Introduction
Bladder cancer is one of the most common
genitourinary malignancies in the world (1). Complete resection is the primary
treatment strategy for patients with bladder cancer; however, a
study in Germany reported that the 5-year overall survival rate of
patients with bladder cancer after radical cystectomy was <60%
(2).
Ribosome-binding protein 1 (RRBP1) is an endoplasmic
reticulum (ER) membrane protein that is critical for ribosome
binding and the transportation of nascent proteins (3). RRBP1 inhibition could cause ER stress
and significantly reduce cell tumorigenicity (4) RRBP1 was reported to be overexpressed in
lung, colorectal, endometrial and prostate cancer (4–7). High
RRBP1 expression could promote esophageal and colorectal cancer
progression, and can be used to predict patient prognosis (5,8).
However, to the best of our knowledge, the expression of RRBP1 in
bladder cancer has not been previously reported.
C-C chemokine receptor type 7 (CCR7) is a cell
surface chemokine receptor (9).
Increasing evidence has demonstrated that CCR7 is involved in the
development and progression of bladder (10), lung (11), breast (12) and colorectal (13) cancer. Therefore, it was hypothesized
that downregulation of RRBP1 may reduce CCR7 expression leading to
inhibition of bladder cancer migration and invasion.
The present study aimed to investigate the role of
RRBP1 in bladder cancer. The Cancer Genome Atlas (TCGA) database
was searched and analyzed to compare RRBP1 mRNA expression levels
between bladder cancer and healthy tissues, and the overall
survival according to these levels. RRBP1-knockdown was used to
analyze the expression levels of RRBP1 and different cell functions
in bladder cancer cell lines. Finally, the association between
RRBP1 and CCR7 was investigated by comparing CCR7 expression
between control and RRBP1-knockdown cells.
Materials and methods
Bioinformatics analysis
RRBP1 mRNA expression and overall survival in
bladder cancer tissues and healthy tissues in TCGA were analyzed
using the University of Alabama Cancer Database (ualcan.path.uab.edu/analysis.html)
(14). Subsequently, RRBP1 mRNA
expression at different tumor and lymph node stages was analyzed
using LinkedOmics (www.linkedomics.org/admin.php) (15).
Immunohistochemistry (IHC)
The present study was approved by the Ethics
Committee of Beijing Chaoyang Hospital (approval no. 2020-3-18-85).
All subjects provided written informed consent after oral consent
to participate in the present study. The clinical data of the
patients are presented in Table SI.
All surgical specimens were fixed with 10% formalin (10:1 ratio of
formalin to tissue) at room temperature for 24 h and then embedded
in paraffin. The bladder cancer tissues were pathologically
confirmed and para-carcinoma tissues were obtained ~2 cm from the
tumor margin. The section thickness was 3–4 µm. Deparaffinization
and rehydration were performed according to standard procedures
(16). After blocking with 5% bovine
serum albumin (cat. no. G5001; Wuhan Servicebio Technology Co.,
Ltd.) at room temperature for 30 min, sections were incubated
overnight at 4°C with an anti-RRBP1 primary antibody (1:500; cat.
no. ab95983; Abcam) and then an HRP-conjugated secondary antibody
(1:2,000; cat. no. sc-2004; Santa Cruz Biotechnology, Inc.) for 50
min at room temperature. Chromogen detection was performed using
DAB reagent (cat. no. G1211; Wuhan Servicebio Technology Co., Ltd.)
for 3–5 min. Subsequently, the slices were rinsed with running
water and counterstained with haematoxylin (Wuhan Servicebio
Technology Co., Ltd.) for 3 min at room temperature. The expression
levels were evaluated by two pathologists blinded to the clinical
characteristics of the patients according to proportion of cell
staining (0, 0%; 1, ≤25%; 2, 26–50%; 3, 51–75%; and 4, >75%) and
the staining intensity (0, no staining; 1, weak staining; 2,
moderate staining; and 3, strong staining) using a light microscope
(magnification, ×400). The final score was calculated by
multiplying the cell staining proportion score and the staining
intensity score. If the final score was >9, protein expression
was considered high, but if the final score was ≤8, protein
expression was considered low.
Cell lines and cell culture
Bladder cancer cell lines (5637, T24 and UM-UC-3)
and an immortalized urothelial cell line (SV-HUC-1) were obtained
from China Infrastructure of Cell Line Resources, Institute of
Basic Medical Sciences, Chinese Academy of Medical Sciences. All
cells were cultured at 37°C with 5% CO2 and 95%
O2. SV-HUC-1 cells were cultured in a 1:1 mixture of
DMEM and Ham's F12 medium (both HyClone; Cytiva). 5637, T24 and
UM-UC-3 cells were cultured in RPMI-1640 medium (HyClone; Cytiva)
supplemented with 10% FBS (HyClone; Cytiva), 100 U/ml penicillin G
and 100 µg/ml streptomycin (Lonza Group, Ltd.).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from cells using
TRIzol® reagent (cat. no. 3101-100; Shanghai Pufei
Biotechnology Co., Ltd.). Total RNA was reverse transcribed into
cDNA using M-MLV reverse transcriptase (cat. no. M1705; Promega
Corporation). The Oligo dT was provided by Sangon Biotech Co., Ltd.
(cat. no. B0205) and the dNTPs (10 mM) were provided by Promega
Corporation (cat. no. U1240). After heat treatment of RNA/primer
mixture at 70°C for 10 min and cooling immediately on ice for 10
min, the RT reaction was processed at 42°C for 1 h and 70°C for 10
min. Subsequently, qPCR was performed using ExTaq (Takara Bio,
Inc.). The SYBR Master Mixture was provided by Takara Bio, Inc.
