Introduction
Anesthetics are important chemical drugs that allow
patients to undergo operations involving severe pain where the
patient must not move, such as dental treatment (1,2).
Propofol (2,6-diisopropylphenol) is one of the most commonly used
intravenous anesthetics globally as the depth of anesthesia induced
by propofol can be controlled to a greater degree compared with
other anesthetics, such as midazolam, etomidate, thiopental sodium
and ketamine (1,3–5). At the
same time, propofol possesses a number of non-anesthetic effects,
including antitumor function, which has been widely reported
(6,7).
Lung cancer is a leading cause of mortality
worldwide and accounts for >1,000,000 deaths every year
(6,8,9). The
5-year survival rate of patients with lung cancer is <17%
(6). Different anticancer strategies
have been developed and used in clinical treatment of lung cancer,
including surgery, chemotherapy, immunotherapy and targeted therapy
(10–13). However, these strategies do not
effectively improve the long-term survival rate of patients with
lung cancer, so novel effective therapeutic interventions and
targets are urgently needed. Propofol suppresses growth, migration
and invasion of human lung adenocarcinoma A549 cells by
upregulation of microRNA (miR)-1284 and downregulation of miR-372
(6,14). Considering propofol is widely used in
clinical practice, it is important to explore the association
between propofol and lung cancer, as well as the underlying
molecular mechanisms.
Autophagy is a conserved complex process which
maintains the normal function and structure of cells (15,16).
Autophagy is involved in the occurrence and progression of lung
cancer. For example, Xue et al (17) demonstrated that apoptosis stimulating
protein of p53 promotes tumor growth by increasing autophagic flux
in human non-small cell lung cancer, whereas HECT domain and
ankyrin repeat containing E3 ubiquitin protein ligase 1 (HACE1)
acts as a tumor suppressor by ubiquitinating optineurin (OPTN) and
activating selective autophagy (18). Autophagy is also involved in lung
cancer therapy, chemotherapy induces tumor cell autophagy, and
inhibiting autophagy enhances the sensitivity of lung cancer cells
to chemotherapy (19).
The association between propofol and autophagy is
complex. For example, propofol attenuates
hypoxia/reoxygenation-induced autophagy in HK-2 cells, but induces
autophagy in C2C12 cells (16). The
present study aimed to elucidate the antitumor molecular mechanism
of propofol on human lung adenocarcinoma cells and its potential
application on lung cancer therapy.
Materials and methods
Plasmid construction
Short hairpin (sh)RNAs for MBD3 and
HACE1 were designed and inserted into pLKO.1 plasmid
purchased from Sigma-Aldrich (Merck KGaA); their specific sequences
are provided in Table I.
 | Table I.Sequences of shRNAs for MBD3
and HACE1. |
Table I.
Sequences of shRNAs for MBD3
and HACE1.
| shRNA | Target site sequence
(5′→3′) |
|---|
| MBD3 |
|
|
Scramble |
GCGCGATAGCGCTAATAATTT |
|
shMBD3-1 |
AGCAACAAGGTCAAGAGCGAC |
|
shMBD3-2 |
GACCTGAGCACCTTCGACTTC |
|
shMBD3-3 |
GCCGGTGACCAAGATTACCAA |
| HACE1 |
|
|
shHACE1-1 |
CCAGAAATTGATGTGAGTGAT |
|
shHACE1-2 |
GCTGTGCCATATACTCCAAAT |
Cell culture and transfection
Human A549 and H1299 cell lines were purchased from
American Type Culture Collection and cultured in DMEM (Thermo
Fisher Scientific, Inc.) supplemented with 10% FBS, 100 U/ml
penicillin and 100 mg/ml streptomycin (all Gibco; Thermo Fisher
Scientific, Inc.) in a 37°C humidified atmosphere of 5%
CO2. The plasmids containing MBD3 or HACE1 shRNA (3 µg)
were transfected into A549 cells using Lipofectamine®
2000 (Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions, and screened using puromycin (5 µg/ml,
Thermo Fisher Scientific, Inc.) for 48 h after transfection.
Propofol and cycloheximide (CHX)
treatment
Pure propofol was purchased from Sigma-Aldrich
(Merck KGaA) and stock solution of propofol (21 mmol/l) was
prepared in DMSO (Sigma-Aldrich; Merck KGaA). The propofol
concentrations used were as previously described (20). The stock solution of propofol was
diluted to 0.21, 0.18, 0.15, 0.12, 0.09, 0.06 and 0.03 mmol/l with
DMSO (<1%) before addition to DMEM medium supplemented with 10%
FBS, 100 U/ml penicillin and 100 mg/ml streptomycin (1:100). A549
and H1299 cells treated with the indicated concentrations of
propofol were diluted from stock solution and an equal volume of
DMSO was added to the controls (A549 or H1299 cells that did not
receive propofol treatment). CHX was purchased from Sigma-Aldrich
(Merck KGaA), and stock solution of CHX (100 µg/ml) was prepared in
DMSO. For protein stability experiments, A549 cells were treated
with CHX (100 µg/ml) as well as propofol at the indicated time
point (0, 2 or 4 h) at 37°C.
Cell proliferation assay
A total of 3,000 cells was seeded into 96-well
plates and treated in the presence or absence of propofol. The 0 h
time point was defined as 6 h after cells were seeded. After 0, 24,
48 or 72 h, the cells were incubated with MTT solution (cat. no.
C0009; Beyotime Institute of Biotechnology) for 4 h at 37°C, then
the product (formazan) was dissolved in DMSO and quantified
spectrophotometrically at a wavelength of 570 nm using a Microplate
Reader (Bio-Rad Laboratories, Inc.). Experiments were conducted
with six replicates and repeated three times.
Colony formation assay
A total of 1,000 cells was seeded into 6-well plates
and treated in the presence or absence of propofol. After 7 days,
plates were fixed with 4% paraformaldehyde (Merck KGaA) at room
temperature for 30 min, stained with 0.1% crystal violet (cat. no.
C0121; Beyotime Institute of Biotechnology) at room temperature for
30 min and washed three times with PBS buffer. Images were captured
using a camera, the number of colonies were counted manually and
the average number were calculated.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was extracted from cells using a total RNA
kit (Tiangen Biotech Co., Ltd.). Complementary DNA was synthesized
using ReverTra Ace qPCR RT Master Mix (Toyobo Life Science) at 37°C
for 15 min, and 95°C for 5 min, according to the manufacturer's
protocol. RT-qPCR was performed on an ABI 7500 fast real-time PCR
system (pre-denaturation at 95°C for 2 min; denaturation at 95°C
for 30 sec, annealing/extension at 60°C for 34 sec, 40 cycles;
Applied Biosystems; Thermo Fisher Scientific, Inc.) to assess the
relative abundance of HACE1 and MBD3 mRNA using
specific primers (Table II) with
staining by SYBR Green (Toyobo Life Science). The relative
abundance of HACE1 and MBD3 was normalized to that of
GAPDH using the 2−ΔΔCq method (21,22). A
total of three independent experiments was performed.
 | Table II.Sequences of primers used for
RT-qPCR, ChIP-qPCR and bisulfite DNA sequencing. |
Table II.
