Introduction
Lung cancer is the most common malignancy and the
leading cause of death among all types of cancer worldwide, with
11.6% of the total cancer and 18.4% of the total cancer deaths
according to the Global Cancer Statistics 2018. Lung cancer
contains multiple subtypes, such as small cell lung cancer (SCLC),
squamous cell carcinoma and lung adenocarcinoma (LUAD). LUAD is the
most common type of lung cancer (1,2). In
recent decades, an increased understanding of the underlying
molecular mechanisms of LUAD has been obtained, and notable
progress has been made in analysis of LUAD prognosis, especially in
terms of tumour markers (1). For
example, in a previous study, epidermal growth factor receptor
(EGFR) expression was high in 62% of patients with non-small cell
lung cancer (NSCLC), and when treated with inhibitors of EGFR,
these patients had an improved prognosis compared with patients
without EGFR upregulation (3).
Therefore, targeted therapy is becoming increasingly popular, and
new and more efficient diagnostic and therapeutic targets in LUAD
are urgently needed.
Growing evidence has confirmed that the expression
of certain genes to which the cancer has become ‘addicted’ or
‘dependent on’ is necessary for tumour growth (4,5).
Silencing these genes or inhibiting the activity of the proteins
they encode may promote cancer apoptosis (4,5).
Maternal embryonic leucine zipper kinase (MELK), an adenosine
monophosphate-activated protein kinase-related kinase, is
upregulated in multiple malignant tumours, including breast cancer,
melanoma, glioblastoma, hepatocellular carcinoma, acute leukaemia
and ovarian cancer (6–11). In addition, some cancer types have
been reported to depend on the upregulation of MELK, such as breast
cancer, glioma and melanoma, and MELK serves as an oncogenic driver
gene inhibiting increased cell apoptosis and suppressed growth of
tumours (4). OTSSP167, a selective
small molecule inhibitor of MELK, is currently being evaluated in
phase I/II clinical research in patients with breast cancer,
glioblastoma and acute leukaemia (8,11–13).
Notably, bioinformatics screening studies conducted using data from
Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA),
among others, have demonstrated that MELK is one of the most
frequently identified hub genes in lung cancer, and high expression
of MELK is associated with worse overall survival (OS) time
compared with low expression of MELK in patients with NSCLC
(1,14,15).
This suggests that MELK may be a key biomarker for diagnosis and
therapy; however, further clinical data, especially from clinical
samples, are needed to increase the accuracy and credibility of
such findings.
Therefore, in the present study, the differential
expression of MELK in patients with LUAD and its association with
prognosis were assessed. Oncomine, Gene Expression Profiling
Interactive Analysis (GEPIA) and TCGA data were used to determine
the levels of MELK mRNA in malignant tumour and normal lung
tissues. In addition, immunohistochemical staining was used to
detect the expression of MELK protein in LUAD and adjacent normal
tissue. Kaplan-Meier plotter, GEPIA and TCGA survival analyses were
conducted to compare the prognostic outcomes in patients with LUAD
with different levels of MELK.
Materials and methods
Bioinformatics mining methods
Data from the Oncomine (https://www.oncomine.org) (16) and TCGA (https://tcga-data.nci.nih.gov/tcga/) databases
(17,18), and GEPIA (http://gepia.cancer-pku.cn) (18,19),
were used to estimate the expression of MELK in LUAD and adjacent
normal tissues (16–19). The search parameters in Oncomine were
as follows: Analysis type, cancer vs. normal; data source, public;
cancer type, selected specific cancer type; sample type, clinical
specimen and; data type, mRNA. Fold-change (FC) >2, P<0.05
and a gene rank in the top 10% were set as the thresholds for
selecting the datasets. The results of differential expression
analyses, including subgroup, are also from TCGA and GEPIA. FC
>2 and P<0.05 were set as the thresholds of gene
upregulation. Among them, Oncomine and TCGA databases provide the
number of samples in different groups; however, GEPIA does not
provide the number of samples in different groups. Kaplan-Meier
plotterfrom dataset ID 204825 (http://kmplot.com/analysis), GEPIA and TCGA were used
to analyse the prognosis of patients with differential expression
of MELK (20). Briefly, the patient
samples were divided into 2 groups according to transcripts per
million (TPM) value. The data with TPM greater than upper quartile
was assigned to a high expression group and the others with TPM
below upper quartile belonged to low/medium expression group. Then
the gene expression data were put into R software to obtain the
survival plots, in which the number-at-risk was shown below the
main plot.
Patients
A total of 75 cancer tissue and 35 normal adjacent
tissue samples were obtained from patients diagnosed with LUAD who
underwent surgical resection were recruited from Jiangxi Provincial
People's Hospital (Nanchang, China) between September 2018 and
August 2019. The distance between normal adjacent tissue and tumour
tissue was 3 cm. The inclusion/exclusion criteria were as follows:
i) Patients who underwent primary surgical resection and had
histopathologically confirmed LUAD; ii) patients who were ≥18 years
old, iii) patients who had no pulmonary metastases or other
concomitant cancers; and iv) patients for whom informed consent was
provided for the use of tissues. Detailed patient clinical
information was collected retrospectively, including age (46 males
and 29 females), sex (mean age, 61.6 years; range 40–83 years),
tumour location, tumour size, histological differentiation grade,
lymph node metastasis, and Tumour-Node-Metastasis (TNM) stage. All
patients were staged according to the 8th edition of the TNM
staging system for lung cancer (21). There was missing data in tumour
location, tumour size, histological differentiation, T stage, N
stage, M stage and TNM stage. The research was approved by the
Ethical Committee of Jiangxi Provincial People's Hospital (Jiangxi,
China) (approval no. 2018070). Informed consent was provided by the
clinicians and obtained from the patients who provided written
informed consent for the usage of the tissues for research.