(cat. no. DRR041B). The following primer were used for the qPCR:
RRBP1 forward, 5′-GAGATGGCGAAAACTCACCAC-3′ and reverse,
5′-CTCGAAGGAGGACAGTCACAT-3′; CCR7 forward,
5′-TTCATCGGCGTCAAGTTCC-3′ and reverse, 5′-AAGGTGGTGGTGGTCTCG-3′;
and GAPDH forward, 5′-TGACTTCAACAGCGACACCCA-3′ and reverse,
5′-CACCCTGTTGCTGTAGCCAAA-3′. The following thermocycling conditions
were used for the qPCR: Initial denaturation at 95°C for 30 sec;
followed by 40 cycles of 95°C for 5 sec; and final extension at
60°C for 30 sec. mRNA expression levels were quantified using the
2−ΔΔCq method (17) and
normalized to the internal reference gene GAPDH.
Western blotting
Total protein was extracted from cells using RIPA
lysis buffer (cat. no. P0013B; Beyotime Institute of Technology).
Protein concentration was determined using a BCA Protein Assay kit
(cat. no. P0010S; Beyotime Institute of Biotechnology). Proteins
(50 µg) were separated via 10% SDS-PAGE and transferred to PVDF
membranes, which were blocked with 5% non-fat milk powder. The
membranes were incubated overnight at 4°C with primary antibodies
targeted against the following: RRBP1 (1:500; cat. no. ab95983;
Abcam), CCR7 (1:1,000; cat. no. ab32527; Abcam) and GAPDH (1:2,000;
cat. no. ab8245; Abcam). Subsequently, they were incubated with
HRP-conjugated anti-rabbit IgG (1:2,000; cat. no. sc-2004; Santa
Cruz Biotechnology, Inc.) and anti-mouse IgG (1:2,000; cat. no.
sc-2005; Santa Cruz Biotechnology, Inc.) secondary antibodies.
Protein bands were visualized using an ECL kit (cat. no.
M3121/1859022; Thermo Fisher Scientific, Inc.). GAPDH was used as
the loading control. Image J (version 1.51n; National Institutes of
Health) was used to quantify blots.
Lentivirus construction and
infection
Lentivirus RRBP1 short hairpin RNA (shRNA) and
negative control shRNA both containing GFP were purchased from
Shanghai GeneChem Co., Ltd. The target sequences of the shRNAs were
as follows: shRNA-1, 5′-GCAAGCCAGGATGGATATTTA-3′; shRNA-2,
5′-GCCAAGAAGAAGTCTGGTTCA-3′; and negative control shRNA,
5′-TTCTCCGAACGTGTCACGT-3′. T24 cells were seeded in a six-well
plate at 30% confluence, the medium was removed and replaced with 2
ml complete medium containing Polybrene (cat. no. sc-134220; Santa
Cruz Biotechnology, Inc.) at a final concentration of 5 µg/ml. T24
cells were transfected with negative control or RRBP1 shRNA
(MOI=10) according to the manufacturer's instructions (Shanghai
GeneChem Co., Ltd). The volume and concentration of control,
shRNA-1 and shRNA-2 lentivirus were 2 µl 1×109
transduction units (TU)/ml, 3.33 µl 6×108 TU/ml and 3.33
µl 6×108 TU/ml, respectively. After 16 h of infection,
the medium was changed to complete medium. At 72 h
post-transfection, transfection efficiency was assessed via RT-qPCR
and western blotting.
Migration and invasion assays
Cell invasion was assessed using Matrigel-coated
Transwell chambers (cat. no. 354480; Corning Inc.) and cell
migration was assessed using Transwell chambers. Cells were seeded
(1×105 cells/well) in DMEM without FBS into the upper
chamber. DMEM (600 µl) supplemented with 30% FBS was plated into
the lower chambers. Following incubation at 37°C for 16 h,
non-invading/migratory cells were removed using cotton swabs and
invading/migratory cells were fixed with 4% methanol for 30 min at
room temperature. Following staining with 0.5% crystal violet at
room temperature for 5 min, invading/migratory cells were counted
using a light microscope (magnification, ×100 and ×200).
For the wound healing assay, T24 cells
(5×104) were seeded into a 96-well plate and at 90%
confluence, a single scratch was made in the middle of each well.
GFP-transfected cells were incubated in DMEM supplemented with 1%
FBS at 37°C. Cell migration was observed using a Celigo Image
cytometer (Nexcelom Bioscience LLC) at 0, 8 and 24 h, and the
migration distance was automatically measured. The Celigo Image
cytometer identified and scanned GFP-positive cells. The images
captured by the image cytometer were equivalent to 100× the same
resolution of the microscope.
Statistical analysis
Statistical analyses were performed using Stata
software (version 14.0; StataCorp LP) and GraphPad Prism software
(version 7; GraphPad Software, Inc.). Data are presented as the
mean ± SD. The unpaired Student's t-test was used to analyze
comparisons between two groups. The Kruskal-Wallis test was used to
analyze comparisons between different tumor stages and lymph node
metastasis status, followed by the Nemenyi test for individual
comparisons among multiple groups of tumor stages. One-way ANOVA
with Bonferroni test was used to analyze comparisons among multiple
groups in the cell experiments. P<0.05 was considered to
indicate a statistically significant difference.
Results
RRBP1 is highly expressed in bladder
cancer and is correlated with patient prognosis
To investigate whether RRBP1 served a role in
bladder cancer, the TCGA database (14) was searched and LinkedOmics was used
to analyze multi-omics data (15).