Sequences of primers used for
RT-qPCR, ChIP-qPCR and bisulfite DNA sequencing.
| A, RT-qPCR |
|---|
|
|---|
| Target gene | Forward primer
(5′→3′) | Reverse primer
(5′→3′) |
|---|
| GAPDH |
AGGTGAAGGTCGGAGTCAACG |
CTCAGCCTTGACGGTGCCAT |
| HACE1 |
TGCCAGAACGGTCACAAGACG |
CTGTGCTGTATCTCTCTGACCATGA |
| Methyl-CpG
binding domain protein 3 |
GAGAGGGAAGAAGTGCCCAGAAG |
GGAAGTCGAAGGTGCTCAGGTC |
|
| B,
ChIP-qPCR |
|
| Target
gene | Forward primer
(5′→3′) | Reverse primer
(5′→3′) |
|
| F1R1 |
TTTGCTTCCCACCCATTTCCTG |
CTTCTGTGGCCCAGACAGTTTCAAC |
| F2R2 |
AAGAGGCCAAGCAAGACTGGAAC |
CATTGCACTCCAGCCTGGGT |
| F3R3 |
AAGTGTTCAACTTCTGTGCAGAGC |
GACACAGCCTAGTGGGAAATCCA |
|
| C, Bisulfite DNA
sequencing |
|
| Target
gene | Forward primer
(5′→3′) | Reverse primer
(5′→3′) |
|
| HACE1
promoter |
ATAGGGATATAATATAGTTTAA |
AAAAACTATAATTTCCAACTA |
Bisulfite DNA sequencing
Genomic DNA (gDNA) was extracted from A549 cells
treated in the presence or absence of propofol using the standard
phenol-chloroform extraction method (23). Then, gDNA was treated with bisulfite
using the CpGenome Turbo Bisulfite Modification kit (EMD Millipore)
according to the manufacturer's instructions (24). The modified DNA was amplified using
Platinum Taq DNA Polymerase (Thermo Fisher Scientific, Inc.) with
the respective primer sets (Table
II) that recognize bisulfite-modified DNA only. Then, PCR
products were cloned into the pMD18-T vector (Takara Bio, Inc.) and
Sanger sequencing was performed by an external company (BioSune
Bio, Inc; www.biosune.com).
Immunoprecipitation and
immunoblotting
For immunoprecipitation, cells were lysed in RIPA
buffer [50 mM Tris-HCl (pH 7.6), 150 mM NaCl, 5 mM EDTA, 0.1% SDS
and 1% NP-40] supplemented with a protease inhibitor cocktail, cell
lysates were centrifuged at 4°C with 12,000 × g for 10 min,
incubated with OPTN antibody (1:100; cat. no. 10837-1-AP;
ProteinTech Group, Inc.) or normal rabbit IgG (1:100; cat. no.
2729; Cell Signaling Technology, Inc.), and Protein G agarose beads
(Merck KGaA) overnight at 4°C, washed three times with COIP buffer
at 4°C. The immunoprecipitates were enriched and denatured at 100°C
for 10 min in 2X SDS-PAGE loading buffer. The inputs,
immunoprecipitates and cell lysates (10 µl/lane) were then
subjected to SDS-PAGE (10%) and transferred to PVDF membranes
(Bio-Rad Laboratories, Inc.) with 200 mA for three hours as
previously described (25). The
membrane was blocked with 5% non-fat milk at room temperature for 1
h and incubated with appropriate antibodies against GAPDH (1:5,000;
cat. no. 60004-1-Ig; ProteinTech Group, Inc.), HACE1 (1:1,000; cat.
no. ab133637; Abcam), ubiquitin (1:500; cat. no. sc-47721; Santa
Cruz Biotechnology, Inc.), OPTN (1:2,000), microtubule-associated
protein 1A/1B-light chain 3 (LC3) (1:500; cat. no. L7543;
Sigma-Aldrich; Merck KGaA), MBD3 (1:1,000; cat. no. 14258-1-AP;
ProteinTech Group, Inc.), tet methylcytosine dioxygenase (TET)1
(1:1,000; cat. no. 61444; Active Motif, Inc.), TET2 (1:1,000; cat.
no. 21207-1-AP; ProteinTech Group, Inc.), metastasis-associated 1
family member 2 (MTA2) (1:1,000; cat. no. 66195-1-Ig; ProteinTech
Group, Inc.) or TET3 (1:800; cat. no. 61395; Active Motif, Inc.)
overnight at 4°C, washed three times with TBST (50 mM Tris-HCl, 150
mM NaCl and 0.1% Tween-20, pH 7.6), and then incubated with
secondary antibodies [HRP-conjugated Affinipure Goat Anti-Mouse IgG
(H+L), cat. no. SA00001-1, 1:5,000 dilution; HRP-conjugated
Affinipure Goat Anti-Rabbit IgG (H+L), cat. no. SA00001-2, 1:5,000
dilution.] at room temperature for one hour, washed three times
with TBST, the signals were visualized with high-sig ECL Western
Blotting Substrate (cat. no. 180–5001, Tanon Science and Technology
Co., Ltd.) using a Tanon 5200 Imaging System (Tanon Science and
Technology Co., Ltd.). Gray values of protein bands were quantified
using ImageJ software (version 1.52; National Institutes of Health)
and calculated.
Chromatin immunoprecipitation
(ChIP)
ChIP experiments were performed as previously
described (26). Briefly, A549 cells
treated in the presence or absence of propofol were crosslinked in
1% formaldehyde (Sigma-Aldrich; Merck KGaA) for 10 min at room
temperature, followed by quenching in 125 mM glycine, then washed
three times with ice-cold PBS, and resuspended with 270 µl lysis
buffer [50 mM Tris-Cl (pH 8.0), 10 mM EDTA, 1% SDS and protease
inhibitor]. Following incubation on ice for 5 min, cells were
sonicated with Bioruptor (Diagenode SA) for 15 cycles of 30 sec on,
30 sec off at high setting. Samples were centrifuged at 13,000 × g
for 10 min at 4°C. Then, 100 µl supernatant was diluted 10 times
with ChIP dilution buffer [20.00 mM Tris-Cl (pH 8.0), 0.01% SDS,
1.10% Triton X-100, 1.10 mM EDTA and 167.00 mM NaCl] and incubated
with 5 µg control rabbit IgG (cat. no. 2729; Cell Signaling
Technology, Inc.) or anti-MBD3 (cat. no. 14258-1-AP; ProteinTech
Group, Inc.) antibody at 4°C overnight. Samples were further
incubated with 40 µl Protein G beads at 4°C for 2 h. The beads were
washed three times with low salt wash buffer [20.0 mM Tris-Cl (pH
8.0), 150.0 mM NaCl, 0.1% SDS, 1.0% Triton X-100, 2.0 mM EDTA],
three times with high salt wash buffer [20.0 mM Tris-Cl (pH 8.0),
500.0 mM NaCl, 1.0% NP-40, 0.1% SDS, 2.0 mM EDTA], three times with
LiCl wash buffer [20 mM Tris-Cl (pH 8.0), 500 mM LiCl, 1% NP-40, 1
mM EDTA, 1% deoxycholate] and three times with TE buffer [100 mM
Tris-Cl (pH 8.0), 1 mM EDTA]. The washed beads were resuspended
with 500 µl fresh elution buffer (1.0% SDS and 0.1 M sodium
bicarbonate) and incubated at 65°C for 30 min. Eluted DNA was
adjusted to 300 mM NaCl and incubated at 65°C for 4 h, followed by
incubation at 55°C for 1 h with 50 µg proteinase K. DNA was
purified using the phenol-chloroform method and subjected to the
same qPCR analysis as aforementioned (26). Primer sequences are presented in
Table II.