Immunohistochemistry (IHC)
Immunohistochemical staining was performed as
described previously (22). Briefly,
the tissue was fixed with 4% formaldehyde at room temperature for
24 h and then embedded with paraffin and tissue specimens were cut
into 5-µm serial sections. Then, the sections were dewaxed and
dehydrated in a xylene and different concentration of alcohol
solution (100, 95, 90, 80%). Heat mediated antigen retrieval was
performed with Tris/EDTA buffer pH 9.0. The endogenous peroxidase
activity was then blocked using 0.3% hydrogen peroxide for 10 min
at room temperature. The sections were cooled and blocked by
incubating with normal goat serum for 1 h at room temperature. The
sections were then incubated overnight at 4°C with rabbit
anti-human MELK antibody (cat. no. ab129373; 1:200; Abcam). The
sections were next incubated with biotinylated secondary goat
anti-rabbit polyclonal antibody (cat. no. ab6720; 1:800; Abcam) for
30 min at room temperature, followed by incubation with
streptavidin horseradish peroxidase complex. Finally, sections were
visualized by 3,3′-diaminobenzidine staining. The results of
immunohistochemical staining were calculated according to the
immunoreaction score (IRS) and evaluated by 2 pathologists in a
double-blind manner as described previously (23). Briefly, the range of the IRS score
was from 0–12, which was calculated as staining intensity (SI) ×
percentage of positive cells (PP). High expression of MELK was
represented by IRS >4, while low expression of MELK was
represented by IRS ≤4. For haematoxylin and eosin staining
(H&E), which was also performed in a double-blind manner, 5-µm
sections from the paraffin blocks were stained with H&E for 5
min at room temperature by one pathologist, and the other
pathologist observed the results under an optical microscope
(Olympus Corporation).
Statistical analysis
Oncomine, TCGA and GEPIA data were analysed using an
unpaired Student's t-test for the comparison of 2 groups or ANOVA
followed by a post hoc Tukey's honest significance difference (HSD)
test for multiple comparisons conducted to compare the differential
mRNA and protein expression of MELK. The patients' clinical data
are presented as the mean ± S.E.M. SPSS 19.0 software (IBM Corp.)
was used to analyse the clinical data using the unpaired t-test and
χ2 tests as appropriate. Kaplan-Meier plotter, GEPIA and
TCGA analyses were performed to assess the effect of MELK on the
overall survival time of patients with LUAD. Log-rank P-values and
HRs with 95% CIs were calculated and displayed on the webpage.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression levels of MELK in multiple
cancer types
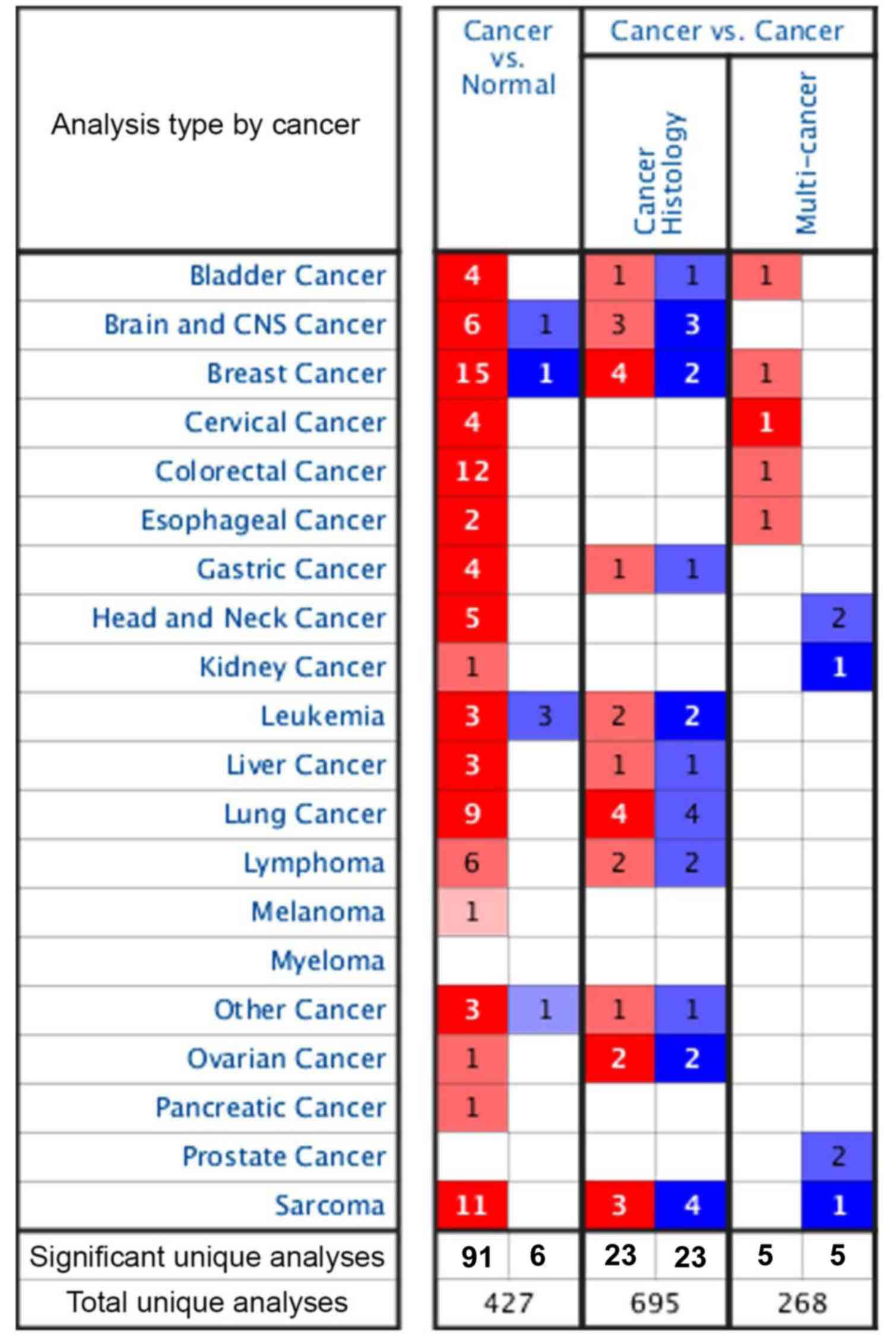
MELK expression in different cancer types was
assessed using the Oncomine database (Fig. 1). In the cancer vs. cancer
comparison, there were 695 cancer histology comparisons and 268
multi-cancer comparisons, and both had the same number of
significant unique samples regardless of if they were in the MELK
high or low expression groups. In addition, in the cancer vs.
normal adjacent comparison, there were 427 unique comparisons for
MELK expression in various tumours and 97 comparisons were unique.