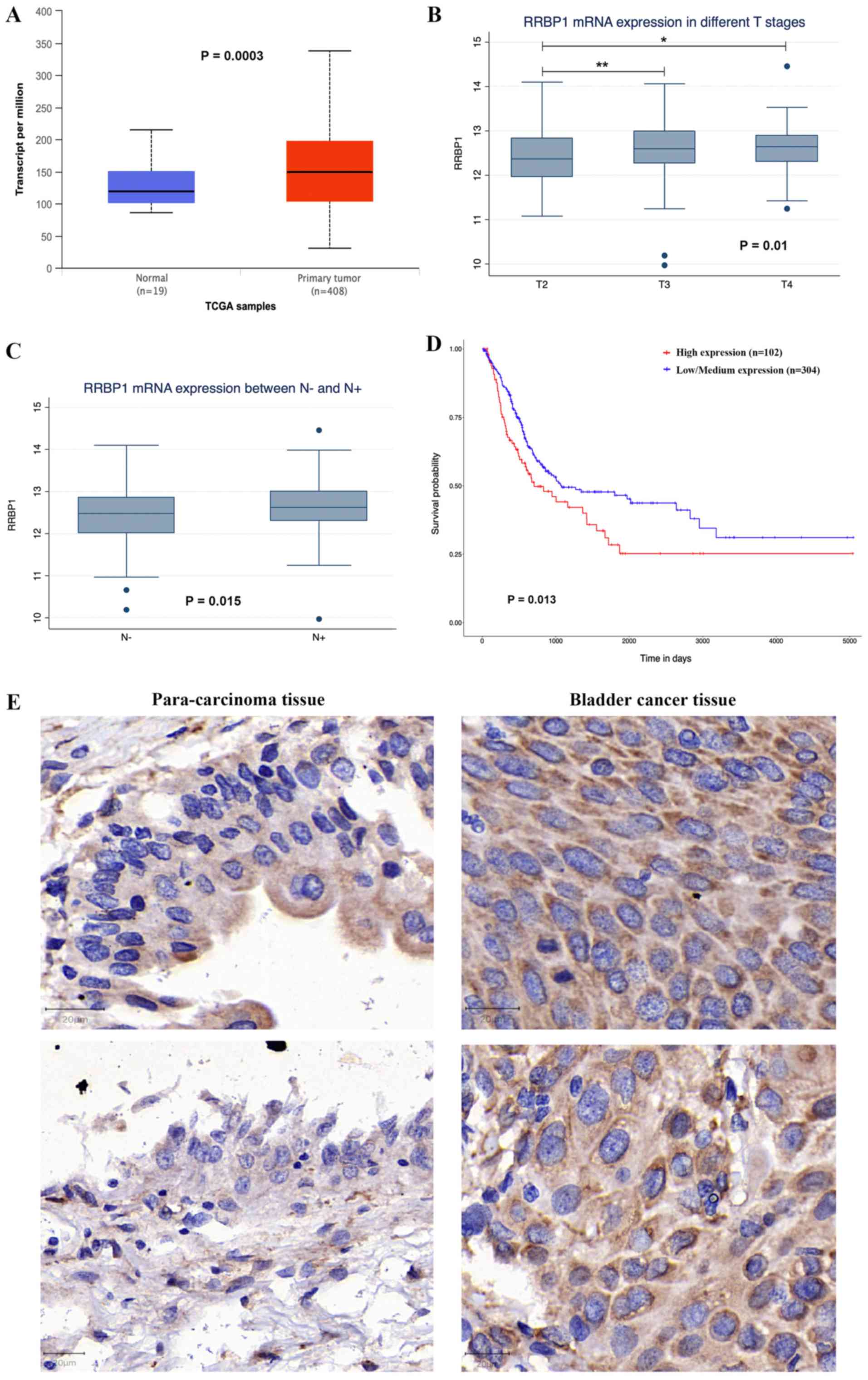
RRBP1 expression was significantly upregulated in bladder cancer
tissues compared with healthy bladder tissues (P=0.0003; Fig. 1A). High RRBP1 expression was
associated with tumor stage (P=0.01) and lymph node metastasis
(P=0.015; Fig. 1B and C). The IHC
results indicated that RRBP1 protein expression was higher in
bladder cancer tissues compared with para-carcinoma tissues
(Figs. 1E and S1). Moreover, patients with high RRBP1
expression displayed shorter overall survival compared with
patients with low and medium RRBP1 expression in the UALCAN
database (P=0.013; Fig. 1D).
RRBP1 overexpression in bladder cancer
cells
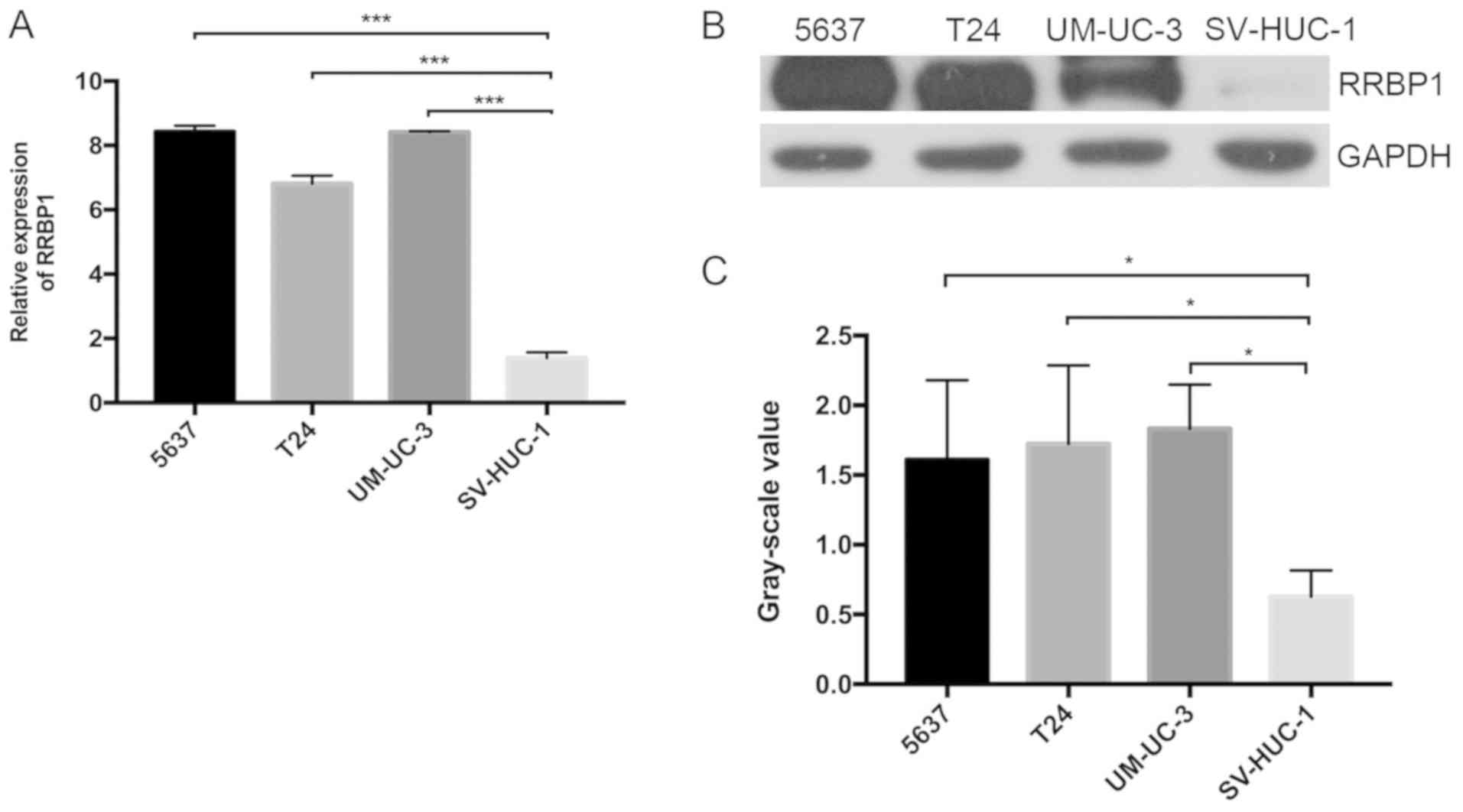
RRBP1 expression was also detected in bladder cancer
cell lines (5637, T24 and UM-UC-3) and SV-HUC-1 cells via RT-qPCR.
SV-HUC-1 is an epithelial SV40 cell line that originated from the
ureter. RRBP1 mRNA expression was significantly higher in 5637