Statistical analysis
Data are presented as the mean ± SD of ≥3
independent repeats. One-way ANOVA was performed with Tukey's post
hoc multiple comparisons test using GraphPad Prism software version
5.0 (GraphPad Software, Inc.). P<0.05 and P<0.01 were
considered to indicate a statistically significantly
difference.
Results
Propofol inhibits proliferation of
human A549 cells
In order to detect the effect of propofol on human
non-small cell lung cancer, A549 cells were treated in the presence
or absence of propofol (0, 30, 60, 90, 120, 150, 180 and 210
µmol/l). MTT assay indicted that propofol inhibited proliferation
of A549 cells in a dose-dependent manner (Fig. 1A). Propofol exhibited significant
cell proliferation inhibition at 60, 90 and 120 µmol/l (Fig. 1A). Propofol at >120 µmol/l
resulted in little further inhibition; therefore, concentrations of
60 and 120 µmol/l were selected for use in further experiments
(Fig. 1A). Then, the viability of
A549 and H1299 cells was detected at different time points (0, 24,
48 and 72 h) following treatment in the presence or absence of
propofol (60or 120 µmol/l), which demonstrated that propofol
exhibited an inhibitory effect in a time-dependent manner (Fig. 1B). Furthermore, a colony formation
assay was performed; decreased colony numbers were observed in
propofol-treated groups compared with the control group (Fig. 1C). These results demonstrated that
propofol inhibited proliferation of A549 cells.
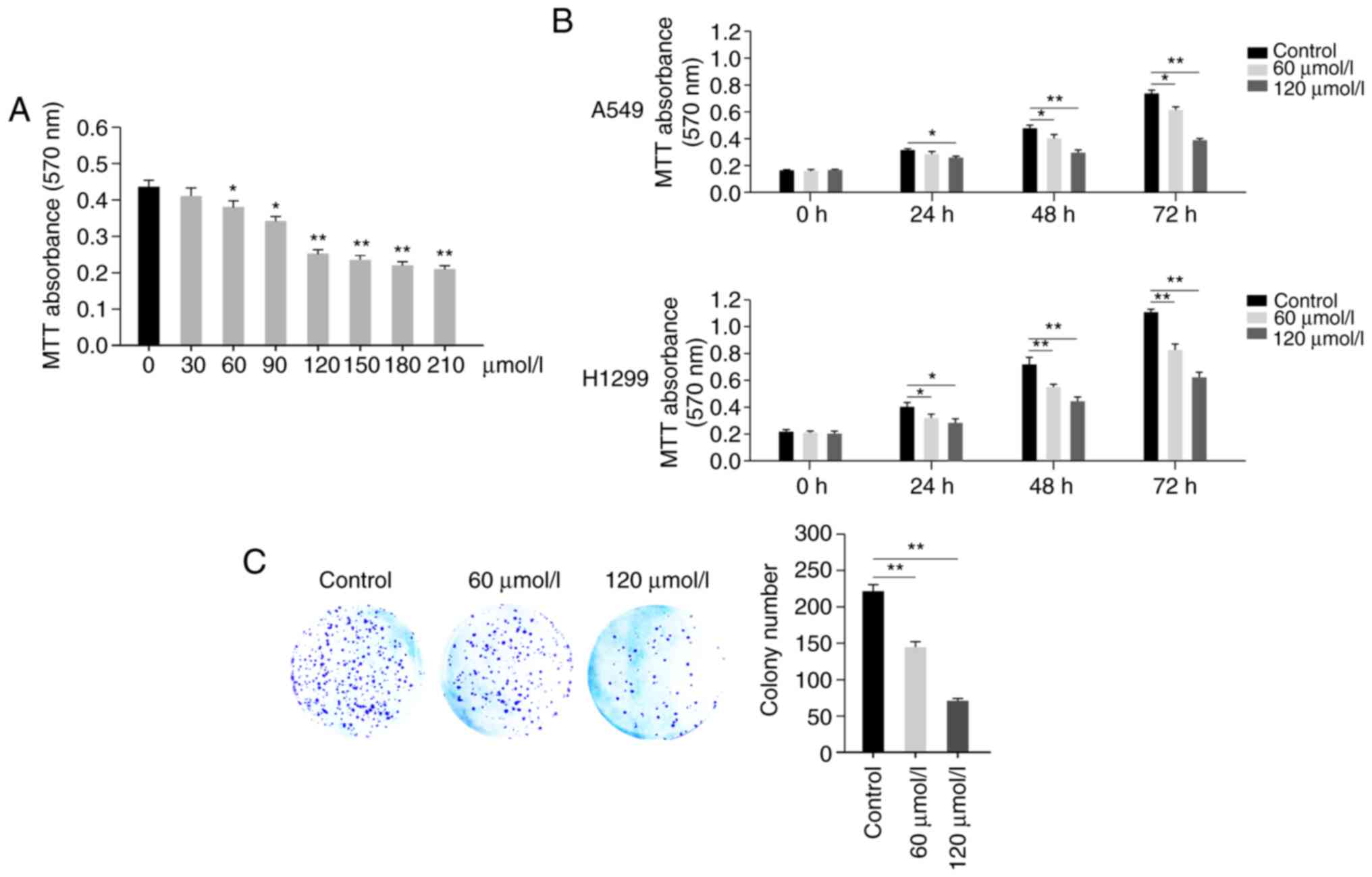 | Figure 1.Propofol inhibits proliferation of
human A549 cells. (A) Propofol inhibits proliferation of A549 cells
in a dose-dependent manner. Viability of A549 cells was detected by
MTT assay at 72 h after treatment with propofol (0, 30, 60, 90,
120, 150, 180 and 210 µmol/l). (B) Propofol inhibits proliferation
of A549 and H1299 cells in a time-dependent manner. The viability
of A549 or H1299 cells was detected by MTT assay at different time
points (0, 24, 48 and 72 h) following treatment with propofol (60
or 120 µmol/l). The 0 h time point was 6 h after cells were seeded.