Among them, 91 analyses demonstrated higher expression of MELK in
18 kinds of malignant tumours compared with adjacent normal
tissues, including bladder, brain and central nervous system (CNS),
breast, cervical, colorectal, oesophageal, gastric, head and neck,
kidney, liver, lung, ovarian and pancreatic cancer types, as well
as melanoma, lymphoma, leukaemia, sarcoma and other cancer types.
The next 6 analyses, including analyses in brain and CNS cancer,
breast cancer, leukaemia and other cancer types, showed lower
expression of MELK (Fig. 1). In
addition, there were numerous more significant comparisons in 4
types of tumours, namely, breast cancer (15 unique analyses),
colorectal cancer (12 unique analyses), sarcoma (11 unique
analyses) and lung cancer (9 unique analyses), compared with other
types of tumours, wherein MELK expression in tumours was higher
compared with that in normal tissues. Lung cancer is the leading
cause of cancer incidence and mortality worldwide, and LUAD is the
most prevalent subtype (2);
therefore, the role of MELK in LUAD was further evaluated.
Expression of MELK in LUAD in
different databases
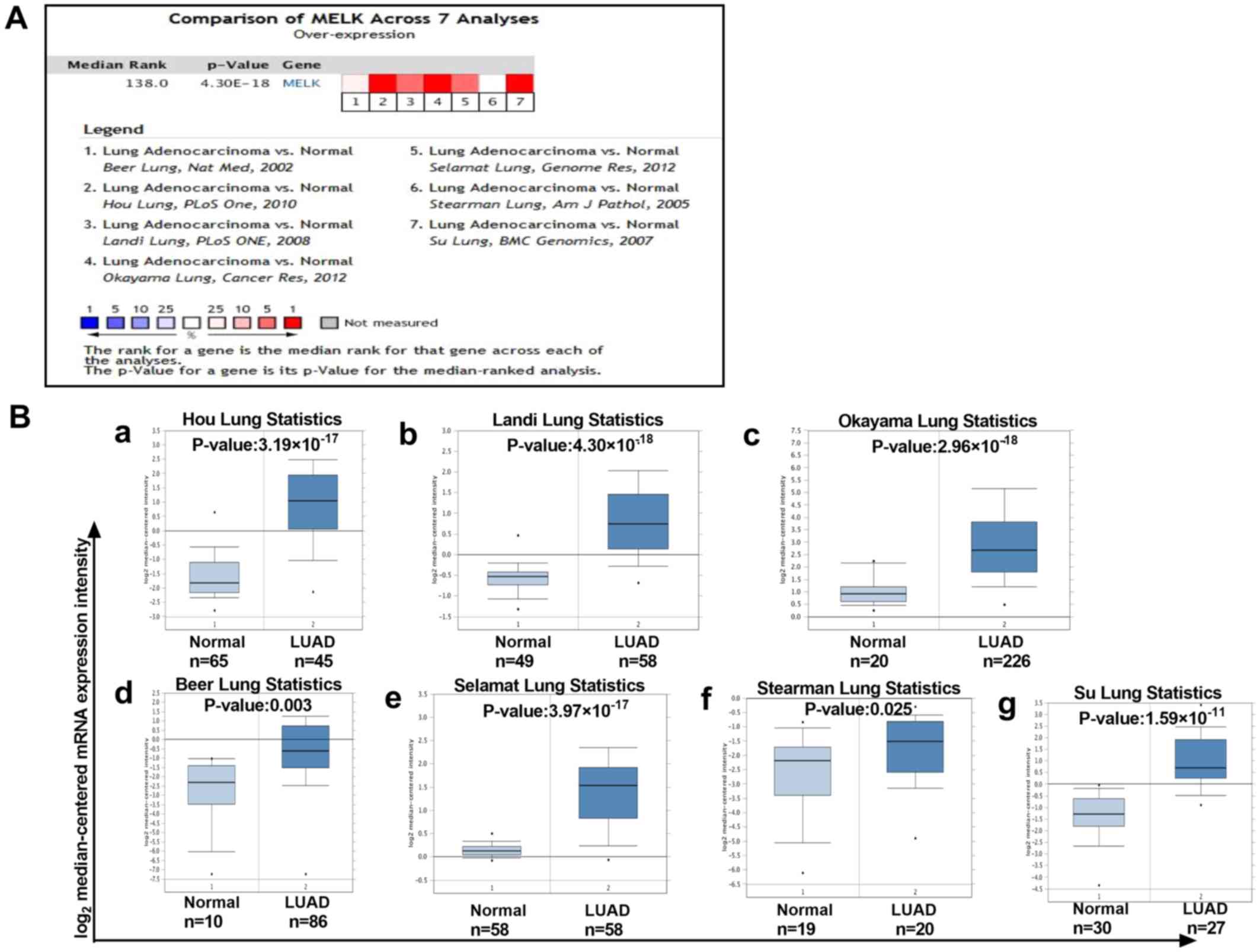
A meta-analysis of 7 studies of MELK mRNA levels in
LUAD was performed using the Oncomine database. As demonstrated,
there was a higher level of MELK in LUAD tissues compared with that
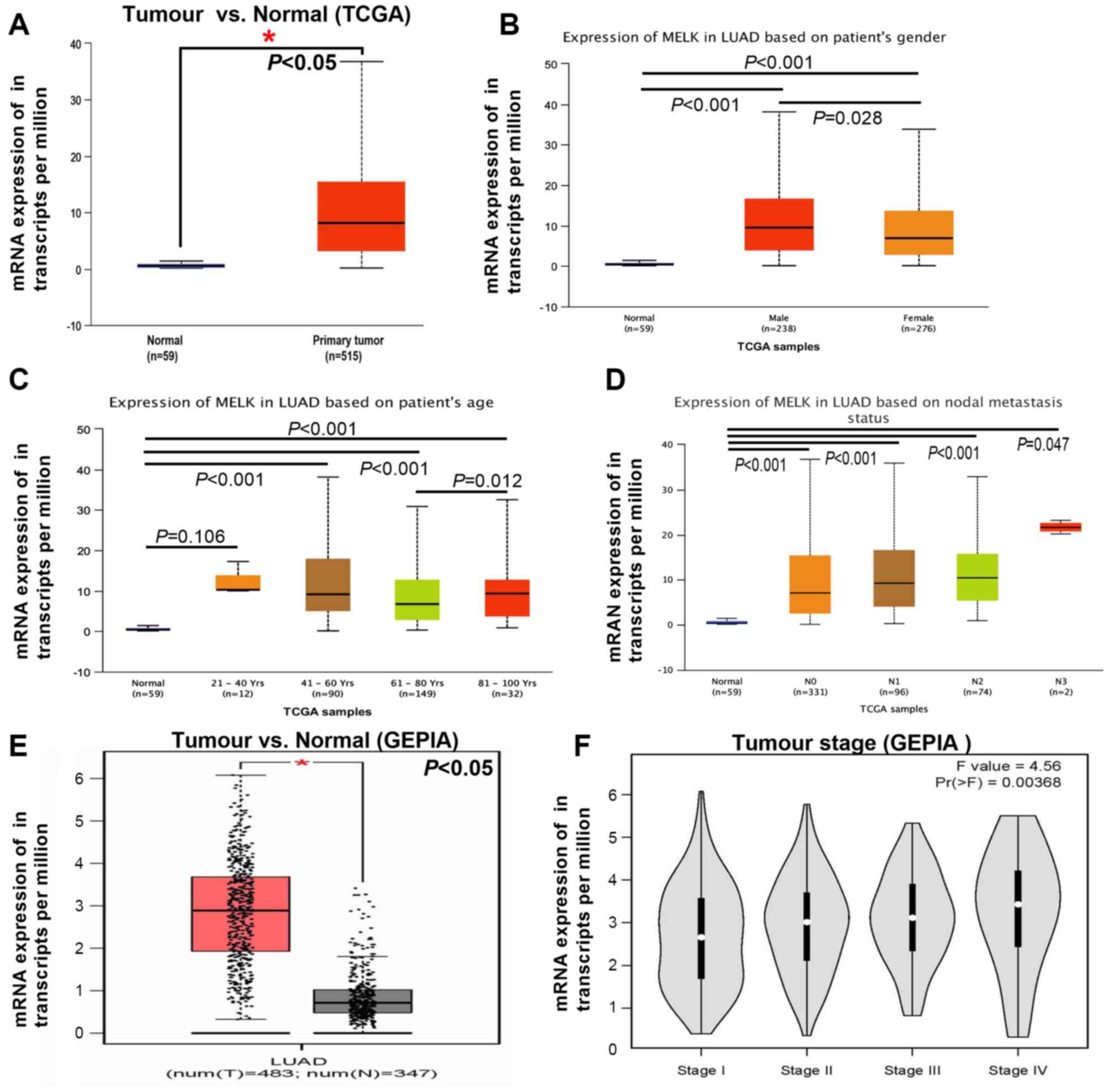
in normal tissues in different datasets (Fig. 2A and B). To verify the aforementioned
results, TCGA and GEPIA datasets were used to analyse MELK
expression in patients with LUAD. As expected, higher levels of
MELK were observed in LUAD compared with those in adjacent normal
tissues both in TCGA and GEPIA datasets (Fig. 3A and E). In addition, the mRNA
expression of MELK was assessed with regard to the different
clinical parameters of LUAD, including sex, age, lymph node
metastasis and tumour stage. MELK expression levels were
significantly higher both in male and female patients with LUAD
compared with those in normal tissues, and higher levels of MELK
mRNA expression were found in male compared with female patients
with LUAD (Fig. 3B). Notably, the
upregulation of MELK in LUAD only occurred in patients >40 years
old (Fig. 3C). There was no
significant difference in the expression of MELK between patients
<40 years old and normal tissue (Fig.
3C). In addition, the expression of MELK in LUAD tissues had no
correlation with lymph node metastasis, but all were higher than
normal group (Fig. 3D). The mRNA
expression of MELK in different stages of LUAD was analysed and the
results demonstrated that the levels of MELK increased gradually
with tumour grade, representing a possible role of MELK in cancer
progression and invasion (Fig. 3F).
Overall, the aforementioned findings were consistent, suggesting
that the expression of MELK in tumours was higher compared with
that in normal lung tissues.