(P<0.01), T24 (P<0.01) and UM-UC-3 cells (P<0.01) compared
with SV-HUC-1 cells (Fig. 2A).
Similarly, RRBP1 protein expression levels were significantly
higher in bladder cancer cell lines compared with SV-HUC-1 cells
(P<0.05; Fig. 2B and C). The
results indicated that RRBP1 was highly expressed in bladder
cancer, which may indicate that it serves a role in
tumorigenesis.
RRBP1 knockdown in a high-expressing
bladder cancer cell
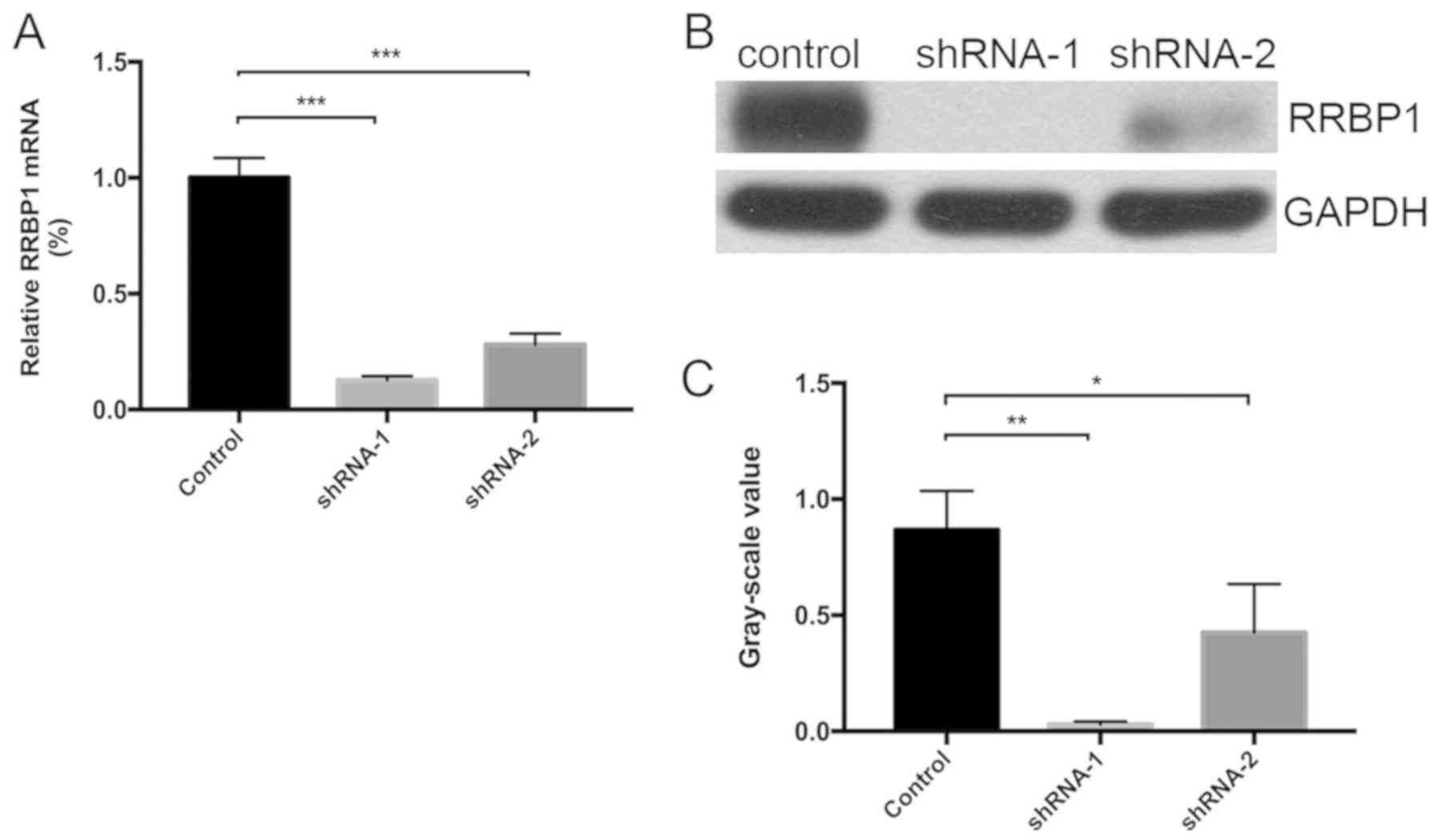
T24 cells were infected with RRBP1 shRNA-containing
lentiviruses, shRNA-1 or shRNA-2. RRBP1 knockdown efficiency was
assessed via western blotting and RT-qPCR. Compared with the
control group, shRNA-1 significantly inhibited RRBP1 expression by
87.3% at the mRNA level (P<0.001) and by 96.55% at the protein
level (P=0.002), whereas shRNA-2 significantly inhibited RRBP1
expression by 72.0% at the mRNA level (P<0.001) and by 50.57% at
the protein level (P=0.038) (Fig.