(C) Propofol inhibits the colony formation of A549 cells. A total
of 1,000 A549 cells were seeded into 6-well plates and treated with
propofol for 7 days. Plates were fixed, stained and colony numbers
were counted and calculated. Scale bar, 1 cm. Data are expressed as
the mean ± SD and analyzed using one-way ANOVA with Tukey's post
hoc test. *P<0.05, **P<0.01. |
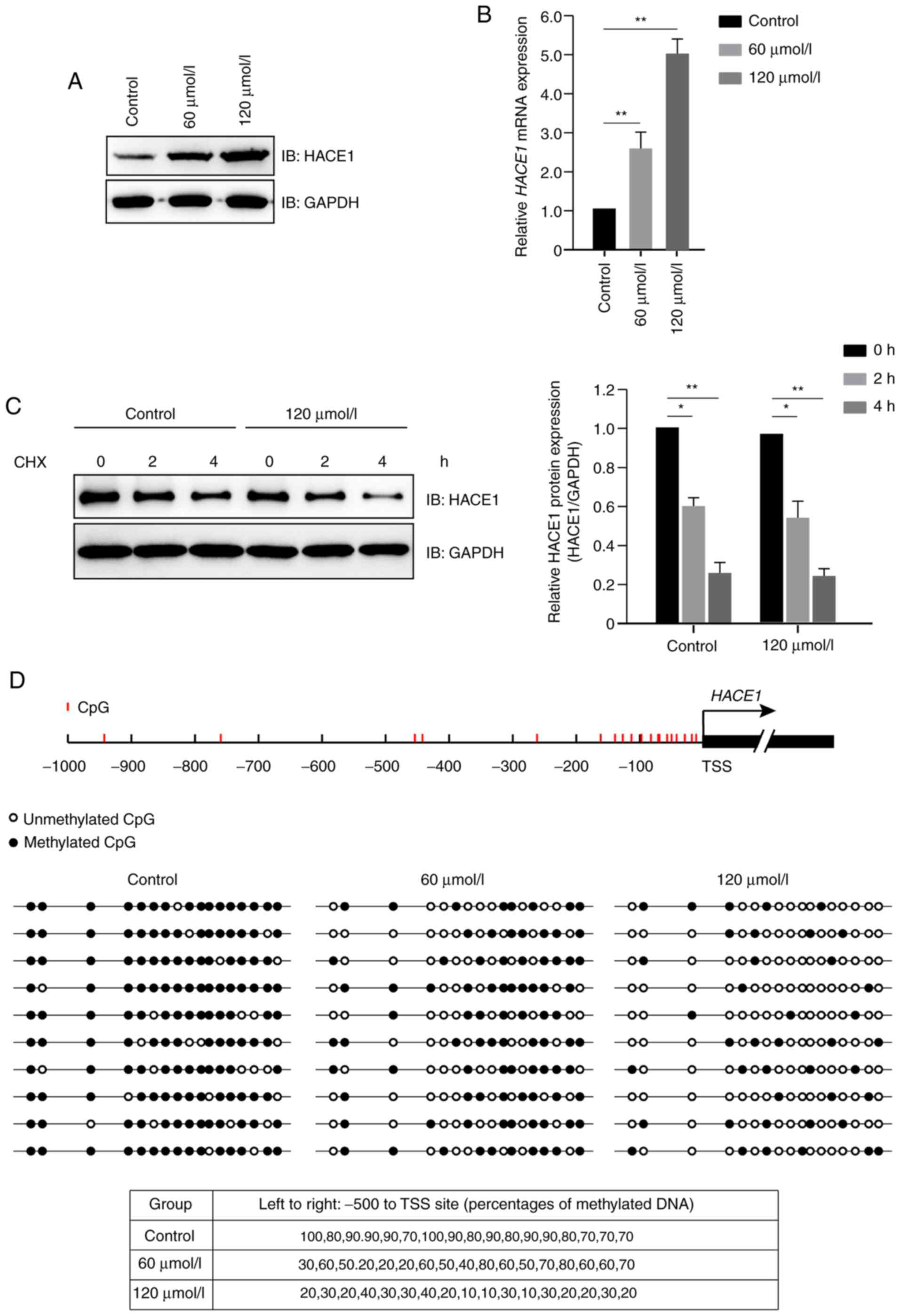
Propofol promotes demethylation of
HACE1 gene promoter in human A549 cells
Propofol increased protein expression levels of
HACE1 (Fig. 2A); further study
indicated that propofol increased the expression levels of HACE1
primarily at the transcriptional, but not translational, level
(Fig. 2B and C). Subsequently, a DNA
methylation detection experiment was performed, which demonstrated
that propofol promoted demethylation of HACE1 gene promoter
in a dose-dependent manner in A549 cells (Fig. 2D).
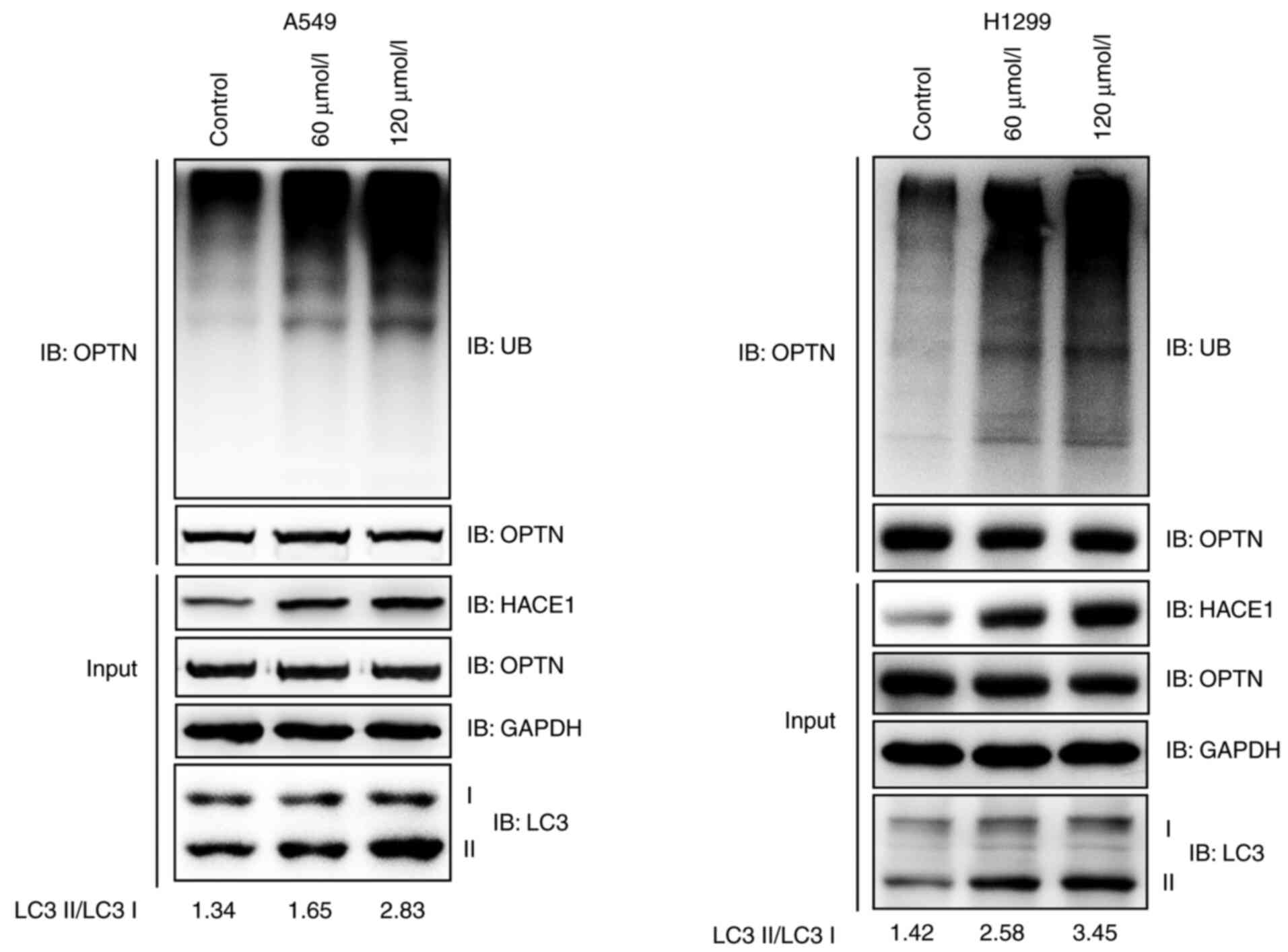
Propofol activates HACE1-OPTN
axis-mediated autophagy in human A549 and H1299 cells
Ubiquitination of the autophagy receptor OPTN by
HACE1 has previously been shown to activate selective autophagy,
resulting in tumor suppression in lung cancer (18). It was therefore investigated whether
propofol activated HACE1-OPTN axis-mediated autophagy. The
ubiquitination of OPTN notably increased when A549 or H1299 cells
were treated with propofol (Fig.