MELK protein expression is upregulated
in tumour tissue compared with normal tissue from patients with
LUAD
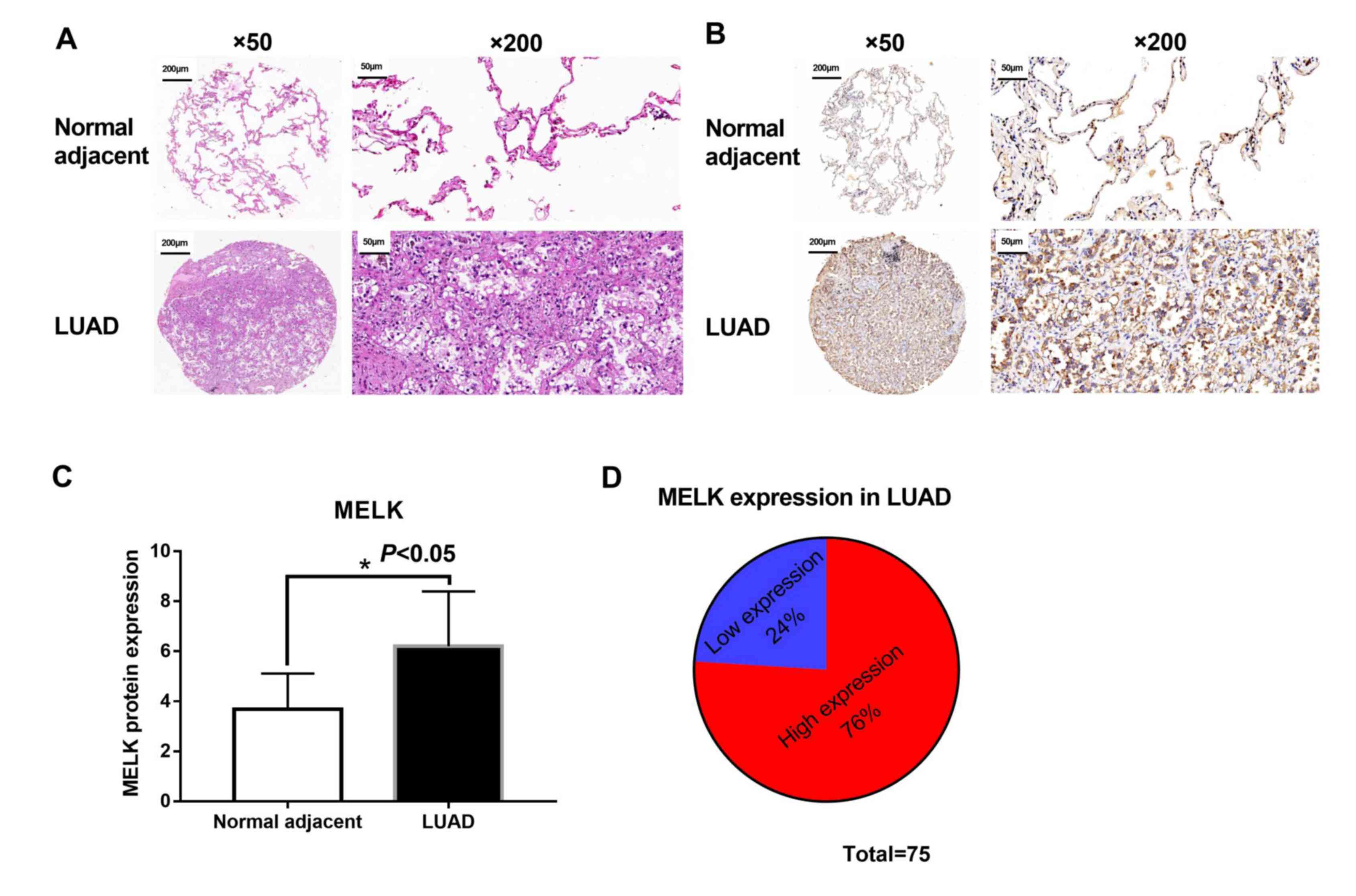
Tumour and matched adjacent normal tissues were
collected from 75 patients with LUAD to evaluate the protein
expression of MELK. Morphological changes were observed between
tumour and matched normal tissues from patients with LUAD using
H&E staining. Matched normal tissues showed normal histological
status and consolidation and less alveoli were presented in LUAD
tissues (Fig. 4A). Subsequently, the
levels of MELK protein in these tissues were assessed using IHC.
The analysis suggested that MELK protein was increased in patients
with LUAD (Fig. 4B and C), and the
high expression rate of MELK was 76% (57/75; Fig. 4D). The association between MELK
protein expression and clinicopathological features in 75 patients
with LUAD was compared, and the results demonstrated that a high
level of MELK was associated with M stage and TNM stage (Table I). However, no significant difference
was found between MELK protein expression and age, sex,
histological differentiation, T stage and N stage.
 | Table I.Association between MELK protein
expression and clinicopathological features in patients with lung
adenocarcinoma (n=75). |
Table I.
Association between MELK protein
expression and clinicopathological features in patients with lung
adenocarcinoma (n=75).
|
|
| MELK levels, n
(%) |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
features | Patients, n (%) | Low | High | P-value |
|---|
| Age, years |
|
|
| >0.999 |
|
<60 | 32 (42.7) | 7 (38.9) | 25 (43.9) |
|
|
≥60 | 43 (57.3) | 11 (61.1) | 32 (56.1) |
|
| Sex |
|
|
| 0.257 |
|
Male | 46 (61.3) | 9 (50.0) | 37 (64.9) |
|
|
Female | 29 (38.7) | 9 (50.0) | 20 (35.1) |
|
| Tumour
location |
|
|
| 0.480 |
|
Left | 28 (37.8) | 7 (41.2) | 21 (36.8) |
|
|
Right | 46 (62.2) | 10 (58.8) | 36 (63.2) |
|
| Tumour size,
cm |
|
|
| 0.611 |
|
<5 | 31 (59.6) | 9 (60.0) | 22 (59.5) |
|
| ≥5 | 21 (40.4) | 6 (40.0) | 15 (40.5) |
|
| Histological
differentiation |
|
|
| 0.088 |
| No | 2 (5.0) | 1 (7.7) | 1 (3.7) |
|
|