3).
RRBP1 knockdown impairs T24 cell
migration and invasion
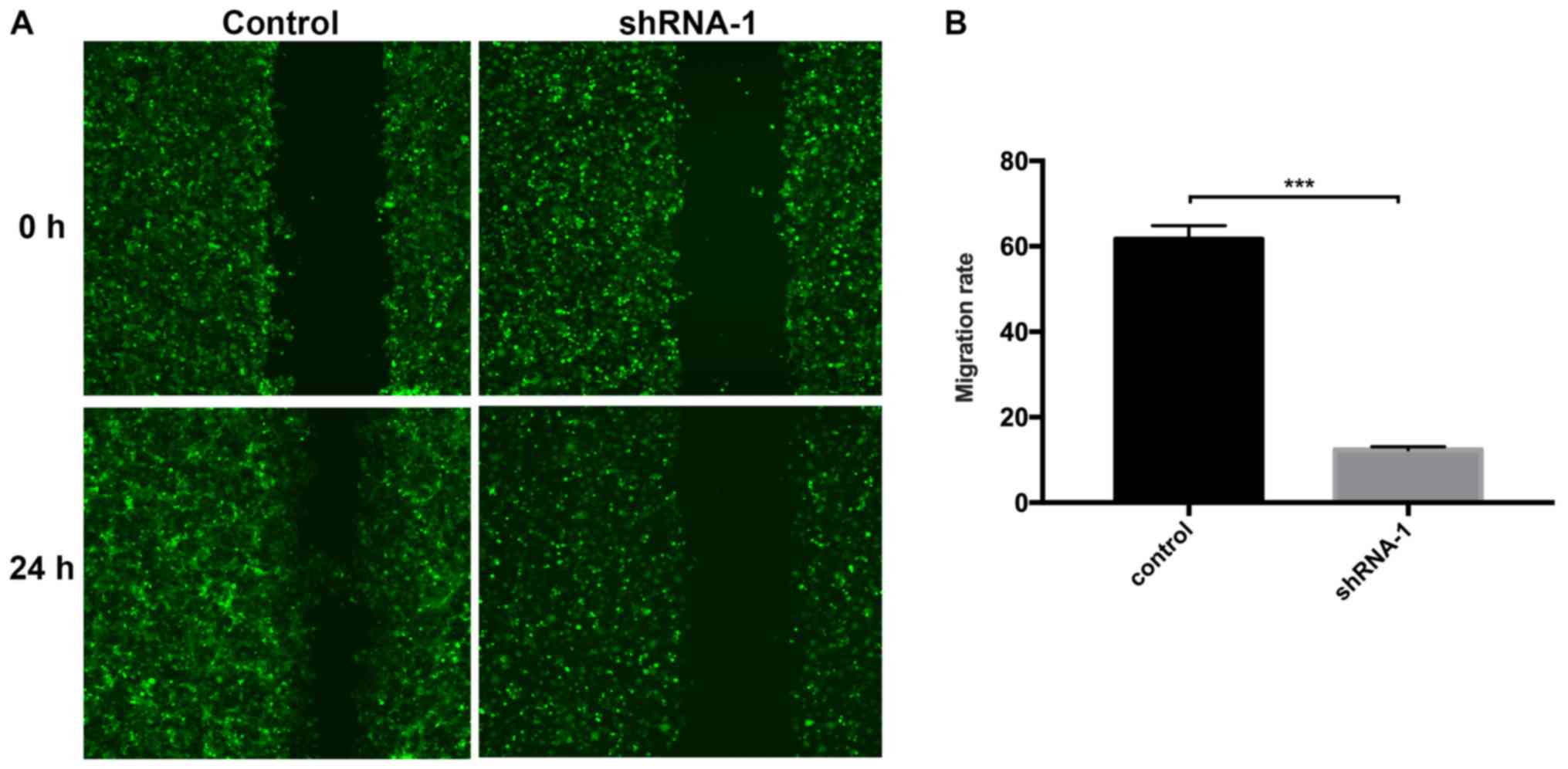
Subsequently, the effect of RRBP1 on T24 cell
migration and invasion was assessed. The wound healing assay
indicated that T24 cell migration was significantly reduced
following shRNA-1 transfection compared with the control group
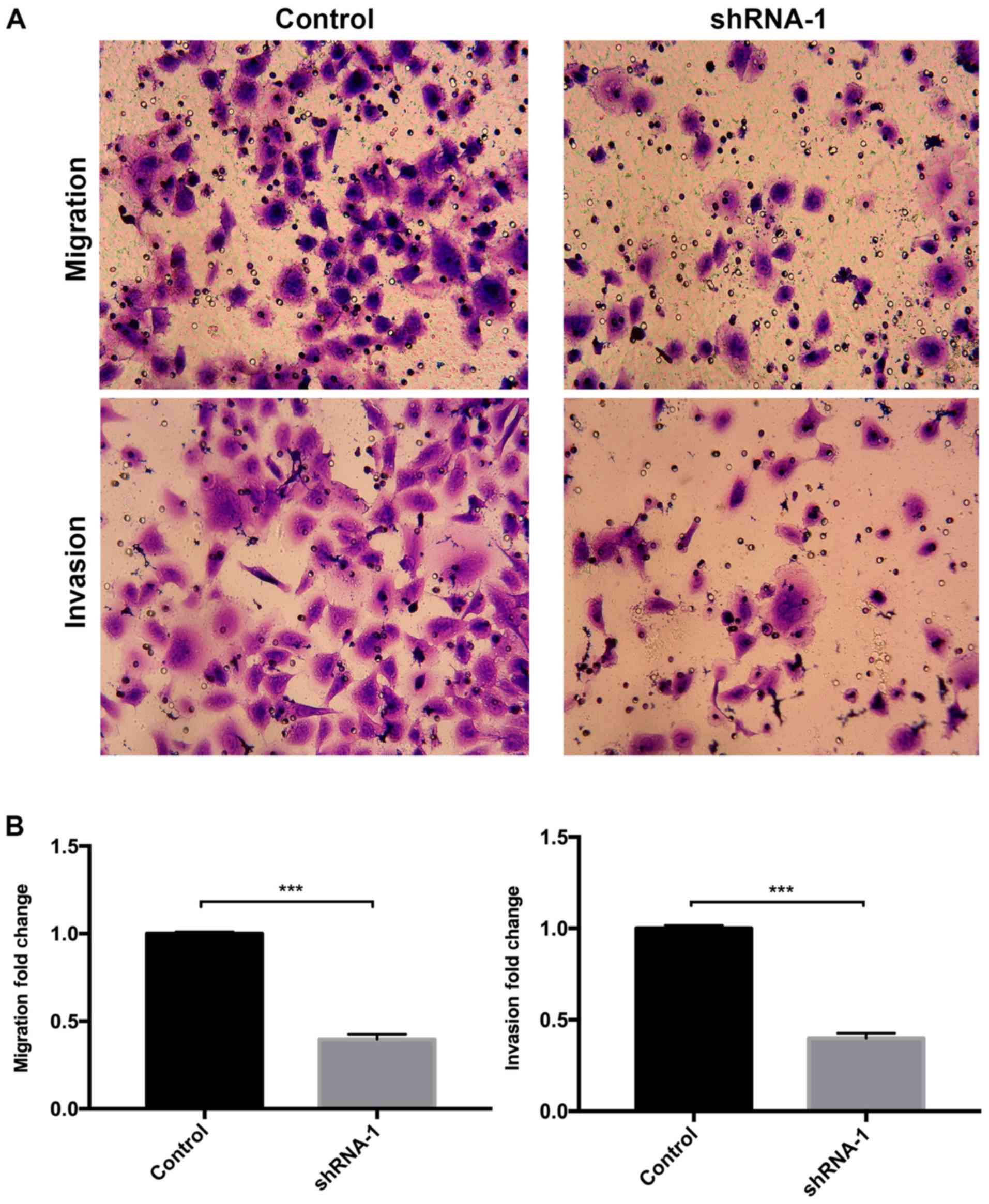
(Fig. 4). Similarly, the Transwell
assay results suggested that RRBP1 knockdown by shRNA-1
significantly inhibited T24 cell migration compared with the
control group (Fig. 5A and B). In
addition, RRBP1 knockdown by shRNA-1 significantly decreased T24
cell invasion compared with the control group (Fig. 5A and B).
RRBP1 knockdown suppresses T24 cell
migration and invasion via suppression of CCR7
Previous studies indicated that CCR7 could enhance
bladder cancer proliferation, migration and invasion (18–20).
Therefore, the association between RRBP1 and CCR7 was investigated.
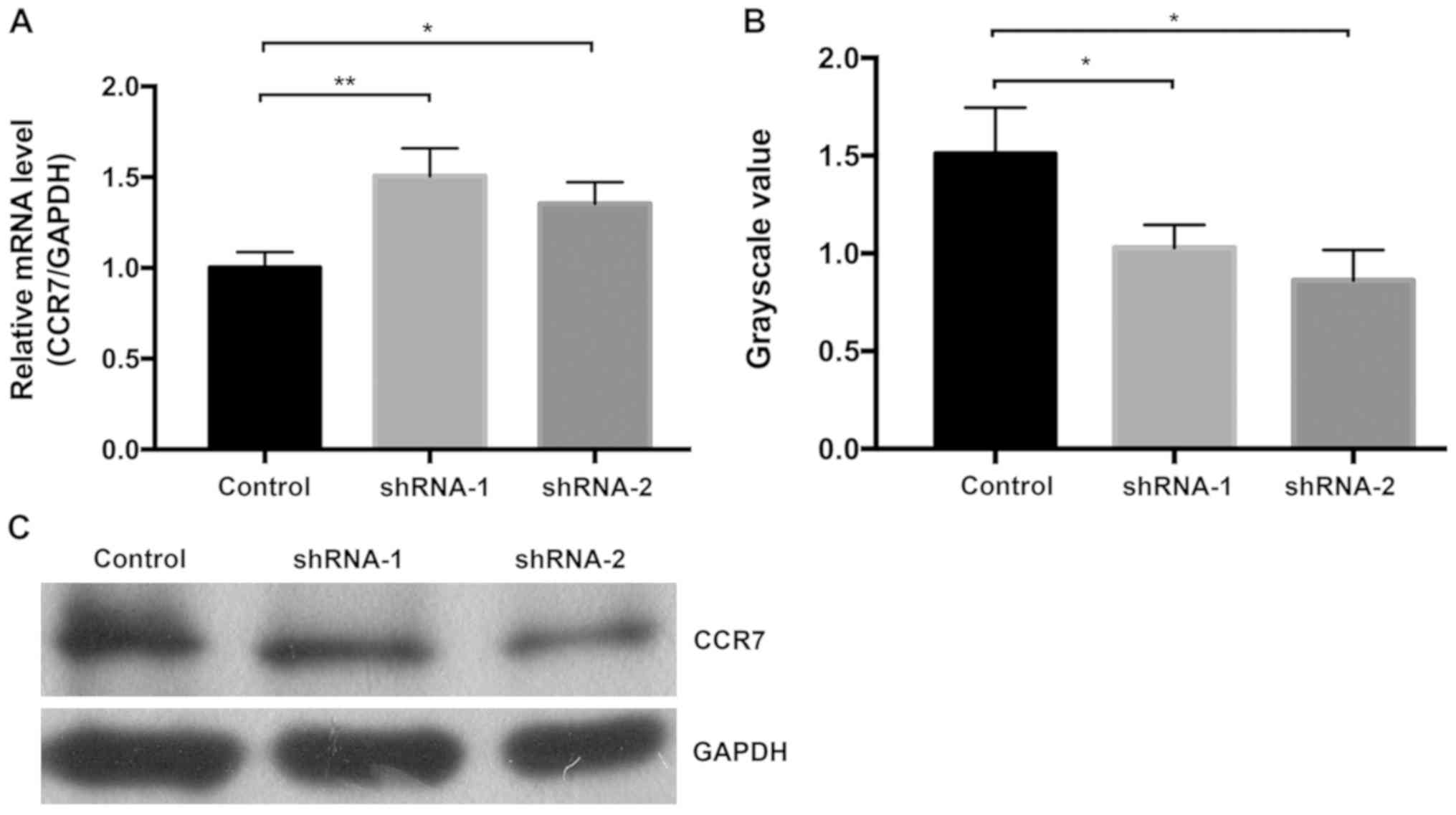
Compared with the control group, the mRNA expression levels of CCR7
were significantly increased after RRBP1 knockdown (P=0.007 vs.
shRNA-1 and P=0.036 vs. shRNA-2; Fig.
6A); however, the protein expression levels of CCR7 were
significantly decreased after RRBP1 knockdown (P<0.05; Fig. 6B and C). The results indicated that
RRBP1 knockdown might suppress CCR7 mRNA translation and inhibit
bladder cancer cell migration and invasion.
Discussion
To the best of our knowledge, the present study was
the first to investigate RRBP1 expression in bladder cancer. To
investigate the association between RRBP1 expression and bladder
cancer, the TCGA database was searched. RRBP1 mRNA expression was