3A). LC3 is the most commonly used marker of autophagosomes
(18); the ratio of LC3 II to LC3 I
notably increased in propofol-treated groups compared with the
control group (Fig. 3A). These data
indicated that propofol activated HACE1-OPTN axis-mediated
autophagy.
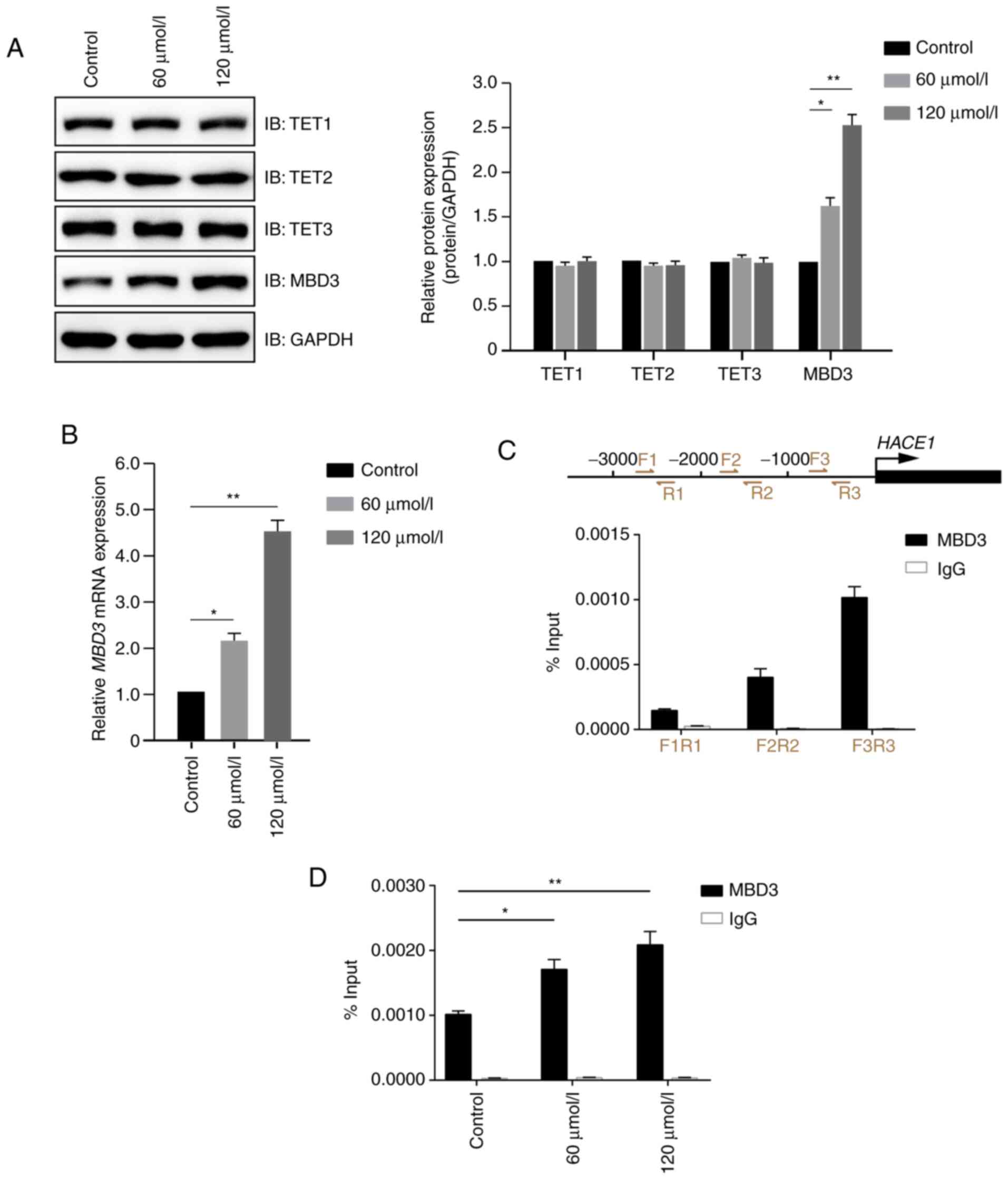
Propofol promotes expression levels of
MBD3 and binding to HACE1 gene promoter
As propofol promoted demethylation of HACE1
gene promoter (Fig. 2C), the
underlying molecular mechanism was investigated.
Demethylation-associated molecules, including TET1, TET2, TET3,
MBD3 and MTA2), were detected by immunoblotting in A549 cells
treated in the presence or absence of propofol. Propofol exhibited
no notable effect on the protein expression levels of TET1, TET2,
TET2 and MTA2, but significantly increased MBD3 protein expression
levels compared with the control group (Figs. 4A and S1). Further study demonstrated that
propofol increased the expression levels of MBD3 primarily at the
transcriptional level (Fig. 4B). As
MBD3 is a transcription factor, it was then determined whether MBD3
could bind to the promoter of HACE1. The present study
demonstrated that MBD3 preferentially bound the −1000 to −1 bp
region of HACE1 promoter (Fig.
4C) in a dose-dependent manner (Fig.
4D). These data indicated that propofol promoted demethylation
of HACE1 promoter by regulating MBD3 expression levels and
binding to HACE1 promoter.