Low | 16 (40.0) | 2 (15.4) | 14 (51.9) |
|
|
Moderate | 14 (35.0) | 5 (38.5) | 9 (33.3) |
|
|
High | 8 (20.0) | 5 (38.5) | 3 (11.1) |
|
| T stage |
|
|
| 0.057 |
|
T1-2 | 30 (55.6) | 12 (75.0) | 18 (47.4) |
|
|
T3-4 | 24 (44.4) | 4 (25.0) | 20 (52.6) |
|
| N stage |
|
|
| 0.382 |
| N0 | 26 (50.0) | 9 (56.3) | 17 (47.2) |
|
|
N1-3 | 26 (50.0) | 7 (43.8) | 19 (52.8%) |
|
| M stage |
|
|
| 0.032a |
| M0 | 37 (50.7) | 13 (72.2) | 24 (43.) |
|
| M1 | 36 (49.3) | 5 (27.8) | 31 (56.4) |
|
| TNM stage |
|
|
| 0.045a |
|
I–II | 25 (62.5) | 11 (84.6) | 14 (51.9) |
|
|
III–IV | 15 (37.5) | 2 (15.4) | 13 (48.1) |
|
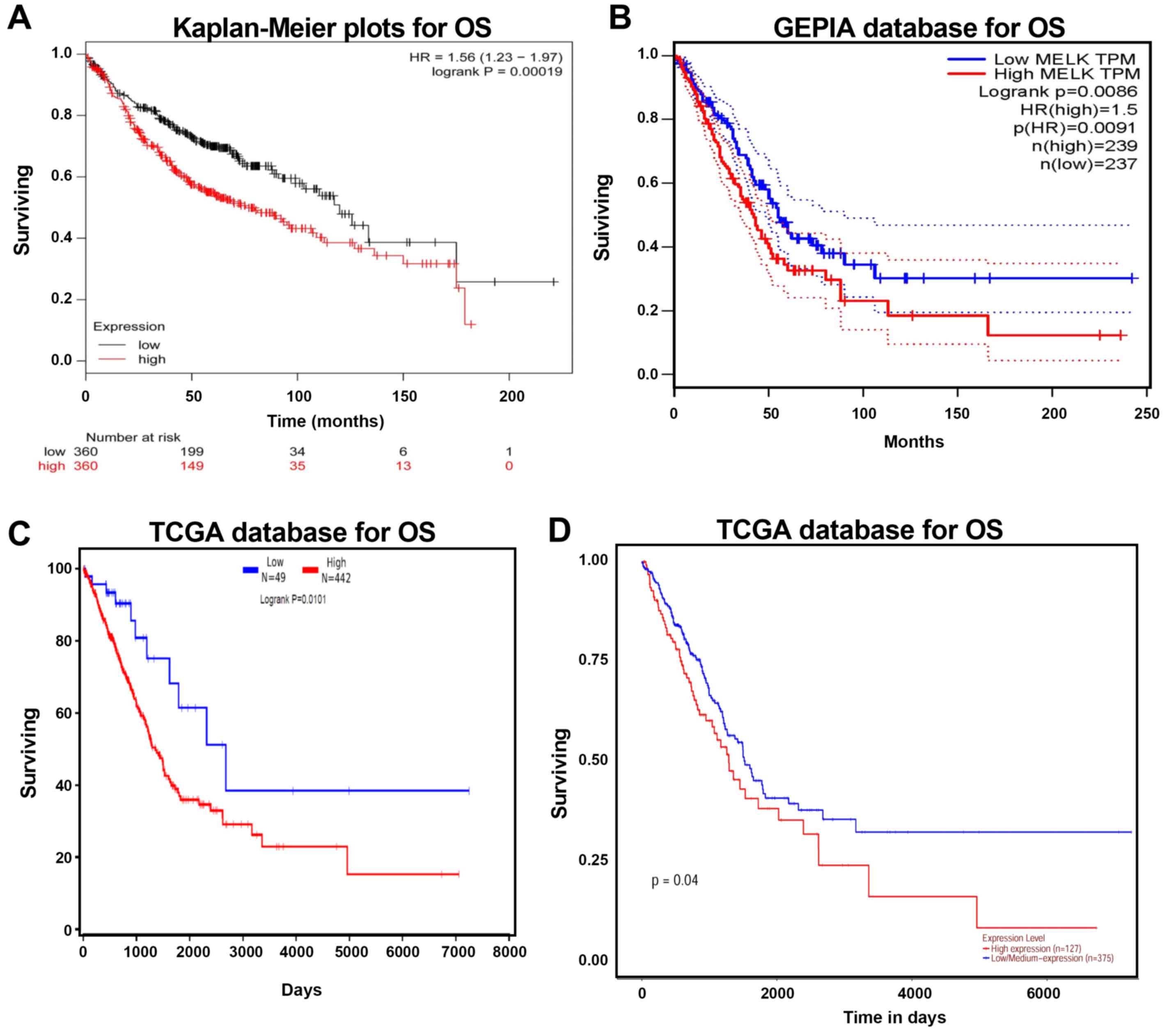
High MELK mRNA expression in patients
with LUAD is associated with a poor prognosis
Kaplan-Meier plotter, GEPIA and TCGA were used to
analyse the prognosis of patients with LUAD with high and low MELK
expression levels. The association between MELK mRNA level and OS
in LUAD patients was analysed using the Kaplan-Meier plotter. The
analysis demonstrated that high MELK expression was negatively
associated with OS time (Fig. 5A).
Similar trends were observed in the GEPIA and TCGA datasets, and
upregulation of MELK increased the risk of mortality (Fig. 5B-D). Overall, these results suggested
that MELK upregulation decreases survival in patients with
LUAD.
Discussion
MELK is a highly conserved serine/threonine kinase
that was originally cloned from mice and is expressed in a wide
range of early embryonic cellular stages (9). In recent years, MELK has been
identified as a modulator of intracellular signalling and it
mediates various cellular and biological processes, including the
cell cycle, cell proliferation, apoptosis, cell renewal, gene
expression and oncogenesis (15). In
the present study, it was found that the expression of MELK was
higher in tumour tissues compared with that in normal lung tissues,
and that increased MELK expression was associated with a poor
prognosis in patients with LUAD. The findings of the present study
are consistent with those of previous studies, indicating that MELK
may be a promising therapeutic target in LUAD (1,13,24).
