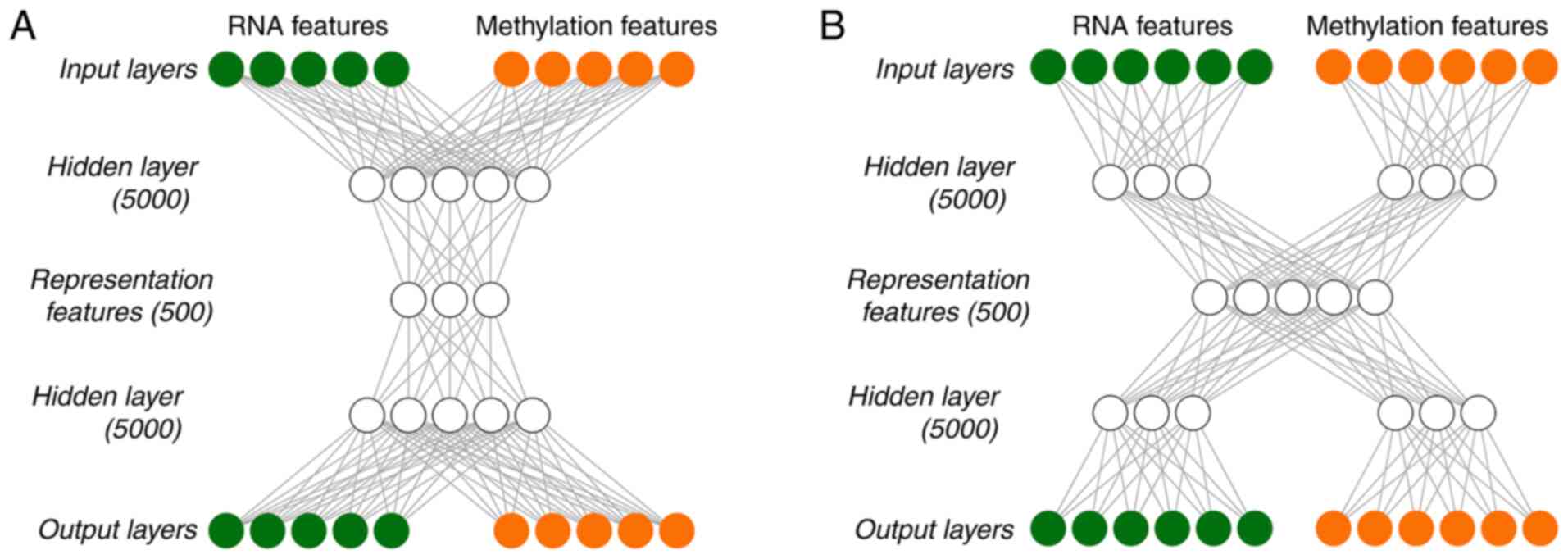
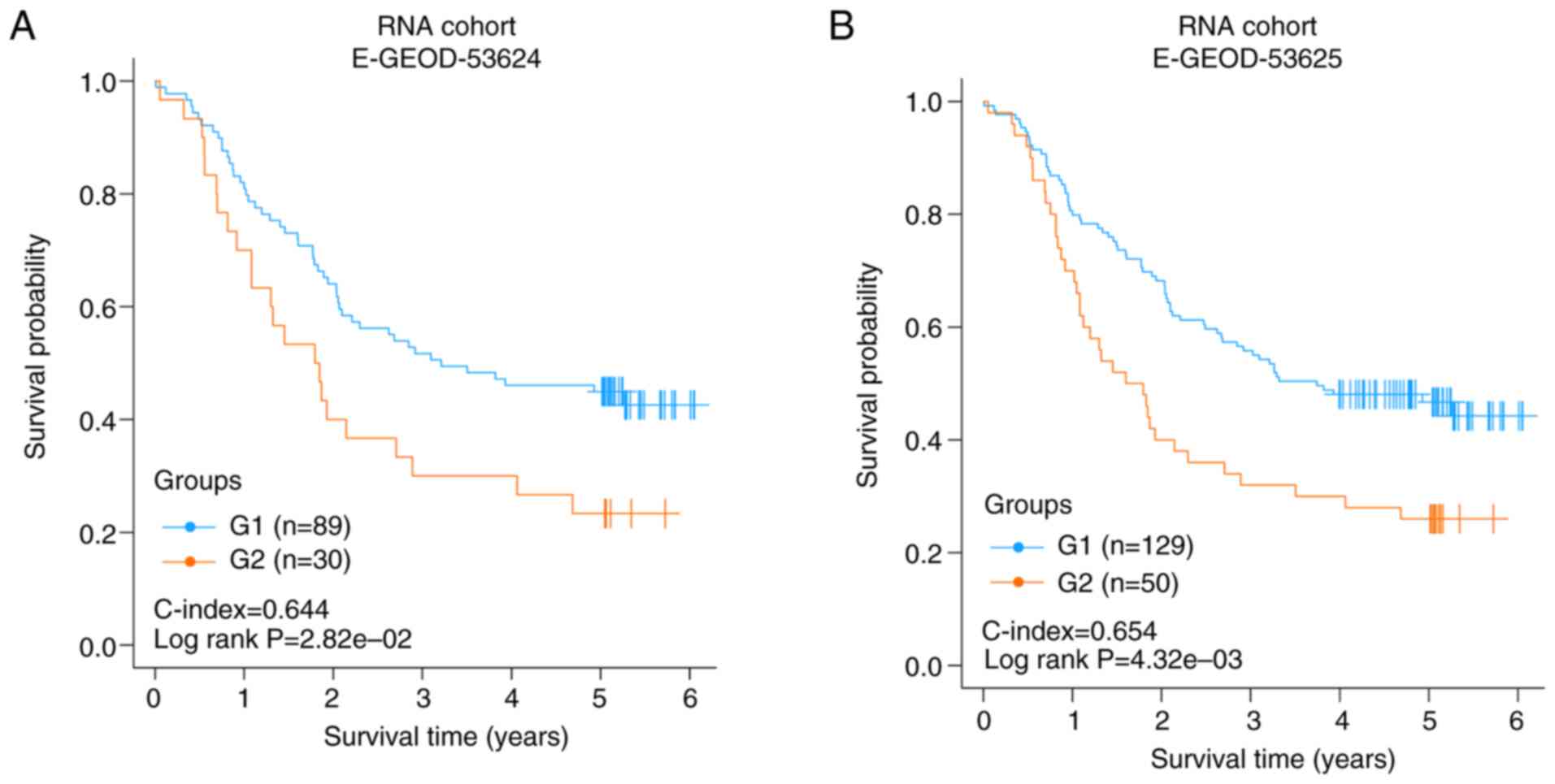
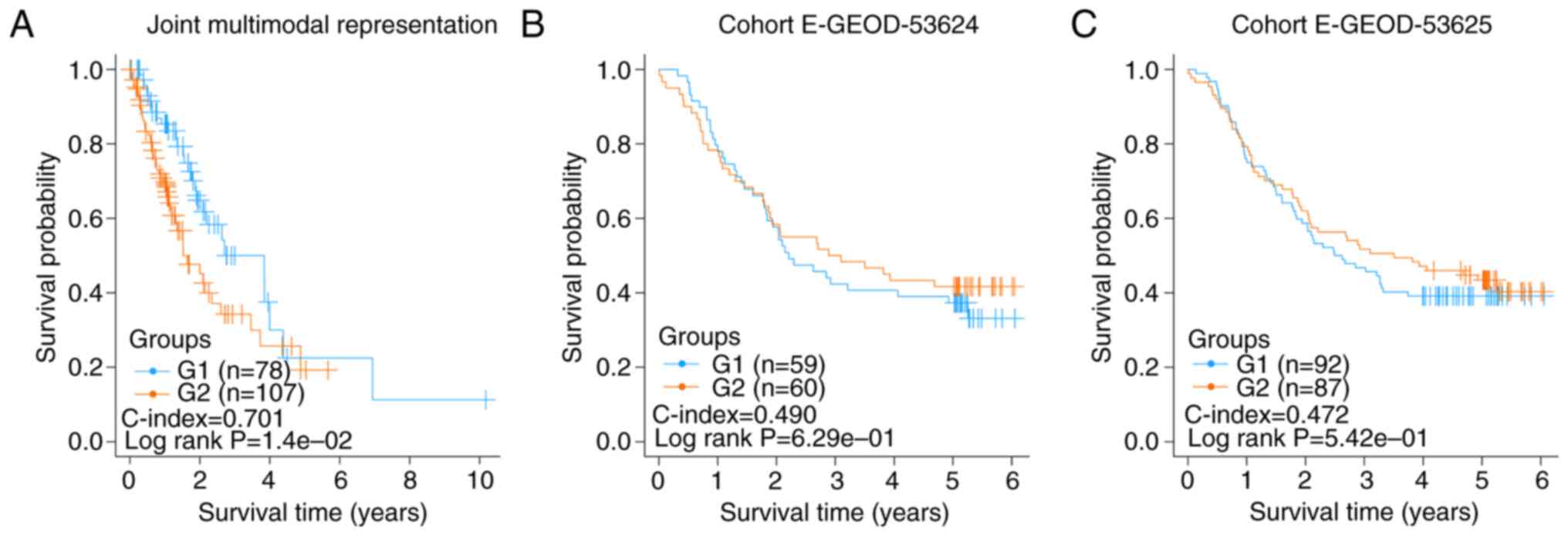
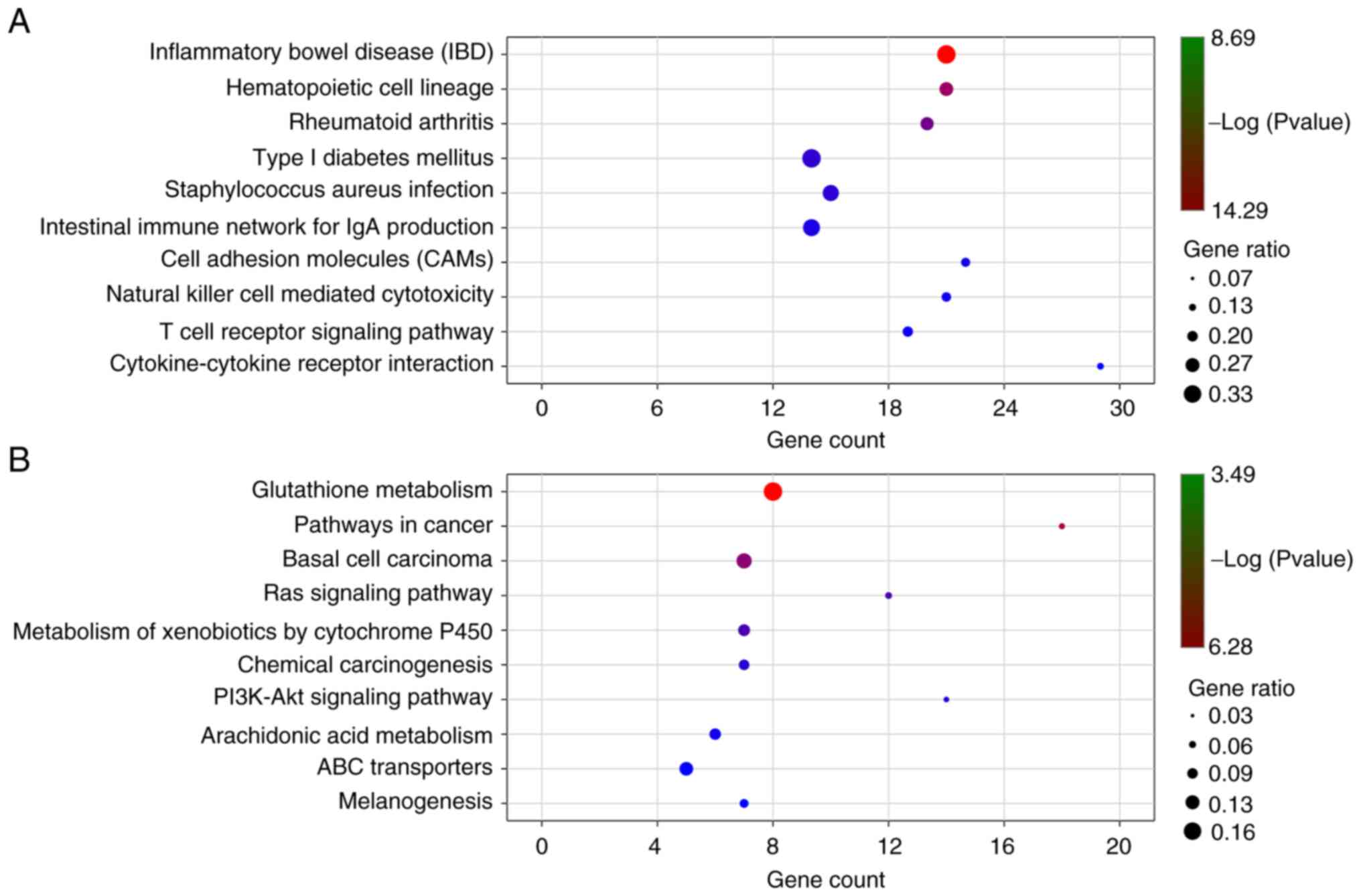
|
1
|
Lin DC, Wang MR and Koeffler HP: Genomic
and epigenomic aberrations in esophageal squamous cell carcinoma
and implications for patients. Gastroenterology. 154:374–389. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhao J, He YT, Zheng RS, Zhang SW and Chen
WQ: Analysis of esophageal cancer time trends in China, 1989–2008.