Propofol inhibits proliferation of
human A549 cells in a MBD3-dependent manner
A total of three shRNAs for MBD3 were designed and
transfected into A549 cells; immunoblotting analysis indicated that
both the protein and mRNA expression levels of MBD3 were
significantly decreased in cells transfected with shMBD3-1 or
shMBD3-2 compared with cells transfected with scramble (Fig. 5A and B). The shRNAs for MBD3
(shMBD3-1 and sh shMBD3-2) were selected for further study. The
present results also indicated that MBD3 knockdown decreased the
protein expression levels of HACE1 (Fig.
5A). Furthermore, MTT and colony formation assays indicated
that MBD3 knockdown abolished propofol-mediated inhibition of cell
proliferation (Fig. 5C and D). These
results demonstrated that propofol inhibited proliferation of human
A549 cells in a MBD3-dependent manner.
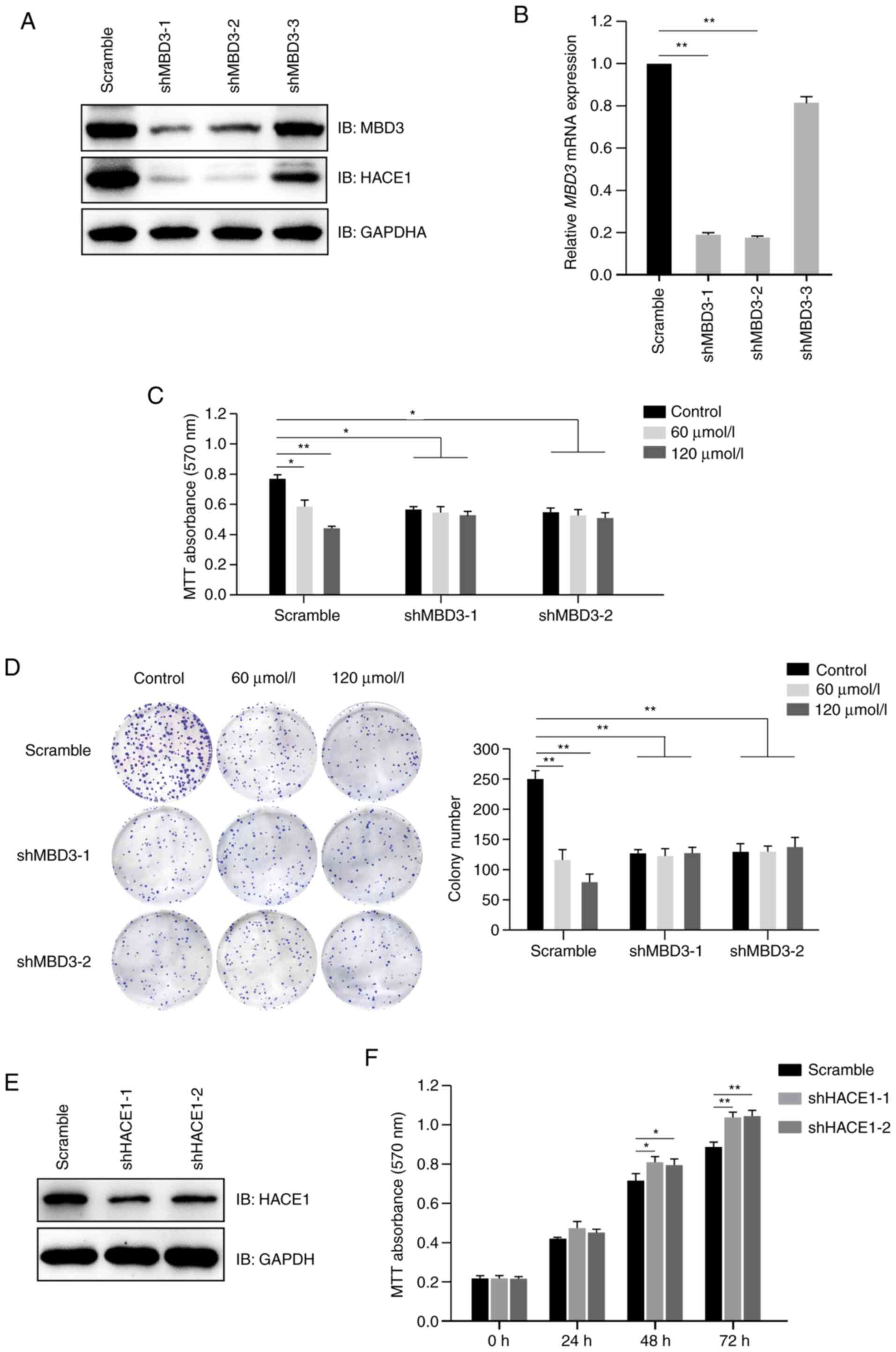 | Figure 5.Propofol inhibits proliferation of
human A549 cells in a MBD3-dependent manner. (A) MBD3 knockdown
decreased the expression levels of HACE1. A549 cells were
transfected with shRNAs (scramble control, 1, 2 or 3) for MBD3, and
then subjected to screening for puromycin resistance to establish
stably expressed cell lines. Protein expression levels of MBD3 and
HACE1 were detected by immunoblotting. (B) Assessment of knockdown
efficiency of MBD3 shRNAs by RT-qPCR. Total RNA was extracted from
A549 cells stably transfected with MBD3 shRNAs, then complementary
DNA was synthesized, and mRNA expression levels of MBD3 were
detected by qPCR. A549 cells stably transfected with MBD3 shRNAs
(scramble, 1 or 2) were treated in the presence or absence of
propofol. (C) MBD3 knockdown abolished the inhibitory effect of
propofol on cell proliferation, as detected by MTT assay. (D) MBD3
knockdown abolished the inhibitory effect of propofol on cell
colony formation. The viability of A549 cells was detected by MTT
assay. Scale bar, 1 cm. (E) Assessment of knockdown efficiency of
shRNAs for HACE1 by immunoblotting. A549 cells were transfected
with shRNAs (scramble, 1 or 2) for HACE1, and then subjected to
screening for puromycin resistance to establish stably expressed
cell lines. The protein expression levels of HACE1 were detected by
immunoblotting. (F) HACE1 knockdown promotes proliferation of A549
cells. The viability of A549 cells stably transfected with HACE1
shRNAs were detected by MTT assay. Data are expressed as the mean ±
SD of three independent experiments and analyzed using one-way
ANOVA with Tukey's post hoc test. *P<0.05, **P<0.01. MBD3,
methyl-CpG binding domain protein 3; HACE1, HECT domain and ankyrin
repeat containing E3 ubiquitin protein ligase 1; sh, short hairpin;
RT-q, reverse transcription-quantitative; IB, immunoblot. |
Downregulation of HACE1 promotes
proliferation of A549 cells
In order to investigate the effect of HACE1 on cell
proliferation, two HACE1 shRNAs were designed and tested in
A549 cells. Immunoblotting analysis indicated that HACE1
significantly decreased in cells transfected with shHACE1-1 or
shHACE1-2 compared with cells transfected with scramble (Fig. 5E). MTT assay demonstrated that HACE1
knockdown promoted proliferation of A549 cells (Fig. 5F).