The expression of MELK is negatively associated with
the survival rate of patients with cancer (7–9,11,25). In
addition, the selective MELK inhibitor OTS167 has been shown to
inhibit proliferation and promote apoptosis in a variety of
tumours, leading to improved prognosis (8,9). Hence,
MELK has been identified as a novel marker for predicting cancer
outcomes and as a potential therapeutic target for some cancer
types. Notably, MELK is 1 of 16 hub genes from the GEO database
that were significantly associated with a worse 5-year OS rate of
patients with lung cancer analysed using GEPIA (1). In addition, Zang et al (15) found that MELK mRNA expression level
in tumour tissues was significantly higher compared with that in
normal/benign tissue analysed using data from the TCGA and GEO
databases, and that it was negatively associated with prognosis in
patients with LUAD when subjected to Kaplan-Meier analysis. Inoue
et al (24) confirmed that
inhibition of the expression of MELK increased apoptosis and
decreased the proliferation of SCLC cells. To further identify the
role of MELK in LUAD and approve its use as a biomarker, additional
data are needed for validation, including data from different
databases and clinical samples. In the present study, the mRNA and
protein expression levels of MELK in LUAD were analysed using
Oncomine, TCGA and GEPIA, and immunohistochemistry, respectively.
In the present study, results from the bioinformatics and
immunohistochemistry analyses suggested that the MELK expression
was higher in LUAD tissues compared with that in normal lung
tissues. In the present study, no association was found between
MELK expression and sex, age, tumour location, tumour size,
histological differentiation, T stage or N stage in clinical
samples. However, in TCGA samples, males had higher expression of
MELK compared with females and there was no difference observed
between normal tissue and tumour tissue from patients with LUAD
21–40 years old. In GEPIA samples, the levels of MELK increased
gradually with tumour grade. These difference between clinical
samples and databases may be due to the small sample size. In
addition, in the present study, Kaplan-Meier, GEPIA and TCGA
survival analyses were all used to evaluate prognosis and the
result suggested that high expression of MELK was associated with
poor OS probability in patients with LUAD. The findings of the
present study and those of previous studies are consistent, and all
demonstrate that MELK may serve as a potential diagnostic biomarker
or therapeutic target for lung cancer (1,13,15,24).
MELK has been reported to participate in tumour
progression via the JNK, p53, Bcl-G and forkhead box protein M1
(FOXM1) signalling pathways, which are all extremely important in
multiple human cancer types (8,25–28). Gu
et al (28) indicated that
MELK regulates glioma cell growth via the JNK/p53 pathway. MELK
triggers carcinogenesis through inhibition of the pro-apoptotic
protein Bcl-G in breast cancer and hepatocarcinoma (25,27).
Inhibition of MELK induces G2/M arrest and reduces cell
proliferation by decreasing FoxM1 phosphorylation and upregulating
p53 expression in chronic lymphocytic leukaemia (8). In addition, MELK is also reported to
have critical roles in the formation or maintenance of cancer stem
cells, which are associated with cancer recurrence (11,12).
Researchers have also demonstrated that high levels of MELK
contribute to cancer progression and stem cell maintenance in SCLC,
and that inhibition of MELK increases apoptosis and suppresses the
growth of cancer cells by reducing the activity of FoxM1 (24). However, the molecular mechanism of
MELK in the progression of LUAD remains unclear. Whether the action
of MELK in LUAD is the same as that in SCLC and other cancer types
needs further investigation.
The present study has several limitations. Firstly,
only 75 LUAD samples were used and a larger sample size is needed
for future research. Secondly, only bioinformatics analysis was
used to calculate the survival rate of patients with LUAD with high
and low expression of MELK and further studies are required to
verify the findings of the present study and increase accuracy.
Thirdly, the present study mainly discussed MELK expression and
prognosis in patients with LUAD, and the detailed molecular
mechanism of MELK involvement in LUAD was not explored. All these
limitations need to be investigated in future studies.
In summary, the present study demonstrated that MELK
is highly expressed in LUAD and that there is a negative
association between MELK expression and prognosis in affected
patients. MELK may serve as a potential diagnostic marker and
therapeutic target in LUAD; however, more studies are needed to
investigate the potential mechanisms of MELK in this disease.
Acknowledgements
Not applicable.
Funding
This study was supported by the Science and
Technology Plan Project of Jiangxi Administration of Traditional
Chinese Medicine, China (grant no. 2019A067).
Availability of data and materials
The three datasets generated and/or analyzed during
the current study are available in the Oncomine databases
(https://www.oncomine.org), The Cancer Genome
Atlas database (https://tcga-data.nci.nih.gov/tcga/), the GEPIA
database (http://gepia.cancer-pku.cn) and
Kaplan-Meier Plotter database (http://kmplot.com/analysis).
Authors' contributions
SC and ZX designed the study. ZL, XC, XW, HT and LY
collected the data. SC, LY, and HT analysed the data. SC, LY and ZX
drafted and revised the manuscript for important intellectual
content. All authors have read and approved the manuscript.
Ethics approval and consent to
participate
The research was approved by the Ethical Committee
of Jiangxi Provincial People's Hospital (Nanchang, China) (approval
no. 2018070), informed consent was provided by the clinicians and
obtained from the patients who provided written informed consent
for the usage of the tissues for research.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
LUAD
|
lung adenocarcinoma
|
|
NSCLC
|
non-small cell lung cancer
|
|
MELK
|
maternal embryonic leucine zipper
kinase
|
|
GEPIA
|
Gene Expression Profiling Interactive
Analysis
|
|
TCGA
|
The Cancer Genome Atlas
|
References
|
1
|
Li Z, Sang M, Tian Z, Liu Z, Lv J, Zhang F
and Shan B: Identification of key biomarkers and potential
molecular mechanisms in lung cancer by bioinformatics analysis.
Oncol Lett. 18:4429–4440. 2019.PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sharma SV, Bell DW, Settleman J and Haber
DA: Epidermal growth factor receptor mutations in lung cancer. Nat
Rev Cancer. 7:169–181. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Giuliano CJ, Lin A, Smith JC, Palladino AC
and Sheltzer JM: MELK expression correlates with tumor mitotic
activity but is not required for cancer growth. Elife. 7:2018.
|
|
5
|
Luo J, Solimini NL and Elledge SJ:
Principles of cancer therapy: Oncogene and non-oncogene addiction.
Cell. 136:823–837. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jiang P and Zhang D: Maternal embryonic
leucine zipper kinase (MELK): A novel regulator in cell cycle
control, embryonic development, and cancer. Int J Mol Sci.