Asian Pac J Cancer Prev. 13:4613–4617. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liang H, Fan JH and Qiao YL: Epidemiology,
etiology, and prevention of esophageal squamous cell carcinoma in
China. Cancer Biol Med. 14:33–41. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang GQ, Jiao GG, Chang FB, Fang WH, Song
JX, Lu N, Lin DM, Xie YQ and Yang L: Long-term results of operation
for 420 patients with early squamous cell esophageal carcinoma
discovered by screening. Ann Thorac Surg. 77:1740–1744. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Abnet CC, Arnold M and Wei WQ:
Epidemiology of esophageal squamous cell carcinoma.
Gastroenterology. 154:360–373. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gao YB, Chen ZL, Li JG, Hu XD, Shi XJ, Sun
ZM, Zhang F, Zhao ZR, Li ZT, Liu ZY, et al: Genetic landscape of
esophageal squamous cell carcinoma. Nat Genet. 46:1097–1102. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Baylin SB: DNA methylation and gene
silencing in cancer. Nat Clin Pract Oncol. 2 (Suppl 1):S4–S11.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yamada N, Yasui K, Dohi O, Gen Y, Tomie A,
Kitaichi T, Iwai N, Mitsuyoshi H, Sumida Y, Moriguchi M, et al:
Genome-wide DNA methylation analysis in hepatocellular carcinoma.
Oncol Rep. 35:2228–2236. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Riley RD, Hayden JA, Steyerberg EW, Moons
KG, Abrams K, Kyzas PA, Malats N, Briggs A, Schroter S, Altman DG,
et al: Prognosis research strategy (PROGRESS) 2: Prognostic factor
research. PLoS Med. 10:e10013802013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Iida M, Ikeda F, Hata J, Hirakawa Y, Ohara
T, Mukai N, Yoshida D, Yonemoto K, Esaki M, Kitazono T, et al:
Development and validation of a risk assessment tool for gastric
cancer in a general Japanese population. Gastric Cancer.
21:383–390. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pavlou M, Ambler G, Seaman SR, Guttmann O,
Elliott P, King M and Omar RZ: How to develop a more accurate risk
prediction model when there are few events. BMJ. 351:h38682015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hoffman MM, Ernst J, Wilder SP, Kundaje A,
Harris RS, Libbrecht M, Giardine B, Ellenbogen PM, Bilmes JA,
Birney E, et al: Integrative annotation of chromatin elements from
ENCODE data. Nucleic Acids Res. 41:827–841. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bengio Y: Learning Deep Architectures for
AI. Foundations and Trends® in Machine Learning, 2009.
simplehttps://doi.org/10.1561/2200000006
|
|
14
|
Papazafeiropoulos G, Vu QV, Truong VH,
Luong MC and Pham VT: Prediction of buckling coefficient of
stiffened plate girders using deep learning algorithm. CIGOS 2019,
Innovation for Sustainable Infrastructure Springer. Ha-Minh C, Dao
D, Benboudjema F, Derrible S, Huynh D and Tang A: Lecture Notes in
Civil Engineering. Vol 54. Springer; Singapore: simplehttps://doi.org/10.1007/978-981-15-0802-8_183
|
|
15
|
Chaudhary K, Poirion OB, Lu L and Garmire
LX: Deep learning-based multi-omics integration robustly predicts
survival in liver cancer. Clin Cancer Res. 24:1248–1259. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Poirion OB, Chaudhary K, Huang S and
Garmire LX: Multi-omics-based pan-cancer prognosis prediction using
an ensemble of deep-learning and machine-learning models. medRxiv.
doi: https://doi.org/10.1101/19010082.
|
|
17
|
Xie G, Dong C, Kong Y, Zhong JF, Li M and
Wang K: Group lasso regularized deep learning for cancer prognosis
from multi-omics and clinical features. Genes (Basel). 10:2402019.