Discussion
Besides anesthetic properties, propofol possesses
numerous non-anesthetic effects (6).
For example, Hsing et al (27) showed propofol decreases reactive
oxygen species generation, thus inhibiting endotoxic inflammation.
Cui et al (28) demonstrated
that propofol prevents oxygen or glucose deprivation-induced
autophagy in PC-12 cells, as well as cerebral ischemia-reperfusion
injury in rats. The association between propofol and tumors has
been extensively studied, revealing that propofol serves as a tumor
suppressor or promoting factor depend on the type of cancer
(29,30). The present study demonstrated that
propofol inhibited proliferation of human A549 and H1299 cells.
Propofol has been shown to suppress growth, migration and invasion
of A549 cells by upregulation of miR-1284 and downregulation of
miR-372 (14,31), which is consistent with the results
of present study. In the present study, propofol >120 µmol/l
exhibited little inhibition; however, the specific underlying
mechanism requires further investigation, although it was
hypothesized that the concentration of propofol reached saturation
at 120 µmol/l.
HACE1 is frequently downregulated or lost in
numerous types of tumor, such as lung and liver cancer, and acts as
a tumor suppressor by ubiquitinating OPTN and activating selective
autophagy (18,24). The study found that propofol promoted
HACE1 expression levels by demethylating HACE1 gene
promoter, which activated HACE1-OPTN axis-mediated autophagy.
In mammalian cells, DNA methylation and
demethylation are critical for regulating gene expression levels
and serve important roles in physiological and pathological
processes, such as mammalian puberty and cancer development
(32). MBD3 induces gDNA
demethylation at specific targets and is also involved in
maintaining the demethylated and active state of numerous genes,
including progonadoliberin-1, serine/threonine-protein kinase Chk2
and 39S ribosomal protein L32, mitochondrial (26,33–35). The
present findings indicated that propofol promoted expression levels
of MBD3 and enhanced its binding to the HACE1 gene promoter.
This may be due to low antibody titer or weak binding of MBD3 to
DNA. Further investigation is required to determine whether MBD3
promotes demethylation of HACE1 promoter or maintains the
demethylated state. The effect of propofol on mRNA expression
levels of MBD3 and its specific underlying mechanism also requires
further study. The present study hypothesized that propofol affects
mRNA expression levels of MBD3 either by demethylating MBD3
gene promoter or by regulating transcription of MBD3.
Selective autophagy is involved in removal of
damaged or superfluous organelles from the cytosol, which is
necessary to maintain homeostasis and cell function (36–39). In
the present study, propofol activated selective autophagy of A549
and H1299 cells by increasing HACE1 expression levels, indicating
that propofol may be a powerful therapeutic drug for lung cancer;
this remains to be assessed in an animal model.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by research grants
from Science and Technology Department of Yunnan Province, and
Kunming Medical University Joint Special Project [grant nos.
2018FE001-(070) and 2019FE001-(248)].
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
ZW and SL conceived and designed the experiments.
SL, HY, MZ, LG, YW, ZL and YQ performed the experiments, collected
the data and analyzed the results. ZW and SL wrote the paper. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kodama M, Higuchi H, Ishii-Maruhama M,
Nakano M, Honda-Wakasugi Y, Maeda S and Miyawaki T: Multi-drug
therapy for epilepsy influenced bispectral index after a bolus
propofol administration without affecting propofol's
pharmacokinetics: A prospective cohort study. Sci Rep.
10:15782020.PubMed/NCBI
|
|
2
|
Sona Khan M, Trenet W, Xing N, Sibley B,
Abbas M, Al-Rashida M, Rauf K and Mandyam CD: A novel sulfonamide,
4-FS, reduces ethanol drinking and physical withdrawal associated
with ethanol dependence. Int J Mol Sci. 21:44112020.
|
|
3
|
Yoon HK, Jun K, Park SK, Ji SH, Jang YE,
Yoo S, Kim JT and Kim WH: Anesthetic agents and cardiovascular
outcomes of noncardiac surgery after coronary stent insertion. J
Clin Med. 9:4292020.
|
|
4
|
Kang Y, Saito M and Toyoda H: Molecular
and regulatory mechanisms of desensitization and resensitization of
GABAA receptors with a special reference to propofol/barbiturate.
Int J Mol Sci. 21:5632020.
|
|
5
|
Cho YJ, Nam K, Kim TK, Choi SW, Kim SJ,
Hausenloy DJ and Jeon Y: Sevoflurane, propofol and carvedilol block
myocardial protection by limb remote ischemic preconditioning. Int
J Mol Sci. 20:2692019.
|
|
6
|
Sun H and Gao D: Propofol suppresses
growth, migration and invasion of A549 cells by down-regulation of
miR-372. BMC Cancer. 18:12522018.PubMed/NCBI
|
|
7
|
Vasileiou I, Xanthos T, Koudouna E, Perrea
D, Klonaris C, Katsargyris A and Papadimitriou L: Propofol: A
review of its non-anaesthetic effects. Eur J Pharmacol. 605:1–8.
2009.PubMed/NCBI
|
|
8
|
Bode AM, Dong Z and Wang H: Cancer
prevention and control: Alarming challenges in China. Natl Sci Rev.
3:117–127. 2016.PubMed/NCBI
|
|
9
|
Xia T, Zhu Y, Mu L, Zhang ZF and Liu S:
Pulmonary diseases induced by ambient ultrafine and engineered
nanoparticles in twenty-first century. Natl Sci Rev. 3:416–429.
2016.PubMed/NCBI
|
|
10
|
Zanoaga O, Braicu C, Jurj A, Rusu A, Buiga
R and Berindan-Neagoe I: Progress in research on the role of
flavonoids in lung cancer. Int J Mol Sci. 20:42912019.
|
|
11
|
Loong HH, Kwan SS, Mok TS and Lau YM:
Therapeutic strategies in EGFR mutant non-small cell lung cancer.
Curr Treat Options Oncol. 19:582018.PubMed/NCBI
|
|
12
|
Xue J, Yang J, Luo M, Cho WC and Liu X:
MicroRNA-targeted therapeutics for lung cancer treatment. Expert
Opin Drug Discov. 12:141–157. 2017.PubMed/NCBI
|
|
13
|
Sarne V, Huter S, Braunmueller S, Rakob L,
Jacobi N, Kitzwögerer M, Wiesner C, Obrist P and Seeboeck R:
Promoter methylation of selected genes in non-small-cell lung
cancer patients and cell lines. Int J Mol Sci. 21:45952020.
|
|
14
|
Wang Q, Liu S, Zhao X, Wang Y, Tian D and
Jiang W: MiR-372-3p promotes cell growth and metastasis by
targeting FGF9 in lung squamous cell carcinoma. Cancer Med.