14:21551–21560. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Speers C, Zhao SG, Kothari V, Santola A,
Liu M, Wilder-Romans K, Evans J, Batra N, Bartelink H, Hayes DF, et
al: Maternal embryonic leucine zipper kinase (MELK) as a novel
mediator and biomarker of radioresistance in human breast cancer.
Clin Cancer Res. 22:5864–5875. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang Y, Zhou X, Li Y, Xu Y, Lu K, Li P
and Wang X: Inhibition of maternal embryonic leucine zipper kinase
with OTSSP167 displays potent anti-leukemic effects in chronic
lymphocytic leukemia. Oncogene. 37:5520–5533. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kohler RS, Kettelhack H,
Knipprath-Mészaros AM, Fedier A, Schoetzau A, Jacob F and
Heinzelmann-Schwarz V: MELK expression in ovarian cancer correlates
with poor outcome and its inhibition by OTSSP167 abrogates
proliferation and viability of ovarian cancer cells. Gynecol Oncol.
145:159–166. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xia H, Kong SN, Chen J, Shi M, Sekar K,
Seshachalam VP, Rajasekaran M, Goh BKP, Ooi LL and Hui KM: MELK is
an oncogenic kinase essential for early hepatocellular carcinoma
recurrence. Cancer Lett. 383:85–93. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ganguly R, Hong CS, Smith LGF, Kornblum HI
and Nakano I: Maternal embryonic leucine zipper kinase: Key kinase
for stem cell phenotype in glioma and other cancers. Mol Cancer
Ther. 13:1393–1398. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chung S and Nakamura Y: MELK inhibitor,
novel molecular targeted therapeutics for human cancer stem cells.
Cell Cycle. 12:1655–1656. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chung S, Suzuki H, Miyamoto T, Takamatsu
N, Tatsuguchi A, Ueda K, Kijima K, Nakamura Y and Matsuo Y:
Development of an orally-administrative MELK-targeting inhibitor
that suppresses the growth of various types of human cancer.
Oncotarget. 3:1629–1640. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li Y, Tang H, Sun Z, Bungum AO, Edell ES,
Lingle WL, Stoddard SM, Zhang M, Jen J, Yang P and Wang L:
Network-based approach identified cell cycle genes as predictor of
overall survival in lung adenocarcinoma patients. Lung Cancer.
80:91–98. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zang X, Qian C, Ruan Y, Xie J, Luo T, Xu B
and Jiang J: Higher maternal embryonic leucine zipper kinase mRNA
expression level is a poor prognostic factor in non-small-cell lung
carcinoma patients. Biomark Med. 13:1349–1361. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bornstein S, Schmidt M, Choonoo G, Levin
T, Gray J, Thomas CJ Jr, Wong M and McWeeney S: IL-10 and integrin
signaling pathways are associated with head and neck cancer
progression. BMC Genomics. 17:382016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dastsooz H, Cereda M, Donna D and Oliviero
S: A comprehensive bioinformatics analysis of UBE2C in cancers. Int
J Mol Sci. 20:22282019. View Article : Google Scholar
|
|
19
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu L, Chen S, Bao H, Zhang W, Liao M,
Liang Q and Cheng X: The role of lncRNA CASC2 on prognosis of
malignant tumors: A meta-analysis and bioinformatics. Onco Targets
Ther. 11:4355–4365. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Detterbeck FC, Chansky K, Groome P,
Bolejack V, Crowley J, Shemanski L, Kennedy C, Krasnik M, Peake M
and Rami-Porta R; IASLC Staging and Prognostic Factors Committee,
Advisory Boards, Participating Institutions, : The IASLC Lung
Cancer Staging Project: Methodology and validation used in the
development of proposals for revision of the stage classification
of NSCLC in the forthcoming (eighth) edition of the tnm
classification of lung cancer. J Thorac Oncol. 11:1433–1446. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dadhania V, Zhang M, Zhang L, Bondaruk J,
Majewski T, Siefker-Radtke A, Guo CC, Dinney C, Cogdell DE, Zhang
S, et al: Meta-analysis of the luminal and basal subtypes of
bladder cancer and the identification of signature
immunohistochemical markers for clinical use. EBioMedicine.
12:105–117. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hu DD, Li PC, He YF, Jia W and Hu B:
Overexpression of coiled-coil domain-containing protein 34 (CCDC34)
and its correlation with angiogenesis in esophageal squamous cell
carcinoma. Med Sci Monit. 24:698–705. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Inoue H, Kato T, Olugbile S, Tamura K,
Chung S, Miyamoto T, Matsuo Y, Salgia R, Nakamura Y and Park JH:
Effective growth-suppressive activity of maternal embryonic
leucine-zipper kinase (MELK) inhibitor against small cell lung
cancer. Oncotarget. 7:13621–13633. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hiwatashi K, Ueno S, Sakoda M, Iino S,
Minami K, Yonemori K, Nishizono Y, Kurahara H, Mataki Y, Maemura K,
et al: Expression of maternal embryonic leucine zipper kinase
(MELK) correlates to malignant potentials in hepatocellular
carcinoma. Anticancer Res. 36:5183–5188. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu S, Chen X, Hu C, Wang J, Shen Y and
Zhong Z: Up-regulated maternal embryonic leucine zipper kinase
predicts poor prognosis of hepatocellular carcinoma patients in a
Chinese Han population. Med Sci Monit. 23:5705–5713. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lin ML, Park JH, Nishidate T, Nakamura Y
and Katagiri T: Involvement of maternal embryonic leucine zipper
kinase (MELK) in mammary carcinogenesis through interaction with
Bcl-G, a pro-apoptotic member of the Bcl-2 family. Breast Cancer
Res. 9:R172007. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gu C, Banasavadi-Siddegowda YK, Joshi K,
Nakamura Y, Kurt H, Gupta S and Nakano I: Tumor-specific activation
of the C-JUN/MELK pathway regulates glioma stem cell growth in a
p53-dependent manner. Stem Cells. 31:870–881. 2013. View Article : Google Scholar : PubMed/NCBI
|