View Article : Google Scholar
|
|
18
|
Lu T, Chen D, Wang Y, Sun X, Li S, Miao S,
Wo Y, Dong Y, Leng X, Du W and Jiao W: Identification of DNA
methylation-driven genes in esophageal squamous cell carcinoma: A
study based on the cancer genome atlas. Cancer Cell Int. 19:522019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hansen KD:
IlluminaHumanMethylation450kanno. ilmn12. hg19: Annotation for
illumina's 450k methylation arrays. R package version 0.2 12016.
doi:
10.18129/B9.bioc.IlluminaHumanMethylation450kanno.ilmn12.hg19.
|
|
20
|
Hastie T, Tibshirani R, Narasimhan B and
Chu G: Impute: Impute: Imputation for microarray data. R package
version. 2019.
|
|
21
|
Malika C, Ghazzali N, Boiteau V and
Niknafs A: NbClust: An R package for determining the relevant
number of clusters in a data set. J Stat Software. 61:1–36.
2014.
|
|
22
|
Schröder MS, Culhane AC, Quackenbush J and
Haibe-Kains B: Survcomp: An R/Bioconductor package for performance
assessment and comparison of survival models. Bioinformatics.
27:3206–3208. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Becker N, Werft W, Toedt G, Lichter P and
Benner A: PenalizedSVM: A R-package for feature selection SVM
classification. Bioinformatics. 25:1711–1712. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Brembilla-Perrot B, de la Chaise Terrier
A, Skeik L, Cherrier F and Pernot C: Factors predicting the
response to an antiarrhythmic during an electrophysiologic study
for ventricular tachycardia. Arch Mal Coeur Vaiss. 80:1497–1503.
1987.(In French). PubMed/NCBI
|
|
27
|
Chen J, Kwong DL, Cao T, Hu Q, Zhang L,
Ming X, Chen J, Fu L and Guan X: Esophageal squamous cell carcinoma
(ESCC): Advance in genomics and molecular genetics. Dis Esophagus.
28:84–89. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Song G, Xu J, He L, Sun X, Xiong R, Luo Y,
Hu X, Zhang R, Yue Q, Liu K and Feng G: Systematic profiling
identifies PDLIM2 as a novel prognostic predictor for oesophageal
squamous cell carcinoma (ESCC). J Cell Mol Med. 23:5751–5761. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Y, Yao H and Zhao S: Auto-encoder
based dimensionality reduction. Neurocomputing. 184:232–242. 2016.
View Article : Google Scholar
|
|
30
|
Xiao Y, Wu J, Lin Z and Zhao X: A
semi-supervised deep learning method based on stacked sparse
auto-encoder for cancer prediction using RNA-seq data. Comput
Methods Programs Biomed. 166:99–105. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Qiao Y, Luo X, Li C, Tian H and Ma J:
Heterogeneous graph-based joint representation learning for users
and POIs in location-based social network. Information Processing
Management. 57:1021512020. View Article : Google Scholar
|
|
32
|
Wang F, Yan Z, Lv J, Xin J, Dang Y, Sun X,
An Y, Qi Y, Jiang Q, Zhu W, et al: Gene expression profiling
reveals distinct molecular subtypes of esophageal squamous cell
carcinoma in Asian populations. Neoplasia. 21:571–581. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cancer Genome Atlas Research Network;
Analysis Working Group: Asan University; BC Cancer Agency; Brigham
and Women's Hospital; Broad Institute; Brown University; Case
Western Reserve University; Dana-Farber Cancer Institute; Duke
University, et al, . Integrated genomic characterization of
oesophageal carcinoma. Nature. 541:169–174. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu W, Snell JM, Jeck WR, Hoadley KA,
Wilkerson MD, Parker JS, Patel N, Mlombe YB, Mulima G, Liomba NG,
et al: Subtyping sub-saharan esophageal squamous cell carcinoma by
comprehensive molecular analysis. JCI Insight. 1:e887552016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Albelda S: Role of integrins and other
cell adhesion molecules in tumor progression and metastasis. Lab
Invest. 68:4–17. 1993.PubMed/NCBI
|
|
36
|
Chen YZ, Xue JY, Chen CM, Yang BL, Xu QH,
Wu F, Liu F, Ye X, Meng X, Liu GY, et al: PPAR signaling pathway
may be an important predictor of breast cancer response to
neoadjuvant chemotherapy. Cancer Chemother Pharmacol. 70:637–644.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dong C, Wang X, Xu H, Zhan X, Ren H, Liu
Z, Liu G and Liu L: Identification of a cytokine-cytokine receptor
interaction gene signature for predicting clinical outcomes in
patients with colorectal cancer. Int J Clin Exp Med. 10:9009–9018.
2017.
|
|
38
|
Ertel A, Verghese A, Byers SW, Ochs M and
Tozeren A: Pathway-specific differences between tumor cell lines
and normal and tumor tissue cells. Mol Cancer. 5:552006. View Article : Google Scholar : PubMed/NCBI
|