significantly higher in bladder cancer tissues compared with
healthy bladder tissues, and RRBP1 expression was associated with
tumor stage, lymph node metastasis and overall survival. RRBP1 was
also upregulated in bladder cancer cells compared with normal
urothelial cells. RRBP1 knockdown inhibited bladder cancer cell
migration and invasion compared with the control group. CCR7 mRNA
expression levels were significantly increased following RRBP1
knockdown compared with the control group. By contrast, the
expression of CCR7 protein was significantly decreased by RRBP1
knockdown compared with the control group, which indicated that
RRBP1 knockdown might suppress CCR7 mRNA translation and inhibit
bladder cancer cell migration and invasion.
A number of studies have demonstrated that RRBP1
overexpression correlated with the progression and prognosis of
various types of cancer. Pan et al (5) examined RRBP1 expression via IHC in
colorectal tissues and reported that RRBP1 was aberrantly
overexpressed in colorectal cancer. Compared with patients with low
RRBP1 expression, patients with high RRBP1 expression had shorter
disease-specific survival. RRBP1 was also highly expressed in
esophageal carcinoma tissues and was associated with T stage, lymph
node metastasis, TNM stage and survival (8). Similar studies were performed in
endometrial endometrioid adenocarcinoma and prostate cancer, and
RRBP1 was recognized as a potential protein marker (6,7).
However, the association between RRBP1 and bladder cancer is not
completely understood. In the present study, RRBP1 was upregulated
in bladder cancer tissues compared with para-carcinoma tissues,
which was associated with bladder cancer cell migration and
invasion.
