance. J Cancer. 8:3641–3647.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Behrens C, Solis LM, Lin H, Yuan P, Tang
X, Kadara H, Riquelme E, Galindo H, Moran CA, Kalhor N, et al: EZH2
protein expression associates with the early pathogenesis, tumor
progression, and prognosis of non-small cell lung carcinoma. Clin
Cancer Res. 19:6556–6565. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Shi R, Li M, Raghavan V, Tam S, Cabanero
M, Pham NA, Shepherd FA, Moghal N and Tsao MS: Targeting the
CDK4/6-Rb pathway enhances response to PI3K inhibition in
PIK3CA-mutant lung squamous cell carcinoma. Clin Cancer Res.
24:5990–6000. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chen L and Pan J: Dual cyclin-dependent
kinase 4/6 inhibition by PD-0332991 induces apoptosis and
senescence in oesophageal squamous cell carcinoma cells. Br J
Pharmacol. 174:2427–2443. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Ma Y, Lin D, Sun W, Xiao T, Yuan J, Han N,
Guo S, Feng X, Su K, Mao Y, et al: Expression of targeting protein
for xklp2 associated with both malignant transformation of
respiratory epithelium and progression of squamous cell lung
cancer. Clin Cancer Res. 12:1121–1127. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Mollinari C, Kleman JP, Jiang W, Schoehn
G, Hunter T and Margolis RL: PRC1 is a microtubule binding and
bundling protein essential to maintain the mitotic spindle midzone.
J Cell Biol. 157:1175–1186. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhan P, Xi GM, Liu HB, Liu YF, Xu WJ, Zhu
Q, Zhou ZJ, Miao YY, Wang XX, Jin JJ, et al: Protein regulator of
cytokinesis-1 expression: Prognostic value in lung squamous cell
carcinoma patients. J Thorac Dis. 9:2054–2060. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Huang J, Zhou Y, Thomas GS, Gu Z, Yang Y,
Xu H, Tricot G and Zhan F: NEDD8 inhibition overcomes CKS1B-induced
drug resistance by upregulation of p21 in multiple myeloma. Clin
Cancer Res. 21:5532–5542. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhan F, Colla S, Wu X, Chen B, Stewart JP,
Kuehl WM, Barlogie B and Shaughnessy JD Jr: CKS1B, overexpressed in
aggressive disease, regulates multiple myeloma growth and survival
through SKP2- and p27Kip1-dependent and -independent mechanisms.
Blood. 109:4995–5001. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Romero Arenas MA, Whitsett TG, Aronova A,
Henderson SA, LoBello J, Habra MA, Grubbs EG, Lee JE, Sircar K,
Zarnegar R, et al: Protein expression of PTTG1 as a diagnostic
biomarker in adrenocortical carcinoma. Ann Surg Oncol. 25:801–807.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Guo XC, Li L, Gao ZH, Zhou HW, Li J and
Wang QQ: The long non-coding RNA PTTG3P promotes growth and
metastasis of cervical cancer through PTTG1. Aging (Albany NY).
11:1333–1341. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Huang JL, Cao SW, Ou QS, Yang B, Zheng SH,
Tang J, Chen J, Hu YW, Zheng L and Wang Q: The long non-coding RNA
PTTG3P promotes cell growth and metastasis via up-regulating PTTG1
and activating PI3K/AKT signaling in hepatocellular carcinoma. Mol
Cancer. 17:932018. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
You H, Lin H and Zhang Z: CKS2 in human
cancers: Clinical roles and current perspectives (Review). Mol Clin
Oncol. 3:459–463. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Hauge S, Naucke C, Hasvold G, Joel M,
Rødland GE, Juzenas P, Stokke T and Syljuåsen RG: Combined
inhibition of Wee1 and Chk1 gives synergistic DNA damage in S-phase
due to distinct regulation of CDK activity and CDC45 loading.
Oncotarget. 8:10966–10979. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Jensen JB, Munksgaard PP, Sørensen CM,
Fristrup N, Birkenkamp-Demtroder K, Ulhøi BP, Jensen KM, Ørntoft TF
and Dyrskjøt L: High expression of karyopherin-α2 defines poor
prognosis in non-muscle-invasive bladder cancer and in patients
with invasive bladder cancer undergoing radical cystectomy. Eur
Urol. 59:841–848. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Goto Y, Kurozumi A, Arai T, Nohata N,
Kojima S, Okato A, Kato M, Yamazaki K, Ishida Y, Naya Y, et al:
Impact of novel miR-145-3p regulatory networks on survival in
patients with castration-resistant prostate cancer. Br J Cancer.
117:409–420. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Garnett MJ, Mansfeld J, Godwin C,
Matsusaka T, Wu J, Russell P, Pines J and Venkitaraman AR: UBE2S
elongates ubiquitin chains on APC/C substrates to promote mitotic
exit. Nat Cell Biol. 11:1363–1369. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Akter KA, Hyodo T, Asano E, Sato N,
Mansour MA, Ito S, Hamaguchi M and Senga T: Erratum to: UBE2S is
associated with malignant characteristics of breast cancer cells.
Tumor Biol. 37:69992016. View Article : Google Scholar
|
|
78
|
Ben-Eliezer I, Pomerantz Y, Galiani D,
Nevo N and Dekel N: Appropriate expression of Ube2C and Ube2S
controls the progression of the first meiotic division. FASEB J.
29:4670–4681. 2015. View Article : Google Scholar : PubMed/NCBI
|