6:1323–1330. 2017.PubMed/NCBI
|
|
15
|
Wang X, Li W, Zhang N, Zheng X and Jing Z:
Opportunities and challenges of co-targeting epidermal growth
factor receptor and autophagy signaling in non-small cell lung
cancer. Oncol Lett. 18:499–506. 2019.PubMed/NCBI
|
|
16
|
Wang H, Peng X, Huang Y, Xiao Y, Wang Z
and Zhan L: Propofol attenuates hypoxia/reoxygenation-induced
apoptosis and autophagy in HK-2 cells by inhibiting JNK activation.
Yonsei Med J. 60:1195–1202. 2019.PubMed/NCBI
|
|
17
|
Xue Y, Han H, Wu L, Pan B, Dong B, Yin CC,
Tian Z, Liu X, Yang Y, Zhang H, et al: iASPP facilitates tumor
growth by promoting mTOR-dependent autophagy in human
non-small-cell lung cancer. Cell Death Dis. 8:e31502017.PubMed/NCBI
|
|
18
|
Liu Z, Chen P, Gao H, Gu Y, Yang J, Peng
H, Xu X, Wang H, Yang M, Liu X, et al: Ubiquitylation of autophagy
receptor optineurin by HACE1 activates selective autophagy for
tumor suppression. Cancer Cell. 26:106–120. 2014.PubMed/NCBI
|
|
19
|
Liao SX, Sun PP, Gu YH, Rao XM, Zhang LY
and Ou-Yang Y: Autophagy and pulmonary disease. Ther Adv Respir
Dis. 13:17534666198905382019.PubMed/NCBI
|
|
20
|
Xu YB, Du QH, Zhang MY, Yun P and He CY:
Propofol suppresses proliferation, invasion and angiogenesis by
down-regulating ERK-VEGF/MMP-9 signaling in Eca-109 esophageal
squamous cell carcinoma cells. Eur Rev Med Pharmacol Sci.
17:2486–2494. 2013.PubMed/NCBI
|
|
21
|
Xu X, Li C, Gao X, Xia K, Guo H, Li Y, Hao
Z, Zhang L, Gao D, Xu C, et al: Excessive UBE3A dosage impairs
retinoic acid signaling and synaptic plasticity in autism spectrum
disorders. Cell Res. 28:48–68. 2018.PubMed/NCBI
|
|
22
|
Li C, Han T, Guo R, Chen P, Peng C, Prag G
and Hu R: An integrative synthetic biology approach to
interrogating cellular ubiquitin and ufm signaling. Int J Mol Sci.
21:42312020.
|
|
23
|
Longchamps RJ, Castellani CA, Yang SY,
Newcomb CE, Sumpter JA, Lane J, Grove ML, Guallar E, Pankratz N,
Taylor KD, et al: Evaluation of mitochondrial DNA copy number
estimation techniques. PLoS One. 15:e02281662020.PubMed/NCBI
|
|
24
|
Yu Z, Li Y, Han T and Liu Z: Demethylation
of the HACE1 gene promoter inhibits the proliferation of human
liver cancer cells. Oncol Lett. 17:4361–4368. 2019.PubMed/NCBI
|
|
25
|
Li H, Liang Z, Yang J, Wang D, Wang H, Zhu
M, Geng B and Xu EY: DAZL is a master translational regulator of
murine spermatogenesis. Natl Sci Rev. 6:455–468. 2019.PubMed/NCBI
|
|
26
|
Li C, Lu W, Yang L, Li Z, Zhou X, Guo R,
Wang J, Wu Z, Dong Z, Ning G, et al: MKRN3 regulates the epigenetic
switch of mammalian puberty via ubiquitination of MBD3. Natl Sci
Rev. 7:671–685. 2020.
|
|
27
|
Hsing CH, Lin MC, Choi PC, Huang WC, Kai
JI, Tsai CC, Cheng YL, Hsieh CY, Wang CY, Chang YP, et al:
Anesthetic propofol reduces endotoxic inflammation by inhibiting
reactive oxygen species-regulated Akt/IKKβ/NF-κB signaling. PLoS
One. 6:e175982011.PubMed/NCBI
|
|
28
|
Cui D, Wang L, Qi A, Zhou Q, Zhang X and
Jiang W: Propofol prevents autophagic cell death following oxygen
and glucose deprivation in PC12 cells and cerebral
ischemia-reperfusion injury in rats. PLoS One.
7:e353242012.PubMed/NCBI
|
|
29
|
Zhang L, Wang N, Zhou S, Ye W, Jing G and
Zhang M: Propofol induces proliferation and invasion of gallbladder
cancer cells through activation of Nrf2. J Exp Clin Cancer Res.
31:662012.PubMed/NCBI
|
|
30
|
Du Q, Liu J, Zhang X, Zhang X, Zhu H, Wei
M and Wang S: Propofol inhibits proliferation, migration, and
invasion but promotes apoptosis by regulation of Sox4 in
endometrial cancer cells. Braz J Med Biol Res.
51:e68032018.PubMed/NCBI
|
|
31
|
Liu WZ and Liu N: Propofol inhibits lung
cancer A549 cell growth and epithelial-mesenchymal transition
process by upregulation of MicroRNA-1284. Oncol Res. 27:1–8.
2018.PubMed/NCBI
|
|
32
|
Xu X, Tao Y, Gao X, Zhang L, Li X, Zou W,
Ruan K, Wang F, Xu GL and Hu R: A CRISPR-based approach for
targeted DNA demethylation. Cell Discov. 2:160092016.PubMed/NCBI
|
|
33
|
Brown SE, Suderman MJ, Hallett M and Szyf
M: DNA demethylation induced by the methyl-CpG-binding domain
protein MBD3. Gene. 420:99–106. 2008.PubMed/NCBI
|
|
34
|
Peng L, Li Y, Xi Y, Li W, Li J, Lv R,
Zhang L, Zou Q, Dong S, Luo H, et al: MBD3L2 promotes Tet2
enzymatic activity for mediating 5-methylcytosine oxidation. J Cell
Sci. 129:1059–1071. 2016.PubMed/NCBI
|
|
35
|
Brown SE and Szyf M: Epigenetic
programming of the rRNA promoter by MBD3. Mol Cell Biol.
27:4938–4952. 2007.PubMed/NCBI
|
|
36
|
Lee CW, Wilfling F, Ronchi P, Allegretti
M, Mosalaganti S, Jentsch S, Beck M and Pfander B: Selective
autophagy degrades nuclear pore complexes. Nat Cell Biol.
22:159–166. 2020.PubMed/NCBI
|
|
37
|
Yamasaki A, Alam JM, Noshiro D, Hirata E,
Fujioka Y, Suzuki K, Ohsumi Y and Noda NN: Liquidity is a critical
determinant for selective autophagy of protein condensates. Mol
Cell. 77:1163–1175.e9. 2020.PubMed/NCBI
|
|
38
|
Zhao ZQ, Yu ZY, Li J and Ouyang XN:
Gefitinib induces lung cancer cell autophagy and apoptosis via
blockade of the PI3K/AKT/mTOR pathway. Oncol Lett. 12:63–68.
2016.PubMed/NCBI
|
|
39
|
Ren S, Ding C and Sun Y: Morphology
remodeling and selective autophagy of intracellular organelles
during viral infections. Int J Mol Sci. 21:36892020.
|