RRBP1 was originally identified as a
ribosome-binding protein on the rough ER. RRBP1 has been studied in
yeast, where it is a member of the ER stress response and
associated unfolded protein response (UPR) (21). shRNA-mediated RRBP1 inhibition caused
ER stress and significantly reduced cell tumorigenicity, which was
related to a significant downregulation of glucose-regulated
protein 78 (GRP78) (4). GRP78, a
modulator of UPR, is an antiapoptotic protein, which is widely
upregulated in cancer and serves a key role in chemotherapy
resistance in several types of cancer (22).
RRBP1 promotes the ribosome-independent localization
of a subset of mRNA to the ER (23).
p180 is required for efficient ER-anchoring of bulk poly(A) and
certain transcripts, such as placental alkaline phosphatase and
calreticulin, to the ER (23). The
present study indicated that RRBP1 knockdown increased CCR7 mRNA
and suppressed CCR7 protein expression compared with the control
group, which might be due to RRBP1 knockdown restricting the
combination between CCR7 mRNA and ER. Therefore, further research
should be performed.
The present study had several limitations. Firstly,
the clinicopathological characteristics of RRBP1 were analyzed
using TCGA data. Therefore, the association between RRBP1
expression and the clinicopathological characteristics of patients
requires further investigation. Secondly, multivariate analysis
should be performed to confirm whether high RRBP1 expression is an
independent prognostic factor in the clinical data. Thirdly, a
rescue experiment was not conducted to validate the role of CCR7.
Fourthly, the lack of additional cell lines in the functional
experiments was also a limitation.
In conclusion, the present study indicated that
RRBP1 was associated with bladder cancer cell migration, invasion
and prognosis. RRBP1 may serve as a key player in the maintenance
of tumor tumorigenesis, and CCR7 might serve a role in the process.
The role of RRBP1 in bladder cancer tumorigenesis requires further
investigation.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Kunning Zhang and
Dr Xingran Jiang both from the Department of Pathology of Beijing
Chaoyang Hospital (Beijing, China) for evaluating the IHC
staining.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81772700) and the
Beijing Natural Science Foundation (grant no. 7194271).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HP and NX analyzed the data. MW contributed to
writing the manuscript and performing the statistical analyses. MW,
SL and BZ performed the experiments and collected the data. MW and
JW analyzed and interpreted the data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The present study was approved by Beijing Chaoyang
Hospital's Institutional Research Ethics Board (approval no.
2020-3-18-85). All subjects provided written informed consent after
oral consent to participate in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hautmann RE, de Petriconi RC, Pfeiffer C
and Volkmer BG: Radical cystectomy for urothelial carcinoma of the
bladder without neoadjuvant or adjuvant therapy: Long-term results
in 1100 patients. Eur Urol. 61:1039–1047. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Savitz AJ and Meyer DI: 180-kD ribosome
receptor is essential for both ribosome binding and protein
translocation. J Cell Biol. 120:853–863. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tsai HY, Yang YF, Wu AT, Yang CJ, Liu YP,
Jan YH, Lee CH, Hsiao YW, Yeh CT, Shen CN, et al: Endoplasmic
reticulum ribosome-binding protein 1 (RRBP1) overexpression is
frequently found in lung cancer patients and alleviates
intracellular stress-induced apoptosis through the enhancement of
GRP78. Oncogene. 32:4921–4931. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pan Y, Cao F, Guo A, Chang W, Chen X, Ma
W, Gao X, Guo S, Fu C and Zhu J: Endoplasmic reticulum
ribosome-binding protein 1, RRBP1, promotes progression of
colorectal cancer and predicts an unfavourable prognosis. Br J
Cance. 113:763–772. 2015. View Article : Google Scholar
|
|
6
|
Li T, Wang Q, Hong X, Li H, Yang K, Li J
and Lei B: RRBP1 is highly expressed in prostate cancer and
correlates with prognosis. Cancer Manag Res. 11:3021–3027. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu S, Lin M, Ji H, Ding J, Zhu J, Ma R
and Meng F: RRBP1 overexpression is associated with progression and
prognosis in endometrial endometrioid adenocarcinoma. Diagn Pathol.
14:72019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang L, Wang M, Zhang M, Li X, Zhu Z and
Wang H: Expression and significance of RRBP1 in esophageal
carcinoma. Cancer Manag Res. 10:1243–1249. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Legler DF, Allmen EU and Hauser MA: CCR7:
Roles in cancer cell dissemination, migration and metastasis
formation. Int J Biochem Cell Biol. 54:78–82. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xiong Y, Huang F, Li X, Chen Z, Feng D,
Jiang H, Chen W and Zhang X: CCL21/CCR7 interaction promotes
cellular migration and invasion via modulation of the MEK/ERK1/2
signaling pathway and correlates with lymphatic metastatic spread
and poor prognosis in urinary bladder cancer. Int J Oncol.
51:75–90. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Takanami I: Overexpression of CCR7 mRNA in
nonsmall cell lung cancer: Correlation with lymph node metastasis.
Int J Cancer. 105:186–189. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
An S, Tiruthani K, Wang Y, Xu L, Hu M, Li
J, Song W, Jiang H, Sun J, Liu R and Huang L: Locally trapping the
C-C chemokine receptor Type 7 by gene delivery nanoparticle
inhibits lymphatic metastasis prior to tumor resection. Small.
15:e18051822019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu Z, Zhu C, Chen C, Zong Y, Feng H, Liu
D, Feng W, Zhao J and Lu A: CCL19 suppresses angiogenesis through
promoting miR-206 and inhibiting Met/ERK/Elk-1/HIF-1α/VEGF-A
pathway in colorectal cancer. Cell Death Dis. 9:9742018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46:D956–D963. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wahafu W, Gai J, Song L, Ping H, Wang M,
Yang F, Niu Y and Xing N: Increased H2S and its
synthases in urothelial cell carcinoma of the bladder, and enhanced
cisplatin-induced apoptosis following H2S inhibition in
EJ cells. Oncol Lett. 15:8484–8490. 2018.PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mo M, Zhou M, Wang L, Qi L, Zhou K, Liu
LF, Chen Z and Zu XB: C CL21/CCR7 enhances the proliferation,
migration, and invasion of human bladder cancer T24 cells. PLoS
One. 10:e01195062015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xiong Y, Huang F, Li X, Chen Z, Feng D,
Jiang H, Chen W and Zhang X: CCL21/CCR7 interaction promotes
cellular migration and invasion via modulation of the MEK/ERK1/2
signaling pathway and correlates with lymphatic metastatic spread
and poor prognosis in urinary bladder cancer. Int J Oncol.
51:75–90. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou M, Wang S, Hu L, Liu F, Zhang Q and
Zhang D: miR-199a-5p suppresses human bladder cancer cell
metastasis by targeting CCR7. BMC Urol. 16:642016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hyde M, Block-Alper L, Felix J, Webster P
and Meyer DI: Induction of secretory pathway components in yeast is
associated with increased stability of their mRNA. J Cell Biol.
156:993–1001. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee AS: Mammalian stress response:
Induction of the glucose-regulated protein family. Curr Opin Cell
Biol. 4:267–273. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cui XA, Zhang H and Palazzo AF: p180
promotes the ribosome-independent localization of a subset of mRNA
to the endoplasmic reticulum. PLoS Biol. 10:e10013362012.
View Article : Google Scholar : PubMed/NCBI
|